Abstract
The Ets-related gene (ERG) located on human chromosome 21 encodes a transcription factor and is thought to be causally related to Down syndrome–associated acute megakaryocytic leukemia in childhood. In clinical adult leukemia, however, increased expression of ERG is indicative of poor prognosis in T-cell acute lymphoblastic leukemia and cytogenetically normal acute myeloid leukemia, although the involvement of ERG in the development of adult leukemia remains elusive. Here, we show that forced expression of ERG in adult BM cells alters differentiation and induces expansion of T and erythroid cells and increases frequencies of myeloid progenitors in mouse BM transplantation models. The expanded T cells then develop T-cell acute lymphoblastic leukemia after acquisition of mutations in the Notch1 gene. Targeted expression of ERG into B cells also altered differentiation and promoted growth of precursor B cells. Overall, these findings suggest a general role of ERG in promoting growth of adult hematopoietic cells in various lineages. In line with this, shRNA-mediated silencing of ERG expression attenuated growth of human leukemia cell lines of various lineages. Thus, ERG is capable of promoting the development of leukemia and is crucial for its maintenance.
Introduction
Ets-related gene (ERG), a member of the ETS transcription factor gene family, is essential for establishing normal megakaryopoiesis and definitive hematopoiesis, as well as maintenance of the hematopoietic stem cell.1 ERG is also involved in oncogenesis by generating fusion genes with FUS/TLS in acute myeloid leukemia (AML),2,3 EWS in Ewing sarcoma4 and TMPRSS2 in prostate cancer.5 The FUS/TLS-ERG fusion protein contains a truncated form of ERG that retains the ETS domain and part of the RNA-binding protein FUS/TLS.2,3 The FUS/TLS-ERG fusion protein is able to transform NIH3T3 cells and to block G-CSF–induced differentiation of the L-G mouse myeloid cell line, depending on the functions of the FUS/TLS part of the fusion.2,6 Notwithstanding these activities, including its ability to enhance cell proliferation, the presence of the FUS/TLS-ERG gene itself is insufficient to initiate leukemia but requires additional cooperating genetic events.7,8 Interestingly, the EWS-ERG fusion protein, in which another RNA-binding protein, EWS, is fused to the ETS domain of ERG, is able to induce leukemia when ectopically expressed in the T-cell compartment in mice,9 although it is noteworthy that the fusion is associated with sarcomas, and not leukemia, in humans. In the case of the TMPRSS2-ERG fusion gene, ERG expression is under control of the TMPRSS2 promoter region containing androgen-response elements.5 Unlike the fusion proteins, ERG protein itself is subject to dysregulated expression in prostate and has been shown to contribute to tumorgenesis.10-15 With respect to leukemiagenesis, ERG has recently drawn much attention in terms of being linked to Down syndrome (DS)–associated acute megakaryocytic leukemia (AMKL)16-18 : ERG is located on human chromosome 21 (Hsa21) and children with germline trisomy 21 (DS) have an increased risk of AMKL.19 Ts(1716)65 Dn mice, used as models of DS containing a trisomic chromosomal DS critical region of Hsa21, develop myeloproliferative disorder, and mice manipulated to be disomic for functional Erg show amelioration of the phenotype.20 Megakaryoblasts isolated from clinical samples from almost all patients with DS with AMKL have somatic mutations in the GATA-1 transcription factor gene, resulting in replacement of full-length globin transcription factor 1 (GATA1) by a shorter isoform, GATA1s,21-25 although the GATA1 mutation alone seems insufficient for leukemogenesis because GATA1s causes only transient proliferation of immature fetal megakaryocytic progenitors in mice.26 Studies manipulating mouse fetal hematopoietic cells to express both GATA1s and ERG have shown that the 2 genes have cooperative roles in cell transformation.17,18 These findings have many implications for infantile leukemia, and, although these studies focused on fetal hematopoietic cells as a target of ERG expression, little is known about the way by which increased ERG expression affects adult hematopoietic systems. In clinical adult human leukemia, a high level of ERG expression is associated with poor prognosis in cytogenetically normal AML and T-cell acute lymphoblastic leukemia (T-ALL),27,28 and increased expression of ERG characterizes a subset of myeloid leukemia with complex karyotype,29 raising the possibility that ERG itself, but not its fusion protein, plays a role in the development or progression/maintenance of leukemia in adults. Here, we explored the effects of forced expression of ERG on adult hematopoietic cells with the use of a mouse BM transplantation (BMT) model. Finally, we provide evidence to show that ERG plays an important role in the maintenance of human leukemia.
Methods
Generation of retrovirus
cDNA for human ERG (NM_182918.3; generously provided by Dr T. Yamada, Hirosaki University, Japan)30 was cloned into the MSCV-ires-GFP vector. B cell–selective lentivirus vector pTRIPΔUEμMarLCD19GFP (generously provided by T. Moreau and C. Tonnelle, Institut Paoli-Calmettes, Centre de Thérapie Cellulaire et Génique, Centre Régional de Lutte Contre le Cancer)31 was modified to allow for coexpression of ERG and GFP, and cloned into the CSII-EF1-iresGFP lentivirus vector (obtained from RIKEN BRC; with permission of Dr H. Miyoshi). cDNA for Kusabira-Orange (KO) was obtained from pGCDNsamIRES-KO plasmid (a generous gift of Dr M. Onodera, National Research Institute for Child Health and Development, Japan).32 Target sequences for shRNA were GACTCTTGGGAGGGAGTTA (shRNA–ERG-1),33 CGACATCCTTCTCTCACAT (shRNA–ERG-2),11 and CTTACGCTGAGTACTTCGA (luciferase).
Retroviral infection of cells and transplantation
BM cells isolated from 8- to 10-week-old Ly5.1-C57BL6 mice (a generous gift of Dr T. Takahashi, ex-chancellor of Aichi Cancer Center) were infected with virus, as described,34 and were transplanted into lethally irradiated Ly5.2-C57BL6 mice (Charles River). Pro-B cells (B220+c-Kit+) were isolated from BM and fetal liver, cultured on OP9 cells (RIKEN BRC) in IMDM supplemented with 15% FSC, SCF, Flt3 ligand (FL), IL-7, and 2-ME, and infected with retrovirus for ERG, GFP-only, and KO. Cells were then sorted for GFP or KO and injected into the BM cavity of syngeneic host or NOD-SCID mice (Charles River). In some experiments, thymocytes were intravenously transferred to lethally irradiated syngeneic hosts along with radioprotective fresh BM cells. All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee at the Aichi Cancer Center.
In vitro culture of cells and colony-forming assays
Human leukemia cells were cultured in IMDM supplemented with 10% FCS. Pro-B cells were plated in triplicate in methylcellulose with SCF, FL, and IL-7. Lineage-negative BM cells were plated in triplicate in methylcellulose with SCF, IL-3, IL-6, and GM-CSF.34
Flow cytometry
Antibodies used were as follows: anti-B220 (RA3-6B2), anti-CD19 (1D3), anti-CD3 (145-2C11), anti–TER-119 (TER-119), anti–Gr-1 (RB6-8C5), anti–c-kit (2B8), anti-CD43 (S7), anti-CD45.1 (A20), anti-CD45.2(104) anti-Mac1 (M1/70), anti-CD4 (GK1.5), anti-CD8 (53-6.7), and anti-CD71 (R17217). Biotinylated annexin V (Sigma) was used to monitor apoptosis. Cytometry was performed with the use of a FACSCalibur (BD Bioscience) and FlowJo software (TreeStar). Flow sorting was conducted with a JSAN cell sorter (Bay Bioscience).
Micrographs
Tissues and colonies were examined under microscopes BH-2 and CKX31 (Olympus), respectively, and images were obtained with a C-4040 digital camera (Olympus). The images were then processed by Photoshop Elements software Version 2.0 (Adobe).
Western blot analysis
Antibodies for ERG (C-17; Santa Cruz Biotechnology), FLAG (M2; Sigma), and tubulin (Sigma) were used.
Cell-cycle analysis
Cells were treated with 0.1% Triton X-100/PBS and incubated with RNase and propidium iodide. Incorporation of BrdU was measured with the use of an anti-BrdU antibody (Bu5.1; Progen Biotechnik).
Microarray and bioinformatics analyses
RNA was extracted with Trizol reagent (Invitrogen). Agilent Mouse Whole Genome 4 × 44K microarrays, Agilent Microarray Scanner, and Agilent Feature Extraction were used. Microarray data are available in the ArrayExpress database under accession nos. E-MEXP-2988 and E-MEXP-2986. Gene set enrichment analyses were performed with the use of Gene Set Enrichment Analysis Version 2.0 software (http://www.broad.mit.edu/gsea) with a log2 ratio of classes metric for gene ranking and 1000 data permutations.
A gene set for genes regulated by Notch1 was composed of DTX1, MYC, NOTCH3, P2RX5, HSPC111, LZTFL1, SHQ1, CR2, EVL, FLJ20485, PWP2H, IGF1R, HES1, PCBP3, SRM, MGC2574, ISG20L1, VARS, AKAP13, SLC29A1, TASP1, DHX37, C20orf6, TCOF1, GPATC4, C1orf33, TRMT1, MRPL4, WDR3, RHOU, PPRC1, FKBP4, APRT, TMEPAI, E2F5, MYBBP1A, IFRD2, KIAA0922, MGC15416, FARSLB, and FLJ14827, as described in the literature.35 Primers used for real-time PCR were GTGTCTGTGCCGTGGCCTCC and TGGGACTACGTGGGCCCGAG (Notch1), CTTGGCCAGGTGGACGCCC and ACACGATGCCCTTGCCCGG (Deltex1), ACACCGGACAAACCAAAGAC and ATGCCGGGAGCTATCTTTCT (Hes1), TGAGGAGACACCGCCCACCA and TGGGCTGTGCGGAGGTTTGC (c-Myc), and, TGCGTGACATCAAAGAGAAG and GATGCCACAGGATTCCATA (β-actin).
Southern blot analysis of genomic DNA and detection of mutations in the Notch1 gene
Proviral integration patterns were determined by Southern blot analysis of isolated DNA with the use of the EcoRI restriction enzyme and a [32P] dCTP-labeled EGFP probe. Mutations in the Notch1 gene were detected, as described.36
Results
ERG induces rapid expansion of T cells and erythroblasts in mice
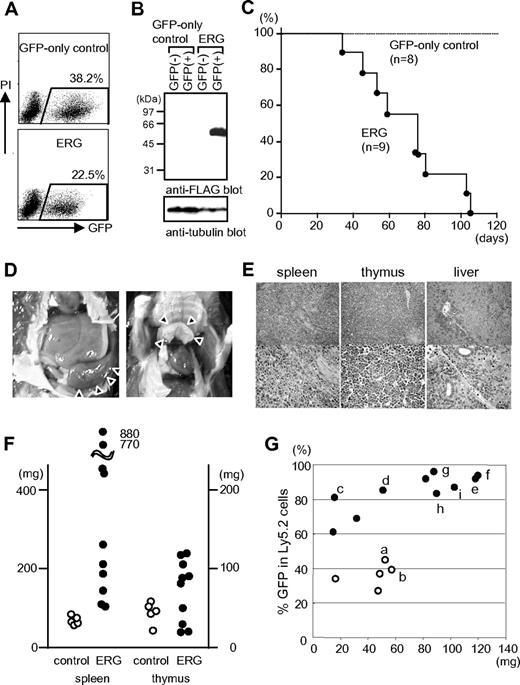
We initially investigated the effects of increased ERG expression on adult mouse hematopoiesis with the use of a mouse BMT model. BM cells harvested from Ly5.2-B6 mice treated with 5-fluorouracil (5-FU) 4 days earlier were infected with a retroviral vector for ERG that coexpresses GFP by virtue of an internal ribosome entry site (ires) element, or a control vector. The infection efficiency ranged from ∼ 20% to 40% (Figure 1A), and the expression of the exogenous ERG was confirmed by Western blot analysis (Figure 1B). Cells were then transplanted into lethally irradiated congenic Ly5.1-B6 mice. Mice that received a transplant with control cells remained healthy (n = 8), but all mice that received a transplant with cells expressing ERG succumbed to leukemia within 110 days (n = 9) (Figure 1C). A second cohort of mice comprising 5 control and 18 ERG mice showed similar results (not shown); all of the control mice and 10 ERG mice were killed between days 50 and 80 after transplantation (8 ERG mice died during this period). Typical ERG mice exhibited an enlarged spleen and thymus (Figure 1D), and histologic examination showed disrupted architecture of the spleen and thymus, and organs (such as the liver and spleen) that were heavily infiltrated by blastic cells (Figure 1E). Although the spleen was invariably enlarged in ERG mice compared with control mice, the thymus from some ERG mice did not show enlargement (Figure 1F). The thymus nevertheless was predominantly populated by GFP+ (therefore ERG-expressing) cells in ERG mice, whereas in control mice GFP+ cells represented only a minor cell population in the thymus (Figure 1G), although ERG- and GFP-only control-infected cells were comparable in regard to percentage of GFP in transplanted cells (Figure 1A). These findings suggest that ERG-expressing cells possess a growth advantage over non–ERG-expressing cells in the thymus.
Mouse BM cells transduced with ERG induce leukemia in hosts that received a transplant. (A) FACS analysis of GFP expression in infected BM cells immediately after infection with GFP-only control and ERG-containing viruses. (B) Western blot analysis of lysates from cells sorted immediately after infection with the use of anti-Flag and antitubulin antibodies. (C) Kaplan-Meier survival curve for cohorts of mice that received a transplant with BM cells transduced with the indicated viruses (n = 8 for vector control; n = 9 for ERG). (D) Splenomegaly (left) and enlarged thymus (right) in mice that received a transplant with cells expressing ERG. (E) Leukemic cell infiltration into organs (spleen, thymus, and liver) was determined with the use of H&E staining. Original magnifications: upper panels, ×40 (SPlan4 4×/0.13 160/-objective); lower panels, ×200 (SPlan20 20×/0.46 160/0.17 objective). (F) Weights of spleen and thymus of mice that received a transplant with cells infected with control and ERG-containing viruses. (G) Percentage of GFP among Ly5.2 donor cells repopulated in thymus of Ly5.1 host mice that received a transplant. Open and filled circles represent control and ERG-transduced cells, respectively. Here, “a” to “i” correspond to mice used for the flow cytometric analysis shown in Figure 2A, Southern blot analysis shown in Figure 3C, and mutational analysis shown in Table 1.
Mouse BM cells transduced with ERG induce leukemia in hosts that received a transplant. (A) FACS analysis of GFP expression in infected BM cells immediately after infection with GFP-only control and ERG-containing viruses. (B) Western blot analysis of lysates from cells sorted immediately after infection with the use of anti-Flag and antitubulin antibodies. (C) Kaplan-Meier survival curve for cohorts of mice that received a transplant with BM cells transduced with the indicated viruses (n = 8 for vector control; n = 9 for ERG). (D) Splenomegaly (left) and enlarged thymus (right) in mice that received a transplant with cells expressing ERG. (E) Leukemic cell infiltration into organs (spleen, thymus, and liver) was determined with the use of H&E staining. Original magnifications: upper panels, ×40 (SPlan4 4×/0.13 160/-objective); lower panels, ×200 (SPlan20 20×/0.46 160/0.17 objective). (F) Weights of spleen and thymus of mice that received a transplant with cells infected with control and ERG-containing viruses. (G) Percentage of GFP among Ly5.2 donor cells repopulated in thymus of Ly5.1 host mice that received a transplant. Open and filled circles represent control and ERG-transduced cells, respectively. Here, “a” to “i” correspond to mice used for the flow cytometric analysis shown in Figure 2A, Southern blot analysis shown in Figure 3C, and mutational analysis shown in Table 1.
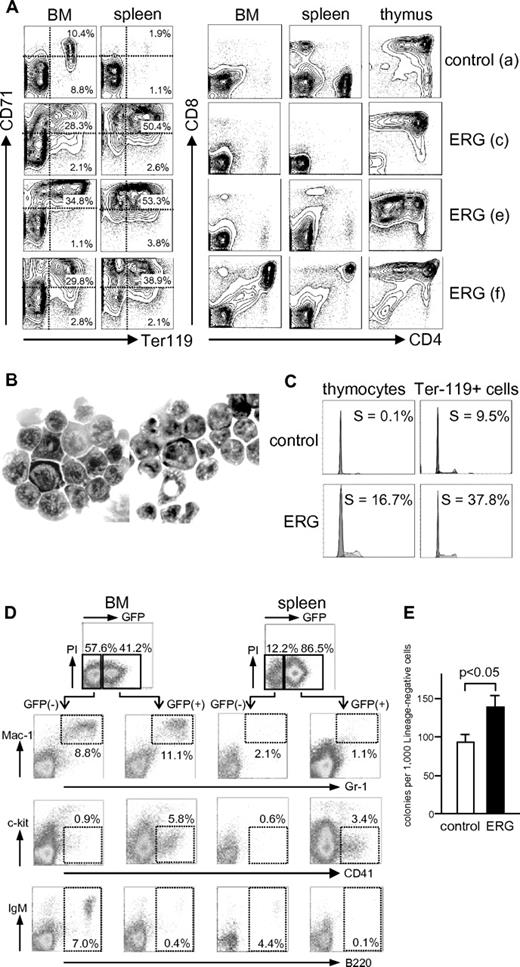
Figure 2A shows flow cytometric analyses of BM, spleen, and thymus of a control mouse (Figure 1G mouse a) and 3 ERG mice (Figure 1G mouse c, mouse e, and mouse f). Results showed increased Ter-119+ erythroblasts in ERG mice, which was accompanied by inhibited differentiation of CD71high immature to CD71low mature erythroblasts37 compared with GFP-only control mice. Analysis of thymus showed an abnormal CD4/CD8 expression profile even in nonenlarged thymus (Figure 1G mouse c), suggestive of an altered T-cell differentiation program. In some ERG mice (Figure 1G f), CD4+CD8+ cells were detected in BM or spleen, whereas they were not detected in control mice. Cytospin preparations of spleen cells (Figure 1G mouse f) showed clusters of morphologically immature erythrocytes (Figure 2B left) and lymphoid cells (Figure 2B right). It is therefore concluded that ERG expression induces rapid expansion of erythroblasts and T cells accompanied by their altered differentiation program in mice. Cell-cycle analysis showed increased proliferation of ERG-expressing erythroblasts and thymocytes compared with normal counterparts taken from mice that received a transplant with GFP-only control-infected BM cells (Figure 2C).
Flow cytometric and morphologic analyses of cells. (A) Typical flow cytometric analyses of cells in BM, spleen, and thymus. Cells were analyzed for expression of the indicated molecules. Typical data are presented for one control mouse that received a transplant with GFP-only–transduced cells and 3 mice that received a transplant with ERG-transduced cells. The “a,“ “c,” “e,” and “f” correspond to mice shown in Figure 1G. (B) Cytospin preparations of spleen cells in an ERG mouse (f) showing clusters of immature erythroid (left) and lymphoid (right) cells. Original magnifications: ×1000 (SPlan100 100×/1.25 oil 160/0.17 objective). (C) Flow cytometric analysis of DNA content of indicated cells in mice that received a transplant with control and ERG-transduced BM cells. Percentage of S phase is presented. Ter-119+ cells were flow-sorted before the analysis. (D-E) Analyses of granulocyte/macrophage, megakaryocytic, and B-cell compartment in ERG mice. Cells obtained from BM and spleen were gated on GFP+ and GFP− fractions for flow cytometric analysis of the indicated molecules. Typical data of 10 mice analyzed are presented (D). Lineage-negative BM cells of control and ERG mice were assayed for colony formation under a granulocyte/macrophage condition, in triplicate. Mean and SD are presented for a typical analysis of 3 independent experiments (E).
Flow cytometric and morphologic analyses of cells. (A) Typical flow cytometric analyses of cells in BM, spleen, and thymus. Cells were analyzed for expression of the indicated molecules. Typical data are presented for one control mouse that received a transplant with GFP-only–transduced cells and 3 mice that received a transplant with ERG-transduced cells. The “a,“ “c,” “e,” and “f” correspond to mice shown in Figure 1G. (B) Cytospin preparations of spleen cells in an ERG mouse (f) showing clusters of immature erythroid (left) and lymphoid (right) cells. Original magnifications: ×1000 (SPlan100 100×/1.25 oil 160/0.17 objective). (C) Flow cytometric analysis of DNA content of indicated cells in mice that received a transplant with control and ERG-transduced BM cells. Percentage of S phase is presented. Ter-119+ cells were flow-sorted before the analysis. (D-E) Analyses of granulocyte/macrophage, megakaryocytic, and B-cell compartment in ERG mice. Cells obtained from BM and spleen were gated on GFP+ and GFP− fractions for flow cytometric analysis of the indicated molecules. Typical data of 10 mice analyzed are presented (D). Lineage-negative BM cells of control and ERG mice were assayed for colony formation under a granulocyte/macrophage condition, in triplicate. Mean and SD are presented for a typical analysis of 3 independent experiments (E).
In contrast to erythroid and T cells, B cells were underrepresented in ERG mice compared with control mice. This underrepresentation was most evident when GFP+ and GFP− fractions were analyzed separately (Figure 2D). B cells were barely detected in GFP+ compartments, unlike the case with GFP−, suggesting that the emergence of B cells is inhibited by ERG expression. Although granulocytes/macrophage fractions were not evidently altered with respect to differentiation, the colony-forming ability of lineage-depleted BM cells in a granulocyte/macrophage condition was significantly enhanced in ERG-expressing than in control cells (Figure 2E), suggesting that ERG expression increased the numbers of myeloid progenitors.
It is reported that mice that received a transplant with ERG-expressing fetal hematopoietic cells are predominantly repopulated by CD41+ cells, which account for > 60% of cells in the spleen.17 Although our mice that received a transplant with ERG-expressing adult BM cells exhibited overrepresentation of CD41+ cells in GFP+ compared with GFP− compartments, the cells accounted for only 5.8% and 3.4% of cells in BM and spleen, respectively, suggesting that the effect of ERG expression differs between fetal and adult hematopoietic cells.
T-ALL emerging in ERG mice acquires the Notch1 mutation
In an effort to determine whether the ERG-expressing thymocytes and erythroblasts are leukemogenic, cells were transplanted into syngeneic hosts.
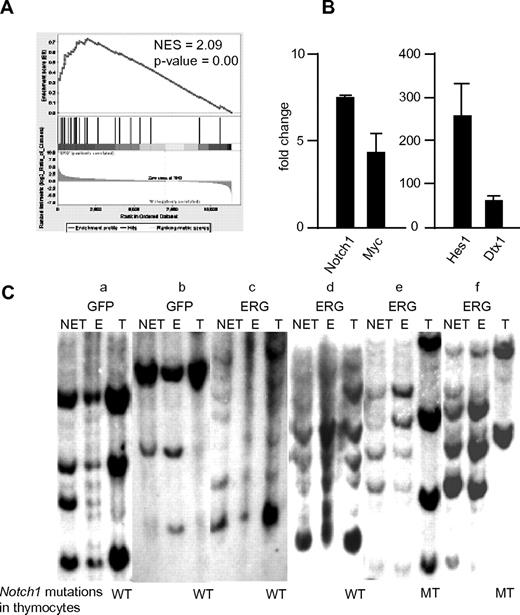
Specifically, all mice that received a transplant with thymocytes taken from enlarged thymi of ERG mice succumbed to leukemic death within 40 days (2 independent experiments each using 3 mice). The leukemic cells massively infiltrated into various organs, including BM, spleen, lymph nodes, kidney, and liver (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The phenotype of the infiltrating cells was similar to that of the transplanted cells (supplemental Figure 1B). Hence, the results showed that ERG-expressing T cells in enlarged thymi are capable of propagating leukemia. To investigate mechanisms underlying the development of T-ALL, we conducted gene expression microarray analysis and compared thymocytes of enlarged thymi of ERG mice with those of control mice. Results showed that genes regulated by Notch1 were specifically enriched in ERG compared with control thymocytes (Figure 3A). Quantitative PCR analysis confirmed this result with Notch1-regulated genes such as Myc, Hes1, Deltex1, and Notch1 itself being up-regulated (Figure 3B). These findings suggest that up-regulation of Notch1 signaling plays important roles in the development of T-ALL.
Gene expression analysis of T cells obtained from enlarged thymi of ERG mice and Southern blot analyses of control and ERG mice. (A) Gene Set Enrichment Analysis showing enrichment of genes regulated by Notch1. (B) Quantitative PCR analysis of transcripts of Notch1-regulated genes, Notch1 itself, Myc, Hes1, and Deltex1 (Dtx1). Results are presented as a fold change compared with control thymocytes after normalization on the basis of transcript levels of β-actin. Results (mean and SD) obtained from 3 mice are shown. (C) Nonerythroid/non-T cells (NET) and erythroid cells (E) were fractionated from BM and spleen of the indicated mice. T cells were obtained from thymi (T). Two GFP-only control and 4 ERG mice were used. Genomic DNA extracted from respective cells was analyzed for virus integration by a GFP probe. The “a” to “f” indicate mice shown in Figure 1G. Mutation status of Notch1 is also shown. WT indicates wild type; MT, mutated.
Gene expression analysis of T cells obtained from enlarged thymi of ERG mice and Southern blot analyses of control and ERG mice. (A) Gene Set Enrichment Analysis showing enrichment of genes regulated by Notch1. (B) Quantitative PCR analysis of transcripts of Notch1-regulated genes, Notch1 itself, Myc, Hes1, and Deltex1 (Dtx1). Results are presented as a fold change compared with control thymocytes after normalization on the basis of transcript levels of β-actin. Results (mean and SD) obtained from 3 mice are shown. (C) Nonerythroid/non-T cells (NET) and erythroid cells (E) were fractionated from BM and spleen of the indicated mice. T cells were obtained from thymi (T). Two GFP-only control and 4 ERG mice were used. Genomic DNA extracted from respective cells was analyzed for virus integration by a GFP probe. The “a” to “f” indicate mice shown in Figure 1G. Mutation status of Notch1 is also shown. WT indicates wild type; MT, mutated.
In an effort to determine the origin of the clones of leukemia in ERG mice, we fractionated T (thymocytes), erythroid and non-T/nonerythroid cells from ERG and control mice (Figure 3C) and subjected them to Southern blot analysis for viral integration. We chose 2 each of ERG mice with and without an enlarged thymus (corresponding to mouse c and mouse d, and mouse e and mouse f in Figure 1G, respectively), as well as control mice (Figure 1G mouse a and mouse b). In control mice that received a transplant with BM cells infected with GFP-only virus (Figure 1G mouse a and mouse b), the virus-infected cells contributed roughly equally to T, erythroid, and non-T/nonerythroid cells. This is also the case for ERG mice without an enlarged thymus (Figure 3C mouse c and mouse d). By contrast, in ERG mice with an enlarged thymus (Figure 3C mouse e and mouse f), whereas the virus-infected cells contributed roughly equally to erythroid and non-T/nonerythroid cells, a subset of clones among those shared by erythroid and non-T/nonerythroid cells dominantly contributed to T cells. These findings suggest that ERG expression promotes growth of T cells relative to the control in thymus that is not yet enlarged (as suggested by Figure 1G; see above-mentioned descriptions), and one or a few clones among the ERG-expressing T cells then become dominant enough to enlarge the thymus, presumably after acquiring additional secondary genetic hits. Given that mutations in the Notch1 gene often accompany the development of leukemia in several mice models of T-ALL,36,38-40 we next searched for such mutations. Results summarized in Table 1 (and also indicated at the bottom of Figure 3C) show that Notch1 was mutated in its PEST domain in cells of enlarged thymus, whereas it was a wild type, or at least under the detection level of the mutation, in cells of nonenlarged thymus of ERG mice. Expression levels of ERG in GFP+ T cells were comparable among nonenlarged thymus of a nonleukemic mouse and enlarged thymus and enlarged spleen of a leukemic mouse (supplemental Figure 2), suggesting that the leukemic transformation was not ascribed to the difference in the levels of ERG expression.
PEST mutations occur at high frequency in ERG-expressing T-cell tumors
| Tumor . | PEST mutation* . | Mutational consequence† . |
|---|---|---|
| e | Deletion AGCAGTCTGCCTGTGCA 7534 | Frameshift, Δ2484 |
| f | Insertion GG/deletion C 7316 | Frameshift, Δ2490 |
| g | Insertion CC/deletion G 7346 | Frameshift, Δ2490 |
| h | Insertion C 7666 | Frameshift, Δ2506 |
| i | Deletion ATGTACAACCGCTGGG 7514 | Frameshift, Δ2446 |
| Tumor . | PEST mutation* . | Mutational consequence† . |
|---|---|---|
| e | Deletion AGCAGTCTGCCTGTGCA 7534 | Frameshift, Δ2484 |
| f | Insertion GG/deletion C 7316 | Frameshift, Δ2490 |
| g | Insertion CC/deletion G 7346 | Frameshift, Δ2490 |
| h | Insertion C 7666 | Frameshift, Δ2506 |
| i | Deletion ATGTACAACCGCTGGG 7514 | Frameshift, Δ2446 |
Tumors e to f correspond to those indicated in Figure 1G. No HD mutations were observed.
Numbers correspond to nucleotide positions in Notch1 cDNA.
Numbers indicate amino acid residues in Notch1 at which mutations occur.
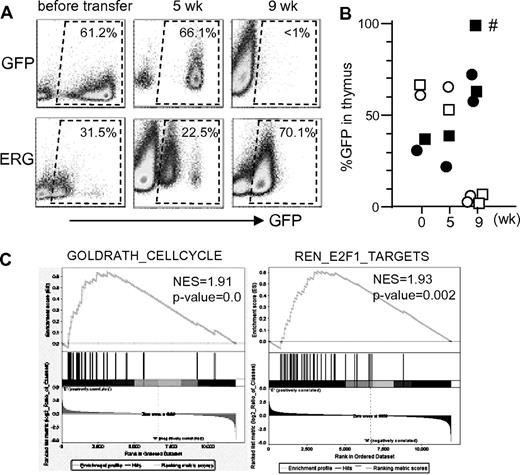
To explore the roles of ERG in T cells before the acquisition of Notch1 mutations, we took thymocytes from GFP-only control and ERG mice without an enlarged thymus and transplanted them into 3 secondary mice (Figure 4A). Thymi of the mice that received a transplant were then analyzed 5 and 9 weeks after the transplantation. In control mice, GFP+ cells were detected 5 weeks but not 9 weeks after the transplantation. In contrast, GFP+ cells were detected even at 9 weeks in mice that received a transplant with ERG-expressing thymocytes. Importantly, Notch1 mutations were not detected from the thymocytes, except those of one mouse with an enlarged thymus at 9 weeks; the other thymi were not enlarged or accompanied by Notch1 mutations (Figure 4B). These findings suggest that ERG expression confers T cells with a competitive growth advantage over control cells. Consistent with this, microarray analysis comparing ERG-expressing (but not mutated for the Notch1 gene) and control thymocytes showed that ERG expression was accompanied by gene sets associated with cell cycle and E2F1-regulated genes (P < .01; Figure 4C) but not by a gene set for Notch1-regulated genes (P = .45; not shown). Overall, these findings suggest that ERG expression promotes growth of T cells, which then become leukemic on acquisition of mutations in the Notch1 gene.
Effects of ERG expression on T-cell repopulation. (A-B) Thymocytes taken from mice that received a transplant with GFP-only control and ERG-transduced BM cells 5 weeks earlier were each transplanted (9 × 106 per mouse) into 3 lethally irradiated syngeneic hosts along with 3 × 105 radioprotective BM cells. Mice were then analyzed for GFP expression in thymus 5 weeks (1 mouse) and 9 weeks (2 mice) after the transplantation. A mouse indicated with an asterisk exhibited Notch1 mutation in thymocytes, whereas all other mice had no detectable Notch1 mutations. (C) Microarray analysis of transcripts in thymocytes taken from mice that received a transplant with GFP-only control and ERG-transduced BM cells 5 weeks earlier. Gene Set Enrichment Analysis showing enrichment of genes associated with cell-cycle and E2F1 target genes. Notch1 mutations were not detected in this experiment.
Effects of ERG expression on T-cell repopulation. (A-B) Thymocytes taken from mice that received a transplant with GFP-only control and ERG-transduced BM cells 5 weeks earlier were each transplanted (9 × 106 per mouse) into 3 lethally irradiated syngeneic hosts along with 3 × 105 radioprotective BM cells. Mice were then analyzed for GFP expression in thymus 5 weeks (1 mouse) and 9 weeks (2 mice) after the transplantation. A mouse indicated with an asterisk exhibited Notch1 mutation in thymocytes, whereas all other mice had no detectable Notch1 mutations. (C) Microarray analysis of transcripts in thymocytes taken from mice that received a transplant with GFP-only control and ERG-transduced BM cells 5 weeks earlier. Gene Set Enrichment Analysis showing enrichment of genes associated with cell-cycle and E2F1 target genes. Notch1 mutations were not detected in this experiment.
In contrast to ERG-expressing T cells, the Ter119+ erythroblasts expanding in ERG mice were unable to propagate leukemia in secondary mice (106 cells were transplanted per mouse). Southern blot analysis shown in Figure 3C showed that ERG-expressing erythroblasts (that were vigorously expanded in mice as shown in Figure 2) were equivalent on the clone basis to non-T/nonerythroid cells, suggesting that the expanding erythroblasts were not clonally selected, in contrast to ERG-expressing T cells that acquired the Notch1 mutation. Collectively, these findings suggest that the ERG-expressing erythroblasts by themselves do not represent transplantable leukemia.
ERG expression promotes growth and inhibits the differentiation of B cells
Although ERG expression appeared to have inhibitory effects on the emergence of B cells in our BMT models, reported microarray analyses of clinical leukemia samples showed higher expression of ERG transcripts in B-cell leukemia than in T-cell and myeloid leukemia,41-44 as well as normal precursor B cells45 (www.oncomine.org/resource/login.html). We then focused on investigating the effect of ERG expression on B cells. B220+c-kit+ pro-B cells were purified with FACS from B6 BM cells and were cultivated on OP9 stroma cells in the presence of cytokines FL, IL-7, and SCF.46 Cells were then infected with ERG- or control virus, sorted for GFP, and assayed for colony formation in semisolid media with the cytokines. Results showed that ERG-expressing pro-B cells yielded significantly more colonies than control pro-B cells (Figure 5A). On replating, ERG-expressing cells yielded even more colonies, whereas control cells yielded fewer colonies than the first plating. In subsequent platings, although control cells failed to form colonies, ERG-expressing cells were still able to form colonies, although the number was markedly lower than the secondary plating (Figure 5B). Experiments that used pro-B cells derived from fetal liver yielded similar results, although the number of colonies formed was significantly greater than that with experiments using BM pro-B cells (supplemental Figure 3). These findings suggested that ERG expression confers on B cells enhanced, although limited, replating activity.
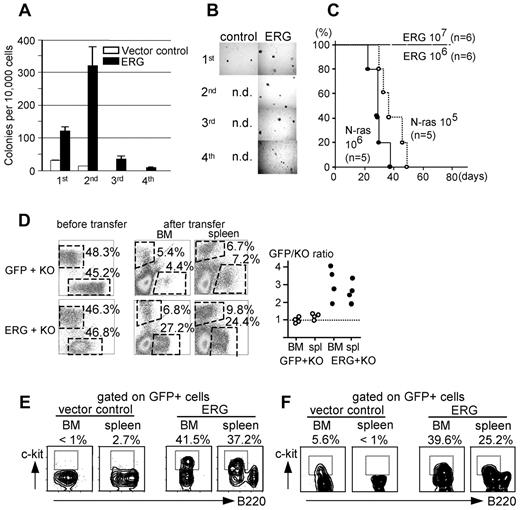
Effects of ERG expression on B cells. (A-B) Colony-forming ability of control and ERG-transduced pro-B cells derived from BM. Number of colonies formed and rounds of plating (A), and photomicrographs of the colonies; original magnification ×40 (UPlan FLN 4×/0.13 PhP objective) (B). (C) Kaplan-Meier survival curve for mice that received a transplant with the indicated number of N-rasG12D- or ERG-expressing pro-B cells. (D) Equal numbers of GFP- and KO-, or ERG- and KO-expressing pro-B cells were transplanted into NOD-SCID mice, and BM and spleen cells were analyzed for expression of GFP and KO after the transplantation. FACS analyses for expression of GFP and KO in pro-B cells before transplantation (left) and in BM and spleen after transplantation (middle). A plot is presented showing the ratio of GFP to KO (right). (E-F) FACS analyses of GFP+ cells in BM and spleen of mice that received a transplant with pro-B cells infected with control or ERG virus (E) and of mice that received a transplant with BM cells infected with a lentivirus containing the CD19 promoter driving GFP-only control and ERG expression in the B-cell compartment (F).
Effects of ERG expression on B cells. (A-B) Colony-forming ability of control and ERG-transduced pro-B cells derived from BM. Number of colonies formed and rounds of plating (A), and photomicrographs of the colonies; original magnification ×40 (UPlan FLN 4×/0.13 PhP objective) (B). (C) Kaplan-Meier survival curve for mice that received a transplant with the indicated number of N-rasG12D- or ERG-expressing pro-B cells. (D) Equal numbers of GFP- and KO-, or ERG- and KO-expressing pro-B cells were transplanted into NOD-SCID mice, and BM and spleen cells were analyzed for expression of GFP and KO after the transplantation. FACS analyses for expression of GFP and KO in pro-B cells before transplantation (left) and in BM and spleen after transplantation (middle). A plot is presented showing the ratio of GFP to KO (right). (E-F) FACS analyses of GFP+ cells in BM and spleen of mice that received a transplant with pro-B cells infected with control or ERG virus (E) and of mice that received a transplant with BM cells infected with a lentivirus containing the CD19 promoter driving GFP-only control and ERG expression in the B-cell compartment (F).
The leukemogenesity of ERG-expressing pro-B cells was then investigated. To this end, the highly efficient infectivity of fetal pro-B cells was used. Cells were infected with virus for ERG and injected into the BM cavity of lethally irradiated syngeneic mice; mice were radioprotected by intravenous transplantation of 2 × 105 fresh BM cells. Virus for N-rasG12D was used as a positive control. Under these experimental conditions, mice injected with N-rasG12D–expressing pro-B cells succumbed to leukemia within 40 (106 injected cells per mouse; n = 5) or 50 (105 injected cells per mouse; n = 5) days. In contrast, mice injected with either 106 or 107 ERG-expressing pro-B cells remained healthy for ≥ 6 months, suggesting that ERG is not competent in inducing leukemia (Figure 5C).
To explore effects of ERG expression in B-cell compartment in more detail, pro-B cells were infected with virus for ERG, GFP-only control, or KO, sorted for GFP or KO, and then mixed so that ERG-expressing (therefore GFP-expressing) cells and KO-expressing cells were equal in number. Similarly, GFP-only–expressing and KO-expressing cells were mixed in a 1:1 ratio. Mixed cells were then transplanted into the BM cavity of NOD-SCID mice. One month later, mice were analyzed for expression of GFP and KO in BM and spleen (Figure 5D). Although the GFP/KO ratio was ∼ 1.0 in mice that received a transplant with the mixture of GFP-control and KO-expressing cells (n = 4), the ratio was 1.8 ∼ 4.0 in mice that received a transplant with the mixture of ERG-expressing and KO-expressing cells (Figure 5D). ERG therefore conferred competitive ability on pro-B cells.
Further FACS analysis showed the inhibitory effects of ERG on B-cell differentiation. Given that the transplanted cells were pro-B cells, these cells express c-kit before transplantation,46 but vector-only–infected pro-B cells lost c-kit expression either in BM and spleen after the transplantation. In contrast, ERG-expressing pro-B cells retained c-kit expression, suggesting that ERG inhibits, albeit incompletely, the differentiation of pro-B cells to pre-B cells (Figure 5E). This inhibited differentiation was further confirmed with the use of a lentivirus driving ERG expression under control of the CD19 promoter (CD19-ERG-iresGFP).31 BM cells were infected with CD19-ERG-iresGFP or vector-only control virus and were transplanted into lethally irradiated syngeneic hosts. In both cases, GFP expression was restricted to the B-cell compartment, as expected. The differentiation of c-kit+ pro-B cells to c-kit− pre-B cells was not apparently disturbed in the GFP+ fraction of mice that received a transplant with vector-only control-infected BM cells (Figure 5F left), whereas c-kit+ cells accounted for 5.6% and < 1% of BM and spleen B220+ B cells, respectively, which is comparable to the percentage of c-kit in normal BM and spleen B cells.46 In contrast, the differentiation of pro-B cells to pre-B cells was inhibited in the GFP+ fraction of mice that received a transplant with CD19-ERG-iresGFP–infected BM cells; c-kit+ cells accounted for 39.6% and 25.2% of BM and spleen B220+ B cells, respectively (Figure 5F right).
Taken together, these findings suggested that ERG expression is not leukemogenic in B cells per se, but it confers on pro-B cells a competitive ability accompanied by inhibited differentiation.
shRNA-mediated silencing of ERG attenuates growth of human leukemia cell lines
Results obtained by our analysis of mice suggest that ERG is able to promote growth of various lineages of hematopoietic cells, but by itself it is insufficient and appears to need additional genetic hits (such as Notch1 mutations in the case of T cells) to elicit leukemia. We therefore finally interrogated if the growth-promoting effects of ERG plays a role in already established leukemia. To this end, human leukemia cell lines were infected with a lentivirus containing shRNA against human ERG and luciferase (control). Two shRNAs against ERG (shERG1 and shERG2) effectively knocked down ERG expression in the cell lines (Figure 6A). Because our lentivirus coexpresses GFP with shRNA, the GFP expression allowed us to follow the virus-infected cells in the bulk liquid culture. Typical results obtained with Reh cells (Figure 6B) showed that, although the percentage of GFP in control (luciferase) shRNA-infected cells was roughly stable, the percentage of GFP in shERG1- and shERG2-infected cells diminished with time. We then investigated the effects of shRNA against ERG with K562, MegA2, HEL, HL60, NB4, Jurkat, JM, Nalm-1, and Nalm-6 cells, in addition to Reh cells. The percentage of GFP expression is shown in Figure 6C as the ratio of the percentage of GFP for ERG shRNA to the percentage of GFP for luciferase (control) shRNA. In all cell lines tested, the ratios diminished over time, suggesting that the silencing of ERG expression has growth-inhibitory effects. In additional experiments that used K562, HL60, HEL, Jurkat, Reh, and MegA2, cells were sorted for GFP after infection with shRNA virus, and cell growth was monitored in liquid culture. The silencing of ERG expression was again found to inhibit cell growth (Figure 6D). The inhibition was not accompanied by cell death in K562, HL60, HEL, and MegA2, suggesting that the inhibition of cell proliferation, and not apoptosis, is the primary cause of the inhibition of cell growth in these cells. The inhibition of cell proliferation was additionally confirmed by decreased uptake of BrdU (supplemental Figure 4A). In contrast, the silencing of ERG expression was accompanied by cell death in Jurkat and Reh cells (Figure 6E; supplemental Figure 4B). These findings suggested that ERG plays pivotal roles in the proliferation or survival or both of leukemia cells even after the leukemia is established; thus, it appears crucial for maintaining human leukemia.
shRNA-mediated silencing of ERG attenuates the growth of human leukemia cell lines. (A) Western blot analysis of ERG expression in cell lines infected with shRNA viruses for luciferase (control) and (ERG(1) and ERG(2). Two different viruses were used for ERG silencing. (B) Typical FACS analysis of cells transduced with the shRNA viruses. Results obtained with Reh cells are presented. Cells were infected with shRNA viruses that coexpress GFP, and percentage of GFP in the bulk liquid culture was monitored over time. (C) Percentage of GFP in the bulk liquid culture of cells infected with the indicated shRNA viruses. K562, NB4, MegA2, Jurkat, JM, Nalm1, Nalm6, and Reh cells were used. The ratio of percentage of GFP for luciferase to percentage of GFP for ERG is presented along with the days elapsed after monitoring began. (D) Growth curves of the indicated cells infected with shRNA viruses for luciferase (○) and ERG [ERG(1), ●; ERG(2), ■]. (E) Growth curves of Reh and Jurkat cells infected with shRNA viruses for luciferase and ERG(1) and ERG(2). The number of viable (●) and dead (○) cells is presented during 5 days of culturing. The initial input cell number was 1 × 105.
shRNA-mediated silencing of ERG attenuates the growth of human leukemia cell lines. (A) Western blot analysis of ERG expression in cell lines infected with shRNA viruses for luciferase (control) and (ERG(1) and ERG(2). Two different viruses were used for ERG silencing. (B) Typical FACS analysis of cells transduced with the shRNA viruses. Results obtained with Reh cells are presented. Cells were infected with shRNA viruses that coexpress GFP, and percentage of GFP in the bulk liquid culture was monitored over time. (C) Percentage of GFP in the bulk liquid culture of cells infected with the indicated shRNA viruses. K562, NB4, MegA2, Jurkat, JM, Nalm1, Nalm6, and Reh cells were used. The ratio of percentage of GFP for luciferase to percentage of GFP for ERG is presented along with the days elapsed after monitoring began. (D) Growth curves of the indicated cells infected with shRNA viruses for luciferase (○) and ERG [ERG(1), ●; ERG(2), ■]. (E) Growth curves of Reh and Jurkat cells infected with shRNA viruses for luciferase and ERG(1) and ERG(2). The number of viable (●) and dead (○) cells is presented during 5 days of culturing. The initial input cell number was 1 × 105.
Discussion
The effects of forced expression of the proto-oncogene ERG on hematopoiesis have been experimentally studied with fetal hematopoietic cells as the target of expression, with particular reference to DS-associated AMKL.17,18 In the study presented here, we used adult hematopoietic cells as the target of ERG expression and found that the effects of ERG differ between adult and fetal hematopoietic cells. Mice that received a transplant with adult BM cells manipulated to express ERG exhibited rapid expansion of T cells and erythroblasts in mice and died as early as 30 days, and none survived beyond 4 months after the transplantation. The lineages affected and phenotypes of the expanded cells are distinct from those of mice that received a transplant with fetal hematopoietic cells overexpressing ERG, whereby leukemia of the CD41+ megakaryocytic lineage emerges.17 The results obtained with adult BM cells treated with 5-FU were reproducible with the use of c-kit+Ter119− cells that are the phenotypical counterparts reported to be used for the viral infection of fetal hematopoietic cells to elicit CD41+ leukemia (not shown).
We observed that thymi of some ERG mice were enlarged, whereas thyme of other ERG mice were not, on elective dissection conducted between 50 and 80 days after transplantation. T cells in the enlarged thymi were phenotypically CD4+CD8+ and CD4−CD8+, reminiscent of leukemia developing in mice in which EWS-ERG expression was targeted to the T-cell compartment.9 The ERG-expressing T cells exhibited increased percentage of S phase on cell-cycle analysis compared with the control, and they rapidly induced leukemia with massive infiltration of leukemic cells in the organs of mice that received a secondary transplant, suggesting that the T cells themselves are capable of propagating leukemia. Gene expression microarray analysis comparing T cells in enlarged thymi with their normal counterparts in thymus showed up-regulation in Notch1-regulated genes, which was then found to be associated with mutations in the PEST domain of the Notch1 gene. Mutations in the Notch1 gene, in particular in its PEST domain, are often found in mouse models of T-ALL; many instances of T-ALL developing in transgenic mice for Tal1, Tal1, and Lmo2, and K-ras are associated with mutation in the PEST domain. However, the mutations in the PEST domain are by themselves weak activators of Notch signaling and are incapable of provoking noticeable changes in proliferation/differentiation of T cells, let alone T-ALL.38 Thus, it is conceivable that Notch1 mutated in the PEST domain works in concert with either one of Tal1, Tal1/Lmo2, and K-ras to elicit leukemia,36,38,40 and here ERG joins the list of murine T-ALL models in which Notch1 mutations play a complementary role.
We then sought to determine the roles of ERG in T cells before they acquire Notch1 mutations. Results presented in Figure 1G and Figure 4 suggested that ERG confers T cells with a growth advantage over control cells. Gene expression analysis comparing ERG-expressing and control thymocytes showed overrepresentation of cell cycle–associated genes in ERG thymocytes. The proliferative nature in T cells evoked by ERG may render T cells prone to acquire Notch1 mutations, as suggested in the case of Tal1/Lmo2.40
In line with the proliferative nature being conferred by ERG in T cells, erythroblasts in our ERG mice also exhibited enhanced proliferation compared with the control (Figure 2C). The ERG-expressing erythroblasts nevertheless were unable to repopulate in animals that received a secondary transplant. Southern blot analysis suggested that the ERG-expressing erythroblasts are derived from clones that are common to nonerythroid/non-T cells and are therefore not clonally selected. ERG expression therefore appears by itself sufficient to promote rapid expansion of erythroblasts but insufficient to elicit transplantable leukemia. Because abnormally expanded erythroblasts and T cells are present in a given mouse, it is not straightforward to evaluate the degree with which abnormal erythroblasts contributed to the lethality of mice. However, abnormal T cells appeared to be confined to the thymus, with BM and spleen being spared in some mice (Figure 2A), suggesting that the expanded erythroblasts are probably not a result of reactive erythropoiesis to the abnormal T cells. The erythroblasts appeared to possess an impaired capacity to produce mature blood cells (Figure 2A-B), making it probable that they made a substantial contribution to the lethality.
In an effort to maximize the cell targets for ERG expression in adult hematopoiesis, we used a strategy that involved a BMT model with a conventional retrovirus to transduce 5-FU–treated BM cells. Because of this strategy, the resulting phenotypes may not necessarily represent those seen in clinical human leukemia; erythroleukemia is a relatively rare disease, and the limited number of cases of erythroleukemia makes strict comparison of erythroleukemia with other types of heterogenous AML not straightforward, but gene expression data available from Gene Expression Omnibus, such as GSE12417 and GSE1729, show that ERG is not uniquely high in erythroleukemia among subsets of AML. Our strategy nevertheless showed a hitherto unrecognized capacity of ERG to promote leukemia of T and possibly erythroid lineages. In our BMT models, however, ERG expression appears to inhibit the emergence of B cells from uncommitted hematopoietic cells, precluding an investigation of how ERG expression affects B cells. Yet, ERG expression is higher in B-cell leukemia than in T-cell leukemia and myeloid leukemia in humans,41-44 raising the possibility that increased ERG expression contributes to B-cell leukemogenesis. To explore this possibility, we used 2 alternative strategies for the targeted expression of ERG in B cells. One was to retrovirally express ERG in pro-B cells isolated from mouse BM or fetal liver. The other complementary approach used a lentivirus vector harboring the CD19 promoter to drive ERG expression exclusively in the B-cell compartment.31 We were thus able to show that forced expression of ERG enhances the growth of pro-B cells in culture, as determined by assaying for colony-forming ability. ERG-expressing pro-B cells also exhibited enhanced competitiveness compared with the control in vivo and inhibition of B-cell differentiation from B220+c-kit+ pro-B to B220+c-kit− pre-B/immature B-cell stages.46 However, ERG expression appears to be incapable of inducing leukemia; although N-RasG12D expression resulted in leukemia in pro-B cells of mice that received a transplant, ERG expression failed to produce the same outcome. Although ERG expression is known to be associated with increased cell migration,12,13 our observation of enhanced competitiveness of ERG-expressing pro-B cells in vivo is unlikely to be associated with increased cell motility because cells were injected directly into the BM cavity to minimize differences in the homing ability of ERG-expressing and control pro-B cells. Analysis of mice that received a transplant with BM cells programmed to express ERG in the B-cell compartment through the use of a lentivirus harboring the CD19 promoter further showed that ERG expression inhibited B-cell differentiation but was insufficient to induce leukemia.
Vigorous expansion of erythroblasts and subsequently of T cells in BM precluded detailed analysis of ERG-expressing granulocyte/macrophages in our BMT mouse model, but results of colony-forming assays (Figure 2E) suggest that ERG promotes growth of progenitors in the granulocytes/macrophage lineage. Overall, these findings suggest growth-promoting effects of ERG in T, erythroid, B, and possibly other lineages of cells, which, coupled with an altered differentiation program (Figures 2A, 5E-F), may contribute to the development of leukemia in various lineages.
We therefore finally sought to determine whether ERG plays a role in already established human leukemia of various lineages. shRNA-mediated silencing of ERG expression attenuated the growth of human leukemia cells (in all 10 cell lines tested) of erythroid, myeloid, megakaryocytic, T-, and B-cell lineages. The attenuated growth observed was ascribed to an inhibition of proliferation or cell death or both, depending on the cell type. Although forced ERG expression attenuated induced apoptosis in NIH3T3 cells,47 we were able to show for the first time here that the silencing of ERG expression induced cell death in leukemia cells. These findings suggest the crucial roles of ERG in maintaining leukemia.
Although our microarray analysis suggested up-regulation of cell cycle–associated genes in ERG-expressing compared with control thymocytes, detailed molecular mechanisms whereby ERG maintains/promotes proliferation and in some cases inhibits cell death are clearly important areas of further investigation. Although ERG has been shown to play important roles in the development of prostate cancer, ERG itself does not seem to elicit noticeable phenotypes in prostate, and must cooperate with other molecules such as PI3K/Akts and possibly androgen responsive genes for prostate cancer to develop.13,15,48,49 However, ERG expression promotes growth of hematopoietic cells of various lineages. Elucidating the underlying mechanisms, which are therefore probably distinct from those involved in prostate cancer, could pave the way toward the development of a new molecular targeting therapy for leukemia, given that silencing of ERG expression attenuates proliferation and in particular induces cell death in a subset of leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Ms Seiko Sato for technical assistance and animal husbandry and Dr Masao Nakagawa for critical reading of the manuscript.
This work was supported by a grant-in-aid for the Second Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health and Welfare (M.S.); grant-in-aid for scientific research from The Ministry of Education, Culture, Sports, Science and Technology (M.S.); and a grant-in-aid for scientific research from the Japan Society for the Promotion of Science (S.T.).
Authorship
Contribution: S.T. designed and performed all experiments and wrote the paper; O.T. conducted histologic examination; and M.S. contributed to the discussion.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shinobu Tsuzuki, Division of Molecular Medicine, Aichi Cancer Center Research Institute, 1-1 Kanokoden, Chikusa-ku, Nagoya, Aichi, 464-8681, Japan; e-mail: stsuzuki@aichi-cc.jp.

![Figure 6. shRNA-mediated silencing of ERG attenuates the growth of human leukemia cell lines. (A) Western blot analysis of ERG expression in cell lines infected with shRNA viruses for luciferase (control) and (ERG(1) and ERG(2). Two different viruses were used for ERG silencing. (B) Typical FACS analysis of cells transduced with the shRNA viruses. Results obtained with Reh cells are presented. Cells were infected with shRNA viruses that coexpress GFP, and percentage of GFP in the bulk liquid culture was monitored over time. (C) Percentage of GFP in the bulk liquid culture of cells infected with the indicated shRNA viruses. K562, NB4, MegA2, Jurkat, JM, Nalm1, Nalm6, and Reh cells were used. The ratio of percentage of GFP for luciferase to percentage of GFP for ERG is presented along with the days elapsed after monitoring began. (D) Growth curves of the indicated cells infected with shRNA viruses for luciferase (○) and ERG [ERG(1), ●; ERG(2), ■]. (E) Growth curves of Reh and Jurkat cells infected with shRNA viruses for luciferase and ERG(1) and ERG(2). The number of viable (●) and dead (○) cells is presented during 5 days of culturing. The initial input cell number was 1 × 105.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/14/10.1182_blood-2010-11-320515/4/m_zh89991169230006.jpeg?Expires=1769104108&Signature=avULK9USW4WTWMRcHzxUw6~ZpSC~O3UEXwxuhAEjMksOiSwfgSuCe4yvS8RwjMYhMk6YuC5p7l7TZgcQhWXymjW4xI4DWDVykpCB~98ZRLOp-YatjsWXja9KiplNrixqa9KQIOBMwefruU9y6ChYo2zfCNd9IUMqzTHcFTDG69jbd-3Nj8auNxOYZgldPxTI~5n3v4iENEi40PF44qO5pud5kl1yiGfw2cSPX6vGexccL67S4EsVqquhYu5tYnKeUNLOthXs0keuYCvntEOW77TAy2yz-QjDj7xEzLpWVMKpBzx8hjJIaXL6953FCx-qUVsj-7mCacin9dp9vTRlEQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal