Abstract
With the aim of finding small molecules that stimulate erythropoiesis earlier than erythropoietin and that enhance erythroid colony-forming unit (CFU-E) production, we studied the mechanism by which glucocorticoids increase CFU-E formation. Using erythroid burst-forming unit (BFU-E) and CFU-E progenitors purified by a new technique, we demonstrate that glucocorticoids stimulate the earliest (BFU-E) progenitors to undergo limited self-renewal, which increases formation of CFU-E cells > 20-fold. Interestingly, glucocorticoids induce expression of genes in BFU-E cells that contain promoter regions highly enriched for hypoxia-induced factor 1α (HIF1α) binding sites. This suggests activation of HIF1α may enhance or replace the effect of glucocorticoids on BFU-E self-renewal. Indeed, HIF1α activation by a prolyl hydroxylase inhibitor (PHI) synergizes with glucocorticoids and enhances production of CFU-Es 170-fold. Because PHIs are able to increase erythroblast production at very low concentrations of glucocorticoids, PHI-induced stimulation of BFU-E progenitors thus represents a conceptually new therapeutic window for treating erythropoietin-resistant anemia.
Introduction
Anemia associated with chronic renal insufficiency can often be successfully treated with recombinant erythropoietin (Epo). Other forms of anemia that are caused by insufficient numbers of Epo-sensitive erythroid colony-forming unit (CFU-E) cells do not respond well to Epo. A drug that stimulates erythropoiesis by increasing the number of CFU-E cells could therefore allow treatment of Epo-resistant anemia and bone marrow failure syndromes.
To identify compounds with the potential to enhance CFU-E regeneration, we studied the mechanism by which glucocorticoids (GCs) promote production of Epo-responsive CFU-E progenitors in vitro. This process, which also requires stem cell factor (SCF), is similar to the physiologic mechanisms of stress erythropoiesis (SE) that replenish CFU-E cells during severe or chronic anemia.1-3
Both in vitro proliferation of fetal liver erythroblasts and SE in vivo require GC receptor (GCR) and is disrupted by GCR mutations that abolish dimerization and transactivation but not transrepression.2-4 Thus, GCs probably stimulate erythroblast production during SE by gene activation rather than by repression.3,4 Although more detailed knowledge about how GCs stimulate SE could lead to better treatment for anemia, such studies have been limited because the cell type that responds to GCs has not been identified.
Here, we used cultured CFU-E and erythroid burst-forming unit (BFU-E) progenitors, highly purified from mouse fetal liver by a new technique, to demonstrate that BFU-E and not CFU-E progenitors respond to GCs by generating more daughter BFU-E cells, that is, by enhancing BFU-E self-renewal. As a consequence, over time this increases the number of CFU-E cells and thus the number of erythroblasts formed from each BFU-E > 10-fold.
To our surprise, we found that promoter regions of many genes regulated by GCR activation in BFU-E cells contain binding sites for hypoxia-induced factor 1α (HIF1α), suggesting that HIF1α activation would enhance expression of these genes and possibly enhance the biologic function of GCR activation.
Transcriptional activation by HIF1α is partly regulated by oxygen-dependent HIF prolyl hydroxylases (Egln1, Egln2, and Egln3).5 These enzymes sense intracellular oxygen tension and use dioxygen as a substrate to hydroxylate a proline residue in HIF1α, which leads to its polyubiqutination by von Hippel–Lindau protein and degradation by the 26S proteasome. Specific prolyl hydroxylase inhibitors (PHIs) have been developed that inhibit HIF1 prolyl hydroxylation. These drugs are able to induce HIF activation in kidneys and to induce Epo production, and they are thus promising erythropoiesis-stimulating drugs. Here, we use dimethyloxalylglycine (DMOG), a commercially available PHI, to show that, as suggested from the enrichment of HIF1α sites in the promoter regions, DMOG enhances the expression of a significant number of genes that are also up-regulated by dexamethasone (Dex). Importantly, the addition of DMOG together with Dex results in a synergistic biologic effect on BFU-E proliferation and self-renewal, leading to 300-fold total increase in production of erythroblasts, 7-fold greater than achieved by Dex alone. We thus show that the mechanism of CFU-E regeneration during SE can be pharmacologically stimulated by PHIs in combination with low GC concentrations.
We propose that the clinical potential of PHIs goes beyond the use as an oral replacement for Epo analogues. In addition to the effect on kidney cells, PHIs intrinsically stimulate BFU-E cells to undergo self-renewal and thus to enhance production of Epo-sensitive CFU-E progenitors. PHIs may therefore have an effect on Epo-resistant anemia and bone marrow failure syndromes such as Diamond-Blackfan anemia (DBA).
Methods
Enrichment of fetal liver erythroid progenitors
Embryonic day 14.5 (E14.5) to E15.5 fetal liver cells were incubated with a cocktail of biotin-labeled lineage antibodies (mouse lineage panel, anti–mouse Ter119; CD16/CD32; Sca-1, and CD41) After magnetic depletion of positive cells, a pure fetal liver erythroid progenitor population is obtained. BFU-E and CFU-E cells were separated from the Kit+ fraction of these cells by flow cytometry. The CFU-E fraction is the 20% highest CD71- and/or CD24a-expressing part of the Kit+ fraction, and the BFU-E fraction is the 10% lowest CD71- and/or CD24a-expressing part. All animal procedures were approved by and performed according to the guidelines of the Massachusetts Institute of Technology Committee on Animal Care.
Serum-free erythroid liquid expansion progenitor growth assay
Our serum-free erythroid liquid expansion (SFELE) medium is modified from a report by Dolzning et al6 and consists of 100 ng/mL recombinant murine stem cell factor (rmSCF), 40 ng/mL recombinant murine insulinlike growth factor-1 (rmIGF-1), and 2 U/mL recombinant human erythropoietin (rhEPO), in StemSpan serum-free expansion medium, with or without Dex or DMOG.
Colony-forming assays
For CFU-E colony-forming assays we used MethoCult M3234 (StemCell Technologies) containing 10 U rhEPO, with or without 100nM Dex. The BFU-E assays were performed in MethoCult M3234 containing 10 U rhEPO, 20 ng/mL rm interleukin-3, 20 ng/mL recombinant murine interleukin-6, and 50 ng/mL rmSCF, (PeproTech), with or without 100nM Dex. CFU-Es were scored at day 3, and BFU-E and myeloid colonies were scored after 8-9 days after staining with 2,7-diaminofluorene (Sigma Chemical).
Cytospin preparations and histologic staining
In vitro cultured cells were centrifuged onto poly-lysine–coated slides for 3 minutes at 500 rpm (Cytospin 3; Thermo Shandon), air dried, and fixed in −20°C methanol for 2 minutes and stained with May-Grünewald-Giemsa according to the manufacturer's recommendations (Sigma Chemical).
Next-generation mRNA sequencing
Samples for Paired-End mRNA-Seq were prepared with the use of the Solexa kit according to the manufacturer's instructions with the exception that we extracted 300-base pair bands before and after the polymerase chain reaction step (Illumina). Images acquired from the Solexa sequencer were processed through the bundled Solexa image extraction pipeline Version 1.4. mRNA-Seq reads were aligned to mouse National Center for Biotechnology Information (NCBI) build 37 (mm9) with the use of ELAND (Illumina). Briefly, the first 32 bases of a read were used as a seed. Each matched seed was then extended ≤ 36 bases and scored to break any ties between multiple matches. For mRNA expression counts, unique reads in the genome that landed within any exons of NCBI gene models (v37.1) were counted. The counts were normalized by the mRNA length to get the final RPKM values (ie, reads per kilobase of exon per million mapped reads).
All sequence data have been uploaded to the NCBI Gene Expression Omnibus database under accession number GSE26086.
Motif enrichment analysis
Whole Genome rVISTA7 tool was used to identify transcription factor binding sites that are conserved between species and enriched in upstream regions of genes unregulated in BFU-E with Dex versus without Dex. In addition, THEME algorithm implemented in the web tool webMOTIFS8 was used to identify significant motifs present in the genes unregulated in BFU-E with Dex versus without Dex.
Results
Purification of BFU-E and CFU-E cells from fetal liver by flow cytometry
Although it is well known that GCs increase formation of CFU-E progenitors in vitro and that the GCR is required for SE in vivo,1-3,6,9,10 it has not been determined whether this mechanism involves increased self-renewal of CFU-E progenitors or earlier BFU-E progenitors. Initial observations that adding Dex increases the size of BFU-E but not of CFU-E colonies in methyl cellulose assays (supplemental Figure 1) led us to formulate the hypothesis that BFU-E and not CFU-E progenitor cells are stimulated by GCs.
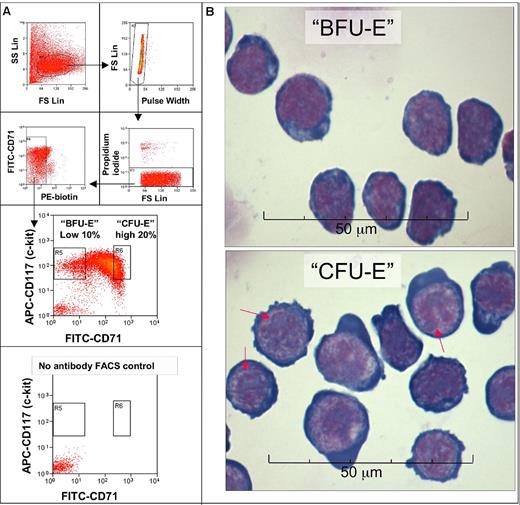
To test this hypothesis we developed techniques to purify BFU-E and CFU-E cells from mouse fetal liver by flow cytometry. A > 90% pure mix of BFU-E and CFU-E cells was obtained by negative selection for Ter119, B220, Mac-1, CD3, Gr-1, Sca-1, CD16/CD32, CD41, and CD34 (Figure 1A; supplemental Tables 1-2, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Within the Kit+ fraction of this cell population the level of CD71 expression was used to separate morphologically distinct and BFU-E (CD7110%low) and CFU-E (CD7120%high) progenitors (Figure 1B).11,12 Eighty-six percent of colonies formed by these BFU-E cells were BFU-E colonies, and 97% of CFU-E cells formed CFU-E colonies (supplemental Table 2).
Enrichment of BFU-E and CFU-E cells by flow cytometric cell sorting. (A) Day 14.5-15.5 mouse fetal liver cells stained with biotin-conjugated antibodies against murine Ter119, B220, Mac-1, CD3, Gr-1, CD32/CD16, Sca-1, CD41, and CD34 were first depleted by magnetic beads (supplemental Table 1). The enriched negative fraction (FLEP) was then stained with streptavidin-phycoerythrin (PE), CD71 fluorescein isothiocyanate (FITC), and CD117 allophycocyanin (APC) antibodies, and the phycoerythrin-negative cells were sorted by FACS into 2 fractions, called BFU-E and CFU-E. The BFU-E fraction is the 10% lowest CD71-expressing part of the Kit (CD117)+ fraction, which has a CD71 signal intensity that is similar to or slightly higher than that of unstained cells. The CFU-E fraction is the 20% highest CD71-expressing part of the Kit+ fraction. Eighty-six percent of colonies formed by sorted BFU-E cells were BFU-E colonies, 11% CFU-E colonies, and 3% other myeloid colonies. Thus, the purity of these sorted BFU-E cells is ∼ 86%. The purity of CFU-E cells is slightly higher because they form 97% CFU-E colonies, 3% late-BFU-E colonies, and no other myeloid colonies (supplemental Table 2). The same FACS setup was later used to separate BFU-E and CFU-Es with higher purities with the use o f CD24a and a combination of CD24a and CD71 (supplemental Table 2). (B) Micrographs of sorted CFU-E (Kit+ CD7120%high) cells and BFU-E (Kit+ CD710%low) cells stained with May-Grünewald-Giemsa. BFU-E cells have a high nuclear/cytoplasmic ratio and very fine nuclear chromatin. CFU-E cells are larger than BFU-E cells with a lower nuclear/cytoplasmic ratio and more regions of heterochromatin. CFU-E cells have multiple large, well-defined nucleoli (red arrows) in the nuclei. The CFU-E cytoplasm is very basophilic and sometimes bulges out from the cell. The morphology and relatively smaller size of BFU-E cells compared with CFU-E cells agree with previous studies on less pure populations of CFU-E and BFU-E cells.11,12 FS indicates forward scatter; SS, side scatter.
Enrichment of BFU-E and CFU-E cells by flow cytometric cell sorting. (A) Day 14.5-15.5 mouse fetal liver cells stained with biotin-conjugated antibodies against murine Ter119, B220, Mac-1, CD3, Gr-1, CD32/CD16, Sca-1, CD41, and CD34 were first depleted by magnetic beads (supplemental Table 1). The enriched negative fraction (FLEP) was then stained with streptavidin-phycoerythrin (PE), CD71 fluorescein isothiocyanate (FITC), and CD117 allophycocyanin (APC) antibodies, and the phycoerythrin-negative cells were sorted by FACS into 2 fractions, called BFU-E and CFU-E. The BFU-E fraction is the 10% lowest CD71-expressing part of the Kit (CD117)+ fraction, which has a CD71 signal intensity that is similar to or slightly higher than that of unstained cells. The CFU-E fraction is the 20% highest CD71-expressing part of the Kit+ fraction. Eighty-six percent of colonies formed by sorted BFU-E cells were BFU-E colonies, 11% CFU-E colonies, and 3% other myeloid colonies. Thus, the purity of these sorted BFU-E cells is ∼ 86%. The purity of CFU-E cells is slightly higher because they form 97% CFU-E colonies, 3% late-BFU-E colonies, and no other myeloid colonies (supplemental Table 2). The same FACS setup was later used to separate BFU-E and CFU-Es with higher purities with the use o f CD24a and a combination of CD24a and CD71 (supplemental Table 2). (B) Micrographs of sorted CFU-E (Kit+ CD7120%high) cells and BFU-E (Kit+ CD710%low) cells stained with May-Grünewald-Giemsa. BFU-E cells have a high nuclear/cytoplasmic ratio and very fine nuclear chromatin. CFU-E cells are larger than BFU-E cells with a lower nuclear/cytoplasmic ratio and more regions of heterochromatin. CFU-E cells have multiple large, well-defined nucleoli (red arrows) in the nuclei. The CFU-E cytoplasm is very basophilic and sometimes bulges out from the cell. The morphology and relatively smaller size of BFU-E cells compared with CFU-E cells agree with previous studies on less pure populations of CFU-E and BFU-E cells.11,12 FS indicates forward scatter; SS, side scatter.
GCs stimulate CFU-E regeneration by allowing more BFU-E cell divisions to occur before differentiation into CFU-Es
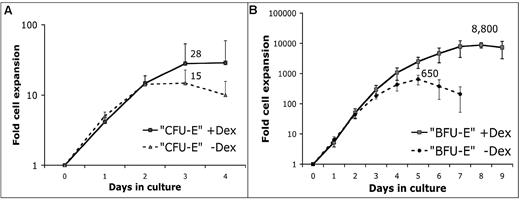
The effect of 100nM Dex on proliferative potential of CFU-E and BFU-E cells was determined by liquid culture in SFELE medium containing SCF, IGF-1, and Epo (see “Methods”). Adding Dex only slightly increased the average number of erythroblasts formed from each CFU-E cell (Figure 2A). In contrast, the proliferative capacity of BFU-E cells was dramatically increased by adding Dex. Although there was no significant difference in the rate of BFU-E cell proliferation during the first 3 days, adding Dex allowed an average BFU-E cell to divide 13 instead of 9 times (Figure 2B).
GCs stimulate stress erythropoiesis by enhancing erythroblast output predominantly from BFU-E progenitors. We determined whether adding 100nM Dex changes the maximum number of erythroblasts that sorted CFU-E and BFU-E cells produce in SFELE medium with or without 100nM Dex. (A) Production of erythroblasts from sorted CFU-E (CD7120%high) cells peaks at day 3 with 28- and 15-fold expansion with and without Dex, respectively (n = 4). Error bars show 1 SD. (B) BFU-E (CD7110%low) cells cultured with 100nM Dex expand 8800-fold (13 cell divisions) with a peak at day 8 and expand 650-fold (9 cell divisions) without Dex with the peak at day 5 (n = 5). At the endpoint of the BFU-E cell cultures cells were routinely stained with May-Grünewald-Giemsa stain, which consistently showed that virtually all cells were erythroblasts equivalent to the cells depicted in Figure 6C (data not shown). Error bars show 1 SD.
GCs stimulate stress erythropoiesis by enhancing erythroblast output predominantly from BFU-E progenitors. We determined whether adding 100nM Dex changes the maximum number of erythroblasts that sorted CFU-E and BFU-E cells produce in SFELE medium with or without 100nM Dex. (A) Production of erythroblasts from sorted CFU-E (CD7120%high) cells peaks at day 3 with 28- and 15-fold expansion with and without Dex, respectively (n = 4). Error bars show 1 SD. (B) BFU-E (CD7110%low) cells cultured with 100nM Dex expand 8800-fold (13 cell divisions) with a peak at day 8 and expand 650-fold (9 cell divisions) without Dex with the peak at day 5 (n = 5). At the endpoint of the BFU-E cell cultures cells were routinely stained with May-Grünewald-Giemsa stain, which consistently showed that virtually all cells were erythroblasts equivalent to the cells depicted in Figure 6C (data not shown). Error bars show 1 SD.
This was confirmed in cultures of single BFU-E cells. As in all SFELE cultures of BFU-E cells, at the end of the culture single BFU-Es exclusively form erythroblasts similar to those presented in Figure 6C and supplemental Figure 6B and D (data not shown). Only 3 of 180 single BFU-Es gave rise to > 1000 erythroblasts without Dex after 8 days of culture. In the presence of Dex approximately one-third of the BFU-Es gave rise to > 1000 and several generated > 100 000 erythroblasts (supplemental Figure 2C-D).
The apparent 2-fold stimulatory effect of Dex on CFU-E cells may be explained by the fact that the CFU-E population contains 3% late BFU-E cells (supplemental Table 2) that are able to respond to Dex. Nonetheless, these results show that Dex enhances formation of erythroblasts mainly by stimulating BFU-E proliferation.
GCs regulate transcription of a small number of mRNAs in BFU-E cells
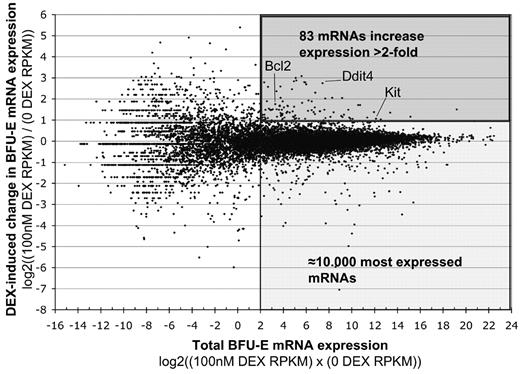
To determine at the molecular level how GCs stimulate BFU-E progenitors, we used next-generation sequencing to determine the difference in relative expression of individual messenger RNAs (mRNAs) in BFU-E cells cultured 4 hours with or without 100nM Dex (Figure 3). To minimize the risk of false-positive GC target genes we chose to compare only highly expressed genes (≈10 000 genes with a RPKM count > 4).13 We defined GC target genes as those whose mRNA expression levels differed ± 2-fold between BFU-E cells cultured 4 hours with and without DEX. By these stringent criteria only 195 genes were differentially expressed, of which 83 were up-regulated and 112 were down-regulated (Figure 3; supplemental Table 3).
The effect of GCs on mRNA expression in BFU-E cells determined by next-generation sequencing. Sorted CD7110%low BFU-E cells were cultured as detailed in Figure 1 for 4 hours either in the absence or presence of 100nM Dex, after which mRNA was extracted and subjected to Illumina next-generation sequencing. mRNA expression was normalized with the RPKM method, which determines the relative expression of each gene, normalized to the total transcriptome, by giving each gene an RPKM count. The mRNA expression R-I plot shows on the y-axis the log2 ratio of expression of individual mRNAs in Dex-stimulated versus nonstimulated cells. Positive values mean higher expression in BFU-E cells treated with 100nM Dex (+1 = 100% up; +2 = 400% up; −1 = 50% down; −2 = 75% down, etc). The x-axis shows the average expression of individual mRNAs, plotted as the log2 of the product of expression of the mRNA in stimulated and nonstimulated cells.
The effect of GCs on mRNA expression in BFU-E cells determined by next-generation sequencing. Sorted CD7110%low BFU-E cells were cultured as detailed in Figure 1 for 4 hours either in the absence or presence of 100nM Dex, after which mRNA was extracted and subjected to Illumina next-generation sequencing. mRNA expression was normalized with the RPKM method, which determines the relative expression of each gene, normalized to the total transcriptome, by giving each gene an RPKM count. The mRNA expression R-I plot shows on the y-axis the log2 ratio of expression of individual mRNAs in Dex-stimulated versus nonstimulated cells. Positive values mean higher expression in BFU-E cells treated with 100nM Dex (+1 = 100% up; +2 = 400% up; −1 = 50% down; −2 = 75% down, etc). The x-axis shows the average expression of individual mRNAs, plotted as the log2 of the product of expression of the mRNA in stimulated and nonstimulated cells.
In addition, we performed small RNA sequencing but detected no significant (± 2-fold) effects of Dex on expression of any micro-RNAs (data not shown).
BFU-E cells respond to GCs by increased expression of genes with promoter regions containing evolutionarily conserved HIF1α binding sequences rather than GC response elements
On the basis of previous studies, functionally relevant GCR target genes in BFU-E cells probably respond to Dex by increased rather than decreased expression.2,4 To find novel strategies to pharmacologically stimulate the 83 up-regulated genes, we computationally analyzed the promoter regions of these 83 genes with the use of Whole Genome rVISTA to determine whether any transcription factor binding sites conserved from mouse to human were overrepresented in 5000 bases of upstream genomic regions (supplemental Figure 3). Surprisingly, there was no overrepresentation of GC response elements (GREs) but instead highly significant enrichment of predicted HIF1α (P = 1 × 10−27) and MYC (P = 1 × 10−24) binding sites. This motif enrichment analysis was repeated with 395 genes up-regulated only ≥ 50% by Dex and with a multiplied RPKM count greater than zero. Again, in this larger set of less significantly enriched GC-target genes, the promoter segments were enriched for HIF1α (P = 1 × 10−12) and MYC (P = 1 × 10−9) but not for GRE sites (data not shown).
To confirm that the significant enrichment is not merely explained by the fact that highly expressed genes in BFU-E cells in general are enriched for HIF1α binding sites, the enrichment analysis was repeated with 2847 well-expressed (multiplied RPKM count > 4) genes that change < 7.2% (log2 [.1]). Neither HIF1α nor any other transcription factor binding motifs were significantly enriched in these promoter regions (data not shown).
We confirmed the results by motif enrichment analysis of −2000- to 200-base pair promoter segments of 83 Dex-induced genes with the use of Bayesian motif discovery approach implemented in webMOTIFS.12 Again, no significant enrichment of GRE motifs was found, whereas the most significantly enriched motif (caCGTGgc) is a HIF1α binding site (supplemental Figure 4). In addition, 3 of the 83 up-regulated genes (Egln3, Pfkfb4, and Spry1) that were not computationally predicted to be HIF1α targets are experimentally validated HIF1α target genes.15-17
Dex and the PHI DMOG regulate overlapping genes in BFU-E cells
The enrichment of HIF1α motifs in the promoter regions of Dex-induced genes prompted us to test whether a compound that enhances HIF1α activation would alter expression of the same genes in the same direction as Dex. Because none of the recently developed HIF-specific PHIs are commercially available, we used the prototype PHI, DMOG, to test our hypothesis.5,18
For this and the following experiments we used an improved fluorescence-activated cell sorting (FACS) method by which BFU-E cells were purified on the basis of 10% low expression of both CD71 and CD24a (supplemental data; supplemental Table 2). Because, similar to CD71, CD24a is expressed at very low levels in BFU-E cells and at more than a 7-fold higher level in CFU-E cells (supplemental Table 4), and because good antibodies were available, we used CD24a in the purification. By combining CD71 and CD24a we increased the purity of the progenitor populations so that BFU-E cells formed 94% BFU-E colonies and CFU-E cells formed 95% CFU-E colonies at cloning efficiencies of 55%-70% (+ Dex in supplemental Table 2).
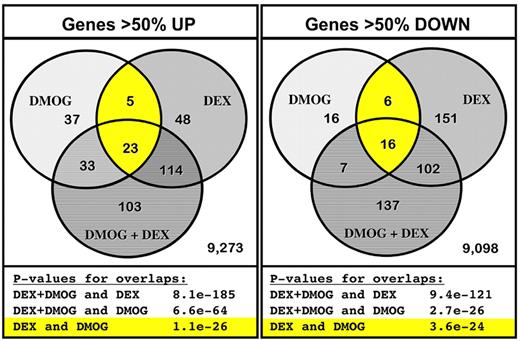
We first determined the change in mRNA expression by next-generation sequencing in BFU-E cells cultured 4 hours in SFELE medium with 100nM Dex, with 333μM DMOG, or with both 100nM Dex and 333μM DMOG. Of ≈9500 highly expressed genes, expression of 98 (DMOG), 190 (Dex), and 273 (DMOG + Dex) genes increased > 50% compared with BFU-E cells cultured 4 hours in SFELE medium only. Expression of 45 (DMOG), 275 (Dex), and 262 (DMOG + Dex) genes decreased > 50%. We tested our hypothesis by comparing the overlap of up-regulated and down-regulated genes in the 3 conditions. Figure 4 shows that, indeed, there was a statistically significant overlap of the genes regulated by Dex and DMOG. Of the 98 genes up-regulated by DMOG alone, 28 were among the 190 genes up-regulated by Dex added alone (P = 1 × 10−26). Similarly, of the 45 genes down-regulated by DMOG added alone 22 were among the 275 genes down-regulated by Dex added alone (P = 4 × 10−24). As expected genes up-regulated by adding both DMOG and Dex to the cultured BFU-E cells overlap with cells treated with DMOG alone (P = 7 × 10−64) and Dex alone (P = 8 × 10−185; supplemental Table 5). Similarly, genes down-regulated by adding both DMOG and Dex overlap with genes down-regulated in cells treated with DMOG alone (P = 3 × 10−26) and Dex alone (P = 9 × 10−121; supplemental Table 6).
The PHI DMOG and Dex have overlapping effects on gene expression in BFU-E cells. Next-generation mRNA sequencing was performed on mRNA extracted from BFU-E (CD71 and CD24a10%high) cells cultured 4 hours in SFELE medium with 100nM Dex, with 333mM DMOG; or with both 100nM Dex and 333mM DMOG. Expression of the 9636 most highly expressed genes (multiplied RPKM > 0 from the 4 groups) from cells treated with Dex, DMOG, and DEX + DMOG was compared with cells cultured in SFELE medium only. The left Venn diagram shows the overlap of the genes that increased > 50%, whereas the Venn diagram to the right shows overlap of genes that decreased to < 50% of that in unstimulated BFU-Es. Significance of the overlapping genes was computed with the use of hypergeometric distribution over all genes detected in any of the samples. The statistical significance of each respective overlap is presented below each diagram. The individual genes in each overlapping group are listed in supplemental Tables 5 and 6.
The PHI DMOG and Dex have overlapping effects on gene expression in BFU-E cells. Next-generation mRNA sequencing was performed on mRNA extracted from BFU-E (CD71 and CD24a10%high) cells cultured 4 hours in SFELE medium with 100nM Dex, with 333mM DMOG; or with both 100nM Dex and 333mM DMOG. Expression of the 9636 most highly expressed genes (multiplied RPKM > 0 from the 4 groups) from cells treated with Dex, DMOG, and DEX + DMOG was compared with cells cultured in SFELE medium only. The left Venn diagram shows the overlap of the genes that increased > 50%, whereas the Venn diagram to the right shows overlap of genes that decreased to < 50% of that in unstimulated BFU-Es. Significance of the overlapping genes was computed with the use of hypergeometric distribution over all genes detected in any of the samples. The statistical significance of each respective overlap is presented below each diagram. The individual genes in each overlapping group are listed in supplemental Tables 5 and 6.
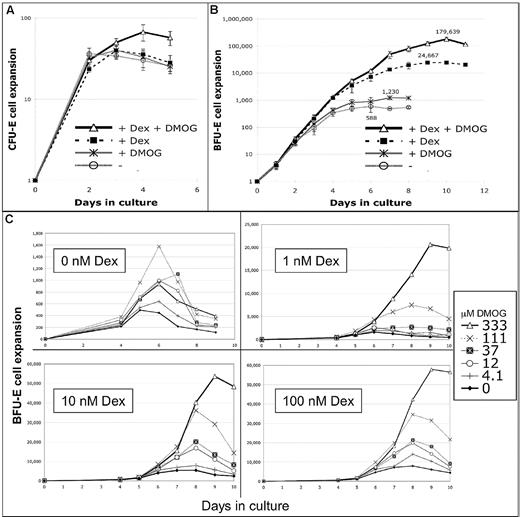
The PHI DMOG synergizes with Dex to increase BFU-E self-renewal and CFU-E production
The striking overlapping effects of Dex and DMOG on gene expression in BFU-E cells prompted us to investigate if the GC effect on BFU-E cells could be enhanced or replaced by DMOG. In vitro culture experiments indeed show that DMOG synergizes with Dex to increase the erythroid output of BFU-E cells, whereas CFU-E cell proliferation is minimally affected (Figure 5A).
DMOG synergizes with Dex to increase the number of erythroblasts formed from a BFU-E cell 300-fold. The experimental protocol was similar to that used in Figure 2A and B except that the CFU-E and BFU-E cells were purer (CD71andCD24a20%high and CD71andCD24a10%low, respectively). The improved method provided a BFU-E population that formed 94% BFU-E, 5% CFU-E, and 1% other myeloid colonies in colony-forming assays with a plating efficiency of 70% (supplemental Table 2). Cultures contained or not 100nM Dex and/or 333μM DMOG. (A) Proliferation of sorted CFU-E cells. Cells do not increase proliferation in response to Dex or DMOG alone, whereas a combination of both Dex and DMOG increase proliferation 1.7-fold (P < .05; n = 6). Error bars show 1 SD. (B) Proliferation of sorted BFU-E cells (n = 4). The maximum increase in proliferation (compared with day 6 with no Dex or DMOG) was 2-fold with DMOG (day 7), 42-fold with 100nM Dex (day 9), and 306-fold with both Dex and DMOG (day 10). The synergistic effect is shown by the fact that DMOG increases the stimulatory effect of Dex on proliferation 7.3-fold (more than the additive 1.7-fold increase). Error bars show 1 SD. (C) BFU-E cells were cultured in SFELE medium containing 0nM, 1nM. 10nM, or 100nM Dex with different concentrations of DMOG. Cells were counted from day 4 until the day cell counts dropped. The y-axis shows the average expansion of several thousand BFU-E cells plated in each experiment. Without Dex 333μM DMOG had little effect on BFU-E proliferation, whereas adding 1nM Dex allows 333μM DMOG to enhance maximum BFU-E proliferation 12-fold. (n = 4) At the endpoint of the BFU-E cell cultures, 95% of cells were erythroblasts (Figure 6C; supplemental Figure 6B,D).
DMOG synergizes with Dex to increase the number of erythroblasts formed from a BFU-E cell 300-fold. The experimental protocol was similar to that used in Figure 2A and B except that the CFU-E and BFU-E cells were purer (CD71andCD24a20%high and CD71andCD24a10%low, respectively). The improved method provided a BFU-E population that formed 94% BFU-E, 5% CFU-E, and 1% other myeloid colonies in colony-forming assays with a plating efficiency of 70% (supplemental Table 2). Cultures contained or not 100nM Dex and/or 333μM DMOG. (A) Proliferation of sorted CFU-E cells. Cells do not increase proliferation in response to Dex or DMOG alone, whereas a combination of both Dex and DMOG increase proliferation 1.7-fold (P < .05; n = 6). Error bars show 1 SD. (B) Proliferation of sorted BFU-E cells (n = 4). The maximum increase in proliferation (compared with day 6 with no Dex or DMOG) was 2-fold with DMOG (day 7), 42-fold with 100nM Dex (day 9), and 306-fold with both Dex and DMOG (day 10). The synergistic effect is shown by the fact that DMOG increases the stimulatory effect of Dex on proliferation 7.3-fold (more than the additive 1.7-fold increase). Error bars show 1 SD. (C) BFU-E cells were cultured in SFELE medium containing 0nM, 1nM. 10nM, or 100nM Dex with different concentrations of DMOG. Cells were counted from day 4 until the day cell counts dropped. The y-axis shows the average expansion of several thousand BFU-E cells plated in each experiment. Without Dex 333μM DMOG had little effect on BFU-E proliferation, whereas adding 1nM Dex allows 333μM DMOG to enhance maximum BFU-E proliferation 12-fold. (n = 4) At the endpoint of the BFU-E cell cultures, 95% of cells were erythroblasts (Figure 6C; supplemental Figure 6B,D).
Although adding 333μM DMOG alone has little effect on BFU-E proliferation, 333μM DMOG synergizes with 100nM Dex and enhances BFU-E proliferation 7-fold over that seen by Dex alone, resulting in a total 300-fold increase in erythroblast formation over that of cultures not containing either substance (Figure 5B). Figure 5C shows that DMOG stimulates BFU-E expansion even in the presence of very low concentrations of Dex. Adding 333μM DMOG enhanced BFU-E proliferation 1.7-fold without Dex, 12-fold with 1nM Dex, and 10-fold with 10nM Dex, compared with 7-fold with 100nM Dex. The results in Figure 5C are replotted in supplemental Figure 5 to more clearly show the synergistic effects of Dex and DMOG. These DMOG and Dex dose-response experiments clearly show that DMOG is a potent enhancer of erythroblast production from BFU-E cells, particularly in the presence of low concentrations of Dex.
DMOG synergize with Dex to promote BFU-E self-renewal and to prevent BFU-E cell exhaustion, allowing more CFU-E cells to be formed
To clarify how DMOG and Dex can stimulate BFU-E cells to make 300 times more erythroblasts, we determined how DMOG and Dex alter the fate of proliferating BFU-E cells. Therefore, daily colony assays were performed during the culture of the cells described in Figure 5B.
Tables 1 and 2 show that adding Dex alone increased the maximum number of CFU-E cells formed 25-fold (28 000 CFU-Es at day 5 per 100 plated BFU-E vs 1100 CFU-Es formed at day 3 in control cultures). Because we previously showed that Dex does not promote CFU-E proliferation or self-renewal (Figure 5A), the 25-fold increase in CFU-E cells probably originates from increased formation of BFU-E cells, each of which in turn produces multiple CFU-E cells. Indeed, Dex alone increased generation of late, “small” BFU-E cells 4.2-fold (maximum of 1000 per 100 plated BFU-E cell at day 5 vs 240 in control cultures at day 2). Adding Dex alone also allowed early “large” BFU-Es to remain undifferentiated during 3 days of in vitro culture (117% of the early BFU-E cells remaining at day 3 with Dex [40 per 100 plated BFU-E vs 42 in the uncultured cells] vs 36% [12 BFU-Es in contrast to 42 in the uncultured cells without Dex]). Further evidence that erythroid progenitors are maintained in cultures with Dex comes from FACS analysis of cultured BFU-E cells at day 3, showing that ≈55% still expressed the early progenitor marker Kit when cultured with Dex, compared with ≈25% without Dex (supplemental Figure 2A).
DMOG and Dex synergize to prevent BFU-E cell exhaustion and enhance CFU-E regeneration: total cell expansion from 100 BFU-E cells
| Dex/DMOG . | Day . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | |
| Nothing added | 100 | 400 | 3100 | 9210 | 34 000 | 50 000 | 58 000* | 49 000 | 54 000 | |||
| DMOG | 100 | 420 | 2600 | 13 000 | 43 000 | 82 000 | 89 000 | 120 000* | 120 000 | |||
| Dex | 100 | 390 | 3000 | 20 000 | 120 000 | 350 000 | 720 000 | 1 400 000 | 2 000 000 | 2 500 000* | 2 500 000 | 2 000 000 |
| DMOG + Dex | 100 | 410 | 3700 | 23 000 | 130 000 | 520 000 | 1 200 000 | 4 900 000 | 8 000 000 | 12 000 000 | 18 000 000* | 12 000 000 |
| Dex/DMOG . | Day . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | |
| Nothing added | 100 | 400 | 3100 | 9210 | 34 000 | 50 000 | 58 000* | 49 000 | 54 000 | |||
| DMOG | 100 | 420 | 2600 | 13 000 | 43 000 | 82 000 | 89 000 | 120 000* | 120 000 | |||
| Dex | 100 | 390 | 3000 | 20 000 | 120 000 | 350 000 | 720 000 | 1 400 000 | 2 000 000 | 2 500 000* | 2 500 000 | 2 000 000 |
| DMOG + Dex | 100 | 410 | 3700 | 23 000 | 130 000 | 520 000 | 1 200 000 | 4 900 000 | 8 000 000 | 12 000 000 | 18 000 000* | 12 000 000 |
Sorted BFU-E (CD24 and CD7110%low) cells were cultured in SFELE medium with or without 100nM Dex and with or without 333μM DMOG (Figure 5B). Although the average of 4 experiments is shown, CFU-E colony-forming assays at days 7 and 8 were performed only twice. Data reported are number of cells formed in culture from 100 BFU-E cells. Results show that a combination of DMOG and Dex reduces differentiation and increases self-renewal of BFU-E cells, which over time results in increased CFU-E colony formation.
Number of erythroblasts reached its highest value that day.
DMOG and Dex synergize to prevent BFU-E cell exhaustion and enhance CFU-E regeneration: total colony numbers from 100 BFU-E cells
| Dex/DMOG . | Day . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | |
| CFU-E, 1 cluster | |||||||||
| Nothing added | 3 | 98 | 630 | 1100* | 1000 | 450 | 50 | ||
| DMOG | 3 | 64 | 600 | 2400 | 2800* | 2700 | 1000 | ||
| Dex | 3 | 42 | 1100 | 5100 | 14 000 | 28 000* | 25 000 | 18 000 | 6600 |
| DMOG + Dex | 3 | 19 | 870 | 8000 | 27 000 | 94 000 | 150 000 | 190 000* | 160 000 |
| Small BFU-E, 5-20 clusters | |||||||||
| Nothing added | 26 | 130 | 240* | 200 | 53 | ||||
| DMOG | 26 | 130 | 320 | 440* | 100 | ||||
| Dex | 26 | 180 | 580 | 830 | 1000* | ||||
| DMOG + Dex | 26 | 150 | 790 | 2000 | 3800* | ||||
| Large BFU-E, > 20 clusters | |||||||||
| Nothing added | 42* | 28 | 15 | 12 | 0 | ||||
| DMOG | 42 | 43* | 22 | 17 | 0 | ||||
| Dex | 42 | 45 | 49* | 40 | 0 | ||||
| DMOG + Dex | 42 | 72 | 120 | 160* | 8 | ||||
| Dex/DMOG . | Day . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | |
| CFU-E, 1 cluster | |||||||||
| Nothing added | 3 | 98 | 630 | 1100* | 1000 | 450 | 50 | ||
| DMOG | 3 | 64 | 600 | 2400 | 2800* | 2700 | 1000 | ||
| Dex | 3 | 42 | 1100 | 5100 | 14 000 | 28 000* | 25 000 | 18 000 | 6600 |
| DMOG + Dex | 3 | 19 | 870 | 8000 | 27 000 | 94 000 | 150 000 | 190 000* | 160 000 |
| Small BFU-E, 5-20 clusters | |||||||||
| Nothing added | 26 | 130 | 240* | 200 | 53 | ||||
| DMOG | 26 | 130 | 320 | 440* | 100 | ||||
| Dex | 26 | 180 | 580 | 830 | 1000* | ||||
| DMOG + Dex | 26 | 150 | 790 | 2000 | 3800* | ||||
| Large BFU-E, > 20 clusters | |||||||||
| Nothing added | 42* | 28 | 15 | 12 | 0 | ||||
| DMOG | 42 | 43* | 22 | 17 | 0 | ||||
| Dex | 42 | 45 | 49* | 40 | 0 | ||||
| DMOG + Dex | 42 | 72 | 120 | 160* | 8 | ||||
CFU-E and BFU-E colony-forming assays were performed at 24-hour intervals in the presence of 100nM Dex. Shown are the number of different types of colonies formed per 100 “BFU-E” cells plated at 0 hours.
Number of erythroblasts or colony-forming cells reached its highest value that day.
Adding DMOG increased the maximum number of CFU-E cells, formed 2.5-fold compared with culture without Dex (2800 at day 4 vs 1100 in control cultures at day 3) and 6.8-fold compared with the culture with Dex alone (190 000 per 100 plated BFU-E cells at day 7 vs 28 000 at day 5). Because DMOG has little effect on CFU-E proliferation (Figure 5A), the dramatic increase in CFU-E cells in response to DMOG must derive from increased self-renewal of earlier progenitors. Consistent with this notion, DMOG increases the maximum formation of late BFU-Es 1.8-fold in the absence of Dex (440 per 100 plated BFU-E cells at day 3 vs 240 in control cultures at day 2) and 3.8-fold in presence of Dex (3800 at day 5 vs 1000 at day 5).
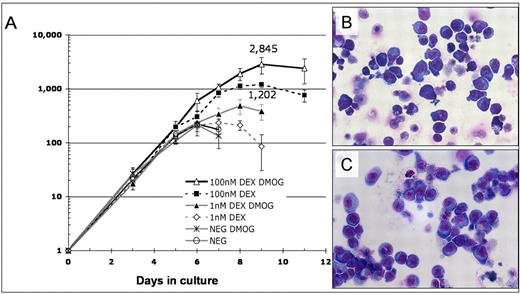
DMOG and Dex synergistically enhance proliferation of adult erythroid progenitors
To exclude the possibility that DMOG synergize with Dex exclusively in fetal liver we determined the effect of DMOG and Dex on proliferation of adult hematopoietic progenitors. To this end mouse bone marrow progenitor cells (Lin−, Kit+, Sca-1−) were cultured in SFELE medium with and without 1nM or 100nM Dex together or not with 333μM DMOG. Figure 6 shows that DMOG increased the output of erythroid cells 2.1-fold when added with 1nM Dex and 2.4-fold with 100nM Dex. More than 85% of cells formed after 11 days of culture with 100nM Dex and 333μM DMOG were erythroblasts, as shown by the Benzidine-Giemsa staining and FACS plots in supplemental Figure 6A,C.
Proliferation of erythroid progenitors in the Lin−, Sca-1−, Kit+ mouse bone marrow progenitor population is synergistically enhanced by DMOG and Dex. (A) Lin−, Kit+, Sca-1− mouse bone marrow cells were cultured in SFELE medium with no additions (control), 333μM DMOG, 1nM or 100nM Dex, 333μM DMOG plus 1nM Dex, or 333μM DMOG plus 100nM Dex. Total cell number was counted daily and normalized to the number of cells added to the culture. Error bars show 1 SD. (B) May-Grünewald-Giemsa staining of bone marrow Lin−, Kit+, Sca-1− cells after 11 days of culture in medium with 333μM DMOG plus 100nM Dex. The erythroid morphology of these cells is further confirmed by FACS and benzidine-Giemsa staining (supplemental Figure 6A,C). (C) Fetal liver BFU-E cells after 10 days of culture in the same medium as panel B. The erythroid morphology is further confirmed by FACS and benzidine-Giemsa (supplemental Figure 6B,D). By these assays 80%-95% of cells are erythroid.
Proliferation of erythroid progenitors in the Lin−, Sca-1−, Kit+ mouse bone marrow progenitor population is synergistically enhanced by DMOG and Dex. (A) Lin−, Kit+, Sca-1− mouse bone marrow cells were cultured in SFELE medium with no additions (control), 333μM DMOG, 1nM or 100nM Dex, 333μM DMOG plus 1nM Dex, or 333μM DMOG plus 100nM Dex. Total cell number was counted daily and normalized to the number of cells added to the culture. Error bars show 1 SD. (B) May-Grünewald-Giemsa staining of bone marrow Lin−, Kit+, Sca-1− cells after 11 days of culture in medium with 333μM DMOG plus 100nM Dex. The erythroid morphology of these cells is further confirmed by FACS and benzidine-Giemsa staining (supplemental Figure 6A,C). (C) Fetal liver BFU-E cells after 10 days of culture in the same medium as panel B. The erythroid morphology is further confirmed by FACS and benzidine-Giemsa (supplemental Figure 6B,D). By these assays 80%-95% of cells are erythroid.
Importantly, DMOG also enhances the effect of Dex in human erythroid progenitors, allowing formation of 10 times more erythroblasts than in cultures with Dex alone (supplemental Figure 7).
Discussion
Here, we first describe a method to obtain > 90% pure erythroid progenitor cells from mouse E14.5-E15.5 fetal liver by magnetic depletion (supplemental Table 1; supplemental text). We next isolate CFU-E and BFU-E progenitor cells from this population to a high degree of purity on the basis of the expression of Kit, the transferrin receptor CD71, and the heat-stable antigen CD24a (Figure 1; supplemental Table 2). The isolated BFU-E (CD24a/CD7110%low) and CFU-E (CD24a/CD7120%high) fractions contain < 10% contamination of other colony-forming cells and have a cloning efficiency of 40%-70% in methyl cellulose assays (supplemental Table 2). For the purpose of this study we needed very pure BFU-E and CFU-E cell populations and, therefore, did not mind that ∼ 70% of these progenitors were lost during sorting. Although there are several reports describing isolation of CFU-E19,20 and BFU-E11,12 cells, our method enables both simultaneous isolation of BFU-E and CFU-Es and purer cell populations.
We next used purified BFU-E and CFU-E cells to determine that the stimulatory effect of GCs is intrinsic to BFU-E cells, with only a minimal effect on Epo-dependent CFU-E cell proliferation (Figures 2 and 5). Early BFU-E cells are particularly responsive to GC stimulation and divide > 6 additional times in the presence of Dex (supplemental Figure 2C-D).
After establishing that BFU-Es are the principal GC target cells, we determined that 4 hours of treatment with 100nM Dex alters expression of ∼ 200 mRNAs > 2-fold (Figure 3), a time point showing maximum gene up-regulation in other cell types.21 In contrast, Dex did not significantly alter expression of any micro-RNAs in BFU-E cells (data not shown).
Because GCR dimerization3,4 and a functional AF2 transactivation domain2 are required to induce production of CFU-E progenitors in vitro and in vivo during SE, we focused our attention to the 83 up-regulated genes. Some of the Dex-induced genes are noteworthy because they inhibit hematopoietic progenitor differentiation, such as the SCF receptor protein tyrosine kinase Kit22 and the acute myelogenous leukemia–associated factor Trib2.23 We also found it interesting that the antiapoptotic protein Bcl2 is up-regulated. Bcl2 is down-regulated in a DBA cell line model,24,25 and it is possible that part of the therapeutic effect of prednisone treatment in DBA is through Bcl2 up-regulation.
Although we were mostly interested in studying up-regulated genes, we note that gene ontology analysis of the genes down-regulated by Dex as well as by DMOG shows a highly significant (P 1 × 10−9 to × 10−20) enrichment of ribosomal protein genes and genes involved in nucleolar processes (data not shown). Because ribosome biogenesis is regulated by mammalian target of rapamycin, the shutdown of ribosomal biogenesis could be secondary to the 3.5- to 10-fold up-regulation of Ddit4 (supplemental Table 3), which is a potent inhibitor of mammalian target of rapamycin signaling.26,27 Note that expression of the transcription factors friend of GATA 1 (globin transcription factor 1) and globin transcription factor 2 decrease on Dex stimulation (supplemental Table 4).
We did not attempt biologic evaluation of the individual GC target genes, but instead we turned to computational evaluation of the promoter regions to find alternative pathways to regulate the GC target genes as a group. Surprisingly, the Dex-induced genes in BFU-E cells were not enriched for GRE sites in their promoter regions but instead for HIF1 (P = 1 × 10−27) and other transcription factor binding sites (supplemental Figures 3-4). The reason our method failed to detect enrichment of GRE sites is probably explained by the fact that GCR regulates transcription by chromatin interaction outside of the promoter regions of the target genes.28 Remarkably, in A549 lung epithelial carcinoma cells 52% of Dex-induced genes and 92% of Dex-repressed genes show no GR binding within 10 kb of the transcription start site.21 Here, we further confirmed that GC-regulated genes do not contain proximal GRE motifs by analyzing genes up-regulated > 2-fold by Dex in different types of cells in 6 publications of others. Supplemental Table 7 shows that GRE motifs were not among the 10 most enriched motifs in promoter regions of the up-regulated genes in any of the cell types analyzed. Furthermore, HIF1α was only highly enriched in 2 of the 6 cell types, and in no other cell type than BFU-E cells was HIF1α the most significantly enriched transcription factor binding motif.
The fact that Dex induces expression of genes whose promoters are significantly enriched for HIF1α binding motifs suggests these pathways overlap in BFU-E cells, and that HIF1α activation would increase the expression of several of Dex-induced genes. Indeed, when we compared genes that change expression ± 50% after a 4-hour treatment of Dex versus treatment with the HIF1α activator DMOG, we found a significant overlap between the up-regulated (P = 1 × 10−26) and down-regulated (P = 4 × 10−24) genes (Figure 4). It is possible that HFI1α and GCR protein cross talk leads to reciprocal enhanced activity, which could explain some of the overlapping effects on gene expression.29,30 Alternatively, the 2 pathways could functionally converge in BFU-E cells and induce expression of genes such as Ddit4 that have both proximal HIF1α binding sites and more distal GREs, which would not be detected by our computational analyses.21
The significant Dex and DMOG overlaps on gene expression led us to investigate if DMOG could enhance or replace the effect of Dex on BFU-E proliferation. We found that, although adding DMOG alone had some intrinsic stimulatory biologic effect on BFU-E progenitors, it was not sufficient to replace the effect of Dex in stimulating BFU-E self-renewal or formation of CFU-Es or erythroblast progeny (Tables 1–2). In the presence of 1, 10, and 100nM Dex, however, DMOG enhanced the erythroblast production from both mouse fetal liver and bone marrow BFU-Es 7- to 12-fold. The experiments in Table 1 show that, during a given BFU-E cell division, the presence of DMOG and Dex increases the probability that a BFU-E will divide and form daughter BFU-E cells (“self-renew”) rather than form more mature CFU-E cells (“differentiate”). Thus, over time, the presence of DMOG and Dex supports formation of more CFU-E progeny than the addition of either alone and subsequently generation of more erythroblasts.
Although activation of either Hif1α or Hif2α could account for the DMOG-induced expression of genes with hypoxia response elements, mRNA sequencing shows that HIF1α is highly expressed in purified BFU-Es but that expression of HIF2α is undetectable (supplemental Table 4). This agrees with previous findings that HIF2α is mainly expressed in endothelial cells31 and suggests that HIF1α and not HIF2α intrinsically enhance BFU-E self-renewal. Although DMOG inhibits HIF prolyl hydroxylases as well as collagen prolyl 4-hydroxylases, the stimulatory effects on CFU-E production are most probably not related to increased collagen prolyl hydroxylation, in part because collagen prolyl 4-hydroxylase α subunits are expressed at low levels in BFU-Es (supplemental Table 4).
There are previous reports that support our findings that PHIs can be used to stimulate erythropoiesis, not only by up-regulating Epo expression but also through intrinsic erythroid progenitor stimulation. In colony-forming assays of spleen cells from phenylhydrazine-treated mice, the addition of SCF to the medium only increased the size of “stress BFU-E” colonies when performed in 20% oxygen. In contrast, when performed at 2% oxygen the addition of SCF increased both the size and number of BFU-Es.32 This suggests that hypoxia cooperates with SCF to promote BFU-E self-renewal and that PHIs possibly could do the same. Hypoxia (HIF activation) is further known to result in a higher proportion of fetal hemoglobin positive (F+) red blood cells,33,34 which in primates is considered a clinical sign of ongoing SE. Furthermore, monkeys receiving PHI treatment induce production of F+ cells to higher levels than can be explained by the level of Epo induction.35 Because PHIs and GCs have synergistic effects on BFU-E self-renewal and because both PHIs and GCs alone induce production of F+ cells,36,37 it will be interesting to evaluate if there is a synergistic effect of PHIs and GCs also on human fetal hemoglobin induction, possibly in combination with hydroxyurea or SCF.
A group of patients who directly could benefit from the CFU-E–promoting effect of PHIs are those with the red cell progenitor disorder DBA.38,39 Importantly, our dose-response curves with the use of Dex, which has a longer half-life and is a ≈30-fold more potent GCR agonist than cortisol,40 are readily converted to physiologic cortisol levels. Although daytime-free cortisol levels normally reach 14-15nM (corresponding to 0.43-0.45nM Dex), levels increase to 98nM (∼ 3.0nM Dex) after surgery41 and ≤ 120nM (∼ 3.6nM Dex) during septic shock.42 Our dose response curves (in the absence of DMOG) show that normal GC concentrations (0.45nM Dex) hardly increase CFU-E formation, whereas a clear effect is seen at GC levels detected during stress (> 1nM Dex) (Figure 5B-C). Because DMOG enhances the effect of 1nM Dex > 10-fold, it is therefore probable that. although DMOG has little effect without Dex in vitro, DMOG may enhance the effect of endogenous cortisol and promote CFU-E production even without additional GC treatment.
Although increased HIF activation in other tissues theoretically could lead to adverse effects, including tumorigenesis, the therapeutic potential of activating intrinsic HIF1α in BFU-Es represents a new concept for treatment of anemia. Our results suggest that PHIs stimulate erythropoiesis not only by enhancing Epo production from kidneys and liver43 but also by intrinsically stimulating BFU-E self-renewal. The stimulation of BFU-E self-renewal depends on SCF and low levels of GCs and allows over time increased numbers of divisions of individual BFU-E cells and thus increased numbers of CFU-E cells and, subsequently, additional erythroblasts to be formed from each BFU-E.
We conclude by proposing that PHIs should be evaluated not only as a substitute for recombinant Epo treatment but also for the potential to intrinsically induce BFU-E self-renewal in Epo-resistant anemias such as bone marrow failure syndromes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Evelyn Wang; Drs Colin Sieff, Anupama Narla, Gregory Hyde, Song Chou, Beiyan Zhou, Peng Ji, George Bell, David Bartel, Vijay Sankaran, and Shilpa Hattangadi of the Whitehead Institute; and Dr David Bryder of Lund University for advice in experiments. We thank the Daniella Maria Arturi Foundation for promoting discussions and collaborations in the field of Diamond-Blackfan anemia and bone marrow failure syndromes.
This work was supported in part by the National Institutes of Health (grant P01 HL 32262; H.F.L.), by a grant from the Diamond-Blackfan Anemia Foundation (J.F.), a fellowship from the Swedish Research Council (J.F.), and stipends from Maja och Hjalmar Leanders Stiftelse and The Sweden-America Foundation (J.F.).
National Institutes of Health
Authorship
Contribution: J.F. designed, performed, and interpreted all of the experiments and wrote and edited the paper; H.F.L. helped with data interpretation, experimental design, and writing the paper; V.R.E. assisted in obtaining BFU-E and CFU-E cells from mouse fetal liver; C.S. made the small RNA libraries; and S.G. assisted in bioinformatical analysis.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Harvey F. Lodish, Whitehead Institute for Biomedical Research, Nine Cambridge Center, Cambridge, MA 02142; e-mail: lodish@wi.mit.edu.