Abstract
Activating mutations in codon D816 of the tyrosine kinase receptor, KIT, are found in the majority of patients with systemic mastocytosis. We found that the transcription factor, microphthalmia-associated transcription factor (MITF), is highly expressed in bone marrow biopsies from 9 of 10 patients with systemic mastocytosis and activating c-KIT mutations. In primary and transformed mast cells, we show that KIT signaling markedly up-regulates MITF protein. We demonstrate that MITF is required for the proliferative phenotype by inhibiting colony-forming units with sh-RNA knockdown of MITF. Furthermore, constitutively active KIT does not restore growth of primary MITF-deficient mast cells. MITF mRNA levels do not change significantly with KIT signaling, suggesting posttranscriptional regulation. An array screen from mast cells identified candidate miRNAs regulated by KIT signaling. We found that miR-539 and miR-381 are down-regulated by KIT signaling and they repressed MITF expression through conserved miRNA binding sites in the MITF 3′-untranslated region. Forced expression of these miRNAs suppressed MITF protein and inhibited colony-forming capacity of mastocytosis cell lines. This work demonstrates a novel regulatory pathway between 2 critical mast cell factors, KIT and MITF, mediated by miRNAs; dysregulation of this pathway may contribute to abnormal mast cell proliferation and malignant mast cell diseases.
Introduction
KIT is a member of the type III receptor tyrosine kinase family encoded by the proto-oncogene, c-kit. Its ligand, stem cell factor (SCF), binds to KIT, resulting in receptor dimerization and autophosphorylation.1 KIT activation leads to a variety of cellular responses, including mast cell proliferation, survival, differentiation, mediator release, adhesion, and chemotaxis.2 Activating mutations within the kinase domain or internal juxtamembrane deletions of of KIT have been identified in patients with systemic mastocytosis as well as several hematologic malignancies.3
Systemic mastocytosis is characterized by an abnormal accumulation of mast cells in various organs, most commonly the bone marrow. In more than 80% of patients with systemic mastocytosis, a point mutation that results in a substitution of valine for aspartic acid in codon 816 (D816V) is demonstrated.4 A functionally identical mutation (D814) is found in transformed mast cell lines from rodents.5,6 This genetic change results in constitutive activation of KIT signaling and plays a major role in the proliferative phenotype. Although tyrosine kinase inhibitors that target the kinase domain of KIT are clinically available, such as imatinib, the D816V mutation is relatively resistant.7 Furthermore, the transcriptional events downstream of KIT signaling that participate in establishment of the transformed phenotype remain to be elucidated.
The microphthalmia-associated transcription factor (MITF) is a basic helix-loop helix leucine zipper transcription factor that is critical for the development of mast cells, as well as melanocytes.8 It is expressed in normal human skin mast cells, as well as human mastocytosis; however, it is not highly expressed in human monocytes or granulocytes.9,10 Mice with a deficiency of MITF display an absence of mast cells, and MITF-deficient cultured mast cells appear immature and do not express genes critical for mast cell function.11 The in vivo and in vitro mast cell and melanocyte phenotype of Mitf mutant mice is strikingly similar to that of SCF or KIT-deficient mice.12,13 In melanocytes, 2 kinases, MAP kinase and p90 rsk, are activated downstream of ras in response to SCF and target MITF for phosphorylation.14,15 The MAP kinase, ERK-2, phosphorylates MITF at its amino-terminus and results in the recruitment of the coactivator, p300/CBP. This event increases the transcriptional activity of MITF on a melanocyte gene target promoter.16 SCF treatment also activates p90 rsk, which phosphorylates MITF at its carboxyl-terminus. This coupled phosphorylation targets the transcription factor for proteosome-mediated destruction.15 Thus, SCF stimulation appears to activate MITF while shortening its half-life. In melanocytes, KIT signals may activate MITF-dependent transcription of critical melanocyte genes; however, the functional link between KIT and MITF in mast cells has not been determined.
MicroRNAs (miRNAs) are a class of small, noncoding RNA nucleotides that regulate protein expression in several fundamental biologic processes and are expressed in a tissue-specific and developmentally regulated fashion. Long primary transcripts of miRNAs are processed to mature double-stranded miRNAs of 18 to 24 bp by the RNases Drosha and Dicer. The fully processed, mature miRNA can regulate protein expression by inhibiting the stability and translation of target mRNAs by pairing with short complementary sequences within the 3′-untranslated portion of the target mRNA. There may be considerable mismatch for the rest of the molecule, however; and a single miRNA potentially may regulate numerous mRNA transcripts. Furthermore, multiple miRNAs may be coordinately regulated, either processed from a single primary RNA transcript or expressed under the control of common cis-regulatory elements. Thus, miRNAs appear to function as negative regulators of protein expression and may have multiple targets.
We sought to determine the mechanisms of KIT-dependent proliferation and transformation of mast cells. We found that protein expression of MITF was increased in patients with systemic mastocytosis and that this up-regulation was dependent on KIT signals in normal and malignant mast cells. We identified miRNAs that were repressed by KIT signaling that had predicted binding to the 3′-untranslated region (UTR) of the MITF mRNA. We found that these KIT-regulated miRNAs specifically repressed MITF expression, requiring the phylogenetically conserved predicted binding sites in the MITF 3′-UTR. Our studies thus demonstrate a novel miRNA regulatory pathway that links MITF and KIT, 2 factors essential for mast cell function.
Methods
Animals
C57/BL6 wild-type mice and Mitf−/− mice were maintained in the Johns Hopkins University Animal Facilities in accordance with institutional guidelines. All mice experiments were approved by the Johns Hopkins University Animal Care and Use Committee. The Mitf−/− mice are bred on a C57/BL6 background and harbor a transgene of the vasopressin-β-galactosidase that disrupts the 5′ genomic locus of the Mitf gene.8 Six- to 10-week (Mitf−/−) and wild-type (Mitf+/+) mice were used to obtain splenocytes and bone marrow.
Cells
Bone marrow-derived mast cells (BMMCs) were cultured from splenocytes and bone marrow from 4- to 8-week-old C57/B6 wild-type and Mitf−/− mice as described.11 The HMC-1.1 cell line (KIT V560G mutant provided by Dr Shau-Ku Huang, Johns Hopkins University) and the HMC-1.2 cell line (KIT D816V and V560G mutant provided by Dr Alasdair Gilfillan) were maintained in RPMI with 10% fetal bovine serum (FBS) and 2mM l-glutamine, 100 units/mL penicillin, and 100 μg/mL of streptomycin. The P815, NIH 3T3, 293, and the Phoenix cell lines (provided by Dr Gary Nolan, Stanford University) were maintained in Dulbecco modified Eagle medium (DMEM) with 10% FBS and 2mM l-glutamine, 100 units/mL penicillin, and 100 μg/mL of streptomycin. The C57 murine mast cell line was maintained in DMEM with 10% FBS and 2mM l-glutamine, 100 units/mL penicillin, 100 μg/mL of streptomycin, and 5 × 10−5M β-mercaptoethanol. The 293GPG cell line was grown in DMEM with 10% FBS and 2mM L-glutamine, 100 units/mL penicillin, and 100 μg/mL of streptomycin with 2 μg/mL puromycin, 300 μg/mL G418, and 2 μg/mL doxycyline.17 For SCF treatment of BMMCs, cells were washed free of cytokine and maintained in DMEM 10% FBS with or without SCF at 100 ng/mL overnight. For imatinib and midostaurin treatment of mastocytoma lines, cells were treated with various concentrations of drug overnight.
Metabolic labeling
BMMCs untreated or treated with 100 ng/mL SCF for 24 hours were washed twice with cold phosphate-buffered saline and preincubated for 30 minutes at 37°C in L-methionine + L-cysteine-free DMEM labeling medium. Cells were then metabolically labeled for 2 hours with 200 μCi of 35S protein labeling mix (PerkinElmer Life and Analytical Sciences), washed free of radioactive amino acids, and incubated in prewarmed complete medium for the indicated chase times. At the indicated time points, the cell extracts were prepared and immunoprecipitated with antibodies to MITF (C5), run on sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and analyzed by autoradiography. The MITF half-life was determined by direct quantification of the band intensities in the autoradiographs by image analysis (AlphaImager 3400).
Patient bone marrow specimens, c-KIT mutation analysis, immunohistochemistry, and Western blotting
Western blots were performed with antibodies to MITF (C5) and α-tubulin as described.18 Relative quantification of protein expression was performed with densitometry measurement of ECL signal using AlphaImager 3400 analysis software. Bone marrow biopsies were obtained from patients with systemic mastocytosis at the National Institutes of Health after informed consent in accordance with the Declaration of Helsinki following a National Institutes of Health Institutional Review Board-approved protocol. Deidentified bone marrow biopsies were obtained from patients with other hematologic diseases at Johns Hopkins University on an Institutional Review Board exempt protocol. Detection of the D816V mutation in c-KIT was performed using reverse-transcribed polymerase chain reaction (RT-PCR)/restriction fragment length polymorphism analysis as described.19 Immunohistochemistry was performed by Paragon Bioservices. The C5 antibody was used for MITF staining,18 and an antitryptase antibody (Dako United Kingdom) was used for tryptase staining. Assessment of MITF and tryptase staining of bone marrow samples was performed by a reviewer (P.N.) who was blinded to the diagnoses.
Plasmids
The MITF 3′-UTR was amplified by PCR from first strand of cDNA from RNA obtained from the C57 murine mast cell line. An EcoR1 site was engineered 3′ of the Xba1 site of pGL4 SV40 pro to allow directional cloning of PCR fragments into the EcoR1/Fse1 site downstream of the luciferase gene. Fragments extending from exon 9 of MITF into the 3′-UTR of various lengths (940, 1580, and 2000 bp) were cloned into the Xba1/Fse1 site of pGL4 SV40pro. Deletions of the miRNA seed sequences for miR-539 and miR-381 were engineered into the 2000-bp 3′-UTR of MITF using the QuikChange site-directed mutagenesis kit according to the manufacturer's recommendations (Stratagene). Constructs were verified by sequencing. The sh-RNA-expressing lentiviral vector pLL3.7 was provided by Dr Van Parijs (Massachusetts Institute of Technology).20 A short hairpin loop targeting a conserved sequence of the murine and human MITF was subcloned into the Xho1/Hpa1 site. The pSICO-dicer lentiviral construct targeting human Dicer was obtained from the Tyler Jacks Laboratory (Massachusetts Institute of Technology).21 The pMMP-IRES-puro retroviral vector has been described.11 pMMP-IRES-puro MITF was constructed by subcloning the murine MITF-A isoform cDNA into the pMMP-IRES-puro backbone. The retroviral construct-expressing MITF-A resistant to sh-RNA silencing (pMMP-IRES-puro MITF rescue) was constructed by site-directed mutagenesis (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). pMMP-IRES-puro KIT was constructed by subcloning the murine c-Kit cDNA into the pMMP-IRES-puro backbone. The pMMP-IRES-puro Kit-D814V was engineered by site-directed mutagenesis. The miRNA-expressing constructs were cloned into the pMSCVpuro retroviral backbone with a puromycin resistance cassette.22 Primers were designed to amplify a genomic fragment that spanned approximately 100 bp upstream of the miR-381 site and 100 bp downstream of the miR-539 site. The fragment was cloned into the Xho1 site of pMSCVpuro. Primer sequences for cloning are described in the Supplemental data.
miRNA arrays
RNA was harvested from BMMCs with and without SCF treatment and from the c-Kit mutant P815 cell line with and without imatinib treatment. RNA was labeled with Cy3 using RNA ligase and hybridized to custom microarrays containing 474 human, mouse, and rat miRNAs as described.23 Ratios of signals were then calculated, and miRNAs that were up-regulated or down-regulated at least 2-fold by KIT signaling were identified. The complete array results are included in the Supplemental data. The microarray data have been deposited into the Gene Expression Omnibus under accession number GSE26783.
Quantitative real-time PCR and RT-PCR
RNA was harvested from cells with Trizol (Invitrogen). cDNA was made with the first strand synthesis kit (Invitrogen) and used for quantitative PCR using a SYBR Green mix (Bio-Rad) on an iQ5 multicolor Real-time PCR Detection System (Bio-Rad). Quantification of MITF mRNA relative to β-actin was determined by the 2−ΔΔCt method.24 mir-RT PCR was performed with the QuantiMir RT kit from System Biosciences according to the manufacturer's instructions. Quantitation of miRNA for human miR539 and miR381 was performed with the TaqMan MicroRNA Assay (Applied Biosystems) according to the manufacturer's recommendations and normalized to miR-17 expression. PCR primers and conditions are described in the Supplemental data.
Reporter assays
HMC-1.1 human mast cell line transfections were performed with the Superfect reagent (QIAGEN) according to the manufacturer's recommendations and as described.18 A total of 4 mL of Superfect solution was used with 2 μg of DNA and 0.05 μg of Sea Pansey luciferase for 3 × 105 cells. Lysates were harvested after 2 days. NIH 3T3 cell line transfections were performed using FuGENE 6 (Invitrogen) according to the manufacturer's recommendations with 0.5 μg of DNA and 0.05 mg of Sea Pansey luciferase. The driver-to-reporter ratio used for transfections was 1:4. Lysate was assayed for luciferase activity using the dual luciferase system (Promega) and normalized to Sea Pansey luciferase activity.
Colony-forming assay
For colony-forming assays, a 1% methylcellulose (StemCell Technologies) was prepared with culture media (RPMI with 2% FBS for HMC cells, and DMEM with 10% FBS for P815 cells). A total of 2.5 × 103 HMC cells and 2 × 104 P815 cells were vortexed thoroughly in 2.5 mL of methylcellulose mixture and placed into 6-well plates. Cells were incubated at 37°C in 5% CO2 for 10 to 12 days. Individual colonies containing more than 50 cells were identified visually and were enumerated with an inverted microscope.
Cell proliferation assay BrdU incorporation and XTT assay
Cell viability and growth of BMMCs in response to SCF and drug treatment were measured with XTT using the CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (Promega) according to the manufacturer's recommendations. A total of 5 × 104 cells were resuspended in 100 μL of DMEM with 10% FBS with SCF or drug treatment for 72 hours. Optical density (OD) readings were done at 550 nm. Cell proliferation of mastocytosis lines were performed with the BrdU Cell Proliferation ELISA kit (Roche Diagnostics) according to the manufacturer's recommendations. A total of 5 × 103 cells were cultured for 2 days and then labeled for 4 hours with bromodeoxyuridine (BrdU). OD readings were done at 450 nm and normalized to vector treated cells.
Retroviral and lentiviral transduction
pMMP-IRES-puro retrovirus was produced as described.11,17 BMMCs were cultured from Mitf+/+ mice and Mitf−/− mice as described.11 After 1 to 2 weeks in culture, 2 × 106 cells/mL from Mitf−/− mice were placed on retronectin-coated plates at 20 μg/mL (RetroNectin)25 and transduced with concentrated retrovirus (100-200 μL) supplemented with interleukin-3 at 5 ng/mL and SCF at 25 ng/mL overnight. Retroviral transduction was repeated twice. After 2 to 3 days, 2 μg/mL of puromycin was added to the culture to select for stably transduced cells.
pMSCVpuro retrovirus was produced as described.22 Constructs were transfected into the amphotrophic Phoenix packaging line, and retroviral supernatants were collected at 48 and 72 hours. One- to 2-week cultures of BMMCs were placed in retronectin-coated plates at 20 μg/mL (RetroNectin)25 and transduced with retroviral supernatant supplemented with interleukin-3 at 5 ng/mL and SCF at 25 ng/mL overnight. Retroviral transductions were repeated twice. After 3 to 5 days in culture, 2 μg/mL of puromycin was added and maintained in culture to select for stably transduced cells. For production of pSICO and pLL3.7 lentivirus, 293 clone T17 cells were transfected with FUGENE with the viral packaging constructs, pRRE, pRSV, and pVSG as described.20,21 Murine (P815) and human (HMC) mastocytoma lines were transduced with polybrene at 8 μg/mL overnight and then repeated. Transduction efficiency of the pLL3.7 virus was more than 90% as assessed by green fluorescent protein positivity. pSICO transduced cells were selected with puromycin.
Results
MITF is expressed in systemic mastocytosis
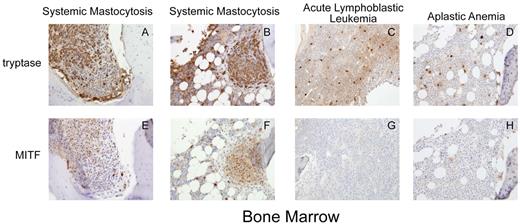
We examined MITF expression by immunohistochemistry on bone marrow samples obtained from 10 adult patients with the diagnosis of systemic mastocytosis. Eight of these patients were found to harbor the D816V c-KIT mutation, whereas 2 patients had other c-KIT activating mutations (Table 1). We found that all 8 patients with D816V c-KIT mutations showed strongly positive nuclear staining for MITF. These cells were often found in aggregates and stained strongly for tryptase, characteristic of mastocytosis lesions (Figure 1A-B,E-F). These patients were diagnosed with indolent systemic mastocytosis, the most common variant of systemic mastocytosis. Bone marrow from the patient with the F522C mutation also showed strong MITF staining.26 This mutation lies within the transmembrane region of c-KIT. Another patient with the diagnosis of aggressive systemic mastocytosis was found to have a D816Y mutation in c-Kit, which is predicted to result in constitutive activation of KIT. However, the bone marrow from this patient did not show strong MITF staining. We also examined MITF staining in bone marrow biopsies from patients with other hematologic disorders associated with increased mast cells. Seven patients had the diagnosis of idiopathic aplastic anemia, one with Fanconi anemia, one with pre-B acute lymphoblastic leukemia, and one with hemophagocytic lymphohistiocytosis (Table 1). However, mast cells in these conditions did not aggregate in clusters, nor did they stain strongly for MITF expression (Figure 1C-D,G-H). These conditions are not commonly associated with mutations in c-KIT.
Tryptase and MITF are coexpressed in patients with systemic mastocytosis and c-KIT mutations
| Patient no. . | Diagnosis . | . | Staining . | |
|---|---|---|---|---|
| Tryptase . | MITF . | |||
| 1 | Indolent systemic mastocytosis | D816V | ++ | + |
| 2 | Indolent systemic mastocytosis | D816V | ++ | + |
| 3 | Indolent systemic mastocytosis | D816V | ++ | + |
| 4 | Indolent systemic mastocytosis | D816V | ++ | + |
| 5 | Indolent systemic mastocytosis | D816V | ++ | + |
| 6 | Indolent systemic mastocytosis | D816V | ++ | + |
| 7 | Indolent systemic mastocytosis | D816V | ++ | + |
| 8 | Indolent systemic mastocytosis | D816V | ++ | + |
| 9 | Systemic mastocytosis | F522C | ++ | + |
| 10 | Aggressive systemic mastocytosis | D816Y | ++ | − |
| 11 | Aplastic anemia | NT | ++ | − |
| 12 | Aplastic anemia | NT | ++ | − |
| 13 | Aplastic anemia | NT | ++ | − |
| 14 | Aplastic anemia | NT | ++ | − |
| 15 | Aplastic anemia | NT | ++ | − |
| 16 | Aplastic anemia | NT | ++ | − |
| 17 | Aplastic anemia | NT | ++ | − |
| 18 | Fanconi anemia | NT | ++ | − |
| 19 | Acute lymphoblastic leukemia | NT | ++ | − |
| 20 | Hemophagocytic lymphohistiocytosis | NT | ++ | − |
| Patient no. . | Diagnosis . | . | Staining . | |
|---|---|---|---|---|
| Tryptase . | MITF . | |||
| 1 | Indolent systemic mastocytosis | D816V | ++ | + |
| 2 | Indolent systemic mastocytosis | D816V | ++ | + |
| 3 | Indolent systemic mastocytosis | D816V | ++ | + |
| 4 | Indolent systemic mastocytosis | D816V | ++ | + |
| 5 | Indolent systemic mastocytosis | D816V | ++ | + |
| 6 | Indolent systemic mastocytosis | D816V | ++ | + |
| 7 | Indolent systemic mastocytosis | D816V | ++ | + |
| 8 | Indolent systemic mastocytosis | D816V | ++ | + |
| 9 | Systemic mastocytosis | F522C | ++ | + |
| 10 | Aggressive systemic mastocytosis | D816Y | ++ | − |
| 11 | Aplastic anemia | NT | ++ | − |
| 12 | Aplastic anemia | NT | ++ | − |
| 13 | Aplastic anemia | NT | ++ | − |
| 14 | Aplastic anemia | NT | ++ | − |
| 15 | Aplastic anemia | NT | ++ | − |
| 16 | Aplastic anemia | NT | ++ | − |
| 17 | Aplastic anemia | NT | ++ | − |
| 18 | Fanconi anemia | NT | ++ | − |
| 19 | Acute lymphoblastic leukemia | NT | ++ | − |
| 20 | Hemophagocytic lymphohistiocytosis | NT | ++ | − |
+ indicates positive staining ++, strongly positive staining −, no detectable staining and — NT, not tested.
Trypase and MITF are coexpressed in systemic mastocytosis. Immunohistochemistry for tryptase and MITF was performed on bone marrow specimens from patients with systemic mastocytosis and other hematologic diseases. (A-D) Cytoplasmic tryptase staining in bone marrow mast cells in 2 patients with systemic mastocytosis and the D816V KIT mutation (A-B), aplastic anemia (C), and acute lymphoblastic leukemia (D). (E-F) Nuclear MITF staining in mast cells in systemic mastocytosis. Mast cells in aplastic anemia and acute lymphoblastic leukemia (G-H) do not express significant MITF. Mast cells in systemic mastocytosis are spindle shaped and grow in clusters, whereas mast cells in other conditions do not. Photomicrograph images were acquired at room temperature with a Kontron ProgRes 3012 digital camera and Roche Image analysis software with a Zeiss Axiophot microscope (original magnification ×20). Images cropped with Photoshop Elements 2.0 (Adobe).
Trypase and MITF are coexpressed in systemic mastocytosis. Immunohistochemistry for tryptase and MITF was performed on bone marrow specimens from patients with systemic mastocytosis and other hematologic diseases. (A-D) Cytoplasmic tryptase staining in bone marrow mast cells in 2 patients with systemic mastocytosis and the D816V KIT mutation (A-B), aplastic anemia (C), and acute lymphoblastic leukemia (D). (E-F) Nuclear MITF staining in mast cells in systemic mastocytosis. Mast cells in aplastic anemia and acute lymphoblastic leukemia (G-H) do not express significant MITF. Mast cells in systemic mastocytosis are spindle shaped and grow in clusters, whereas mast cells in other conditions do not. Photomicrograph images were acquired at room temperature with a Kontron ProgRes 3012 digital camera and Roche Image analysis software with a Zeiss Axiophot microscope (original magnification ×20). Images cropped with Photoshop Elements 2.0 (Adobe).
MITF expression is up-regulated with KIT signaling posttranscriptionally
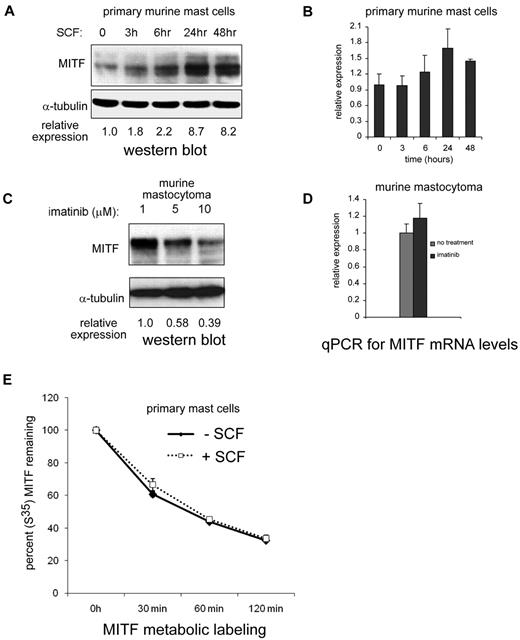
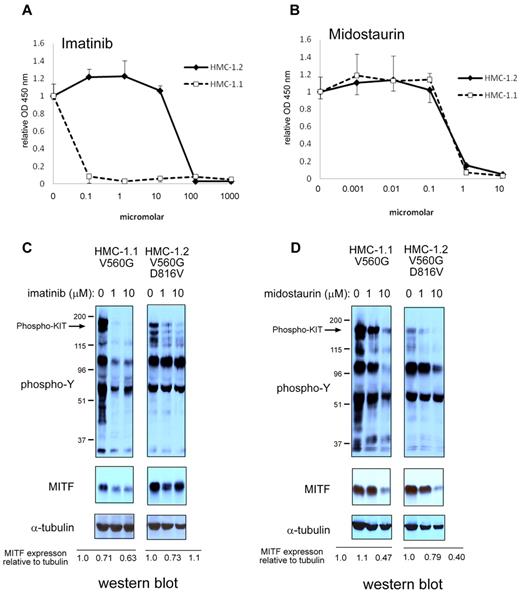
Based on the observation that MITF expression was increased in patients with systemic mastocytosis and c-KIT activating mutations, we investigated whether MITF was regulated by KIT stimulation. BMMCs treated with the KIT ligand SCF markedly up-regulated MITF expression (> 8-fold increase) over 24 to 48 hours (Figure 2A). We then examined whether MITF mRNA levels were regulated by KIT signaling by quantitative PCR. In contrast to the robust increase in protein levels, MITF mRNA increased only to a maximum of 1.8-fold at 24 hours in BMMCs treated with SCF (Figure 2B). We also found that murine and human mastocytoma cell lines with activating c-KIT mutations have high MITF protein expression in the absence of SCF. In P815 cells (which harbor the murine D814V c-KIT mutation), we found a dose-dependent decrease in MITF protein with imatinib treatment (Figure 2C). We found no significant decrease of MITF mRNA levels in P815 mastocytoma cells treated with imatinib (Figure 2D). Given the discordance between the magnitude of protein and mRNA changes with SCF treatment, we considered the possibility that MITF protein was stabilized with SCF treatment. BMMCs were metabolically pulse-labeled with S35 and then stimulated with SCF. As shown in Figure 2E, the half-life of the MITF protein did not change significantly with SCF treatment. The HMC-1.1 cell line harbors the V560G mutation in c-Kit; a similar decrease in MITF protein expression was observed with imatinib treatment. Whereas the HMC-1.1 cell line is sensitive to imatinib, the HMC-1.2 cell line, which has both V560G and D816V c-KIT mutations in c-KIT, is relatively resistant (Figure 3A-B). No significant reduction in MITF protein was seen with imatinib treatment in the 10nM range. The decrease in MITF protein correlated with inhibition of tyrosine phosphorylation (Figure 3C). Both HMC lines are sensitive to the protein kinase C inhibitor midostaurin; treatment resulted in reduced MITF protein, which was concordant with tyrosine phosphorylation inhibition (Figure 3D). The proteosome inhibitor bortezomib also has activity against mastocytosis,27 in part because of inhibition of KIT expression.28 Although both HMC lines are sensitive to bortezomib, MITF expression was only significantly reduced in the HMC-1.2 line, which was concordant with reduction in KIT protein (supplemental Figure 2). This suggests cytotoxic pathways that are MITF independent.
KIT activation up-regulates MITF protein expression. (A) Western blot shows robust increase of MITF protein expression by 24 to 48 hours in primary murine BMMCs treated with SCF (100 ng/mL). Relative expression is calculated by taking the ratio of the densitometry signal for MITF to tubulin and normalizing to 1.0 for the zero time point. (B) Real-time PCR shows MITF mRNA levels with SCF treatment in BMMCs. Signal quantitated with SYBR Green and normalized to β-actin. Relative expression is normalized to the signal of the zero time point. (C) Pharmacologic inhibition of KIT reduces MITF expression. Western blot shows that increasing the concentrations of the KIT inhibitor imatinib results in a dose-dependent decrease in MITF expression in murine (P815). Relative expression is calculated as described for BMMCs. (D) Real-time PCR shows no significant change in MITF mRNA in murine mastocytoma (P815) with imatinib treatment (10μM). (E) MITF protein half-life does not change significantly with KIT activation. Metabolically labeled BMMCs (primary mast cells) with and without SCF treatment were immunoprecipitated with an MITF antibody, and MITF protein was quantitated at various time points after SCF treatment with densitometry of signal intensity of bands on autoradiograph. There was no significant difference in MITF half-life with SCF treatment.
KIT activation up-regulates MITF protein expression. (A) Western blot shows robust increase of MITF protein expression by 24 to 48 hours in primary murine BMMCs treated with SCF (100 ng/mL). Relative expression is calculated by taking the ratio of the densitometry signal for MITF to tubulin and normalizing to 1.0 for the zero time point. (B) Real-time PCR shows MITF mRNA levels with SCF treatment in BMMCs. Signal quantitated with SYBR Green and normalized to β-actin. Relative expression is normalized to the signal of the zero time point. (C) Pharmacologic inhibition of KIT reduces MITF expression. Western blot shows that increasing the concentrations of the KIT inhibitor imatinib results in a dose-dependent decrease in MITF expression in murine (P815). Relative expression is calculated as described for BMMCs. (D) Real-time PCR shows no significant change in MITF mRNA in murine mastocytoma (P815) with imatinib treatment (10μM). (E) MITF protein half-life does not change significantly with KIT activation. Metabolically labeled BMMCs (primary mast cells) with and without SCF treatment were immunoprecipitated with an MITF antibody, and MITF protein was quantitated at various time points after SCF treatment with densitometry of signal intensity of bands on autoradiograph. There was no significant difference in MITF half-life with SCF treatment.
Pharmacologic inhibition of KIT signaling results in MITF protein reduction. (A) D816V mutation confers resistance to imatinib treatment. XTT assay shows sensitivity of HMC-1.1 (KIT D816V-negative) and relative resistance of HMC-1.2 (KIT D816V-positive) to imatinib. (B) D816V-negative and -positive HMC cell lines are sensitive to midostaurin. XTT assays were performed in triplicate and absorbance measured at OD 450. Relative OD 450 value is calculated by normalizing OD 450 reading to OD 450 with no treatment. (C) MITF protein expression is reduced with inhibition of KIT signaling. Western blot shows that both phosphotyrosine signal and MITF protein are reduced with imatinib treatment in HMC-1.1. In HMC-1.2, phosphotyrosine signal is only mildly reduced and MITF protein is not significantly repressed. (D) Midostaurin treatment results in inhibition of phosphotyrosine in both HMC-1.1 and HMC-1.2. Western blot for α-tubulin shows equivalent loading. Relative expression is calculated by taking the ratio of the densitometry signal for MITF to tubulin and normalizing to 1.0 for the zero time point.
Pharmacologic inhibition of KIT signaling results in MITF protein reduction. (A) D816V mutation confers resistance to imatinib treatment. XTT assay shows sensitivity of HMC-1.1 (KIT D816V-negative) and relative resistance of HMC-1.2 (KIT D816V-positive) to imatinib. (B) D816V-negative and -positive HMC cell lines are sensitive to midostaurin. XTT assays were performed in triplicate and absorbance measured at OD 450. Relative OD 450 value is calculated by normalizing OD 450 reading to OD 450 with no treatment. (C) MITF protein expression is reduced with inhibition of KIT signaling. Western blot shows that both phosphotyrosine signal and MITF protein are reduced with imatinib treatment in HMC-1.1. In HMC-1.2, phosphotyrosine signal is only mildly reduced and MITF protein is not significantly repressed. (D) Midostaurin treatment results in inhibition of phosphotyrosine in both HMC-1.1 and HMC-1.2. Western blot for α-tubulin shows equivalent loading. Relative expression is calculated by taking the ratio of the densitometry signal for MITF to tubulin and normalizing to 1.0 for the zero time point.
Mast cells with activating c-Kit mutation require MITF expression for mast cell proliferation
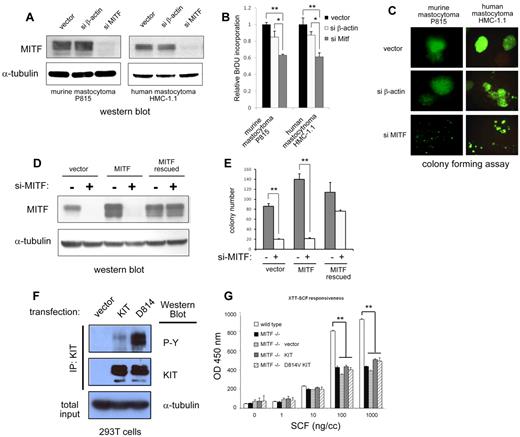
To address the requirement of MITF for KIT dependent proliferation, we suppressed MITF expression in c-KIT mutant mast cell lines with sh-RNA-expressing lentivirus. This sh-RNA efficiently silences MITF protein expression in the c-KIT mutant human HMC-1.1 and murine P815 lines (Figure 4A,D). We first examined the effect of MITF on cell growth. We found that BrdU uptake was modestly decreased in cells with silenced MITF expression compared with wild-type cells over 48 hours (Figure 4B). We then examined the role of MITF on the colony formation of mastocytoma cells in methylcellulose. We found that knockdown of MITF expression in both murine and human mastocytoma cells significantly impaired their ability to form colonies in methylcellulose (Figure 4C). Cells transduced with a control lentivirus targeting β-actin (si-β actin) did not significantly differ from vector alone in proliferative capacity or colony formation. To ascertain that the effect of MITF knockdown was not the result of sh-RNA off-target effects, we constructed an MITF cDNA sequence that was insensitive to sh-RNA silencing. This cDNA contained a MITF sequence with mutations in the wobble sequences in the si-RNA binding site and expressed wild-type MITF protein but was insensitive to MITF sh-RNA silencing (MITF rescued, supplemental Figure 1). As shown the Western blot in Figure 3D, both endogenous MITF and transduced MITF protein were silenced by sh-RNA, but the mutated MITF (MITF rescued) was resistant. Figure 4E shows that forced expression of MITF resistant to the RNAi restored colony-forming capacity, whereas forced expression of wild-type MITF did not, demonstrating a role for MITF in mastocytoma colony formation. We found that MITF knockdown in D816V KIT mutant-positive HMC-1.2 cells also impaired colony/foci growth similar to MITF knockdown in D816V-negative HMC-1.1 cells (supplemental Figure 3).
MITF knockdown inhibits cell growth in c-KIT mutant cells. (A) Western blot shows efficient knockdown of MITF protein expression with sh-RNA-expressing lentivirus (si-MITF) in murine (P815) and human (HMC-1) mastocytoma lines. Control lentivirus targeting β-actin (si-β-actin) shows no effect on MITF expression. (B) MITF repression results in decrease in cell proliferation. BrdU incorporation of both murine and human mastocytoma cells with si-MITF–expressing lentivirus is decreased approximately 40% of control. *P < .5. **P < .005. (C) MITF repression significantly impairs colony-forming capacity of mastocytoma lines. Representative images of mastocytoma colonies transduced with lentiviral empty vector, si-β-actin, and si-MITF in methylcellulose. Murine (P815) and human (HMC-1.1) cells with knockdown of MITF (si-MITF) are impaired in colony-forming capacity. Lentiviral vector expresses green fluorescent protein, and green fluorescent protein–expressing cells are visualized (original magnification ×10). Fluorescent images were obtained with a Nikon fluorescent microscope with excitation filter set at 488 nm. Images were cropped with Photoshop Elements 2.0 (Adobe Systems). (D) MITF resistant to silencing (MITF rescued) restores colony-forming capacity. For controls for potential off-target effects of sh-RNA and lentivirus, an MITF-expressing retrovirus with and without the si-RNA target site was constructed. Western blot shows human mastocytoma cells (HMC-1.1) expressing MITF with si-RNA binding site mutated (MITF rescued) is resistant to silencing, but the wild-type MITF-expressing cells (MITF) are not. (E) Human mastocytoma cells (HMC-1.1) are dependent on MITF expression for colony formation. Knockdown of MITF with the si-MITF-expressing lentivirus impairs colony formation MITF-expressing cells (vector and MITF, white bars), but cells with MITF resistant to silencing (MITF rescued) maintain colony-forming capacity. **P < .005. (F) Constitutive tyrosine phosphorylation of mutant D814V KIT. 293T cells were stably transduced with vector, wild-type KIT, and mutant D814V retrovirus and then immunoprecipitated with an anti-KIT antibody. Western blots performed for phosphotyrosine and KIT expression. Both wild-type and D814V KIT-transduced cells show expression of KIT, but only D814V KIT-transduced cells show significant constitutive tyrosine phosphorylation. Total input from immunoprecipitated lysates was probed for α-tubulin expression and showed roughly equivalent starting material. (G) D814V requires MITF expression for mast cell growth. Cell proliferation of BMMCs in response to SCF (72 hours incubation) was measured with XTT assay (measured at OD 450 nm). Wild-type BMMCs (white bars) proliferate in response to SCF; MITF−/− BMMCs (black bars) do not proliferate as well to high doses of SCF. MITF−/− BMMCs transduced with vector (horizontal bars), KIT (gray bars), or mutant D814V KIT (diagonal bars) retrovirus do not restore proliferative capacity of MITF−/− cells. **P < .005. P values are calculated with Student t test; all experiments performed in triplicate.
MITF knockdown inhibits cell growth in c-KIT mutant cells. (A) Western blot shows efficient knockdown of MITF protein expression with sh-RNA-expressing lentivirus (si-MITF) in murine (P815) and human (HMC-1) mastocytoma lines. Control lentivirus targeting β-actin (si-β-actin) shows no effect on MITF expression. (B) MITF repression results in decrease in cell proliferation. BrdU incorporation of both murine and human mastocytoma cells with si-MITF–expressing lentivirus is decreased approximately 40% of control. *P < .5. **P < .005. (C) MITF repression significantly impairs colony-forming capacity of mastocytoma lines. Representative images of mastocytoma colonies transduced with lentiviral empty vector, si-β-actin, and si-MITF in methylcellulose. Murine (P815) and human (HMC-1.1) cells with knockdown of MITF (si-MITF) are impaired in colony-forming capacity. Lentiviral vector expresses green fluorescent protein, and green fluorescent protein–expressing cells are visualized (original magnification ×10). Fluorescent images were obtained with a Nikon fluorescent microscope with excitation filter set at 488 nm. Images were cropped with Photoshop Elements 2.0 (Adobe Systems). (D) MITF resistant to silencing (MITF rescued) restores colony-forming capacity. For controls for potential off-target effects of sh-RNA and lentivirus, an MITF-expressing retrovirus with and without the si-RNA target site was constructed. Western blot shows human mastocytoma cells (HMC-1.1) expressing MITF with si-RNA binding site mutated (MITF rescued) is resistant to silencing, but the wild-type MITF-expressing cells (MITF) are not. (E) Human mastocytoma cells (HMC-1.1) are dependent on MITF expression for colony formation. Knockdown of MITF with the si-MITF-expressing lentivirus impairs colony formation MITF-expressing cells (vector and MITF, white bars), but cells with MITF resistant to silencing (MITF rescued) maintain colony-forming capacity. **P < .005. (F) Constitutive tyrosine phosphorylation of mutant D814V KIT. 293T cells were stably transduced with vector, wild-type KIT, and mutant D814V retrovirus and then immunoprecipitated with an anti-KIT antibody. Western blots performed for phosphotyrosine and KIT expression. Both wild-type and D814V KIT-transduced cells show expression of KIT, but only D814V KIT-transduced cells show significant constitutive tyrosine phosphorylation. Total input from immunoprecipitated lysates was probed for α-tubulin expression and showed roughly equivalent starting material. (G) D814V requires MITF expression for mast cell growth. Cell proliferation of BMMCs in response to SCF (72 hours incubation) was measured with XTT assay (measured at OD 450 nm). Wild-type BMMCs (white bars) proliferate in response to SCF; MITF−/− BMMCs (black bars) do not proliferate as well to high doses of SCF. MITF−/− BMMCs transduced with vector (horizontal bars), KIT (gray bars), or mutant D814V KIT (diagonal bars) retrovirus do not restore proliferative capacity of MITF−/− cells. **P < .005. P values are calculated with Student t test; all experiments performed in triplicate.
In wild-type BMMCs, MITF promotes normal proliferation in response to SCF. To determine the role of MITF in mediating the proliferative effect of mutant KIT, MITF−/− BMMCs were transduced with wild-type and mutant D814V KIT-expressing retroviruses. The D814V KIT retrovirus expresses constitutively phosphorylated KIT in the absence of SCF treatment (Figure 4F). In response to high doses of SCF, MITF−/− BMMCs (black bars) show a greater than 50% reduction in growth compared with wild-type BMMCs (white bars, Figure 4G). Forced expression of either wild-type KIT (gray bars) or mutant D814V KIT (diagonal striped bars) did not restore proliferative capacity of the MITF−/− BMMCs in response to SCF. These results demonstrate that MITF promotes the proliferative effects of SCF in normal mast cells as well as mastocytosis.
miRNAs 381 and 539 are repressed with KIT signaling in mast cells
These findings suggested that MITF protein expression may be positively regulated by KIT signals through posttranscriptional mechanisms. We considered the possibility that MITF protein expression may be affected by miRNAs in response to SCF treatment. To investigate this possibility, we examined miRNAs regulated by KIT signaling with expression arrays.23 We compared miRNAs differentially expressed in BMMCs with and without SCF, and in the c-Kit mutant P815 line with and without imatinib. We found that these 2 experimental systems yielded 11 shared miRNAs that were significantly increased (> 2-fold) with KIT signals and 8 shared miRNAs that were significantly decreased with KIT signals (Table 2). Given that miRNAs are frequently negative regulators of protein expression, we focused on the miRNAs that were down-regulated with KIT activation as potential candidates for mediating the increase in MITF expression in response to SCF.
MicroRNAs differentially regulated with KIT signaling in mast cells
| miRNA . |
|---|
| Induced with KIT activation |
| miRNA-568 |
| miRNA-627 |
| miRNA-452 |
| miRNA-466 |
| miRNA-215 |
| miRNA-713 |
| miRNA-703 |
| miRNA-467a |
| miRNA-574 |
| miRNA-467b |
| miRNA-215 |
| Repressed with KIT activation |
| miRNA-617 |
| miRNA-294 |
| miRNA-373 |
| miRNA-134 |
| miRNA-539 |
| miRNA-198 |
| miRNA-202 |
| miRNA-346 |
| miRNA . |
|---|
| Induced with KIT activation |
| miRNA-568 |
| miRNA-627 |
| miRNA-452 |
| miRNA-466 |
| miRNA-215 |
| miRNA-713 |
| miRNA-703 |
| miRNA-467a |
| miRNA-574 |
| miRNA-467b |
| miRNA-215 |
| Repressed with KIT activation |
| miRNA-617 |
| miRNA-294 |
| miRNA-373 |
| miRNA-134 |
| miRNA-539 |
| miRNA-198 |
| miRNA-202 |
| miRNA-346 |
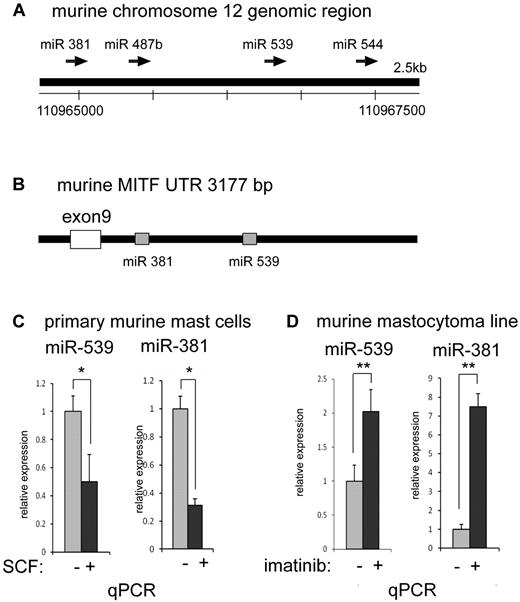
We inspected the down-regulated miRNAs for candidates that potentially targeted MITF. Using the TargetScan S algorithm (www.targetscan.org), we found a putative binding site for miRNA-539 within the 3′-UTR of the MITF gene (Figure 5B); miRNA-539 was identified in our screen as an miRNA that was down-regulated by KIT signaling (Table 2). The putative binding site in the MITF-UTR was phylogenetically conserved among mammals. Inspection of the miRNA-539 genomic region revealed that miRNA-381 and miRNA-487b were in close proximity, suggesting that these miRNAs may be coregulated as a cluster (Figure 5A). In addition, a putative binding site for miRNA-381 was also found in the 3′-UTR of the MITF gene (Figure 5B). Consistent with the microarray results, we found that miRNA-539 was suppressed with KIT signaling by quantitative PCR; miRNA-381 was also decreased with KIT signaling, suggesting coregulation of these miRNAs as a cluster (Figure 5C-D).
miRNAs 381 and 539 are repressed with KIT signaling in mast cells. (A) Schematic representation of the murine genomic region of chromosome 12 with the conserved miRNAs for 381, 487b, 539, and 544. (B) Schematic representation of the murine MITF 3′-UTR depicting phylogenetically conserved binding sites for miR-381 and miR-539. (C) Real-time TaqMan PCR showing the depression of mature miR-381 and miR-539 with activation of KIT in primary BMMCs treated with SCF. *P < .05. (D) Real-time TaqMan PCR showing the increase in mature miR-381 and miR-539 levels with inhibition of KIT in murine mastocytoma (P815) with imatinib. **P < .005. Signals were normalized to miR-16 expression. P values are calculated with Student t test; all experiments performed in triplicate.
miRNAs 381 and 539 are repressed with KIT signaling in mast cells. (A) Schematic representation of the murine genomic region of chromosome 12 with the conserved miRNAs for 381, 487b, 539, and 544. (B) Schematic representation of the murine MITF 3′-UTR depicting phylogenetically conserved binding sites for miR-381 and miR-539. (C) Real-time TaqMan PCR showing the depression of mature miR-381 and miR-539 with activation of KIT in primary BMMCs treated with SCF. *P < .05. (D) Real-time TaqMan PCR showing the increase in mature miR-381 and miR-539 levels with inhibition of KIT in murine mastocytoma (P815) with imatinib. **P < .005. Signals were normalized to miR-16 expression. P values are calculated with Student t test; all experiments performed in triplicate.
Overexpression of miR-381 and 539 repress MITF expression and function
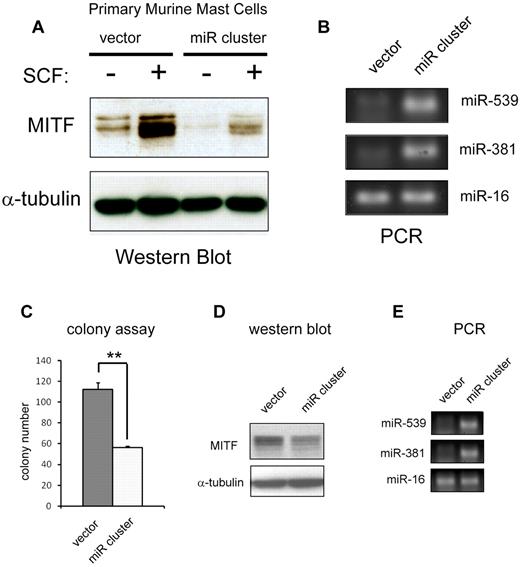
To address whether miRNAs 539 and 381 targeted MITF in vivo, we subcloned the 1.5-kb genomic region that contained these miRNAs into the MSCV-puro retroviral backbone.18 This construct expresses the mature miR-539, miR-381 (Figures 5A-B, 6E), and miR-487b (data not shown). Forced expression of this retrovirus (miR cluster) into BMMCs decreased protein levels of MITF and blunted the up-regulation of MITF protein in response to SCF treatment (Figure 6A). We also assessed the effect of forced expression of the miRNA cluster on colony-forming potential. Overexpression of the miR cluster reduced MITF protein and suppressed colony-forming capacity of HMC-1.1 cells (Figure 6C-D). We also constructed MSCV-puro retroviruses that expressed miR-539 and miR-381 individually. Overexpression of these miRNAs independently did not appreciably affect HMC-1.1 colony-forming potential, nor did it significantly repress MITF protein expression in BMMCs (data not shown). These findings suggest that KIT signals modulate MITF protein levels by regulating this cluster of miRNAs.
Overexpression of miR-381 and miR-539 represses MITF expression and function. (A) Western blot shows an increase in MITF protein with SCF treatment in primary BMMCs. Retroviral overexpression of miR-381 and miR-539 (miR cluster) represses MITF expression at baseline and blunts up-regulation with SCF treatment. (B) PCR analysis shows overexpression of miR-381 and miR-539 in miR cluster-expressing BMMCs. miR-16 expression is not significantly changed. (C) Overexpression of miR-381 and miR-539 in HMC-1 cells (miR cluster) impairs colony-forming capacity. **P < .005. P values are calculated with Student t test; all experiments performed in triplicate. (D) Western blot shows decrease of MITF protein in miR cluster-expressing HMC-1 cells. (E) PCR analysis demonstrates overexpression of miR-539 and miR-381 in HMC cells transduced with the miR cluster compared with vector-transduced cells. miR-16 expression is not significantly changed.
Overexpression of miR-381 and miR-539 represses MITF expression and function. (A) Western blot shows an increase in MITF protein with SCF treatment in primary BMMCs. Retroviral overexpression of miR-381 and miR-539 (miR cluster) represses MITF expression at baseline and blunts up-regulation with SCF treatment. (B) PCR analysis shows overexpression of miR-381 and miR-539 in miR cluster-expressing BMMCs. miR-16 expression is not significantly changed. (C) Overexpression of miR-381 and miR-539 in HMC-1 cells (miR cluster) impairs colony-forming capacity. **P < .005. P values are calculated with Student t test; all experiments performed in triplicate. (D) Western blot shows decrease of MITF protein in miR cluster-expressing HMC-1 cells. (E) PCR analysis demonstrates overexpression of miR-539 and miR-381 in HMC cells transduced with the miR cluster compared with vector-transduced cells. miR-16 expression is not significantly changed.
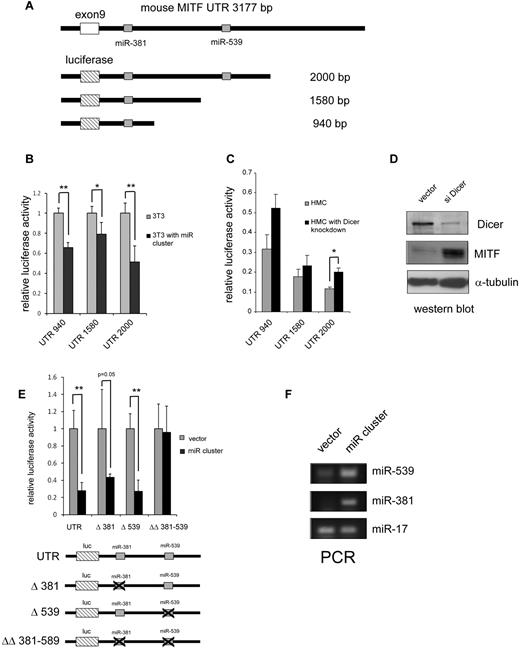
miR-381 and 539 target the 3′-UTR of MITF
To determine whether miR-381 and miR-539 specifically target the 3′-UTR of MITF, we constructed luciferase reporters (pGL4 with the SV40 promoter) linked to the MITF 3′-UTR containing these predicted miRNA binding sites (Figure 7A). Luciferase activity was not significantly different in 3T3 cells with reporter containing the both miR-539 and miR-381 binding sites and truncations containing only the miR-381 binding site (Figure 7B). In 3T3 cells overexpressing miR-381 and miR-539 (miR cluster), the luciferase activity was suppressed for all 3′-UTR reporters (Figure 7B). We next examined the effect of global suppression of miRNAs by knockdown of the miRNA endonuclease Dicer with a sh-RNA containing lentivirus in HMC-1 cells.21 We found that knockdown of Dicer expression increased MITF expression in HMC-1.1 cells and also increased luciferase activity for all MITF 3′-UTR reporters (Figure 7C-D). We next examined whether miR-539 and miR-381 suppressed protein expression specifically through the MITF 3′-UTR. As shown in Figure 7E-F, forced expression of miR-539 and miR-381 (miR cluster) decreases luciferase expression of the reporter containing the wild-type MITF 3′-UTR; it also decreases luciferase expression of reporters with a single miR-381 or miR-539 deleted site. However, the reporter with targeted deletions of both the miR-539 and miR-381 binding sites is resistant to repression by the miR cluster. Taken together, these results are consistent with the conclusion that the 3′-UTR of MITF is specifically regulated by miR-539 and miR-381.
miR-381 and miR-539 target the 3′-UTR of MITF. (A) Schematic representation of the MITF 3′-UTR with the miR-381 and miR-539 binding sites. Luciferase reporter constructs with truncated fragments of the 3′-UTR shown with the lengths depicted. (B) Overexpression of miR-381 and miR-539 (miR cluster) inhibits luciferase activity in 3T3 cells. *P < .05. **P < .005. (C) Knockdown of the miRNA endonuclease, Dicer, results in increased luciferase activity with constructs containing the MITF 3′-UTR. *P < .05. (D) Western blot shows knockdown of Dicer expression with the sh-RNA-expressing lentivirus. MITF protein is increased in Dicer knockdown cells. (E) miR-381 and miR-539 specifically target the miR binding sites in the MITF 3′-UTR. Luciferase reporters with miR-381 and miR-539 binding sites show decreased activity with overexpression of miRs (miR cluster). Luciferase reporters with either miR-381 (Δ381) or miR-539 (Δ539) singly deleted also show decreased activity with overexpression of miRs. Luciferase reporter with and miR-381 and miR-539 binding sites deleted (ΔΔ 381-539) are not repressed by miR cluster overexpression. *P < .05. **P < .005. (F) PCR analysis shows overexpression of miR-539 and miR-381 in miR cluster transduced 3T3 cells. miR-17 is not significantly affected. P values are calculated with Student t test; all experiments are performed in triplicate. Luciferase activity is normalized to Sea Pansey luciferase activity
miR-381 and miR-539 target the 3′-UTR of MITF. (A) Schematic representation of the MITF 3′-UTR with the miR-381 and miR-539 binding sites. Luciferase reporter constructs with truncated fragments of the 3′-UTR shown with the lengths depicted. (B) Overexpression of miR-381 and miR-539 (miR cluster) inhibits luciferase activity in 3T3 cells. *P < .05. **P < .005. (C) Knockdown of the miRNA endonuclease, Dicer, results in increased luciferase activity with constructs containing the MITF 3′-UTR. *P < .05. (D) Western blot shows knockdown of Dicer expression with the sh-RNA-expressing lentivirus. MITF protein is increased in Dicer knockdown cells. (E) miR-381 and miR-539 specifically target the miR binding sites in the MITF 3′-UTR. Luciferase reporters with miR-381 and miR-539 binding sites show decreased activity with overexpression of miRs (miR cluster). Luciferase reporters with either miR-381 (Δ381) or miR-539 (Δ539) singly deleted also show decreased activity with overexpression of miRs. Luciferase reporter with and miR-381 and miR-539 binding sites deleted (ΔΔ 381-539) are not repressed by miR cluster overexpression. *P < .05. **P < .005. (F) PCR analysis shows overexpression of miR-539 and miR-381 in miR cluster transduced 3T3 cells. miR-17 is not significantly affected. P values are calculated with Student t test; all experiments are performed in triplicate. Luciferase activity is normalized to Sea Pansey luciferase activity
Discussion
Activating c-KIT mutations are considered a key contributor to the abnormal proliferation of mast cells in systemic mastocytosis; activating c-KIT mutations can also be detected in other malignancies, such as gastrointestinal stromal tumors and acute myeloid leukemia. Therapy targeted to inhibit KIT signaling may provide benefit in the treatment of some of these diseases. However, relatively little is known about the transcriptional and posttranscriptional pathways that are regulated by KIT signaling. MITF mRNA has been shown previously to be expressed in human mastocytosis and up-regulated with SCF treatment9 ; more recently, forced expression of the D816V KIT mutant in BAF cells was shown to induce MITF mRNA.29 We find that KIT activation results in only modest up-regulation of MITF mRNA in mast cells, but that MITF protein is robustly increased. In this study, we show that colony formation of transformed mast cell lines is significantly dependent on MITF. However, the blunted (but not absent) growth of both primary and transformed mast cells in the absence of MITF suggests KIT-mediated proliferation that is both dependent and independent of MITF function. We have previously shown that, in nonmalignant mast cells, MITF regulates several proteases, transporters, and signaling molecules.11 In addition, MITF-deficient mast cells express lower levels of KIT, and KIT may be a transcriptional target of MITF.11,30 However, we have found that KIT expression is not significantly affected by MITF knockdown transformed mast cells, nor is KIT signaling impaired (supplemental Figure 3). Thus, the critical targets of MITF that may contribute to the transformed phenotype in systemic mastocytosis will require further study.
It is well established that systemic mastocytosis is a clonal disease, However, activating c-KIT mutations alone may not be sufficient for malignant transformation.31,32 In advanced mast cell neoplasms, such as aggressive systemic mastocytosis or mast cell leukemia, activating c-KIT mutations are detected, but additional cooperating or initiating genetic defects may be required for the transformed phenotype.4 Our studies suggest that increased MITF expression in response to KIT activation is an important contributor to proliferation in mastocytosis. However, we found that one patient with aggressive systemic mastocytosis and a D816Y mutation did not have high expression of MITF. This finding illustrates that high MITF expression is not found in all cases of mastocytosis and suggests that other cooperating factors remain to be identified. Furthermore, we expect that other mechanisms probably contribute to MITF regulation, including control of mRNA expression at transcriptional and posttranscriptional levels as well as posttranslational modifications.
Both KIT and MITF are essential for normal mast cell development and function. A functional link between these 2 factors has been elucidated in melanocytes, another cell type dependent on KIT and MITF. In melanocytes, KIT signals appear to activate MITF transcriptional function through targeted phosphorylation of MITF, with subsequent degradation of the protein through ubiquitin-mediated proteolysis.14,15 KIT activation in mast cells results in a rapid and reversible mobility shift of MITF protein, concurrent with ERK and AKT phosphorylation. These findings are consistent with a phosphorylation event (supplemental Figure 4). However, the role of MITF phosphorylation in mediating KIT function in mast cells is yet unexplored. We show here, however, that prolonged or constitutive KIT signals in mast cells results in marked up-regulation of MITF protein expression. These findings suggest that a major biologic effect of KIT signals is to regulate MITF levels in mast cells.
KIT signals mediate several functions in mast cells and biologically important mRNA targets have been identified, including the nuclear factors κB and STAT4.33 The miRNA targets regulated by KIT signals, however, have not been reported. In our array screen, we identified several miRNAs that were either increased or decreased with KIT signaling; these miRNAs potentially regulate multiple protein targets with physiologic significance. Given the shared functions of KIT and MITF in mast cells, however, we focused our studies on 2 KIT-regulated miRNAs, miR-381 and miR-539, which target the 3′-UTR of MITF. The expression of these miRNAs is modulated by KIT signals, but the transcription factors that participate in their regulation are yet to be elucidated. A computational analysis (www.genometrafac.cchmc.org)34 of flanking genomic regions identifies several potential factors (supplemental Figure 5A-B); this question will require further study. We also anticipate that SCF-regulated miRNAs mediate other mast cell functions that are not MITF-dependent. Additional studies will be needed to determine the biologic significance of the other SCF-regulated miRNAs that we have identified.
Taken together, our data elucidate a novel regulatory network that links KIT signaling and MITF expression through miRNAs, and provides a basis for the shared functions of KIT and MITF in mast cells. We find this mechanism of MITF regulation in both normal and malignant mast cells, and it is conserved in murine and human cells. Although KIT signals are anticipated to regulate multiple transcriptional targets and biologic functions, we demonstrate a specific pathway in which the coregulated miRNAs miR-381 and mi-539 miRNAs suppress MITF expression and function. In addition to this miRNA pathway, other mechanisms at the transcriptional and posttranslational level regulate MITF function. However, these findings may provide specific downstream targets to modulate KIT function in mast cell-associated diseases.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by in part by the March of Dimes Birth Defects Foundation (Basil O'Conner Starter Scholar Research Award grant 5-FY04-30; C.M.T.), a Children's Cancer Foundation Grant (C.M.T.), the National Institutes of Health (grant 5R01HL077178, C.M.T.; grants 4R01HL077177 and 2R01HL08111, R.K.), and the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (D.D.M., M.C.). J.T.M. is an HHMI Early Career Scientist.
National Institutes of Health
Authorship
Contribution: Y.-N.L. designed and performed the research, analyzed the data, and edited the paper; S.B. performed the research; P.N. reviewed the patient bone marrow biopsies; E.W. performed the miRNA experiments; J.T.M. performed and analyzed the miRNA experiments and edited the paper; R.K. and M.A.M. analyzed data and edited the paper; M.C. performed research and analyzed data on patients with mastocytosis; D.D.M. performed the research and analyzed data on patients with mastocytosis and edited the paper; and C.M.T. designed and performed the research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Clifford M. Takemoto, Division of Pediatric Hematology, Johns Hopkins University, 720 Rutland Ave, Ross 1125, Baltimore MD, 21205; e-mail: ctakemot@jhmi.edu.