Abstract
Hoxb4 overexpression promotes dramatic expansion of bone marrow (BM) hematopoietic stem cells (HSCs) without leukemic transformation and induces development of definitive HSCs from early embryonic yolk sac and differentiating embryonic stem cells. Knockout studies of Hoxb4 showed little effect on hematopoiesis, but interpretation of these results is obscured by the lack of direct evidence that Hoxb4 is expressed in HSCs and possible compensatory effects of other (Hox) genes. To evaluate accurately the pattern of Hoxb4 expression and to gain a better understanding of the physiologic role of Hoxb4 in the hemato-poietic system, we generated a knock-in Hoxb4–yellow fluorescent protein (YFP) reporter mouse model. We show that BM Lin−Sca1+c-Kit+ cells express Hoxb4-YFP and demonstrate functionally in the long-term repopulation assay that definitive HSCs express Hoxb4. Similarly, aorta-gonad-mesonephrous–derived CD45+CD144+ cells, enriched for HSCs, express Hoxb4. Furthermore, yolk sac and placental HSC populations express Hoxb4. Unexpectedly, Hoxb4 expression in the fetal liver HSCs is lower than in the BM, reaching negligible levels in some HSCs, suggesting an insignificant role of Hoxb4 in expansion of fetal liver HSCs. Hoxb4 expression therefore would not appear to correlate with the cycling status of fetal liver HSCs, although highly proliferative HSCs from young BM show strong Hoxb4 expression.
Introduction
The homeobox genes (Hox) are pivotal for embryonic development. In bone marrow (BM)–derived hematopoietic cells expression of Hoxa and Hoxb gene families is largely restricted to the stem and precursor cell populations and down-regulated on lineage commitment.1,2 Aberrant Hox gene expression is common to many human acute myeloid leukemias and genes known to regulate Hox expression, including the caudal-type Cdx2, which has been shown to be dysregulated in 90% of these cases.3 In acute myeloid leukemia, Hox genes have been identified as sites of genomic instability participating in gene fusion, most commonly with the NUP98 gene transcriptional coactivator, producing potent NUP98-Hox oncogenes. Overexpression of various Hox genes in BM cells can lead to enhanced proliferation and sometimes expansion of hematopoietic stem cells (HSCs) in vivo and in in vitro culture systems. However, overexpression of most Hox genes leads to lymphoproliferative and myeloproliferative disorders and development of leukemias.4,5
Transplantation of BM cells overexpressing Hoxb4 or exposure to soluble cell-penetrating forms of HOXB4 protein promote enhanced reconstitution of all hematopoietic lineages in recipient animals.6-8 Notably, the HSC pool in the BM of irradiated recipients receiving Hoxb4-expanded HSCs returns to but does not exceed normal levels,9,10 even when HSCs are first expanded in vitro.10 Importantly, Hoxb4 overexpression does not lead to leukemia or myeloproliferative disease in murine models, indicating that Hoxb4-directed expansion of HSCs may be clinically applicable, although studies in larger animals have shown that HOXB4 overexpression can increase the risk of leukemogenesis.11 These effects are mediated through the interaction of HOXB4 with DNA and are lost if the DNA-binding domain is mutated.12 It was suggested that Hoxb4 may regulate the response of HSCs to extrinsic signals, by directly regulating the expression of genes involved in various pathways, including Wnt and Notch signaling implicated to be involved in HSC self-renewal.13
Definitive HSCs capable of long-term reconstitution of irradiated adult recipients are not detectable during normal development in the early yolk sac and are not produced by in vitro–differentiating embryonic stem (ES) cells.14,15 Remarkably, Hoxb4 overexpression in both early embryonic yolk sac and in vitro–differentiated ES cells allows the generation of definitive HSCs in culture, although with limited lymphoid differentiation capacity.16 Hematopoietic precursor cells with more balanced functional lymphoid production have been achieved with the use of Hoxb4-transduced ES cells in vitro.17 The level of HOXB4 can influence lineage differentiation as shown with the use of human cord blood CD34+ cells.18 Through cooperation with the Cdx4 homeobox gene, enhanced and balanced myelolymphoid engraftment can also be achieved in Hoxb4-overexpressing ES cells.19
Despite all expectations, Hoxb4 loss of function models have shown no profound defects in hematopoiesis. Hoxb4-null mice show a minor defect in HSC proliferation,20 and Hoxb3−/−/Hoxb4−/− compound knockouts have a reduced number of progenitors.21 Furthermore, deletion of Hoxb4 or most of the Hoxb family locus does not compromise HSC production and does not lead to myelolymphoid dysregulation.22 Many Hox family genes act in concert, and the potential for Hox gene compensation by other members of the Hox family complicates identification of their physiologic significance. Another complicating factor impeding the interpretation of knockout studies is that Hoxb4 expression in HSCs has been inferred from gene expression studies on the HSC-enriched populations,1,2 whereby HSCs constitute only a small fraction of cells, and not directly on HSCs.23
Here, we have targeted a fluorescent reporter to the Hoxb4 locus to accurately identify Hoxb4-expressing cells throughout development. We demonstrate that within the adult BM the Hoxb4 reporter exclusively marks the Lin−Sca1+c-Kit+ (LSK) population, including functionally defined self-renewing HSCs. During development definitive HSCs emerging in the aorta-gonad-mesonephrous (AGM) region24 also express Hoxb4, but on passing through the fetal liver (FL) HSCs can be identified within a broad range of Hoxb4 expression, including low/nonexpressing cells. Because Hoxb4 is up-regulated in highly proliferative HSCs in newborn animals, this indicates that Hoxb4 is expressed in HSCs independently of their cycling status.
Methods
Generation of Hoxb4–yellow fluorescent protein reporter mice
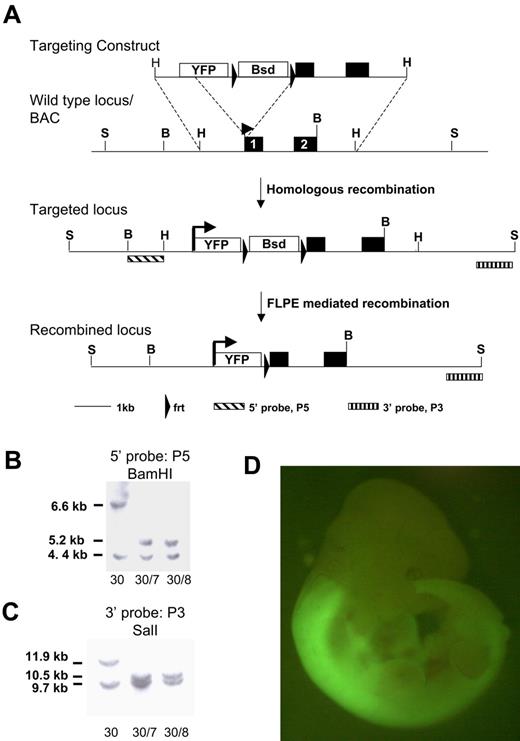
A bacterial artificial chromosome (BAC) clone harboring the Hoxb4 gene was isolated from a murine ES-129/Sv BAC library (CitbCJ7 library; Invitrogen) through polymerase chain reaction (PCR) screening with the use of primers 5′-CCATTGCCAGAGATTTACGG-3′ and 5′-TTCTCCAGCCAAGGTACCAG-3′. This clone was electroporated into the bacterial host DY380. The open reading frame of the improved yellow fluorescent protein (YFP), Venus, was cloned into pBluescript and an frt-flanked PGK/EM7-blasticidin selection cassette inserted downstream. Homology arms of 71 base pairs and 61 base pairs were inserted 5′ of Venus and 3′ of the selection cassette, respectively, to allow targeting to the start codon of Hoxb4 through recombineering. The targeting cassette was electroporated into the DY380/BAC clone, and a correctly targeted BAC was identified. BAC DNA was digested with HindIII and subcloned into pBluescript, using the EM7-blasticidn selection marker to recover the final gene targeting vector shown in Figure 1. ES cells were targeted, and blasticidin-resistant colonies were obtained. A single correctly targeted clone was identified by Southern blotting with use of a 5′ probe (P5) and 3′ probe (P3) on BamHI or SalI DNA digests, respectively. After transient transfection of FLPe recombinase, blasticidin-sensitive clones were harvested and further screened with the use of the above-mentioned restriction digests and probes.
Chimeric mice were generated by injecting the targeted ES cells into C57BL/6 blastocysts, and heterozygous germline-transmitting mice were produced.
Mice
129, C57BL/6, or C57BL/6 × 129 F1 mice were bred in animal facilities at the University of Edinburgh. Donor cells from CD45.2/2 mice or embryos were isolated and transplanted into irradiated C57BL/6, CD45.1/1, or C57BL/6 × 129 F1 CD45.1/2 adult recipients as described previously. Timed matings were scored day 0.5 for embryo stage on discovery of a vaginal plug. Animals were kept in compliance with Home Office regulations, and all animal experiments were approved by the University of Edinburgh Ethical Review Committee. Images were acquired and processed using a 20× objective on an Olympus IX51 microscope with a RETIGA 2000R camera and Volocity 4.2 software.
Cyclophosphamide was administered according to the protocol of Nygren and Bryder.25
Flow cytometry
Antibodies purchased from either eBioscience or PharMingen against the following markers were used: B220-biotin, CD3ϵ-phycoerythrin (PE)/biotin, CD4-PE/biotin, CD4-allophycocyanin (APC)–AF750, CD8a-biotin, CD8a-APC-AF750, CD19-PE cyanine dye 7, CD45-PE, CD45.2-Pacific Blue, CD48-PE, CD49-PE, CD45.2 peridinin chlorophyll protein cyanine dye 5.5, CD150-APC, c-Kit-APC, c-Kit-APC-eF780, GR1-biotin, Ly5.1-PE, Ly5.2–fluorescein isothiocyanate, MAC1-PE/biotin, MAC1-APC, NK1.1-PB, Sca1-PE, Sca1-Pacific Blue, and CD16/32 unconjugated. MoFlo (Dako-Cytomation) and FACSAria (Becton Dickinson) flow cytometers were used for cell sorting. Flow cytometric analysis was performed with a FACScalibur, FACSDiva, or LSRII/Fortessa machines (Becton Dickinson). Data analysis was performed with FlowJo software (TreeStar).
Quantitative reverse-transcription PCR
RNA was prepared from cells with the use of QIAGEN MicroRNA kits and was reverse transcribed with SuperScript III (Invtirogen). Quantitative PCR was performed on a Roche 480 Lightcycler with the Universal Probe Library detection system with the use of the primers/probes as described in supplemental Figure 7 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Long-term repopulation assay
Results
Generation of Hoxb4-YFP reporter mice
The homeobox genes are subject to complex regulation which needs to be considered in gene-targeting strategies.27 We made a knock-in of the enhanced YFP reporter gene, Venus, into the first exon of Hoxb4 to retain all intronic and untranslated region regulatory elements. Hoxb4 is subject to posttranslational regulation,27 and recently Prep1 was shown to modulate translation through binding the 3′ untranslated region28 ; our transgene is therefore designed to be subject to authentic transcriptional and translational control (Figure 1A). Correctly targeted 129Sv (CGR14) ES clones were confirmed by Southern blot analysis and transiently transfected with FLPe recombinase to remove the selection cassette (Figure 1B-C) and then used to generate chimeric animals by blastocyst injection. As shown in Figure 1D, expression of Hoxb4/YFP in the embryo shows the characteristic pattern of expression described previously by anti-HOXB4 immunostaining.27,29 Specifically, strong fluorescence is observed in the neural tube posterior to the rhombomere 6/7 boundary. Both heterozygous and homozygous mice are healthy. In this study we have used heterozygous mice for analysis, thus retaining one functional Hoxb4 allele.
Targeting strategy for introducing the YFP reporter at the Hoxb4 locus. (A) The YFP variant sequence, Venus, and an frt-flanked blasticidin selection cassette was inserted at the start codon of Hoxb4 in a BAC clone. A 5.2-kb HindIII fragment was subcloned from the BAC and used for gene targeting in ES cells. Schematic diagrams of the Hoxb4 locus and of correctly targeted alleles before and after blasticidin selection cassette removal with the use of FLPe-mediated recombination. (B) Southern blot of BamHI-digested DNA probed with 5′ probe (P5). (C) Southern blot of ScaI-digested DNA probed with 3′ probe (P3). Clone 30 is the primary targeted clone. Clones 30/7 and 30/8 were recovered after transient removal of the selection cassette from clone 30. The lower band in both blots represents the wild-type untargeted allele (BamHI, 4.4 kb; ScaI, 9.7 kb). (D) Hoxb4YFP/+ embryo. Note bright YFP expression is in the anterior neural tube; the sharp boundary between rhombomeres 6 and 7 is consistent with previous reports. YFP in open box marks the Venus open reading frame. Bsd in open box marks the PGK/EM7 dual eukaryotic/prokaryotic promoter driven blasticidin selection cassette. B indicates BamHI; H, HindIII; S, ScaI.
Targeting strategy for introducing the YFP reporter at the Hoxb4 locus. (A) The YFP variant sequence, Venus, and an frt-flanked blasticidin selection cassette was inserted at the start codon of Hoxb4 in a BAC clone. A 5.2-kb HindIII fragment was subcloned from the BAC and used for gene targeting in ES cells. Schematic diagrams of the Hoxb4 locus and of correctly targeted alleles before and after blasticidin selection cassette removal with the use of FLPe-mediated recombination. (B) Southern blot of BamHI-digested DNA probed with 5′ probe (P5). (C) Southern blot of ScaI-digested DNA probed with 3′ probe (P3). Clone 30 is the primary targeted clone. Clones 30/7 and 30/8 were recovered after transient removal of the selection cassette from clone 30. The lower band in both blots represents the wild-type untargeted allele (BamHI, 4.4 kb; ScaI, 9.7 kb). (D) Hoxb4YFP/+ embryo. Note bright YFP expression is in the anterior neural tube; the sharp boundary between rhombomeres 6 and 7 is consistent with previous reports. YFP in open box marks the Venus open reading frame. Bsd in open box marks the PGK/EM7 dual eukaryotic/prokaryotic promoter driven blasticidin selection cassette. B indicates BamHI; H, HindIII; S, ScaI.
YFP marks Hoxb4 expression in adult BM HSCs
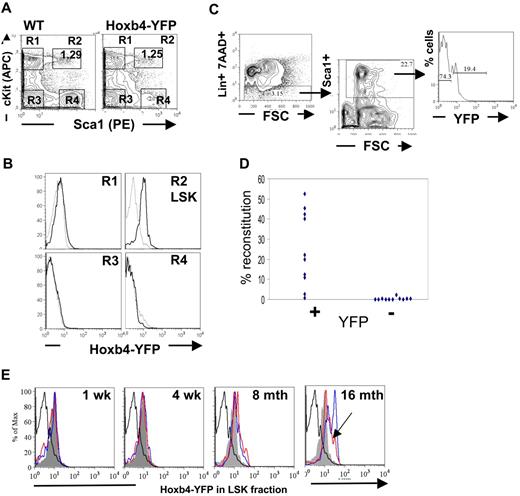
Reverse transcription (RT)–PCR analysis showed that expression of homeobox genes in BM is largely restricted to the hematopoietic stem and early progenitor populations and are generally down-regulated on differentiation.1,2 Analysis of the major hematopoietic cell types within the peripheral blood, BM, and thymic compartments of adult mice by flow cytometry was performed to determine the extent of Hoxb4 expression in Hoxb4YFP/+ mice (supplemental Figures 1-2). Initial assessment of differentiated BM cells with the use of B-lineage markers, B220, CD19, CD43; immunoglobulin D; myeloid markers GR1 and MAC1; and the erythroid lineage markers CD71 and Ter119; or the T-cell markers CD4 and CD8 in thymic lymphocytes showed no significant Hoxb4/YFP expression, which is consistent with the expected down-regulation of Hoxb4 in all mature blood cell lineages (supplemental Figures 1-2; data not shown). In contrast, the BM LSK fraction enriched for HSCs and primitive multipotent progenitors expressed Hoxb4/YFP with almost the entire population showing an increase in fluorescence intensity (Figure 2A-B). Furthermore, using the HSC marker CD150,30 analysis of BM LSK CD48− fractions showed that the Hoxb4/YFP expression could be seen in both CD150− and CD150+ fractions, although the brightest Hoxb4/YFP-expressing cells were observed in the CD150+ population (supplemental Figure 3).
Hoxb4 is expressed in adult BM HSCs. Flow cytometric analysis of BM cells from WT and Hoxb4YFP/+ animals. (A) Gating of the LSK population is shown by 4 gates representing the major populations, marked R1 to R4. (B) Comparison of gated populations R1 to R4 indicated in panel A, showing YFP expression. In the histograms dashed lines indicate WT and solid lines indicate Hoxb4YFP/+ cells. (C) The FACS strategy is outlined, indicating Lin−, Sca1+, and YFP sorting gates. (D) Lin−Sca1+Hoxb4/YFP+ cells in contrast to Lin−Sca1+Hoxb4/YFP− cells effectively reconstitute irradiated recipients. Each symbol indicates one recipient animal and reflects the level of engraftment of donor-derived cells (combined data from 2 separate experiments). (E) BM extracted from WT and Hoxb4YFP/+ mice at 1 week, 4 weeks, 8 months, and 16 months of age were stained with the LSK markers (lacking Mac1 for mice 1 and 4 weeks old). LSK cells were gated (as in panel A) and YFP fluorescence in experimental Hoxb4YFP/+ mice (2 animals at each time point, red- and blue-outlined histograms) compared with LSK cells from WT (thick black-lined histograms) and 8-week-old Hoxb4YFP/+ mice (solid gray histogram). Note that, although the peak of Hoxb4/YFP fluorescence in the red histogram is shifted less than in the blue histogram at 16 months and is similar to that in 8-month-old mice, the entire peak volume is shifted significantly higher (arrowhead).
Hoxb4 is expressed in adult BM HSCs. Flow cytometric analysis of BM cells from WT and Hoxb4YFP/+ animals. (A) Gating of the LSK population is shown by 4 gates representing the major populations, marked R1 to R4. (B) Comparison of gated populations R1 to R4 indicated in panel A, showing YFP expression. In the histograms dashed lines indicate WT and solid lines indicate Hoxb4YFP/+ cells. (C) The FACS strategy is outlined, indicating Lin−, Sca1+, and YFP sorting gates. (D) Lin−Sca1+Hoxb4/YFP+ cells in contrast to Lin−Sca1+Hoxb4/YFP− cells effectively reconstitute irradiated recipients. Each symbol indicates one recipient animal and reflects the level of engraftment of donor-derived cells (combined data from 2 separate experiments). (E) BM extracted from WT and Hoxb4YFP/+ mice at 1 week, 4 weeks, 8 months, and 16 months of age were stained with the LSK markers (lacking Mac1 for mice 1 and 4 weeks old). LSK cells were gated (as in panel A) and YFP fluorescence in experimental Hoxb4YFP/+ mice (2 animals at each time point, red- and blue-outlined histograms) compared with LSK cells from WT (thick black-lined histograms) and 8-week-old Hoxb4YFP/+ mice (solid gray histogram). Note that, although the peak of Hoxb4/YFP fluorescence in the red histogram is shifted less than in the blue histogram at 16 months and is similar to that in 8-month-old mice, the entire peak volume is shifted significantly higher (arrowhead).
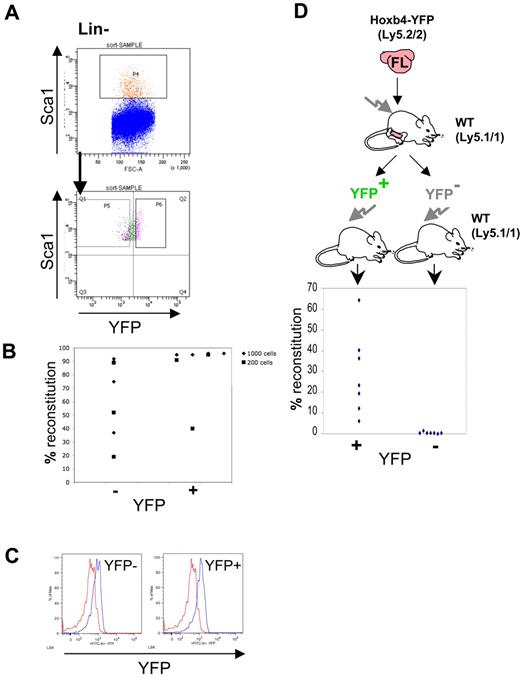
However, because long-term repopulating HSCs constitute only a small fraction of LSK cells, we needed to determine functionally if HSCs express Hoxb4. To this end, Lin−Sca1+ cells purified by fluorescence-activated cell sorting (FACS) from adult BM were further sorted on the basis of YFP expression and transplanted into irradiated CD45.1/1 recipients (Figure 2C). Each recipient mouse received transplants of either 130 Lin−Sca1+Hoxb4+ cells or 850 Lin−Sca1+Hoxb4− cells. The numbers of transplanted cells per recipient were in line with proportional representations of these fractions in BM, and ≤ 10 HSCs were transplanted to minimize the effect of contamination from cell sorting.26 We found that Lin−Sca1+Hoxb4+ cells repopulated all 10 recipients (8 with > 5% and 2 with < 5% donor contribution), whereas Lin−Sca1+Hoxb4− cells reconstituted only 1 animal of 11 recipients at low level (< 5% donor contribution) (Figure 2D) at 16 weeks after transplantation. In all cases short-term reconstitution after 6 weeks was stable and was followed by long-term, multilineage reconstitution at 16 weeks (supplemental Figure 4). Thus, definitive HSCs largely reside within a Hoxb4/YFP-expressing fraction of adult BM cells.
Furthermore, administration of cyclophosphamide enabled us to determine expression of Hoxb4/YFP in the LSK fraction when in cycle because this treatment does not alter HSC phenotype unlike fluorouracil.25 Treated animals expressed Hoxb4/YFP at close, although slightly reduced, levels to those seen in control animals (supplemental Figure 5).
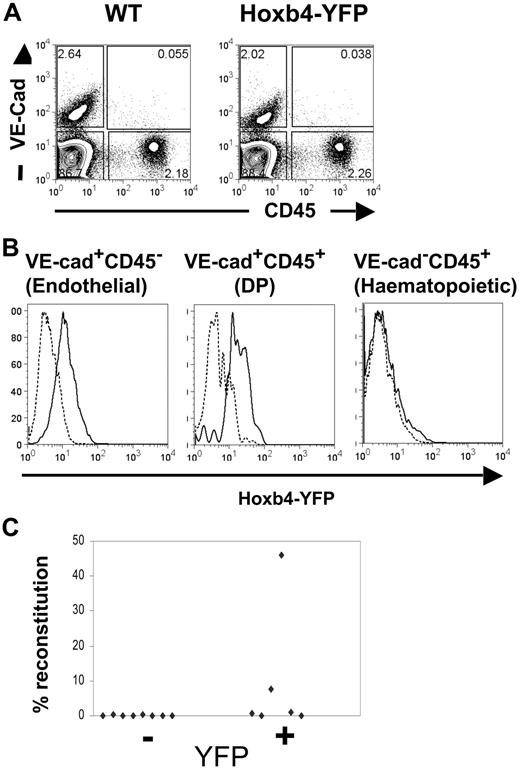
Hoxb4 is expressed in embryonic day 11.5 AGM-definitive HSCs
We and others have shown previously that the first HSCs emerging in the AGM region coexpress hematopoietic and endothelial markers.24,31,32 Specifically, these reside within the double-positive (DP) vascular endothelial (VE)–cad+CD45+ population.24 Embyronic day 11.5 (E11.5) AGM regions were isolated from wild-type and Hoxb4YFP/+ mice; cell suspensions were prepared and stained for VE-cadherin and CD45 as described previously.24 We found that the level of Hoxb4/YFP expression in the DP fraction is similar to that detected in the endothelial (VE-cad+CD45−) fraction (Figure 3A-B), which again underscores the putative lineage relationship between these 2 cell populations.24,32-34 Virtually all the DP population displayed a significant shift in YFP fluorescence (in contrast, the hematopoietic VE-cad−CD45+ population in the AGM region lacked any significant expression of Hoxb4/YFP, although expression may be restricted to a small subset present in the leading foot of the histogram). To confirm this observation functionally, Hoxb4/YFP+ and Hoxb4/YFP− cells from E11.5 AGM were flow sorted and transplanted into irradiated recipients in 2 separate experiments (1.4-2.0 embryo equivalents per recipient). We found that 4 of 8 recipients that received transplants of Hoxb4/YFP+ cells were repopulated (2 at high level and 2 at low level), whereas none of the 8 mice that received a transplant with Hoxb4/YFP− cells showed donor-derived hematopoietic engraftment (Figure 3C). Thus, the first definitive HSCs emerging in the AGM region express Hoxb4.
Hoxb4 is expressed in CD45+CD144+ cells and HSCs from the E11.5 AGM. (A) AGM regions from WT and Hoxb4YFP/+ embryos were analyzed by flow cytometry with the use of CD45 and VE-cadherin markers. (B) Histograms comparing YFP fluorescence are shown for the endothelial, hematopoietic, and double-positive (DP) cell populations. In the histograms dashed lines indicate WT and solid lines indicate Hoxb4YFP/+ cells. (C) Cells from E11.5 AGM region were sorted on the basis of YFP expression and injected into irradiated recipients. Donor contribution to the peripheral blood is displayed (data obtained from 2 independent experiments).
Hoxb4 is expressed in CD45+CD144+ cells and HSCs from the E11.5 AGM. (A) AGM regions from WT and Hoxb4YFP/+ embryos were analyzed by flow cytometry with the use of CD45 and VE-cadherin markers. (B) Histograms comparing YFP fluorescence are shown for the endothelial, hematopoietic, and double-positive (DP) cell populations. In the histograms dashed lines indicate WT and solid lines indicate Hoxb4YFP/+ cells. (C) Cells from E11.5 AGM region were sorted on the basis of YFP expression and injected into irradiated recipients. Donor contribution to the peripheral blood is displayed (data obtained from 2 independent experiments).
In addition, we analyzed E11.5 yolk sacs and E11.5 and E12.5 placenta. In the yolk sac Hoxb4/YFP was observed in DP (VE-cad+CD45+) cells, phenotypically similar to those which are enriched for HSCs in the AGM region, as well as in endothelial (VE-cad+CD45−) cells.24 For the placenta, cell suspensions were stained with CD34 and c-Kit. Here, the CD34+c-Kit+ population, previously shown to harbor HSCs, was seen to express Hoxb4/YFP at both stages (supplemental Figure 6).
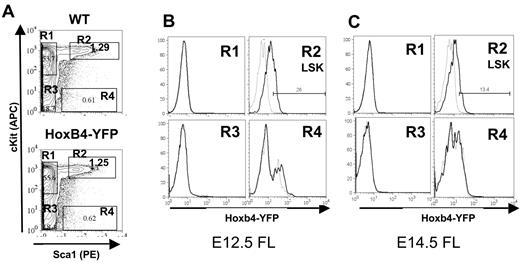
Hoxb4 is broadly expressed in FL-derived definitive HSCs
HSCs in the FL reside within the Lin−c-Kit+Sca1+Mac1+ population.35 Therefore, for characterization of FL HSCs the anti-Mac1 antibody was omitted from the Lin cocktail. E12.5 and E14.5 FLs from Hoxb4YFP/+ embryos were analyzed for Hoxb4/YFP expression by FACS. We found that Hoxb4/YFP expression is slightly reduced in the E12.5 FL LSK fraction than in the HSC (VE-cad+CD45+) fraction in the AGM region (Figure 4A-B) and is further down-regulated in the E14.5 FL LSK fraction (Figure 4A-C). Similar low Hoxb4/YFP levels were seen in E16.5 FL (results not shown). This significant progressive down-regulation of Hoxb4/YFP during FL hematopoiesis was consistently observed across 3 independent experiments (compare Figure 4B with 4C). Further analysis was performed on E14.5 FL with the use of CD15036 along with LSK+CD48− staining. Both CD150+ and CD150− fractions showed similar expression patterns of Hoxb4/YFP. However, in agreement with our earlier analysis expression of Hoxb4/YFP was lower in the CD150+ fraction in the FL compared with adult BM (supplemental Figure 3).
Hoxb4 is down-regulated in the fetal liver. Flow cytometric analysis of FL cells from WT and Hoxb4YFP/+ embryos at E12.5 and E14.5. (A) Gating of the LSK population is shown by 4 gates representing the major populations, marked R1 to R4. (B) Comparison of gated populations R1 to R4 indicated in panel A showing YFP expression at E12.5. In the histograms dashed lines indicate WT and solid lines indicate Hoxb4YFP/+ cells. (C) Labeling is identical to panel B but at E14.5. The fold change in mean fluorescence intensity between WT and Hoxb4YFP/+ at E12.5 is 2.39 (SD = 0.35) and at E14.5 is 1.48 (SD = 0.087); P = .02. Data are representative of 3 independent experiments.
Hoxb4 is down-regulated in the fetal liver. Flow cytometric analysis of FL cells from WT and Hoxb4YFP/+ embryos at E12.5 and E14.5. (A) Gating of the LSK population is shown by 4 gates representing the major populations, marked R1 to R4. (B) Comparison of gated populations R1 to R4 indicated in panel A showing YFP expression at E12.5. In the histograms dashed lines indicate WT and solid lines indicate Hoxb4YFP/+ cells. (C) Labeling is identical to panel B but at E14.5. The fold change in mean fluorescence intensity between WT and Hoxb4YFP/+ at E12.5 is 2.39 (SD = 0.35) and at E14.5 is 1.48 (SD = 0.087); P = .02. Data are representative of 3 independent experiments.
To determine whether other Hox group 4 paralogs could compensate for the reduced Hoxb4 levels in the FL, quantitative RT-PCR analysis was performed on both E14.5 FL and adult BM LSK cells. We observed no signs of up-regulation of other Hoxb4 paralogs in the FL, and in both tissues Hoxb4 was seen to have the highest relative expression level (supplemental Figure 7).
We then tested functionally if FL-derived HSCs express Hoxb4. To this end, Lin−Sca1+ cells from E14.5 Hoxb4YFP/+ FLs (omitting anti-Mac1 and anti-CD4 stains from the Lin cocktail) were sorted on the basis of YFP expression (Figure 5A). For both Lin−Sca1+YFP+ and Lin−Sca1+YFP− fractions either 200 or 1000 sorted cells were transplanted into irradiated recipients in 2 separate experiments (sort purity is shown in supplemental Figure 8).26 After 6 and 16 weeks the contribution to the peripheral blood by donor HSCs was determined by flow cytometry (Figure 5B). We found engraftment within all mice that received a transplant, although reconstitution levels were generally lower with Hoxb4/YFP− phenotype cells whereby only 2 mice achieved 90% reconstitution, whereas most mice (6 of 7) receiving the Hoxb4/YFP+ fraction showed high level (≥ 90%) reconstitution in all but one case (40%). In all cases engraftment was multilineage with no lymphoid/ myeloid bias observed (supplemental Figure 4). Thus, in the FL, transplantable HSCs express Hoxb4/YFP over a wide range.
Hoxb4 is broadly expressed in embryonic HSCs from the E14.5 fetal liver. (A) FL cells were gated using Lin− and Sca1+ markers and sorted on the basis of YFP expression. Recovered cells were tested in long-term reconstitution experiments (200 or 1000 cells injected). (B) Donor contribution to the peripheral blood is displayed from 2 independent sorting experiments. (C) Assessment of Hoxb4/YFP expression in the donor-derived CD45.2+ LSK fraction from recipient CD45.1 mice reconstituted with sorted FL HSCs 16 weeks after transplantation. (D) Irradiated recipients were injected with one E14.5 FL to obtain > 90% peripheral blood reconstitution. BM from reconstituted mice was sorted as in Figure 2C into Lin−Sca1+Hoxb4/YFP+ and Lin−Sca1+ Hoxb4/YFP− populations and injected into irradiated recipients. Contribution to the peripheral blood was determined by flow cytometry. HSCs were only observed in the Hoxb4/YFP-expressing cell fraction. Red histo-grams represent control BM LSK cells, and blue histograms show LSK CD45.2 cells in BM from Hoxb4/YFP-reconstituted CD45.1 recipients. Result from one experiment.
Hoxb4 is broadly expressed in embryonic HSCs from the E14.5 fetal liver. (A) FL cells were gated using Lin− and Sca1+ markers and sorted on the basis of YFP expression. Recovered cells were tested in long-term reconstitution experiments (200 or 1000 cells injected). (B) Donor contribution to the peripheral blood is displayed from 2 independent sorting experiments. (C) Assessment of Hoxb4/YFP expression in the donor-derived CD45.2+ LSK fraction from recipient CD45.1 mice reconstituted with sorted FL HSCs 16 weeks after transplantation. (D) Irradiated recipients were injected with one E14.5 FL to obtain > 90% peripheral blood reconstitution. BM from reconstituted mice was sorted as in Figure 2C into Lin−Sca1+Hoxb4/YFP+ and Lin−Sca1+ Hoxb4/YFP− populations and injected into irradiated recipients. Contribution to the peripheral blood was determined by flow cytometry. HSCs were only observed in the Hoxb4/YFP-expressing cell fraction. Red histo-grams represent control BM LSK cells, and blue histograms show LSK CD45.2 cells in BM from Hoxb4/YFP-reconstituted CD45.1 recipients. Result from one experiment.
This observation is further supported by published microarray data (GSE1559)37 in which there is a 1.8-fold reduction in Hoxb4 expression in the HSC-enriched fractions of FL compared with adult BM.
Hoxb4 is up-regulated in FL HSCs engrafted in the adult BM environment
We next wanted to determine whether donor-derived HSCs from the FL sorting experiments adopted an adult phenotype for Hoxb4 expression in the recipient BM (CD45.1), so we performed FACS analysis to determine YFP expression in the CD45.2+ LSK fraction. In both experiments, donor-derived LSK cells showed Hoxb4/YFP expression similar to that seen in adult BM regardless of their initial YFP expression level (Figure 5C). Our data suggest that during expansion in the FL, HSCs express Hoxb4 over a broad range, although one which does not exceed that seen in adult BM, and on BM colonization up-regulate Hoxb4. Furthermore, analysis of CD45.2+LSKFlt3−CD150+ cells from recipients reconstituted with both YFP− and YFP+ FL HSCs shows that either can sustain LT HSCs at 4 months after transplantation (supplemental Figure 9).
To determine functionally that all FL HSCs up-regulate Hoxb4 expression in BM after circumvention of the embryonic maturation process, whole E14.5 FL was directly transplanted into irradiated animals (∼ 500 HSCs26 ). After 3 months the LSK fraction from BM of highly repopulated animals (≥ 90% contribution to peripheral blood) had up-regulated Hoxb4/YFP to levels identical to those seen in the BM of control Hoxb4YFP/+ animals (not shown). These reconstituted animals were sorted by FACS on the basis of YFP expression in the Lin−Sca1+ fraction as described earlier, and transplanted into secondary recipients (Figure 5D). We found that, although transplantation of Lin−Sca1+HoxB4− cells resulted in none of 7 animals being reconstituted, Lin−Sca1+Hoxb4+ cells reconstituted 6 of 7 recipient animals (> 5% donor contribution) (Figure 5D). This result indicates that up-regulation of Hoxb4 expression occurs in FL-derived definitive HSCs within the adult BM environment and does not require the full maturation period in the FL, also confirming the earlier observation of Hoxb4 expression in BM HSCs.
Progressive up-regulation of Hoxb4 in BM HSCs with age
Recent work has indicated that there is a developmental switch between the fetal to adult-type HSC in the BM within 3-4 weeks after birth.38 Therefore, we decided to test the status of Hoxb4/YFP expression in the LSK fraction during this early period. To this end we isolated BM from mice at 1 and 4 weeks after birth and performed FACS analysis with LSK markers (omitting the anti-Mac1 stain) (Figure 2E). We found that in all cases Hoxb4/YFP expression was greater than that observed in the FL (compare with Figure 3) and is at a level seen in the BM LSK population of 8-week-old mice.
To test whether the level of Hoxb4 expression is stable throughout life, we analyzed expression of Hoxb4 in mice of 8 and 16 months old and found progressively increasing levels of Hoxb4/YFP expression in their BM LSK populations (Figure 2E).
Discussion
Hoxb4 has been shown to promote the expansion of adult HSCs in vitro and in vivo,6,9,10 as well as to promote the emergence of HSCs from ES and yolk sac cells,16 indicating a potentially significant role in both adult and embryonic HSCs. Surprisingly, given the results obtained from overexpression experiments, Hoxb4 knockout studies showed little effect on hematopoiesis.20,21 This may be explained by possible compensation effects exerted by other members of the large Hox gene family which are known to be involved in promoting proliferation of hematopoietic cells. In addition, although previous studies that used RT-PCR analysis indicated that Hoxb4 is expressed predominantly in the LSK fraction,1 to our knowledge, no direct evidence has shown that Hoxb4 is actually expressed in HSCs, which constitute only ∼ 3% of the LSK fraction.23 In the absence of such clear evidence, the interpretation of knockout experiments is ambiguous. Here, we have produced a Hoxb4 knock-in a YFP reporter mouse model with minimal disturbance of the locus to preserve spatiotemporal regulation as much as possible. This model thus allows us to directly question HSC activity in the mouse by facilitating purification of Hoxb4-expressing cells.
Using this reporter system we have demonstrated that the BM LSK population is an exclusive fraction within the adult hematopoietic system that expresses Hoxb4. Furthermore, transplantation of subfractionated BM showed that definitive HSC activity was confined to Hoxb4/YFP-positive cells, supporting the initial conclusion that Hoxb4 is intrinsically expressed in HSCs. More stringent phenotypic analysis with the LSK CD48−CD150+ markers exhibited strong Hoxb4/YFP expression. CD150− cells have been shown to harbor HSCs,39,40 and our data indicate that CD150− cells within the LSK CD48− fraction express Hoxb4, although at lower levels than the CD150+ fraction.
We then focused on embryonic stages and found that the VE-cadherin+CD45+ fraction enriched for the first HSCs emerging in the AGM region24,32 also express Hoxb4. Furthermore, functional analysis showed that long-term reconstitution activity resides within the CD45+VE-cadherin+Hoxb4+ fraction. Of note, significant Hoxb4 expression observed in endothelial and HSCs, but not the CD45+ population, once again points to a possible common origin for some endothelial cells and HSCs.33,34 Extending the analysis to the embryonic yolk sac and placenta, we found expression of Hoxb4 in cell populations known to harbor HSCs. However, Hoxb4 expression in the yolk sac itself is not sufficient to ensure efficient expansion of HSCs in physiologic conditions, because this organ shows limited capacity to generate HSCs.
However, on colonization of the FL, Hoxb4 expression in the HSC-enriched LSKMac1+ fraction is broadened but still lower than that seen in adult BM. Furthermore, long-term transplantation experiments showed that HSCs in the E14.5 FL are found in both Hoxb4 negative/low and positive fractions. We find down-regulation of Hoxb4 in some HSCs in the FL unexpected, given its potency in HSC expansion seen in overexpression studies. In support of this result, not all FL LSK CD48−CD150+ cells express Hoxb4. Taken together, the above data indicate that, although Hoxb4 may contribute to the initiation and primary expansion of HSCs in the AGM region, it is not absolutely required for further expansion of HSCs in the FL. Insulin-like growth factor 2 and angiopoietin-like molecules identified in FL have been shown to expand HSCs ex vivo, suggesting an important role in vivo.41,42 Importantly, mice reconstituted with Hoxb4-negative HSCs up-regulate Hoxb4 once resident in the BM, indicating that Hoxb4 may play a more important role in this location. Our data have not shown any functional differences between Hoxb4low/neg and Hoxb4+ HSCs; however, a difference in self-renewal capacity between the 2 populations has not been ruled out in serial transplantation experiments. Both showed similar lymphoid and myeloid contribution. However, more detailed analysis would be required to show subtle differences in potential.
Consistent with rapid expansion, most HSCs in the FL are in cycle35 and become quiescent in the BM at ∼ 3-4 weeks after birth on gaining an adult phenotype and largely remain quiescent during steady-state adult hematopoiesis.38,43 We initially thought that Hoxb4 down-regulation may be associated with high proliferative activity of HSCs, but we found strong up-regulation of Hoxb4 already in highly proliferative BM HSCs in 1-week-old mice. Furthermore, induction of proliferation via chemotherapeutic stress did not reduce Hoxb4 expression within adult BM LSK cells to levels observed in FL LSK cells. Thus, in contrast to other genes such as MAC1, AA4.1, and Sox17 that are expressed in fetal-type HSCs and down-regulated only at ∼ 3-4 weeks after birth,44 Hoxb4 expression could be regulated directly by the BM microenvironment. If so, the previously proposed role for Hoxb4 in regulating HSC residency in the BM niche45 may occur through regulating c-myc expression.46
We then noticed that in aging mice Hoxb4 is expressed at higher levels than in younger mice. This finding correlates with 2 key effects described in Hoxb4 overexpression studies: HSC expansion and an increased myeloid-to-lymphoid lineage ratio.16,47 Indeed, both an increased HSC pool and a myeloid differentiation bias have been described in aging animals.16,47-49 It would be interesting to investigate if there is any relationship between Hoxb4 and the tumor suppressor gene p16INK4a in influencing HSC function, given their increased expression with age. Although Hoxb4 can promote self-renewal decisions in HSCs, it does not promote sustained self-renewal over an organism's lifespan, and sustained self-renewal is maintained by other genes, such as Bmi1. Furthermore, these observations warrant further investigation, given the interaction between Bmi1 and the Ink4a locus.50
To summarize, we have generated a novel Hoxb4-YFP reporter mouse model that enabled us to directly investigate Hoxb4 expression in HSCs in embryonic and adult tissues. It is especially important for the analysis of the physiologic role of Hoxb4 because knockout studies are potentially confounded by paralog redundancy within the Hox clusters. In addition, this reporter model has, for the first time, allowed direct functional testing of Hoxb4-expressing cells and has shown that, although Hoxb4 is expressed in the first emerging definitive HSCs in the AGM region, it is not required in HSCs undergoing expansion in the FL. In contrast, BM HSCs intrinsically express Hoxb4 even at early stages after birth when HSCs still retain fetal characteristics.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Jan Vrana, Andrew Sanderson, Simon Monard, and Helen Ferry for assistance with cell sorting. We are grateful to John Verth, Yvonne Gibson and Carol Manson for animal management and irradiation. We thank Simon Tomlinson for analyzing the microarray data.
This work was supported by Leukemia Research, the MRC, and the EU FPVI integrated project EuroStemCell.
Authorship
Contribution: D.H. designed and performed the experiments and wrote the article; R.G., S.L., and N.B.-V. performed FACS sorting and transplantation experiments; J.U. performed ES cell culture and chimeric mouse production; S.E.W.J. provided FACS sorting expertise and advice; and A.M. designed the study and wrote the article.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alexander Medvinsky, Ontogeny of Haematopoietic Stem Cells Group, MRC/JDRF Centre in Stem Cell Biology, Institute for Stem Cell Research, University of Edinburgh, King's Buildings, West Mains Rd, Edinburgh, EH9 3JQ, United Kingdom; e-mail: a.medvinsky@ed.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal