Abstract
In multiple myeloma (MM) pathogenesis, hyperdiploidy and nonhyperdiploidy are recognized as 2 major cytogenetic pathways. Here, we assessed the role of hyperdiploidy in 426 patients with monoclonal plasma cell disorders, among them 246 patients with AL amyloidosis (AL), by interphase fluorescence in situ hybridization. Hyperdiploidy was defined by a well-established score requiring trisomies for at least 2 of the 3 chromosomes 5, 9, and 15. The hyperdiploidy frequency in AL was a mere 11% compared with 30% in monoclonal gammopathy of undetermined significance (P < .001) and 46% in AL with concomitant MM I (P < .001). Overall, hyperdiploidy was associated with an intact immunoglobulin, κ light chain restriction, higher age, and bone marrow plasmacytosis, but was unrelated to the organ involvement pattern in AL. Clustering of 6 major cytogenetic aberrations in AL by an oncogenetic tree model showed that hyperdiploidy and t(11;14) were almost mutually exclusive, whereas gain of 1q21 favored hyperdiploidy. Deletion 13q14 and secondary IgH translocations were equally distributed between ploidy groups. We conclude that the interphase fluorescence in situ hybridization–based hyperdiploidy score is also a feasible tool to delineate hyperdiploid patients in early-stage monoclonal gammopathies and that the cytogenetic pathogenetic concepts developed in MM are transferable to AL.
Introduction
In symptomatic multiple myeloma (MM), which is characterized by the expansion of malignant monoclonal plasma cells, karyotypic instability has been detected as a major pathogenetic factor. Numerical and structural chromosome aberrations have permitted to delineate hyperdiploid and nonhyperdiploid forms as 2 major pathogenetic pathways, which are approximately equally distributed.1 The hyperdiploid group is characterized by gains of the odd chromosomes 3, 5, 7, 9, 11, 15, 19, and 21, whereas the nonhyperdiploid group typically displays chromosomal rearrangements and deletions. The biologic validity of hyperdiploid versus nonhyperdiploid subgroups has been substantiated by studies showing different molecular signatures, methylation patterns, and prognosis.2-4
Ploidy is less well studied in 2 other related plasma cell dyscrasia entities: monoclonal gammopathy of undetermined significance (MGUS), a premalignant clonal plasma cell dyscrasia, which can progress into overt MM,5,6 or can lead to systemic light chain amyloidosis (AL), a disease with deposition of immunoglobulin light chains as amyloid fibrils in different organs.7 This paucity of cytogenetic data in these 2 disease entities is the result of their small percentages of monoclonal plasma cells and low proliferative activity in the bone marrow, which has hampered conventional karyotypic analysis. These limitations have been overcome with the development of interphase fluorescence in situ hybridization (iFISH) on sorted CD138+ plasma cells, which allows the assessment of chromosomal abnormalities independent of the low degree of bone marrow plasmacytosis and the low proliferative rate.8-10 iFISH techniques detecting trisomies of odd chromosomes have been established as a surrogate to identify ploidy subtypes. Two scores have been established, with hyperdiploidy defined as the presence of extra copies of at least 2 of the 3 chromosomes 5, 9, and 1511 or 9, 11, and 15.12
Whereas iFISH-based data suggest that the dichotomy of hyperdiploid and nonhyperdiploid categories is also applicable to MGUS,12 previous cytogenetic studies in AL have not addressed hyperdiploidy.13-16 Therefore, the aim of our study was to investigate hyperdiploidy as an underlying cytogenetic pathogenetic mechanism for the development of AL. In detail, we aimed to determine the hyperdiploidy frequency in AL with an underlying monoclonal gammopathy or MM stage I (MM I) compared with a MGUS/MM I control group, to characterize the hematologic and clinical characteristics of the respective ploidy categories, and to identify their clustering with other cytogenetic aberrations in an oncogenetic tree model.
Methods
Patients
We evaluated a series of 246 consecutive AL patients from our institution who were tested for iFISH abnormalities from June 2003 to August 2010. A total of 236 patients were untreated, 10 patients had received prior conventional chemotherapy without achieving a hematologic remission before referral to our center. Patients in hematologic relapse were not considered for this study to exclude a bias regarding clonal evolution. Seventy-seven of the 246 patients had been part of a previous cytogenetic study, which had not addressed hyperdiploidy.16 A total of 180 patients with MGUS or MM I not requiring therapy were used as a control group. Patients with an IgM heavy chain subtype17 or MM stage II or III were not included in this study. For the classification of patients into the MGUS and MM I groups, we applied standard diagnostic criteria except for bone marrow plasmacytosis of 30% instead of 10% as cutoff value between MGUS and MM I, according to the Boston group and Mayo Clinic criteria for AL and our own practice.16,18-21 Thus, we obtained 4 patient groups: (1) 220 AL patients with an underlying monoclonal gammopathy; (2) 26 AL patients with a concomitant MM I (AL + MM I); (3) 151 MGUS “patients”; and (4) 29 MM I patients. Table 1 details the patient characteristics for each of the 4 groups. As expected, patients with AL ± MM I displayed a higher frequency of λ light chain restriction and a lower frequency of intact immunoglobulin detection compared with the MGUS/MM I group (P < .001 each). However, age, sex, and bone marrow plasmacytosis were equally distributed between AL ± MM I and MGUS/MM I (P = .38, P = .24, and P = .50, respectively).
Hematologic and clinical characteristics for each of the patient groups
| . | AL (n = 220) . | AL + MM I (n = 26) . | MGUS (n = 151) . | MM I (n = 29) . | AL ± MM I vs MGUS/MM I, P . |
|---|---|---|---|---|---|
| Median age, y (range) | 63 (36-82) | 57 (41-80) | 59 (29-87) | 64 (40-82) | .38† |
| Sex, no. male/female | 127/93 | 12/14 | 74/77 | 17/12 | .24‡ |
| Light chain, no. κ/λ | 49/170* | 8/18 | 99/51* | 16/13 | < .001‡ |
| Intact immunoglobulin, no. yes/no | 90/129 | 14/12 | 144/7 | 29/0 | < .001‡ |
| Median plasmacytosis, % (range) | 9 (1-29) | 23 (4-44) | 9 (1-28) | 30 (3-50) | .50† |
| . | AL (n = 220) . | AL + MM I (n = 26) . | MGUS (n = 151) . | MM I (n = 29) . | AL ± MM I vs MGUS/MM I, P . |
|---|---|---|---|---|---|
| Median age, y (range) | 63 (36-82) | 57 (41-80) | 59 (29-87) | 64 (40-82) | .38† |
| Sex, no. male/female | 127/93 | 12/14 | 74/77 | 17/12 | .24‡ |
| Light chain, no. κ/λ | 49/170* | 8/18 | 99/51* | 16/13 | < .001‡ |
| Intact immunoglobulin, no. yes/no | 90/129 | 14/12 | 144/7 | 29/0 | < .001‡ |
| Median plasmacytosis, % (range) | 9 (1-29) | 23 (4-44) | 9 (1-28) | 30 (3-50) | .50† |
These patient groups include one patient with a gammopathy biclonal for κ and λ each.
Exact Wilcoxon rank-sum test.
Fisher exact test.
Informed consent was obtained from the patients in accordance with the Declaration of Helsinki. The study was approved by the Ethics Committee of the University of Heidelberg.
Cytogenetic testing
CD138+ bone marrow plasma cells were purified by auto-magnetic-activated cell sorting with anti-CD138 immunobeads as described previously.22 iFISH analyses were performed using a panel of 2-color probe sets for the detection of numerical changes for the chromosome loci 1q21/8p21, 5p15/5q35, 9q34/15q22, 11q23/13q14, 11p15/11cen, and 17p13/19q13 as well as the translocations t(11;14)(q13;q32), t(4;14)(p16;q32), t(14;16)(q32;q23), and an IgH breakapart probe. Following hybridization according to the manufacturer's instructions (Kreatech and Vysis), a total of 100 interphase nuclei per probe were evaluated using a DM RXA fluorescence microscope (Leica) or the ASI automated iFISH spot counting system (Applied Spectral Imaging). Hybridization efficiency was validated on interphase nuclei obtained from the peripheral blood and bone marrow of a healthy donor. The thresholds for gains, deletions, and translocations were uniformly set at 10%.
For the classification of patients into hyperdiploid versus nonhyperdiploid group, we applied the criteria by Wuilleme et al,11 which require trisomies of at least 2 of the 3 chromosomes 5, 9, and 15 to assign a patient into the hyperdiploidy group. This score had been validated by concomitant DNA staining11 and had already been used for ploidy determination by iFISH in MGUS and MM,23-25 although it might underestimate the frequency of hyperdiploidy.23
Confirmation of monoclonal plasma cell purity
To make sure that in the setting of early monoclonal gammopathies no cytogenetic aberrations were missed because of the contamination of a small monoclonal plasma cell population by a substantial number of polyclonal plasma cells, we evaluated the purity of the plasma cell clone by assessment of clonal sizes and by light chain restriction analysis.
As for clonal size in the 2 early gammopathy groups (AL and MGUS), a clonal aberration by iFISH was detected in 210 of 220 (95%) and 138 of 151 (91%) patients, respectively. The largest clonal aberration encompassed 72% and 63% of plasma cells in median, respectively, well above the 10% threshold. In AL, 198 of 210 (94%) patients displayed an aberration encompassing at least 20% of plasma cells, as did 130 of 138 (94%) MGUS “patients.” The remainder, all specimens with a maximum clonal size of less than 20%, were revisited for possible aberrations below the 10% cutoff used in our study, and the very few questionable cases were retested by iFISH staining, leading to a ploidy reassignment in a single patient.
The flow cytometric analysis after plasma cell enrichment was performed in a cohort of 32 patients with AL, which showed a median ratio of involved versus noninvolved light chains of 10:1 (range, 2.4:1 to 177:1, with 31 of 32 patients displaying at least a ratio of 4:1). Taken together, the monoclonal population clearly outweighed possibly contaminating nonmonoclonal plasma cells.
Statistical analysis
To compare frequencies of the aforementioned cytogenetic aberrations and hyperdiploidy between patient groups, Fisher exact test was performed. The P values obtained for the distribution of additional cytogenetic aberrations among hyperdiploidy and nonhyperdiploidy were subsequently adjusted by the Holm method to control the family-wise error rate at 0.05.26
Oncogenetic tree models derived by maximum likelihood estimation were applied as previously described.27 The state of a cytogenetically normal cell is represented by the root of the tree. Models were built using the following 2 subsets: (1) gains of 1q21, 5p15/5q35, 9q34, 11q23, 15q22, 17p13, and 19q13; and (2) gain of 1q21, hyperdiploidy, t(11;14), t(4;14), IgH rearrangement with unknown partner, and deletion of 13q14. Each subset provides the leaves of the corresponding tree. The length of the paths between the root and the corresponding leaves encodes the marginal probabilities of the observed aberrations. The length (horizontal distance) from the chromosomal aberration to the root or the next inner node is the negative logarithm of the conditional probability that this aberration occurs at this node, given that the hidden events corresponding to this node happened. Nonoverlapping paths to the root represent independent events. A nonparametric bootstrap28 is used to assess the uncertainty of the obtained tree models. Confidence values are given for the internal edges of the maximum likelihood trees, based on 500 bootstrap datasets. The tree displays the splits occurring in more than 10% of the bootstrap datasets in the 2 patient groups. The proposed tree structure has to be interpreted with caution, still allowing us to formulate hypotheses about the association between aforementioned chromosomal aberrations.
To compare hematologic and clinical characteristics between hyperdiploid and nonhyperdiploid groups, Fisher exact test for categorical variables and exact Wilcoxon rank-sum test for quantitative variables were performed, respectively. To analyze the number of chromosomal aberrations, the Cochran-Armitage test for trend was performed. The distribution of patients with gain of 11q23 defined by t(11;14) and hyperdiploidy was analyzed by Fisher exact test. Exact 2-sided 95% confidence intervals (CIs) for the probability of gain of 11p15 and 11cen were estimated according to Clopper et al.29
All statistical tests were 2-sided at significance level of 5%. Statistical analysis was carried out using the software environment R Version 2.9.2,30 together with the R packages ”oncomodel,” Version 0.7 and ”coin,” Version 1.0-6. For the Cochran-Armitage tests, StatXact 7.0 (Cytel Inc) was used.
Results
Frequencies of chromosomal gains
The frequencies of extra copies of 1q21, 5p15/5q35, 9q34, 11q23, 15q22, 17p13, and 19q13 are specified for each patient group in Table 2. Applying the score by Wuilleme et al,11 hyperdiploidy was found in 11% of AL patients, whereas it was detected more frequently in MGUS with 30% (P < .001). Interestingly, the transition of monoclonal gammopathy into MM I was paralleled by an increasing hyperdiploidy rate, namely, from 11% to 46% (P < .001) in AL patients and from 30% to 45% (P = .1) in the MGUS/MM I control group. Accordingly, there was no difference between MM I with and without AL (P = 1). When individual trisomies were assessed in the oncogenetic tree model (Figure 1), gains of 17p13 (with the lowest frequency in each patient group) and 19q13, which are not entered into the score by Wuilleme et al,11 closely clustered with the branches 5p15/5q35, 9q34, and 15q22 both in AL ± MM I and MGUS/MM I. Gain of 1q21, known as progression marker in MM rather than a marker of hyperdiploidy,31 clustered apart as expected. Most remarkable was the clustering of gain of 11q23. Particularly in AL ± MM I, gain of 11q23 appeared to represent hyperdiploidy very poorly because it separated even earlier from the classic hyperdiploidy probes than gain of 1q21.
Frequencies of extra copies of odd chromosomes for each of the patient groups
| . | 1q21, % . | 5p15/5q35, % . | 9q34, % . | 11q23, % . | 15q22, % . | 17p13, % . | 19q13, % . | Hyperdiploid,11 % . |
|---|---|---|---|---|---|---|---|---|
| AL (n = 220) | 21 | 9 | 18 | 34 | 13 | 5 | 14 | 11 |
| AL + MM I (n = 26) | 50 | 32 | 58 | 46 | 38 | 15 | 42 | 46 |
| MGUS (n = 151) | 14 | 29 | 32 | 36 | 32 | 5 | 28 | 30 |
| MM I (n = 29) | 48 | 28 | 52 | 41 | 52 | 0 | 38 | 45 |
| . | 1q21, % . | 5p15/5q35, % . | 9q34, % . | 11q23, % . | 15q22, % . | 17p13, % . | 19q13, % . | Hyperdiploid,11 % . |
|---|---|---|---|---|---|---|---|---|
| AL (n = 220) | 21 | 9 | 18 | 34 | 13 | 5 | 14 | 11 |
| AL + MM I (n = 26) | 50 | 32 | 58 | 46 | 38 | 15 | 42 | 46 |
| MGUS (n = 151) | 14 | 29 | 32 | 36 | 32 | 5 | 28 | 30 |
| MM I (n = 29) | 48 | 28 | 52 | 41 | 52 | 0 | 38 | 45 |
The clustering of chromosomal gains (1q21, 5p15/5q35, 9q34, 11q23, 15q22, 17p13, and 19q13) in an oncogenetic tree model of AL ± MM I and MGUS/MM I. The complete dataset with all 7 branches tested is complete in 243 of 246 AL and in 173 of 180 MGUS/MM I patients. At least one of the 7 investigated aberrations was detected in 58% (n = 140) of the evaluable AL patients and in 60% (n = 104) of the MGUS/MM I patients. The length of each horizontal edge e is proportional to −log(pe). Bootstrap confidence values (in percentage) for the inner nodes are given, based on 500 bootstrap samples. In addition to the clusters shown in the graph for gains in the AL group, gains of 15q22 and 17p13 were identified as a separate cluster in 42% of the bootstrap samples and gains of 9q34 and 17p13 built a cluster in 41% of samples; in another 23%, 15q22 was added to the cluster of 9q34 and 17p13. For MGUS/MM I cases, gains of 19q13 and 17p13 were grouped together in 65% of the 500 bootstrap samples, whereas 5p15 and 17p13 were separately clustered in 21% of samples. In 22% of samples, we observed a terminal cluster of 5p15 together with 15q22.
The clustering of chromosomal gains (1q21, 5p15/5q35, 9q34, 11q23, 15q22, 17p13, and 19q13) in an oncogenetic tree model of AL ± MM I and MGUS/MM I. The complete dataset with all 7 branches tested is complete in 243 of 246 AL and in 173 of 180 MGUS/MM I patients. At least one of the 7 investigated aberrations was detected in 58% (n = 140) of the evaluable AL patients and in 60% (n = 104) of the MGUS/MM I patients. The length of each horizontal edge e is proportional to −log(pe). Bootstrap confidence values (in percentage) for the inner nodes are given, based on 500 bootstrap samples. In addition to the clusters shown in the graph for gains in the AL group, gains of 15q22 and 17p13 were identified as a separate cluster in 42% of the bootstrap samples and gains of 9q34 and 17p13 built a cluster in 41% of samples; in another 23%, 15q22 was added to the cluster of 9q34 and 17p13. For MGUS/MM I cases, gains of 19q13 and 17p13 were grouped together in 65% of the 500 bootstrap samples, whereas 5p15 and 17p13 were separately clustered in 21% of samples. In 22% of samples, we observed a terminal cluster of 5p15 together with 15q22.
Based on these results, we added gains of 17p13 and 19q13 to the 3 probes defining the score by Wuilleme et al11 and analyzed the distribution of these 5 chromosomal gain frequencies in the 4 patient groups. This confirmed that the rate of gained chromosomes was lower in AL compared with MGUS (P < .001, frequency distribution; Table 3) and was higher when the plasma cell disorder had reached the stage of MM I (P < .001 for AL; P = .26 for MGUS).
Number of extra copies of the 5 probes 5p15/5q35, 9q34, 15q22, 17p13, and 19q13 for each of the patient groups
| . | No extra copy, % . | 1 extra copy, % . | 2 extra copies, % . | 3 extra copies, % . | 4 extra copies, % . | 5 extra copies, % . |
|---|---|---|---|---|---|---|
| AL (n = 220) | 76 | 8 | 6 | 4 | 3 | 3 |
| AL + MM I (n = 25*) | 44 | 8 | 12 | 8 | 20 | 8 |
| MGUS (n = 144*) | 58 | 7 | 6 | 9 | 17 | 3 |
| MM I (n = 29) | 41 | 7 | 14 | 17 | 21 | 0 |
| . | No extra copy, % . | 1 extra copy, % . | 2 extra copies, % . | 3 extra copies, % . | 4 extra copies, % . | 5 extra copies, % . |
|---|---|---|---|---|---|---|
| AL (n = 220) | 76 | 8 | 6 | 4 | 3 | 3 |
| AL + MM I (n = 25*) | 44 | 8 | 12 | 8 | 20 | 8 |
| MGUS (n = 144*) | 58 | 7 | 6 | 9 | 17 | 3 |
| MM I (n = 29) | 41 | 7 | 14 | 17 | 21 | 0 |
Only patients with all 5 probes available are included.
Dichotomy of patients with gain of 11q23
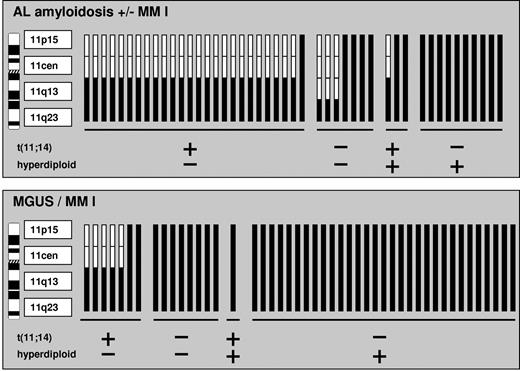
We assessed AL ± MM I patients with gain of 11q23 in more detail and could delineate 2 subgroups (Table 4): they displayed either a concomitant t(11;14) or hyperdiploidy as defined by the score by Wuilleme et al11 (P < .001). Using additional probes for 11p15 and 11cen, we could show that t(11;14)-positive patients with gain of 11q23 generally present a normal diploid status for the loci 11p15 and 11cen, so that the gain of 11q23 is merely related to the t(11;14) rearrangement (Figure 2). Because all patients with gain of 11q23 and concomitant hyperdiploidy on the contrary also displayed extra copies of 11p15 and 11cen, gain of 11q23 in these patients reflected trisomy of the whole chromosome 11. This dichotomy was highly statistically significant (2-sided 95% CIs for the probability of gain of 11p15 and 11cen between the t(11;14)-positive/hyperdiploidy negative group and the t(11;14)-negative/hyperdiploidy positive group were not overlapping; 95% CI, 0.01-0.3 and 0.7-1.0 respectively).
Distribution of patients with gain of 11q23 in subgroups defined by t(11;14) and hyperdiploidy
| . | t (11;14) negative . | t (11;14) positive . |
|---|---|---|
| AL ± MM I (n = 246) | ||
| Hyperdiploidy negative | 13* | 51 |
| Hyperdiploidy positive | 18 | 5† |
| MGUS/MM I (n = 180) | ||
| Hyperdiploidy negative | 13 | 12 |
| Hyperdiploidy positive | 40 | 1† |
| . | t (11;14) negative . | t (11;14) positive . |
|---|---|---|
| AL ± MM I (n = 246) | ||
| Hyperdiploidy negative | 13* | 51 |
| Hyperdiploidy positive | 18 | 5† |
| MGUS/MM I (n = 180) | ||
| Hyperdiploidy negative | 13 | 12 |
| Hyperdiploidy positive | 40 | 1† |
Three patients with a local gain of 11q23 without a concomitant gain of 11q13 (see Figure 2) are included in this number.
In patients with t(11;14) and hyperdiploidy, the clonal size of both aberrations was similar, suggesting that the same clone harbors both aberrations.
Gains of 11p15, 11cen, and 11q13 in patients positive for gain of 11q23 depending on their t(11;14) and hyperdiploidy status. A chromosomal gain is represented by black bars and a normal diploid status is indicated by white bars. Patient samples are grouped according to t(11;14) and hyperdiploidy subgroups.
Gains of 11p15, 11cen, and 11q13 in patients positive for gain of 11q23 depending on their t(11;14) and hyperdiploidy status. A chromosomal gain is represented by black bars and a normal diploid status is indicated by white bars. Patient samples are grouped according to t(11;14) and hyperdiploidy subgroups.
The same dichotomy emerged in our MGUS/MM I patients with gain of 11q23 (P < .001, 2-sided 95% CIs for gains of 11q15 and 11cen not overlapping; 95% CI, 0.04-0.7 and 0.9-1.0, respectively). However, because of the lower t(11;14) frequency and the higher hyperdiploidy rate in this group, the distribution pattern was inversed (Table 4).
Finally, we analyzed our samples with gain of 11q13 without concomitant gain of 11q23. This constellation was invariably associated with t(11;14) (65 of 66 patients in AL ± MM I, 28 of 28 in MGUS/MM I), and accordingly only a single of 60 patients tested for 11p15 and 11cen had gained chromosome 11 as a whole.
Association of hyperdiploidy with hematologic and clinical parameters
As for hematologic parameters in the overall study population, hyperdiploid patients displayed higher frequencies of an intact immunoglobulin (P < .001; Table 5) and κ light chain restriction (P < .001) as well as a higher degree of bone marrow plasmacytosis (P = .007) compared with nonhyperdiploid patients. As for clinical parameters, hyperdiploid patients were diagnosed at a higher age (P = .04), but no sex predilection was observed (P = .5).
Association of hyperdiploidy with clinical and hematologic parameters of the entire study population
| . | Age (median, range), y . | Sex (male vs female), % . | Intact Ig, % . | Ig subtype (IgG vs IgA), % . | Light chain restricition (κ vs λ), % . | Bone marrow plasmacytosis, % (median, range) . |
|---|---|---|---|---|---|---|
| Hyperdiploid (n = 95) | 63 (40-87) | 51-49 | 89 | 79-21 | 57-43 | 12 (1-50) |
| Nonhyperdiploid (n = 331) | 60 (29-84) | 55-45 | 58 | 76-24 | 36-64 | 9 (1-46) |
| P | .04 | .5 | < .001 | .6 | < .001 | .007 |
| . | Age (median, range), y . | Sex (male vs female), % . | Intact Ig, % . | Ig subtype (IgG vs IgA), % . | Light chain restricition (κ vs λ), % . | Bone marrow plasmacytosis, % (median, range) . |
|---|---|---|---|---|---|---|
| Hyperdiploid (n = 95) | 63 (40-87) | 51-49 | 89 | 79-21 | 57-43 | 12 (1-50) |
| Nonhyperdiploid (n = 331) | 60 (29-84) | 55-45 | 58 | 76-24 | 36-64 | 9 (1-46) |
| P | .04 | .5 | < .001 | .6 | < .001 | .007 |
In AL ± MM I, ploidy status proved unrelated to number of involved organs and the organ involvement pattern.
Association of hyperdiploid and nonhyperdiploid phenotypes with other cytogenetic aberrations
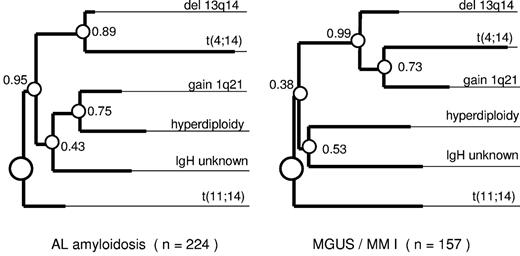
Both in the AL ± MM I and the MGUS/MM I groups, t(11;14) was rarely detected in the hyperdiploid group (Figure 3). This inverse association was highly significant (P < .001 in both entities). All 5 cases with translocation t(14;16) also belonged to the nonhyperdiploid category; however, the very small number excluded its assignment to ploidy groups (P = 1). On the contrary, t(4;14) and IgH translocations with an unknown partner were equally distributed among the ploidy groups (P = 1 in both entities). Both ploidy groups also displayed similar frequencies of deletion 13q14 (P = 1 in both entities). This cytogenetic aberration pattern is also visualized by our oncogenetic tree model, which assigns t(11;14) and hyperdiploidy both in AL ± MM I and MGUS/MM I to separate branches originating right from the root of the tree (Figure 4). In addition to t(11;14) and hyperdiploidy, deletion 13q14, with t(4;14) linked to it, and IgH translocations with an unknown translocation partner emerged as distinct branches, which separated very early from the hyperdiploidy branch. Although the oncogenetic tree models for AL ± MM I and MGUS/MM I were largely overlapping, there was one difference regarding gain of 1q21. Whereas it clustered with the deletion of 13q14/t(4;14) branch in MGUS/MM I, it was associated with hyperdiploidy in AL ± MM I (P = .001; Figure 3).
The frequencies of cytogenetic aberrations in hyperdiploid versus nonhyperdiploid patients in the AL ± MM and MGUS/MM I entities. Statistical significance was reached for the inverse association of hyperdiploidy with t(11;14) in both AL ± MM I (P < .001) and MGUS/MM I (P < .001) as well as for the association of hyperdiploidy with gain of 1q21 in AL ± MM I (P = .02).
The frequencies of cytogenetic aberrations in hyperdiploid versus nonhyperdiploid patients in the AL ± MM and MGUS/MM I entities. Statistical significance was reached for the inverse association of hyperdiploidy with t(11;14) in both AL ± MM I (P < .001) and MGUS/MM I (P < .001) as well as for the association of hyperdiploidy with gain of 1q21 in AL ± MM I (P = .02).
The clustering of the major cytogenetic aberrations (gain of 1q21, hyperdiploidy, t(11;14), t(4;14), IgH split with unknown translocation partner, and deletion of 13q14) in an oncogenetic tree model of AL ± MM I and MGUS/MM I. The complete dataset with all 6 branches tested is complete in 241 of 246 AL and in 176 of 180 MGUS/MM I patients. At least one of the 6 investigated aberrations was detected in 93% (n = 224) of the evaluable AL patients and in 89% (n = 157) of the MGUS/MM I patients. The length of each horizontal edge e is proportional to −log(pe). Bootstrap confidence values (in percentage) for the inner nodes are given, based on 500 bootstrap samples. Besides the clusters shown in this figure, t(4;14) clustered together with gain 1q21, del 13q14, and hyperdiploidy in 46% of the bootstrap samples for AL cases and in 22% for MGUS/MM I cases.
The clustering of the major cytogenetic aberrations (gain of 1q21, hyperdiploidy, t(11;14), t(4;14), IgH split with unknown translocation partner, and deletion of 13q14) in an oncogenetic tree model of AL ± MM I and MGUS/MM I. The complete dataset with all 6 branches tested is complete in 241 of 246 AL and in 176 of 180 MGUS/MM I patients. At least one of the 6 investigated aberrations was detected in 93% (n = 224) of the evaluable AL patients and in 89% (n = 157) of the MGUS/MM I patients. The length of each horizontal edge e is proportional to −log(pe). Bootstrap confidence values (in percentage) for the inner nodes are given, based on 500 bootstrap samples. Besides the clusters shown in this figure, t(4;14) clustered together with gain 1q21, del 13q14, and hyperdiploidy in 46% of the bootstrap samples for AL cases and in 22% for MGUS/MM I cases.
Because the oncogenetic tree clustering is based on the principle of plasma cell clones harboring more than one cytogenetic aberration, we also used the 6 major cytogenetic aberrations considered in the oncogenetic tree model as a measure for the degree of chromosomal instability. The aberration frequencies were alike in the AL and the MGUS groups (P = .49; Table 6). However, within both groups, MM I was associated with an increased number of detected chromosomal aberrations (P < .001 in AL ± MM I, P < .001 in MGUS/MM I).
Chromosomal instability as determined by the number of major cytogenetic aberrations (IgH translocations, gain of 1q21, deletion 13q14, and hyperdiploidy)
| . | No chromosome aberration, % . | 1 chromosome aberration, % . | 2 chromosome aberrations, % . | 3 chromosome aberrations, % . | 4 chromosome aberrations, % . |
|---|---|---|---|---|---|
| AL (n = 217)* | 7 | 54 | 28 | 9 | 2 |
| AL + MM I (n = 26) | 0 | 35 | 23 | 35 | 8 |
| MGUS (n = 149)* | 11 | 51 | 28 | 8 | 2 |
| MM I (n = 29) | 3.5 | 28 | 31 | 34 | 3.5 |
| . | No chromosome aberration, % . | 1 chromosome aberration, % . | 2 chromosome aberrations, % . | 3 chromosome aberrations, % . | 4 chromosome aberrations, % . |
|---|---|---|---|---|---|
| AL (n = 217)* | 7 | 54 | 28 | 9 | 2 |
| AL + MM I (n = 26) | 0 | 35 | 23 | 35 | 8 |
| MGUS (n = 149)* | 11 | 51 | 28 | 8 | 2 |
| MM I (n = 29) | 3.5 | 28 | 31 | 34 | 3.5 |
Only patients with all 4 probes available are included.
Discussion
In this study, we could establish that in AL, as in MGUS and MM I, a portion of patients have a hyperdiploid karyotype as underlying cytogenetic instability assessed by the score of Wuilleme et al.11 So, hyperdiploidy can be detected at an early stage of plasma cell dyscrasia best represented by AL, which becomes rather symptomatic because of the amyloidogenicity of the light chains, its “unlucky protein,”13,16 than because of the malignancy of the plasma cell clone.
The cytogenetic aberration patterns known to go along with ploidy groups in MM could be reproduced both in our AL ± MM I and our MGUS/MM I groups. Accordingly, the oncogenetic trees for both entities largely overlapped. In detail, we could confirm the strong inverse association between “primary” IgH translocations, particularly t(11;14), and hyperdiploidy, which has repeatedly been described in MM11,17,23,32-37 and supports the concept of dichotomy of hyperdiploidy and nonhyperdiploidy.2,36,38 In addition, in line with previous MM studies,36,37 we could show that the inverse association of hyperdiploidy and IgH translocations does not apply to IgH translocations with an unidentified partner, the “secondary” IgH translocations, which were equally distributed between hyperdiploidy and nonhyperdiploidy. Despite its association with nonhyperdiploidy in most MM studies,10,11,24,25,36 deletion of 13q14 in our early-stage monoclonal gammopathy groups was rather equally distributed between hyperdiploidy and nonhyperdiploidy as well. Overall, the recurring overlap of aberration patterns in AL and MGUS highlights the common pathogenetic mechanisms underlying these plasma cell dyscrasia entities.
Despite these shared pathogenetic patterns, the frequencies for individual aberrations widely differed among disease entities. First, hyperdiploidy was rare in AL compared with MGUS and MM I. Vice versa, AL showed its known high prevalence of t(11;14),13,15,16,39 which is considered a very early and possible initiating event of clonal immortalization driving the nonhyperdiploid pathway. However, beyond this AL versus non-AL comparison, the distribution frequencies of karyotypic aberrations also depended on the stage of the plasma cell disorder from MGUS to MM I. Strikingly, we observed a markedly higher prevalence of hyperdiploid karyotypes in AL with concomitant MM I (46%) compared with AL without MM I (11%, P < .001). Of note, this hyperdiploidy frequency of AL patients with concomitant MM I was close to the 51% to 60% range consistently reported for symptomatic MM patients by previous studies using different techniques, such as conventional karyotyping, iFISH analysis, and flow cytometric DNA content measurement.10-12,35,40,41 Vice versa, the frequency of t(11;14) dropped in AL during disease transition to MM I (55% vs 35%, P = .06).
The high prevalence of hyperdiploidy in the AL + MM I group suggests that the hyperdiploid karyotypes are biologically distinct and manifest themselves with a higher plasma cell burden. Accordingly, the hyperdiploid phenotype in our study frequently showed an intact immunoglobulin and a κ light chain restriction, mimicking features known to be characteristic of MM. A second factor may also contribute to the higher hyperdiploidy frequency of 46% in the AL + MM I group: our study demonstrates a growing overall karyotypic instability during progression of the plasma cell dyscrasia (Table 6). It might be therefore speculated that this growing overall karyotypic instability also leads to additional chromosomal gains and thus contributes to the higher hyperdiploidy frequency in the AL + MM I group (Tables 2–3). From a biologic viewpoint, it is probable that chromosomal gains characteristic of hyperdiploidy are gained rather step by step than at once.42 AL represents a particular early stage of monoclonal gammopathy, so inherently hyperdiploid phenotypes may not have expanded and completed their chromosomal gains yet, which contributes to a lower hyperdiploidy frequency in the AL group compared with the AL + MM I group. In our study, the association of hyperdiploidy in AL ± MM I with gain of 1q21 (P < .001; Figure 3), which is a known progression factor in MM,43 supports the concept of hyperdiploidy as an evolving process in an early stage of myelomagenesis. Although the dichotomy concept of hyperdiploidy versus nonhyperdiploidy is well estabilished,44 only a few studies have yet interpreted hyperdiploidy as a possible progression-related event in plasma cell dyscrasias.6,25 The concept of acquiescence of additional chromosomal gains during disease progression is also supported by a small follow-up analysis of MGUS “patients” who acquired slowly gradual chromosome changes.45 Our study, again, was only based on cross-sectional, but not on longitudinal, individual follow-up data. However, given the validation of monoclonal plasma cell purity by light chain staining and assessment of clonal sizes in our study, the increasing hyperdiploidy frequency during disease progression truly reflects a biologic phenomenon and does not merely mirror the higher degree of bone marrow plasmacytosis in advanced gammopathies.
As for the assessment of individual trisomies, the oncogenetic tree model confirmed the strong association of gains of 17p13 and 19q13 with gains of 5p15/5q35, 9q34, and 15q22 used in the score by Wuilleme et al.11 Gain of 11q23, however, had a distinct branching pattern. For both AL ± MM I and MGUS/MM I groups, we could elaborate the dichotomy of 11q23 gains, which split into t(11;14)-positive and hyperdiploid karyotypes. In the first group, gain of 11q23 is the consequence of a complex translocation involving t(11;14). In the hyperdiploid group, gain of 11q23 represents an additional chromosome 11 as confirmed by gains for 11p15 and 11cen. In AL, t(11;14) has a particularly high frequency and hyperdiploidy is rare, so 11q23 is most frequently detected in conjunction with t(11;14), which accounts for their common clustering in the oncogenetic tree model described previously by our group.16 Vice versa, in MGUS/MM I, where hyperdiploidy frequencies prevail more than t(11;14) frequencies, gain of 11q23 truly reflects an additional chromosome 11 in most cases. Therefore, chromosome 11 iFISH probes should be chosen and interpreted cautiously. Beyond that, our chromosome 11 workup again mirrors the role of t(11;14) and hyperdiploidy as antagonizing poles in plasma cell neoplasms.
In conclusion, AL, with its particularly early-stage monoclonal gammopathy, appears to be a suitable model to investigate the biology of MM oncogenesis.
Preliminary data were presented in abstract form at the 51st Annual Meeting of the American Society of Hematology, New Orleans, LA, December 5-8, 2009.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Michaela Brough for excellent technical assistance in iFISH analysis and Katrin Heimlich and Maria Dörner for their excellent technical assistance in plasma cell enrichment.
This work was supported in part by grants from the Tumorzentrum Heidelberg/Mannheim (D.10026942) (A.J., K.N.), the Dietmar-Hopp-Stiftung Heidelberger Konzept zur Optimierung der Diagnostik und Therapie des Multiplen Myeloms (H.G.), and Celgene Germany (S.O.S.).
Authorship
Contribution: T.B., U.H., A.J., and S.O.S. conceived and designed the study; T.B., U.H., K.N., D.H., H.G., and S.O.S. provided study materials/patients; T.B., U.H., A.J., and S.O.S. collected and assembled data; S.P.-Z., D.K., D.H., A.S., C.R.B., H.G., and A.J. conducted plasma cell enrichment and iFISH; T.B., U.H., M.M., A.J., and S.O.S. analyzed and interpreted data; C.H. and A.B. performed statistical analysis; T.B., U.H., C.H., A.B., M.M., A.J., and S.O.S. wrote the manuscript; and T.B., U.H., C.H., A.B., M.M., A.S., K.N., D.H., C.R.B., A.D.H., H.G., A.J., and S.O.S. gave final approval of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stefan O. Schonland, Amyloidosis Center, Department of Internal Medicine, Division of Hematology/Oncology, University of Heidelberg, Im Neuenheimer Feld 410, 69120 Heidelberg, Germany; e-mail: Stefan.Schoenland@med.uni-heidelberg.de.
References
Author notes
T.B. and U.H. contributed equally to this study.
A.J. and S.O.S. are to be considered equal last authors.