Abstract
The histone H3 lysine 36 dimethyl–specific demethylase KDM2b/JHDM1b, which is highly expressed in various human leukemias, was previously found to be important in regulating cell proliferation and cellular senescence. However, its functions in leukemia development and maintenance are unclear. Here, we demonstrate that ectopic expression of Kdm2b/Jhdm1b is sufficient to transform hematopoietic progenitors. Conversely, depletion of Kdm2b/Jhdm1b in hematopoietic progenitors significantly impairs Hoxa9/Meis1-induced leukemic transformation. In leukemic stem cells, knockdown of Kdm2b/Jhdm1b impairs their self-renewing capability in vitro and in vivo. The functions of Kdm2b/Jhdm1b are mediated by its silencing of p15Ink4b expression through active demethylation of histone H3 lysine 36 dimethyl. Thus, our study suggests that Kdm2b/Jhdm1b functions as an oncogene and plays a critical role in leukemia development and maintenance.
Introduction
Previous studies have shown that human leukemic cells from the same patient are composed of heterogeneous cell populations with various proliferation capacities and differentiation status. In the proposed leukemia stem cell (LSC) model, a fraction of LSCs resides at the apex of leukemia cellular hierarchy. Similar to hematopoietic stem cells (HSCs) in normal blood development, LSCs can give rise to the entire cellular hierarchy and sustain leukemia expansion through an unlimited self-renewal capability.1 This model is supported by studies in which LSC-enriched cell populations, such as the CD34+CD38− leukemic cells in human acute myeloid leukemia (AML), transplanted into SCID mice are able to fully recapitulate the process of leukemia development.2,3
LSCs can be derived from different cellular compartments according to the leukemia type and disease stage. In a Junb inactivation-induced chronic myeloid leukemia (CML) murine model, the CML-like disease can only develop from Junb-inactivated HSCs but not progenitor cells, indicating that LSCs may derive from HSCs.4 However, in the accelerated and myeloid blast crisis phases of human CML, only leukemic granulocyte-macrophage progenitors can be expanded and display an aberrant self-renewing capacity in vitro in the methylcellulose replating assay.5 In addition, the fact that certain murine AMLs can be induced by retroviral transduction of oncogenes, such as Mll fusion genes, into the granulocyte-macrophage progenitor population indicates that LSCs can originate from committed progenitor cells directly.6,7 These studies suggest that the “stemness” program of LSCs could be activated by various oncogenic stimuli in different cellular contexts. However, the molecular mechanisms underlying LSC self-renewal is not well understood.8
The leukemic stem cell model implies that epigenetic regulation at certain critical gene loci might be important in determining the phenotypic difference between self-renewing LSCs and their non–self-renewing progeny.8 One example that supports this notion comes from the demonstration that the Ink4a-Arf-Ink4b locus, which encodes 3 tumor suppressors, including p16Ink4a, p15Ink4b, and p19Arf is controlled by the Polycomb repressive complex 1 (PRC1) in both normal HSCs and LSCs.9,10 Biochemical analysis has shown that the PRC1 complex contains an ubiquitin E3 ligase activity and catalyzes the monoubiquitylation of histone H2A at lysine 119, which may serve as an epigenetic mark for the recruitment of other transcriptional repressors to the Ink4a-Arf-Ink4b locus.11,12 Consistently, deletion of BMI-1, a component of the PRC1 complex, in LSCs leads to de-repression of Ink4a-Arf expression and loss of their self-renewal capacity.10 In addition, Somervaille et al13 also found that some epigenetic modifiers, such as chromobox 5 and high mobility group box 3, are up-regulated in LSCs and coordinate each other to maintain the LSC program. However, it is unclear whether the functions of chromobox 5 and high mobility group box 3 in LSC maintenance are mediated through the Ink4/Arf or other gene loci.
In an effort to identify other epigenetic regulators important for LSC maintenance, we analyzed the expression level of all known epigenetic factors in human leukemias with the use of the available databases. Interestingly, we found that KDM2b/JHDM1b, a JmjC-domain containing protein, is highly expressed in human leukemia samples. Kdm2b/Jhdm1b was first identified as a hotspot for proviral insertion in murine tumors generated by random mutagenesis of Moloney murine leukemia virus.14 However, it was shown paradoxically to function as both an oncogene and a tumor suppressor, depending on the screen and analytic methods.14,15 In our previous studies, we have demonstrated that KDM2b/JHDM1b is an histone H3 lysine 36 dimethyl (H3K36me2)–specific demethylase important for maintaining proliferation of murine embryonic fibroblasts (MEFs) because depletion of Kdm2b/Jhdm1b causes premature cellular senescence and defective cellular proliferation,16 supporting an oncogenic function for Kdm2b/Jhdm1b.
The observation that KDM2b/JHDM1b is up-regulated in human leukemias prompted us to investigate its role in leukemogenesis. Here, we demonstrate that enforced expression of Kdm2b/Jhdm1b facilitates proliferation of hematopoietic progenitor cells (HPCs) and induces leukemic transformation. This leukemic property depends on its H3K36me2-demethylase activity and its down-stream target p15Ink4b. In addition, we show that Kdm2b/Jhdm1b is necessary for the development and maintenance of leukemia in a mouse AML model. Our study thus establishes KDM2b/JHDM1b as a critical epigenetic factor for leukemogenesis and raises the possibility that KDM2b/JHDM1b might serve as a potential therapeutic target for the treatment of leukemia.
Methods
Lentiviral vector construction and virus production
Stable knockdown (KD) was achieved with a lentiviral system obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. The mouse U6 promoter was cloned from mouse genomic DNA and was inserted into the NotI site of pTY-EF1α-nLacZ. For the LV-U6 shRNA-Pgk-Pac construct, the Pgk-Pac cassette is inserted between NotI/EcoRI sites to replace the EF1α-nLacZ cassette. The hairpin RNA targeting Kdm2b/Jhdm1b (5′-GCTCCAACTCAGTTACTGT-3′) and the control shRNA (5′-GTTCAGATGTGCGGCGAGT-3′) were cloned into the BbsI/HindIII sites under the control of the U6 promoter.17 To generate Hoxa9-Meis1-GFP expression vector, the Hoxa9, Meis1, and GFP cDNAs were amplified by PCR, ligated to a P2A linker (5′-CAACTGCTGAATTTTGACCTTCTTAAGTTGGCGGGAGACGTCGAGTCCAACCCTGGGCCC-3′) and cloned into the SpeI/EcoRI sites under EF1α promoter. To generate wild-type and mutant Jhdm1b-H211A rescue constructs, the small interfering RNA target site of Jhdm1b was mutated to 5′-ATTGCGTTGAGTTACTGT-3′ by PCR mutagenesis. The small interfering RNA–resistant wild-type, mutant Kdm2b/Jhdm1b and truncated LacZ cDNAs were amplified by PCR and cloned into the SpeI/EcoRI sites of the Pac-2a cassette. To generate lentiviral viruses, the transducing vectors pTY, pHP, and pHEF1α-VSVG were cotransfected into 293T cells. The supernatant was harvested at 24, 36, and 48 hours after transfection, filtered through 0.45-μm membrane, and concentrated with a Spin-column. Concentrated viruses were snap-frozen and saved at −80°C for later use.
Hematopoietic progenitor isolation, culture, and viral transduction
Hematopoietic progenitor cells are isolated from either E14.5 fetal liver or femurs of 4- to 6-week Ly5.2 C57BL/6 mice. The red blood cells in the fetal liver and BMs are lysed by ammonium chloride solution (Stem Cell Technologies 07800) and filtered with a 70-μm nylon filter. The isolated single-cell suspension is labeled with allophycocyanin-conjugated anti–c-kit antibody in staining buffer (1× PBS, 2% FBS) at 4°C for 30 minutes. The c-kit+ HPCs are sorted with the use of a BD FACSAria II flow cytometry. The sorted cells are cultured in DMEM supplemented with 15% (vol/vol) FBS, 1% (vol/vol) penicillin/streptomycin, 1× essential amino acid, 1× sodium pyruvate, 1× GlutaMax, 16 ng/mL murine recombinant IL-3, 20 ng/mL murine recombinant IL-6, and 100 ng/mL SCF. For transduction of HPCs, 5 × 105 cells were divided into 10 wells of a 96-well plate and transduced with 10 μL of concentrated virus (MOI ∼ 10) plus polybrene (8 ng/mL). For each round of transduction, the plates were centrifuged at 2000 rpm for 2 hours at 20°C. Two to 3 rounds of transduction were performed for each experiment.
Serial methylcellulose replating assay and BM transplantation
Approximately 3 × 104 sorted GFP+ cells were mixed with 3 mL of methylcellulose (MethoCult GF M3534) medium supplemented with GM-CSF (10 ng/mL) and evenly distributed into three 25-cm2 plates. After 10-14 days, the colony numbers were counted under a microscope. The colonies were picked up, and cells were pooled and replated (104 cells/plate) onto secondary methylcellulose plates. Three to 4 rounds of replating were performed for each experiment.
For BM transplantation, recipient Ly5.1 C57BL/6 mice were subjected to total body irradiation at a dose of 9.5 Gy with the use of a cesium irradiator. Donor cells (2.5 × 105) and radiation protector cells (2.5 × 105) isolated from BM of Ly5.1 C57BL/6 mice were mixed in 1× PBS and transplanted into the recipient mice through retro-orbital injection. For the transplantation of Kdm2b/Jhdm1b overexpressed HPCs, we transplanted 2.5 × 105 transduced cells and 5.0 × 105 Ly5.1+Ly5.2+ protecting cells into irradiated C57BL/6 Ly5.1+Ly5.2+ recipient mice. The mice were fed with water supplemented with trimethoprim/sulfamethoxazole for 4 weeks after transplantation.
FACS analysis and cell sorting
For FACS analysis and cell sorting, cells were stained with antibodies in staining buffer (1× PBS, 2% FBS) and incubated at 4°C for 30 minutes. The samples were washed once with staining buffer before subjected to FACS analysis with the use of a BD FACSAriaII cell sorter. The antibodies used in this study include anti–Mac-1(eBioscience), anti–Gr-1(eBioscience), anti–c-kit (eBioscience), anti-B220 (eBioscience), anti-CD3 (eBioscience), anti-Ly5.1(eBioscience), and anti-Ly5.2 (eBioscience).
Histologic analysis
Mouse tissues were fixed in 4% paraformaldehyde, dehydrated in 70% ethanol, and paraffin-embedded. H&E staining was performed as previously described.18 For May-Grünwald/Giemsa staining, BM and peripheral blood smears are stained with Jenner solution (EMS; no. 26250-1A) for 6 minutes and Giemsa solution (EMS; no. 26250-02) for 15 minutes. The slides were washed with distilled water before viewed under a microscope. The samples were examined using an Axio Observer Z1 microscope (Carl Zeiss) with a Zeiss EC Plan-NeoFluar 10×/0.3 Ph1 DIC objective lens (Carl Zeiss). The images were captured by an AxioCam MRC5 camera (Carl Zeiss) and processed by AxioVision40 V4.8.2.0 image software (Carl Zeiss).
Quantitative RT-PCR and ChIP assays
RNA was extracted and purified from cells with the use of Qiashredder (QIAGEN) and RNeasy (QIAGEN) spin columns. Total RNA (1 μg) was subjected to reverse transcription with the use of random primers (Promega) and the Superscript II reverse transcriptase (Invitrogen). cDNA levels were assayed by real-time PCR with the use of SYBR GreenER (Invitrogen) and analyzed on the ABI 7300 Real-Time PCR system with DSD software Version 1.3.1. The expression of individual genes is normalized to expression level of Gapdh. Chip assays that used M2 anti-Flag antibody (Sigma) and anti-H3K36me2 antibody13,19 were carried out as reported previously16 with the following modifications: 20 μL of M2 agarose (Sigma) was used in the immunoprecipitation, and chromatin-bound beads were washed 3 times each with TSEI, TSEII, and TESIII followed by 2 washes in 10mM Tris, pH 7.5, 1mM EDTA. Histone modification ChIPs were carried out as previously reported.16 DNA that underwent ChIP was analyzed by quantitative PCR (qPCR), and data are presented as the percentage of input as determined with Applied Biosystems's SDS software Absolute Quantification protocol. Primers for qPCR and ChIP assays are listed in supplemental Tables 1 and 2, respectively (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Bisulfite sequencing analysis
Genomic DNA was isolated and subjected to bisulfite conversion with the use of the EZ DNA methylation-gold Kit (Zymo Research). The high CpG regions of p15Ink4b promoter and first exon reported to be methylated in certain cancers20,21 (318-644 bp, GenBank accession no. U66084) was amplified by PCR and cloned in pST-Blue vector (Novagen). DNA sequences of the PCR primers are forward: 5′-TAAGTTTTAGATTAGGAAATTTAAAGTTTT G-3′; reverse: 5′-TAAAATTACTACAACCTAATCTC-3′. The clones were sequenced by the University of North Carolina Chapel Hill genome analysis facility.
Western blot analysis
Total proteins were extracted with RIPA buffer and separated by electrophoresis with the use of 6%-8% gel. Flag monoclonal M2 antibody (Sigma) was used at the dilution of 1:10 000 for Western blot analysis.
Results
Expression of Kdm2b/Jhdm1b is up-regulated in leukemic stem cells
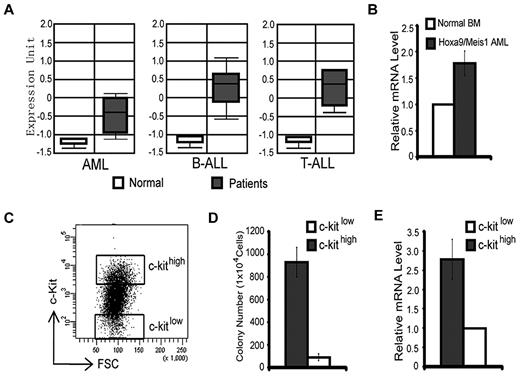
We have previously demonstrated that the H3K36me2-specific demethylase KDM2b/JHDM1b plays an important role in promoting cell proliferation and in preventing cellular senescence in MEFs.16 Interestingly, an analysis of the Oncomine database22 indicated that the expression of Kdm2b/Jhdm1b is up-regulated in samples of human AML, T-cell acute lymphoid leukemia, and B-cell acute lymphoid leukemia compared with healthy controls (Figure 1A). These findings raised the possibility that Kdm2b/Jhdm1b may have a role in leukemogenesis or maintenance of LSCs or both. To address this possibility, we analyzed Kdm2b/Jhdm1b expression in a murine AML model and found that it is expressed at a higher level in the Hoxa9/Meis1-induced leukemia cells than in normal BM (Figure 1B).23 To further analyze the expression of Kdm2b/Jhdm1b in different leukemic cell populations, we sorted the Hoxa9/Meis1-induced AML cells into 2 populations on the basis of their c-kit expression level (c-kithigh vs c-kitlow) (Figure 1C). Methylcellulose colony formation assays showed that the c-kithigh cell population has a higher capability to form colonies than the c-kitlow cell population (Figure 1D), indicating that the c-kithigh cell population is enriched for LSCs, whereas the c-kitlow cell population is depleted of LSCs. RT-qPCR analysis shows that the expression level of Kdm2b/Jhdm1b is ∼ 3-fold higher in the LSC-enriched c-kithigh population than in the LSC-depleted c-kitlow population (Figure 1E). Collectively, these analyses indicate that high Kdm2b/Jhdm2b expression correlates with LSC potential and might play an important role in leukemia development and maintenance.
Expression of Kdm2b/Jhdm1b is up-regulated in leukemic stem cells. (A) KDM2B/JHDM1B is overexpressed at the RNA level in human AML, T-cell acute lymphoid leukemia (T-ALL), and B-cell acute lymphoid leukemia (B-ALL) compared with their normal controls. Data are derived from Oncomine. 22 (B) Kdm2b/Jhdm1b is overexpressed in Hoxa9/Meis1-induced AML samples compared with that of normal BM. Relative mRNA levels are measured by RT-qPCR and normalized to Gapdh level. The level in normal BM is arbitrarily set to 1. (C) Separation of Hoxa9/Meis1-transformed murine AML cells into c-kithigh and c-kitlow populations by FACS. These 2 cell populations are used for methylcellulose colony formation assays. (D) The c-kithigh cell population has higher colony formation capability compared with the c-kitlow population. (E) RT-qPCR analysis shows that the c-kithigh cell population expresses a higher level of Kdm2b/Jhdm1b than the c-kitlow cell population. Relative mRNA levels are measured by RT-qPCR and normalized to Gapdh level. The level of c-kitlow population is arbitrarily set to 1. FSC indicates forward scatter.
Expression of Kdm2b/Jhdm1b is up-regulated in leukemic stem cells. (A) KDM2B/JHDM1B is overexpressed at the RNA level in human AML, T-cell acute lymphoid leukemia (T-ALL), and B-cell acute lymphoid leukemia (B-ALL) compared with their normal controls. Data are derived from Oncomine. 22 (B) Kdm2b/Jhdm1b is overexpressed in Hoxa9/Meis1-induced AML samples compared with that of normal BM. Relative mRNA levels are measured by RT-qPCR and normalized to Gapdh level. The level in normal BM is arbitrarily set to 1. (C) Separation of Hoxa9/Meis1-transformed murine AML cells into c-kithigh and c-kitlow populations by FACS. These 2 cell populations are used for methylcellulose colony formation assays. (D) The c-kithigh cell population has higher colony formation capability compared with the c-kitlow population. (E) RT-qPCR analysis shows that the c-kithigh cell population expresses a higher level of Kdm2b/Jhdm1b than the c-kitlow cell population. Relative mRNA levels are measured by RT-qPCR and normalized to Gapdh level. The level of c-kitlow population is arbitrarily set to 1. FSC indicates forward scatter.
Enforced expression of Kdm2b/Jhdm1b is sufficient for BM transformation
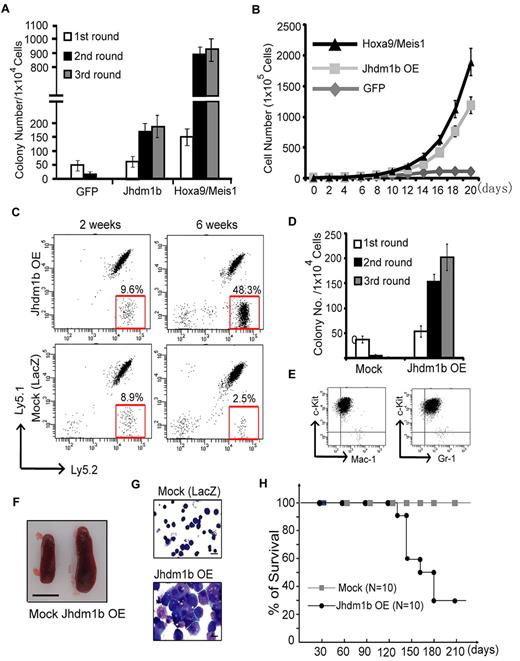
To examine whether overexpression of Kdm2b/Jhdm1b is sufficient to immortalize HPCs in vitro, c-kit+ HPCs were isolated from mouse BM and transduced with lentivirus that expressed Kdm2b/Jhdm1b. As controls, transductions with lentiviruses expressing GFP or Hoxa9/Meis1 were also performed in parallel. The transduction efficiency of HPCs was estimated at ∼ 80%-90% on the basis of the percentage of GFP+ cells analyzed 48 hours after transduction (supplemental Figure 1A). After verifying overexpression of Kdm2b/Jhdm1b in the transduced cells (supplemental Figure 1B), its effect on colony formation was evaluated by methylcellulose assay. Compared with GFP-expressing cells, whose colony formation capacity decreases with each round of replating, enforced expression of Kdm2b/Jhdm1b confers methylcellulose replating capability, although the colony numbers are fewer than that obtained from cells transduced with Hoxa9/Meis1 (Figure 2A). Importantly, cells derived from enforced Kdm2b/Jhdm1b expression can sustain in liquid culture medium supplemented with IL-3, IL-6, and SCF. In contrast, the control GFP-expressing cells were not able to proliferate under the same conditions (Figure 2B). These results suggest that enforced expression of Kdm2b/Jhdm1b alone is sufficient to immortalize c-kit+ cells in vitro, consistent with a potential role of Kdm2b/Jhdm1b in leukemogenesis.
Enforced expression of Kdm2b/Jhdm1b is sufficient for BM transformation. (A) Enforced expression of Kdm2b/Jhdm1b is capable of immortalizing BM cells in vitro. Shown are colony numbers of c-kit+ HPCs transduced with lentiviral vectors expressing GFP, Kdm2b/Jhdm1b, or Hoxa9-Meis1 at each round of methylcellulose replating. (B) Growth curve of c-kit+ hematopoietic progenitors transduced with lentiviral vectors expressing GFP, Kdm2b/Jhdm1b, or Hoxa9-Meis1 in suspension culture. (C) FACS analysis of the genetically modified Ly5.2+ donor cells, LacZ and Jhdm1b overexpression (Jhdm1b OE) in the peripheral blood of recipients. The results show that the percentage of Kdm2b/Jhdm1b-overexpressing Ly5.2+ cells increases gradually after transplantation. (D) Methylcellulose replating assay shows demonstrates that Kdm2b/Jhdm1b-overexpressing BM cells isolated from recipient mice can form increased numbers of colonies. In contrast, mock-transplanted BM cells fail to grow continuously in the methylcellulose replating assay. Colony numbers for each round of replating are shown. (E) Flow cytometric analysis shows that Kdm2b/Jhdm1b-overexpressed colonies express a high level of c-kit and low levels of myeloid lineage markers Mac-1/Gr-1. (F) Splenomegaly is observed in mice that received a transplant with Kdm2b/Jhdm1b-overexpressing BM cells. Shown is a representative picture of spleens harvested from mice 6 weeks after transplantation of BM cells transduced with lentiviral vectors expressing LacZ or Kdm2b/Jhdm1b. Bar size represents 1.0 cm. (G) May-Grünwald/Giemsa staining showing typical leukoblasts in the BM of mice that received a transplant with Kdm2b/Jhdm1b-overexpressing BM cells. Bar size represents 10 μm. (H) Survival curve shows mice that received a transplant with mock cells survived ≥ 210 days after transplantation, whereas most mice that received a transplant with Kdm2b/Jhdm1b-overexpressing cells died within 120-180 days after transplantation.
Enforced expression of Kdm2b/Jhdm1b is sufficient for BM transformation. (A) Enforced expression of Kdm2b/Jhdm1b is capable of immortalizing BM cells in vitro. Shown are colony numbers of c-kit+ HPCs transduced with lentiviral vectors expressing GFP, Kdm2b/Jhdm1b, or Hoxa9-Meis1 at each round of methylcellulose replating. (B) Growth curve of c-kit+ hematopoietic progenitors transduced with lentiviral vectors expressing GFP, Kdm2b/Jhdm1b, or Hoxa9-Meis1 in suspension culture. (C) FACS analysis of the genetically modified Ly5.2+ donor cells, LacZ and Jhdm1b overexpression (Jhdm1b OE) in the peripheral blood of recipients. The results show that the percentage of Kdm2b/Jhdm1b-overexpressing Ly5.2+ cells increases gradually after transplantation. (D) Methylcellulose replating assay shows demonstrates that Kdm2b/Jhdm1b-overexpressing BM cells isolated from recipient mice can form increased numbers of colonies. In contrast, mock-transplanted BM cells fail to grow continuously in the methylcellulose replating assay. Colony numbers for each round of replating are shown. (E) Flow cytometric analysis shows that Kdm2b/Jhdm1b-overexpressed colonies express a high level of c-kit and low levels of myeloid lineage markers Mac-1/Gr-1. (F) Splenomegaly is observed in mice that received a transplant with Kdm2b/Jhdm1b-overexpressing BM cells. Shown is a representative picture of spleens harvested from mice 6 weeks after transplantation of BM cells transduced with lentiviral vectors expressing LacZ or Kdm2b/Jhdm1b. Bar size represents 1.0 cm. (G) May-Grünwald/Giemsa staining showing typical leukoblasts in the BM of mice that received a transplant with Kdm2b/Jhdm1b-overexpressing BM cells. Bar size represents 10 μm. (H) Survival curve shows mice that received a transplant with mock cells survived ≥ 210 days after transplantation, whereas most mice that received a transplant with Kdm2b/Jhdm1b-overexpressing cells died within 120-180 days after transplantation.
To determine whether enforced expression of Kdm2b/Jhdm1b in HPCs is sufficient to induce leukemia in vivo, we transplanted the transduced Ly5.2+ HPCs with normal Ly5.1+Ly5.2+ radioprotective BM cells into lethally irradiated mice. The proliferation rate of transplanted cells was monitored by measuring the percentage of Ly5.2+ cells in the peripheral blood. Compared with control LacZ-transduced HPCs, the percentage of Ly5.2+ cells was greater in mice that received a transplant with Kdm2b/Jhdm1b-overexpressing HPCs (Figure 2C). To determine whether the transplanted cells were transformed, we isolated the Ly5.2+ c-kit+ cells from the BM of recipient mice 4 weeks after transplantation and performed serial methylcellulose replating assay. Compared with the LacZ-transduced cells, which failed to form colonies after a second round of plating, the Kdm2b/Jhdm1b-overexpressing BM cells have increased colony numbers at each round of replating (Figure 2D). FACS analysis of these transformed cells indicates that they express high levels of c-kit and relative low levels of Mac-1/Gr-1 (Figure 2E). The recipient mice in the same group gradually display splenomegaly (Figure 2F), and the typical leukoblasts are also observed in BM smears (Figure 2G). Consequently, mice that received a transplant with Kdm2b/Jhdm1b-overexpressing HPCs died within 4-6 months after transplantation. In contrast, all of the mice in the control group survived in the same period (Figure 2H). Collectively, these results suggest that enforced expression of Kdm2b/Jhdm1b is sufficient to transform hematopoietic progenitors in vivo.
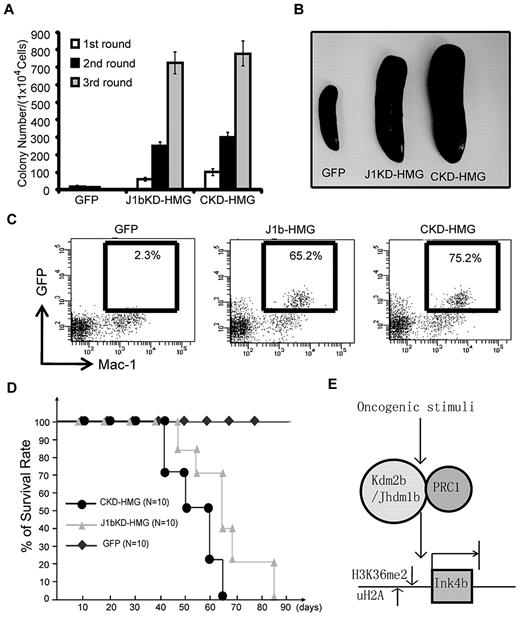
Kdm2b/Jhmd1b is required for Hoxa9/Meis1-induced leukemic transformation in vitro
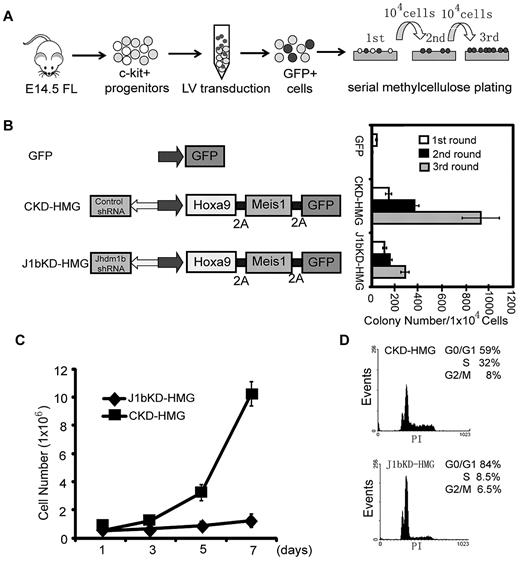
To examine whether Kdm2b/Jhdm1b is necessary for AML development, we used the well-established murine AML model that involved lentiviral transduction of Hoxa9/Meis1 oncogenes into c-Kit+ HPCs followed by methylcellulose replating assays (Figure 3A).10,23 To facilitate isolation and tracing of the genetically modified cells in the study, transduced cells are marked by GFP. HOXA9, MEIS1, and GFP proteins are linked by the foot-and-mouth disease virus peptide 2A, which allows expression of multiple proteins in a single expression vector.16 To examine the role of Kdm2b/Jhdm1b in Hoxa9/Meis1-induced leukemic transformation, a shRNA against all 3 isoforms of Kdm2b/Jhdm1b is delivered into the cells with the use of the same vector. After FACS (supplemental Figure 2A), Kdm2b/Jhdm1b mRNA levels in transduced GFP+ HPCs were analyzed by RT-qPCR. Results shown in supplemental Figure 2B indicate that the Kdm2b/Jhdm1b mRNA level decreased to 15% of the control level on KD. Methylcellulose replating assays show that KD of Kdm2b/Jhdm1b greatly affected the transformation capacity of Hoxa9/Meis1 (Figure 3B). Although colonies can still be derived from the Kdm2b/Jhdm1b KD cells in the third round of replating, these cells failed to proliferate when cultured in liquid media in the presence of IL-3 (Figure 3C). Flow cytometric analysis shows that, compared with control KD cells, Kdm2b/Jhdm1b KD cells have a higher percentage of cells in the G1 phase and a lower percentage of cells in the S phase (Figure 3D), indicating a G1-to-S transition defect. Collectively, these results suggest that Kdm2b/Jhdm1b is necessary for Hoxa9/Meis1-induced leukemic transformation as well as proliferation in vitro.
Kdm2b/Jhdm1b is required for Hoxa9-Meis1-induced leukemic transformation in vitro. (A) Flow chart of experimental procedure. To examine the role of Kdm2b/Jhdm1b in Hoxa9-Meis1-induced leukemic transformation in vitro, c-kit+ hematopoietic progenitors were isolated from E14.5 fetal liver (FL) and transduced with lentiviral (LV) vectors expressing various combinations of proteins and shRNAs. Transduced cells were then plated on methylcellulose to evaluate the effect of Kdm2b/Jhdm1b KD on colony formation and replating capacity. (B) KD of Kdm2b/Jhdm1b in Hoxa9-Meis1-induced leukemic cells impairs their methylcellulose colony replating capacity. Colony numbers for each round of replating are shown. (C) Growth curves indicate KD of Jhdm1b in Hoxa9/Meis1/GFP (J1bKD-HMG)–transformed leukemic cells impairs cell proliferation compared with Hoxa9/MeisI/GFP-transformed cells with control KD (CKD-HMG). Transformed cell colonies were picked after the third round of methylcellulose replating and cultured in liquid medium. (D) KD of Kdm2b/Jhdm1b results in a block at G1-to-S phase transition. Flow cytometric analysis of cell cycle status shows that Kdm2b/Jhdm1b KD (J1bKD-HMG) results in a higher percentage of cells in the G1 phase than that of control KD (CKD-HMG).
Kdm2b/Jhdm1b is required for Hoxa9-Meis1-induced leukemic transformation in vitro. (A) Flow chart of experimental procedure. To examine the role of Kdm2b/Jhdm1b in Hoxa9-Meis1-induced leukemic transformation in vitro, c-kit+ hematopoietic progenitors were isolated from E14.5 fetal liver (FL) and transduced with lentiviral (LV) vectors expressing various combinations of proteins and shRNAs. Transduced cells were then plated on methylcellulose to evaluate the effect of Kdm2b/Jhdm1b KD on colony formation and replating capacity. (B) KD of Kdm2b/Jhdm1b in Hoxa9-Meis1-induced leukemic cells impairs their methylcellulose colony replating capacity. Colony numbers for each round of replating are shown. (C) Growth curves indicate KD of Jhdm1b in Hoxa9/Meis1/GFP (J1bKD-HMG)–transformed leukemic cells impairs cell proliferation compared with Hoxa9/MeisI/GFP-transformed cells with control KD (CKD-HMG). Transformed cell colonies were picked after the third round of methylcellulose replating and cultured in liquid medium. (D) KD of Kdm2b/Jhdm1b results in a block at G1-to-S phase transition. Flow cytometric analysis of cell cycle status shows that Kdm2b/Jhdm1b KD (J1bKD-HMG) results in a higher percentage of cells in the G1 phase than that of control KD (CKD-HMG).
Kdm2b/Jhmd1b is required for Hoxa9/Meis1-induced AML development in vivo
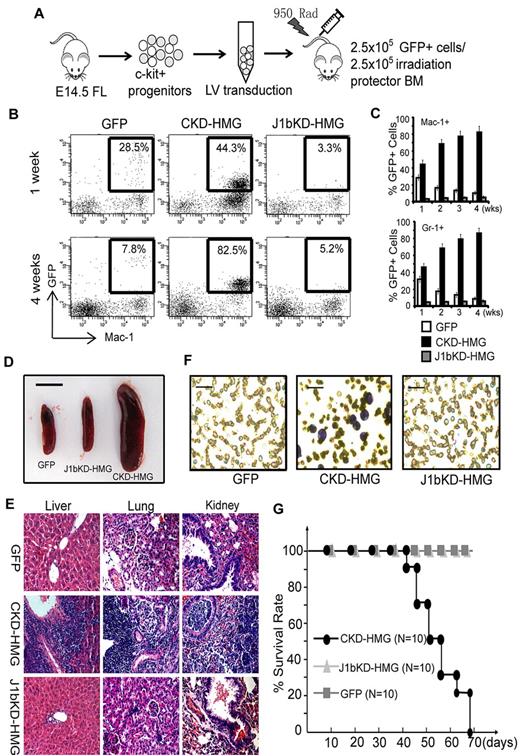
To examine whether Kdm2b/Jhdm1b is required for leukemia development in animals, we cotransplanted 2.5 × 105 various genetically modified GFP+ cells and 2.5 × 105 normal radioprotective BM cells into lethally irradiated syngenic mice (Figure 4A). At different times after transplantation, donor-derived cells in peripheral blood were analyzed by FACS analysis with the use of GFP and lineage markers. Results show that cells transduced with Hoxa9/Meis1/GFP (HMG) and control shRNA (CKD-HMG) can repopulate the recipient by 1 week with 40%-45% of GFP+ cells in the Gr-1+ and Mac-1+ myeloid population (Figure 4B-C). In addition, Hoxa9/Meis1-transformed cells proliferated faster than the radioprotective cells because most of the Mac-1+ and Gr-1+ cells are GFP+ at 4 weeks after transplantation (Figure 4B-C). However, the Kdm2b/Jhdm1b KD cells (J1bKD-HMG) failed to repopulate in vivo as indicated by a persistently low percentage of GFP+ cells after transplantation (Figure 4B-C). Cells transduced with GFP control decreased gradually because of the limited self-renewing capacity of normal HPCs (Figure 4B-C). Consistent with a GFP+-repopulating capability, the mice that received a transplant with CKD-HMG developed typical AML. The mice displayed splenomegaly (Figure 4D) with leukemic cell infiltration in multiple organs, including liver, kidney, and lung (Figure 4E). In addition, typical leukoblasts are found in the peripheral blood and BM smears (Figure 4F; supplemental Figure 3A). FACS analysis indicated that the leukemic cells express both c-kit and myeloid lineage markers such as Mac-1 and Gr-1 (supplemental Figure 3B). The mice also developed other leukemia symptoms such as anemia and bleeding and succumbed to death within 6-10 weeks after transplantation (Figure 4G). In contrast, similar to mice that received a transplant with control GFP cells, the mice that received a transplant with J1bKD-HMG–transduced cells appear healthy and survived > 3 months longer. Collectively, these results suggest that maintaining the expression of Kdm2b/Jhdm1b is important for Hoxa9/Meis1-induced leukemia development in mice.
Kdm2b/Jhdm1 is required for Hoxa9-Meis1-induced AML development in vivo. (A) Flow chart of experimental procedure for BM transplantation assays. To examine the role of Kdm2b/Jhdm1b in Hoxa9-Meis1-induced AML development in vivo, c-kit+ progenitors were isolated from E14.5 fetal liver (FL) of Ly5.2 C57BL/6 embryos. After lentiviral (LV) transduction with vectors expressing various combinations of proteins and shRNAs, the genetically modified cells were mixed with normal irradiation protector cells and were transplanted into lethally irradiated Ly5.1 C57BL/6 mice. (B) FACS analysis of the accumulation kinetics of the genetically modified donor cells (GFP, CKD-HMG, and J1bKD-HMG) in the peripheral blood of recipients. The results indicate that Kdm2b/Jhdm1b KD cells (J1bKD-HMG) failed to repopulate in the Mac-1+ myeloid lineage, whereas the control KD cells (CKD-HMG) can. (C) The percentage of genetically modified cells, marked by GFP, contribute to the myeloid lineage (Gr-1+ and Mac-1+) of peripheral blood at various times after transplantation. The results show that Kdm2b/Jhdm1b KD inhibits repopulating by Hoxa9-Meis1-transduced cells in recipient mice. The percentage of GFP+ cells in a particular lineage is calculated by dividing GFP and lineage marker double-positive cells with the total lineage marker-positive cells. All error bars represent SD (n = 10). (D) Splenomegaly of mice that received a transplant with lentiviral-transduced BM cells expressing CKD-HMG. Shown is a representative picture of spleens harvested from mice 6 weeks after transplantation of BM cells transduced with lentiviral vectors expressing GFP, J1bKD-HMG, or CKD-HMG. Bar size represents 1.0 cm. (E) H&E staining shows leukemic infiltration of multiple organs (spleen, kidney, and lung) in mice that received a transplant with Hoxa9-Meis1-induced leukemic cells (CKD-HMG), whereas there is no obvious leukemic cell infiltration in mice that received a transplant with BM cells transduced with lentiviral vectors expressing GFP or J1bKD-HMG cells. (F) May-Grünwald/Giemsa staining shows typical leukemic cells in the peripheral blood of mice that received a transplant with CKD-HMG cells. Bar size represents 50 μm. (G) Survival curve shows mice that received a transplant with GFP and J1bKD-HMG cells survived ≥ 70 days after transplantation, whereas the mice that received a transplant with CKD-HMG cells all died within 70 days after transplantation.
Kdm2b/Jhdm1 is required for Hoxa9-Meis1-induced AML development in vivo. (A) Flow chart of experimental procedure for BM transplantation assays. To examine the role of Kdm2b/Jhdm1b in Hoxa9-Meis1-induced AML development in vivo, c-kit+ progenitors were isolated from E14.5 fetal liver (FL) of Ly5.2 C57BL/6 embryos. After lentiviral (LV) transduction with vectors expressing various combinations of proteins and shRNAs, the genetically modified cells were mixed with normal irradiation protector cells and were transplanted into lethally irradiated Ly5.1 C57BL/6 mice. (B) FACS analysis of the accumulation kinetics of the genetically modified donor cells (GFP, CKD-HMG, and J1bKD-HMG) in the peripheral blood of recipients. The results indicate that Kdm2b/Jhdm1b KD cells (J1bKD-HMG) failed to repopulate in the Mac-1+ myeloid lineage, whereas the control KD cells (CKD-HMG) can. (C) The percentage of genetically modified cells, marked by GFP, contribute to the myeloid lineage (Gr-1+ and Mac-1+) of peripheral blood at various times after transplantation. The results show that Kdm2b/Jhdm1b KD inhibits repopulating by Hoxa9-Meis1-transduced cells in recipient mice. The percentage of GFP+ cells in a particular lineage is calculated by dividing GFP and lineage marker double-positive cells with the total lineage marker-positive cells. All error bars represent SD (n = 10). (D) Splenomegaly of mice that received a transplant with lentiviral-transduced BM cells expressing CKD-HMG. Shown is a representative picture of spleens harvested from mice 6 weeks after transplantation of BM cells transduced with lentiviral vectors expressing GFP, J1bKD-HMG, or CKD-HMG. Bar size represents 1.0 cm. (E) H&E staining shows leukemic infiltration of multiple organs (spleen, kidney, and lung) in mice that received a transplant with Hoxa9-Meis1-induced leukemic cells (CKD-HMG), whereas there is no obvious leukemic cell infiltration in mice that received a transplant with BM cells transduced with lentiviral vectors expressing GFP or J1bKD-HMG cells. (F) May-Grünwald/Giemsa staining shows typical leukemic cells in the peripheral blood of mice that received a transplant with CKD-HMG cells. Bar size represents 50 μm. (G) Survival curve shows mice that received a transplant with GFP and J1bKD-HMG cells survived ≥ 70 days after transplantation, whereas the mice that received a transplant with CKD-HMG cells all died within 70 days after transplantation.
Enzymatic activity of KDM2b/JHDM1b is required for self-renewal of LSCs
After establishing a role for Kdm2b/Jhdm1b in transformation and development of Hoxa9/Meis1-induced AML, we asked whether KDM2b/JHDM1b and its associated histone demethylase activity are required for self-renewal of LSCs. To this end, GFP+ leukemic cells were isolated from the BM of mice with primary AML and cultured in methylcellulose to select for LSCs. Cells capable of forming colonies were transduced with lentiviral vectors expressing control shRNA or Kdm2b/Jhdm1b shRNA. To determine whether the phenotypes caused by Kdm2b/Jhdm1b KD are related to its H3K36me2 demethylase activity, a mock gene (truncated lacZ), an shRNA-resistant wild-type, or JmjC domain mutant form of Kdm2b/Jhdm1b coupled with Kdm2b/Jhdm1b shRNA is delivered into the cells by the same lentiviral vector (Figure 5A; supplemental Figure 4A). The transduced cells were subjected to serial methylcellulose plating assay as well as leukemia transplantation assay. Methylcellulose plating assays showed that not only the size but also the number of colonies reduced dramatically on Kdm2b/Jhdm1b KD, suggesting that Kdm2b/Jhdm1b expression level in cells is critical for both proliferation of leukemic cells and self-renewal of LSCs. This defect in colony formation can be rescued by the expression of an shRNA-resistant wild-type but not a JmjC-domain mutant KDM2b/JHDM1b protein or LacZ in cells (Figure 5B-C). The differential effect of the wild-type and JmjC-domain mutant is not because of differences in their expression level because similar expression of Kdm2b/Jhdm1b is verified by both RT-qPCR and Western blot analysis (supplemental Figure 4B-C). Consistent with the colony formation assay, leukemia transplantation showed that mice that a received a transplant with control KD or wild-type Kdm2b/Jhdm1b-rescued leukemic cells died within 3-6 weeks after transplantation. In contrast, all mice that received a transplant with Kdm2b/Jhdm1b KD leukemic cells, 7 of 10 mice that received a transplant with JmjC-domain mutant–rescued KD leukemic cells, and 9 of 10 mice that received a transplant with mock-rescued KD leukemic cells survived for ≥ 3 months after transplantation (Figure 5D). These results suggest not only that the KD is specific but also that the enzymatic activity of KDM2b/JHDM1b is responsible for maintaining the leukemic state.
The enzymatic activity of KDM2b/JHDM1b is required for the self-renewal of leukemic stem cells. (A) Flow chart of experimental procedure for analyzing the role of Kdm2b/Jhdm1b in LSC self-renewal. Leukemic cells were isolated from primary AML mice and selected for LSCs through replating on methylcellulose. Cells derived from the colonies were transduced with various lentiviral (LV) vectors, followed by methylcellulose colony formation assay in vitro and secondary transplantation assays in vivo. (B) Photographs of the methylcellulose colony formation assay plates show that Kdm2b/Jhdm1b KD results in a decease of both size and number of colonies. This phenotype can be rescued by wild-type but not a catalytic mutant Kdm2b/Jhdm1b or LacZ. (C) Quantification of the colony numbers derived from the methylcellulose colony replating assays. (D) Survival curve shows prolonged survival of mice that received a transplant of primary Hoxa9-Meis1 leukemia cells with KD of Jhdm1b. This phenotype can be reversed by wild-type but not catalytic mutant Kdm2b/Jhdm1b or LacZ. CFC indicates colony-forming cell; Wt, wild-type; and Mut, mutant.
The enzymatic activity of KDM2b/JHDM1b is required for the self-renewal of leukemic stem cells. (A) Flow chart of experimental procedure for analyzing the role of Kdm2b/Jhdm1b in LSC self-renewal. Leukemic cells were isolated from primary AML mice and selected for LSCs through replating on methylcellulose. Cells derived from the colonies were transduced with various lentiviral (LV) vectors, followed by methylcellulose colony formation assay in vitro and secondary transplantation assays in vivo. (B) Photographs of the methylcellulose colony formation assay plates show that Kdm2b/Jhdm1b KD results in a decease of both size and number of colonies. This phenotype can be rescued by wild-type but not a catalytic mutant Kdm2b/Jhdm1b or LacZ. (C) Quantification of the colony numbers derived from the methylcellulose colony replating assays. (D) Survival curve shows prolonged survival of mice that received a transplant of primary Hoxa9-Meis1 leukemia cells with KD of Jhdm1b. This phenotype can be reversed by wild-type but not catalytic mutant Kdm2b/Jhdm1b or LacZ. CFC indicates colony-forming cell; Wt, wild-type; and Mut, mutant.
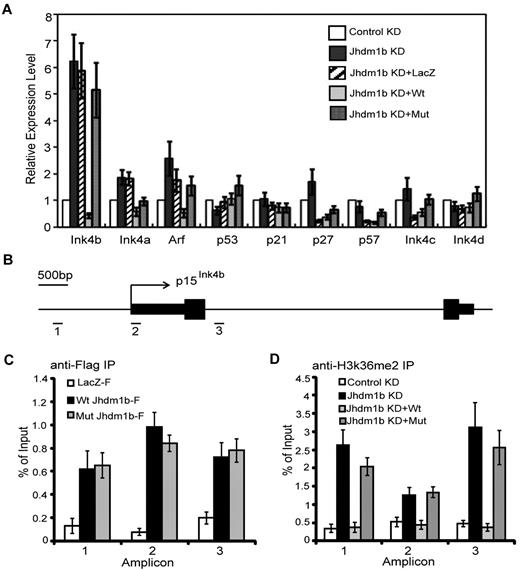
Kdm2b/Jhdm1b directly regulates p15Ink4b expression in leukemic cells
To understand how Kdm2b/Jhdm1b regulates leukemic cell proliferation and LSC self-renewal, we focused on proliferation-related genes. Given that genes with immediate response to Kdm2b/Jhdm1b manipulation are more likely to be its direct targets, we examined the expression of a panel of proliferation-related genes 72 hours after viral transduction with the use of the cells generated in Figure 5A. Consistent with our finding in MEFs,16 p15Ink4b is up-regulated significantly on Kdm2b/Jhdm1b KD. Importantly, p15Ink4b can be re-silenced by introducing an shRNA-resistant wild-type Kdm2b/Jhdm1b, but not a catalytic mutant or irrelevant LacZ (Figure 6A). This result suggests that the H3K36me2 demethylase activity of KDM2b/JHDM1b is involved in p15Ink4b repression. In addition to p15Ink4b, p16Ink4a and p19Arf are also modestly increased in response to Kdm2b/Jhdm1b KD (Figure 6A). However, because p15Ink4b had the most significant changes, we analyzed this effect further.
KDM2b/JHDM1b directly regulates p15Ink4b expression in leukemic cells. (A) RT-qPCR analysis shows that p15Ink4b is significantly up-regulated in response to Kdm2b/Jhdm1b KD. Relative mRNA levels are measured by RT-qPCR and normalized to Gapdh level. (B) Schematic representation of the p15Ink4b locus in mouse, indicating the genomic structure (exons are represented by black boxes), as well as the location of the 3 amplicons analyzed by ChIP assays. (C) ChIP experiments with chromatin prepared from leukemic cells expressing LacZ-Flag, wild-type KDM2b/JHDM1b-Flag, and mutant KDM2b/JHDM1b-Flag were carried out with the use of anti-Flag antibody. KDM2b/JHDM1b-Flag binding was assayed by qPCR at the 3 genomic regions depicted in panel B. (D) ChIP experiments with chromatin prepared from leukemic cells with control KD, Kdm2b/Jhdm1b KD, Kdm2b/Jhdm1b KD reconstituted with wild-type Kdm2b/Jhdm1b or mutant Kdm2b/Jhdm1b were carried out with the use of the anti-H3K36me2 antibody. H3K36me2 level was assayed by qPCR at the 3 genomic regions depicted in panel B.
KDM2b/JHDM1b directly regulates p15Ink4b expression in leukemic cells. (A) RT-qPCR analysis shows that p15Ink4b is significantly up-regulated in response to Kdm2b/Jhdm1b KD. Relative mRNA levels are measured by RT-qPCR and normalized to Gapdh level. (B) Schematic representation of the p15Ink4b locus in mouse, indicating the genomic structure (exons are represented by black boxes), as well as the location of the 3 amplicons analyzed by ChIP assays. (C) ChIP experiments with chromatin prepared from leukemic cells expressing LacZ-Flag, wild-type KDM2b/JHDM1b-Flag, and mutant KDM2b/JHDM1b-Flag were carried out with the use of anti-Flag antibody. KDM2b/JHDM1b-Flag binding was assayed by qPCR at the 3 genomic regions depicted in panel B. (D) ChIP experiments with chromatin prepared from leukemic cells with control KD, Kdm2b/Jhdm1b KD, Kdm2b/Jhdm1b KD reconstituted with wild-type Kdm2b/Jhdm1b or mutant Kdm2b/Jhdm1b were carried out with the use of the anti-H3K36me2 antibody. H3K36me2 level was assayed by qPCR at the 3 genomic regions depicted in panel B.
To determine whether p15Ink4b is a direct target of Kdm2b/Jhdm1b, ChIP assay was carried out with the use of chromatin prepared from leukemic cells harboring wild-type or catalytic mutant rescue Kdm2b/Jhdm1b transgenes (Figure 5A). Both wild-type and mutant KDM2b/JHDM1b-Flag were found to be enriched from the promoter region (amplicon 1) to the first intron (amplicon 3) compared with the LacZ-Flag control (Figure 6B-C), indicating that p15Ink4b is a direct KDM2b/JHDM1b target. Consistent with its H3K36me2-specific demethylase activity, KD of Kdm2b/Jhdm1b results in an increased H3K36me2 levels across the 3 regions compared with control KD (Figure 6B,D). Importantly, reintroduction of an shRNA-resistant wild-type, but not the catalytic mutant, Kdm2b/Jhdm1b results in a decreased level of H3K36me2 comparable to that of control KD (Figure 6D). The change in H3K36me2 level at the p15Ink4b locus appears to be gene specific because no significant increase in the global H3K36me2 level is observed on Kdm2b/Jhdm1b KD (supplemental Figure 5A).
To further substantiate p15Ink4b as a direct target, we measured the p15Ink4b expression by RT-qPCR in mock and Kdm2b/Jhdm1b-overexpressing HPCs that were cultured over 2 weeks. The result shows that overexpression of Kdm2b/Jhdm1b resulted in repression of p15Ink4b (supplemental Figure 5B). Further ChIP analysis with the use of the same cells showed that KDM2b/JHDM1b-Flag is enriched and the H3K36me2 level is reduced at the p15Ink4b locus in Kdm2b/Jhdm1b-overexpressing cells (supplemental Figure 5C). Because previous studies have suggested a link between DNA methylation and p15Ink4b silencing in leukemia,20,21 we asked whether p15Ink4b silencing on Kdm2b/Jhdm1b overexpression uses a similar mechanism. To this end, we analyzed the DNA methylation levels of p15Ink4b promoter and first exon in normal HPCs, Kdm2b/Jhdm1b immortalized HPCs, and Hoxa9/Meis1-transformed cells by bisulfite sequencing. Results shown in supplemental Figure 5D show no significant differences in DNA methylation levels, indicating that changes in histone modifications, but not DNA methylation, are responsible for down-regulation of p15Ink4b in the Kdm2b/Jhdm1b-overexpressing and Hoxa9/Meis1-transformed cells. These results suggest that silencing of p15Ink4b in leukemia may use multiple mechanisms under different conditions. Collectively, these data suggest that p15Ink4b is a KDM2b/JHDM1b direct target and that KDM2b/JHDM1b down-regulates p15Ink4b expression by maintaining low levels of H3K36me2.
p15Ink4b is a critical target mediating the function of Kdm2b/Jhdm1b in Hoxa9/Meis1-induced AML
After showing that p15Ink4b is a direct target of KDM2b/JHDM1b, we asked whether p15Ink4b is a major effector that mediates the function of Kdm2b/Jhdm1b in leukemogenesis. To this end, c-kit+ HPCs were isolated from the BM of p15Ink4b knock-out mice. After transduction with lentiviral vectors expressing control GFP, CKD-HMG control shRNA, and J1bKD-HMG, GFP+ cells were sorted and subjected to serial methylcellulose replating assay and BM transplantation assays. Results in Figure 7A show that Hoxa9/Meis1 can induce transformation of p15Ink4b-null HPCs regardless of whether Kdm2b/Jhdm1b is knocked down or not. This is in direct contrast with the results obtained when wild-type HPCs are used (Figure 3B), indicating that transformation by Hoxa9/Meis1 largely depends on Kdm2b/Jhdm1b-mediated repression of p15Ink4b. Consistent with this in vitro result, mice that received a transplant with J1bKD-HMG p15Ink4b-null cells exhibited the same splenomegaly phenotype as that of mice that received a transplant with CKD-HMG cells, indicating that the transformed cells can proliferate in animals even if Kdm2b/Jhdm1b is depleted (Figure 7B). In addition, FACS analysis shows that GFP+ cells can repopulate and dominate the myeloid lineage cell population in mice 4 weeks after transplantation regardless of whether Kdm2b/Jhdm1b is depleted (Figure 7C). Finally, all mice that received a transplant with J1bKD-HMG p15Ink4b-null cells died of leukemia within 3 months of transplantation, although there is a 2- to 3-week delay compared with mice that received a transplant with CKD-HMG cells (Figure 7D), indicating that other Kdm2b/Jhdm1b targets beside p15Ink4b may also contribute to the effect of Kdm2b/Jhdm1b in leukemogenesis. Nevertheless, the above-mentioned results suggest that p15Ink4b is a main target that mediates the function of Kdm2b/Jhdm1b in leukemogenesis.
p15Ink4b is a critical target mediating the function of KDM2b/JHDM1b in Hoxa9-Meis1-induced AML. (A) Loss of p15Ink4b abrogates the effect of Kdm2b/Jhdm1b KD on Hoxa9-Meis1-induced leukemic transformation. Serial methylcellulose replating assay shows that similar numbers of colonies were obtained from p15Ink4b-null HPCs transduced with Hoxa9-Meis1 regardless of whether Kdm2b/Jhdm1b was knocked down. (B) Splenomegaly is observed in mice that received a transplant with both control KD (CKD/HMG) and Kdm2b/Jhdm1b KD (J1bKD-HMG) Hoxa9-Mesi1-transduced p15Ink4b-null hematopoietic progenitors. (C) Flow cytometric analysis shows that both J1bKD-HMG and CKD/HMG transduced p15Ink4b-null hematopoietic progenitor donor cells can repopulate and dominate peripheral blood of recipient mice 4 weeks after transplantation. (D) Survival curve shows that mice that received a transplant with p15Ink4b-null HPCs transduced with either CKD-HMG or J1bKD-HMG die within 90 days after transplantation. (E) Proposed model for epigenetic regulation of the Ink4b locus in LSCs. In LSCs, p15Ink4b is suppressed by multiple epigenetic modifiers, including KDM2b/JHDM1b and Polycomb group proteins. In this model, Kdm2b/Jhdm1b is up-regulated by oncogenic stimuli and maintains at a high level in LSCs. Demethylation of H3K36 by KDM2b/JHDM1b and concomitant H2A ubiquitylation by the associated Polycomb group of proteins results in silencing of p15Ink4b. Conversely, depletion of Kdm2b/Jhdm1b causes an increase in the H3K36me2 level concomitant with the loss of Polycomb group proteins, leading to de-repression of p15Ink4b, resulting in defects in leukemic cell proliferation and LSC self-renewal.
p15Ink4b is a critical target mediating the function of KDM2b/JHDM1b in Hoxa9-Meis1-induced AML. (A) Loss of p15Ink4b abrogates the effect of Kdm2b/Jhdm1b KD on Hoxa9-Meis1-induced leukemic transformation. Serial methylcellulose replating assay shows that similar numbers of colonies were obtained from p15Ink4b-null HPCs transduced with Hoxa9-Meis1 regardless of whether Kdm2b/Jhdm1b was knocked down. (B) Splenomegaly is observed in mice that received a transplant with both control KD (CKD/HMG) and Kdm2b/Jhdm1b KD (J1bKD-HMG) Hoxa9-Mesi1-transduced p15Ink4b-null hematopoietic progenitors. (C) Flow cytometric analysis shows that both J1bKD-HMG and CKD/HMG transduced p15Ink4b-null hematopoietic progenitor donor cells can repopulate and dominate peripheral blood of recipient mice 4 weeks after transplantation. (D) Survival curve shows that mice that received a transplant with p15Ink4b-null HPCs transduced with either CKD-HMG or J1bKD-HMG die within 90 days after transplantation. (E) Proposed model for epigenetic regulation of the Ink4b locus in LSCs. In LSCs, p15Ink4b is suppressed by multiple epigenetic modifiers, including KDM2b/JHDM1b and Polycomb group proteins. In this model, Kdm2b/Jhdm1b is up-regulated by oncogenic stimuli and maintains at a high level in LSCs. Demethylation of H3K36 by KDM2b/JHDM1b and concomitant H2A ubiquitylation by the associated Polycomb group of proteins results in silencing of p15Ink4b. Conversely, depletion of Kdm2b/Jhdm1b causes an increase in the H3K36me2 level concomitant with the loss of Polycomb group proteins, leading to de-repression of p15Ink4b, resulting in defects in leukemic cell proliferation and LSC self-renewal.
Discussion
The Kdm2b/Jhdm1b gene was first identified as a hot spot for proviral insertion in murine tumors generated by random insertion of Moloney murine leukemia virus.14 Our previous studies have shown that depletion of Kdm2b/Jhdm1b in MEFs results in defective cellular proliferation and premature senescence, implicating that Kdm2b/Jhdm1b has proto-oncogene properties.16 In the current study, we provide 3 lines of evidence that further support that Kdm2b/Jhdm1b functions as an oncogene and plays critical roles in both leukemogenesis and LSC self-renewal. First, overexpression of Kdm2b/Jhdm1b in normal HPCs can increase their methylcellulose replating capability and proliferation in vitro and in vivo (Figure 2), suggesting that an increased level of Kdm2b/Jhdm1b can confer a cell growth advantage and induce leukemic transformation. In addition, the colonies with overexpressed Kdm2b/Jhdm1b maintain a high level of progenitor marker c-kit and express low levels of myeloid markers Mac-1/Gr-1after serial methylcellulose replating, indicating that overexpression of Kdm2b/Jhdm1b is able to restrict cells in the progenitor-like status and to suppress differentiation (Figure 2E). Second, depletion of Kdm2b/Jhdm1b impairs Hoxa9/Meis1-induced leukemogenesis (Figures 3–4). This defect can be largely rescued in p15Ink4b-null HPCs, indicating a block of cellular proliferation because of de-repression of p15Ink4b as a primary cause. Third, depletion of Kdm2b/Jhdm1b in LSCs impairs the capability of LSCs to self-renew, which is manifested by both small size and low number of colonies in the methylcellulose replating assay. This further suggests that high levels of Kdm2b/Jhdm1b might not only promote the proliferation of LSCs but also inhibit their differentiation. In addition, the oncogenic function of Kdm2b/Jhdm1b is supported by the finding that Kdm2b/Jhdm1b is highly expressed in various human leukemias, Hoxa9/Meis1-induced murine AML cells, as well as the LSC-enriched cell population. However, it is currently not clear how Kdm2b/Jhdm1b expression is up-regulated during leukemogenesis and maintained in LSCs but not in non–self-renewing leukemic progeny cells. Further ChIP assays that use oncoprotein-specific antibodies to map their binding sites in the Kdm2b/Jhdm1b regulatory regions will help elucidate the relationship between oncogenic stimuli and Kdm2b/Jhdm1b expression in the initiation and maintenance of leukemia.
The Ink4a-Arf-Ink4b locus plays an important role in regulating cellular proliferation, premature senescence, and apoptosis.24 Mutations and deletions in this locus are frequently found in hematopoietic malignancies.25 Generally, expression of this locus is suppressed in normal adult stem cells and cancer stem cells by various epigenetic regulators. Depletion of Bmi-1, a component in the PRC1, can activate the expression of Ink4a-Arf and result in self-renewal defects in hematopoietic stem cells, neural stem cells, as well as LSCs.9,10,26 Although p15Ink4b and p16Ink4a have a similar mechanism in regulating cell cycle progression and work together in preventing tumorigenesis,27 the transcriptional regulation of p15Ink4b and its role in cancer stem cells is less characterized. In this study, we demonstrate that Kdm2b/Jhdm1b regulates LSC self-renewal mainly through modulating the expression levels of p15Ink4b. We show that the expression of p15Ink4b is up-regulated immediately on Kdm2b/Jhdm1b KD. We also demonstrate that deletion of p15Ink4b can largely rescue the phenotypes caused by Kdm2b/Jhdm1b KD (Figure 7). These results suggest that, although the Ink4a-Arf-Ink4b locus is regulated by multiple epigenetic factors, p15Ink4b and p16Ink4a might have differential regulatory mechanisms. Accordingly, the existence of multiple regulatory mechanisms may allow tight control of the expression of the Ink4a-Arf-Ink4b locus in different conditions. Because the Ink4a-Arf-Ink4b locus also plays an important role in self-renewal of normal adult stem cells, we speculate that Kdm2b/Jhdm1b probably has a similar function as that of Bmi-1 in normal HSCs. Therefore, for therapeutic purposes, it is important to find out whether there is a sensitivity difference between normal HSCs and LSCs in response to Kdm2b/Jhdm1b depletion. Further studies that use Kdm2b/Jhdm1b knockout mice will help answer this question.
Kdm2b/Jdhm1b is a paralog of the first JmjC domain-containing histone demethylase Jhdm1a, which specifically targets H3K36me2 for demethylation.19 We have previously demonstrated that Kdm2b/Jhdm1b also targets H3K36me2 for demethylation.16 In this study, we found that the LSC defects caused by Kdm2b/Jhdm1b depletion can be rescued by wild-type, but not a catalytic mutant, Kdm2b/Jhdm1b, implying that its histone demethylase activity is critical in mediating its function in LSCs (Figure 5). Consistent with the observed defect in LSC self-renewal, we found that the tumor suppressor p15Ink4b is de-repressed on Kdm2b/Jhdm1b KD, which is also rescued by wild-type, but not a catalytic mutant Kdm2b/Jhdm1b. Collectively, these data suggest that H3K36me2 levels are directly linked to p15Ink4b expression (Figure 5). ChIP analysis confirmed that KDM2b/JHDM1b binds to and is responsible for maintaining a low H3K36me2 level at the p15Ink4b locus, resulting in p15Ink4b down-regulation. Our data indicate that the level of H3K36me2 correlates with transcriptional activity, which is consistent with previous studies that showed the association of the H3K36 methyltransferase SET2 with the elongating RNA polymerase II28,29 and that H3K36 methylation is coupled with transcription elongation.29 Given that KDM2b/JHDM1b can actively remove the methyl groups from H3K36, it is not surprising that Kdm2b/Jhdm1b plays an important role in silencing p15Ink4b. Because KDM2b/JHDM1b has been shown to be part of a Polycomb group complex,30,31 it is probable that KDM2b/JHDM1b-mediated H3K36 demethylation is coupled with transcription repression by Polycomb group proteins. Further studies should clarify the exact molecular mechanism underlying p15Ink4b repression in LSCs.
In summary, we propose that KDM2b/JHDM1b is a critical epigenetic factor involved in leukemogenesis and LSC self-renewal (Figure 7E). In response to oncogenic signals, expression of Kdm2b/Jhdm1b is up-regulated during leukemogenesis and is maintained at a high level in LSCs. Binding of KDM2b/JHDM1b to the p15Ink4b locus results in demethylation of H3K36me2 and concomitant repression of p15Ink4b by an associated Polycomb group of proteins. Conversely, depletion of Kdm2b/Jhdm1b causes an increase in H3K36me2 and loss of Polycomb group proteins at the p15Ink4b locus, which leads to de-repression of p15Ink4b and blocks the proliferation of leukemia cells and LSCs. On the basis of this model, developing small molecule inhibitors for KDM2b/JHDM1b enzymatic activity might be a potential strategy for leukemia treatment.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank L. Wolff (National Cancer Institute) for the p15Ink4b-null mice.
This work was partly supported by that National Institutes of Health (grant CA119133) (Y.Z.). J.H. is a fellow of the Leukemia & Lymphoma Society. Y.Z. is an investigator of the Howard Hughes Medical Institute.
National Institutes of Health
Authorship
Contribution: J.H. designed and performed the experiments and contributed data for Figures 1,Figure 2,Figure 3,Figure 4,Figure 5,Figure 6–7; A.T.N. contributed data for Figure 4; Y.Z. oversaw the projects; and J.H. and Y.Z. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yi Zhang, Department of Biochemistry and Biophysics, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599-7295; e-mail: yi_zhang@med.unc.edu.