Abstract
The proteolysis of VWF by ADAMTS13 is an essential step in the regulation of its hemostatic and thrombogenic potential. The cleavage occurs at strand β4 in the structural core of the A2 domain of VWF, so unfolding of the A2 domain is a prerequisite for cleavage. In the present study, we present the crystal structure of an engineered A2 domain that exhibits a significant difference in the α3-β4 loop compared with the previously reported structure of wild-type A2. Intriguingly, a metal ion was detected at a site formed mainly by the C-terminal region of the α3-β4 loop that was later identified as Ca2+ after various biophysical and biochemical studies. Force-probe molecular dynamic simulations of a modeled structure of the wild-type A2 featuring the discovered Ca2+-binding site revealed that an increase in force was needed to unfold strand β4 when Ca2+ was bound. Cleavage assays consistently demonstrated that Ca2+ binding stabilized the A2 domain and impeded its unfolding, and consequently protected it from cleavage by ADAMTS13. We have revealed a novel Ca2+-binding site at the A2 domain of VWF and demonstrated a relationship between Ca2+ and force in the regulation of VWF and primary hemostasis.
Introduction
VWF plays an important role in primary hemostasis by mediating platelet adhesion. VWF is stored in endothelial Weibel-Palade bodies and platelet α-granules as glycosylated multimers consisting of the largest multimeric species, ultra-large VWF.1-4 Upon stimulation, the VWF multimers are released into the circulating blood or subendothelial matrix. Within 2 hours after release into the vessel, the secreted ultra-large VWF multimers are converted by ADAMTS13 proteolysis into smaller multimers ranging from 500-20 000 kDa.5-8 The cleavage site for ADAMTS13 resides at the Tyr1605-Met1606 bond in the A2 domain,1,9 a cryptic site buried in the central β-strand in the intact, folded A2 domain that is inaccessible to ADAMTS13.10
More than a decade ago, it was observed that shear stress promotes the process of VWF proteolysis.11 Since that time, a large body of evidence has supported the notions that the conformation of VWF fibers is altered by shear stress12 and that unfolding of VWF by shear stress leads to enhanced susceptibility of the VWF to ADAMTS13 proteolysis.13-15 Because the hemostatic potential of VWF greatly increases with its length, cleavage by ADAMTS13 provides an important regulatory mechanism for primary hemostasis. The cleavage process is precisely regulated in vivo, and dysfunction of this regulation can lead to severe consequences. For example, mutations in VWF that cause increased susceptibility of VWF to ADAMTS13 lead to a shift of the multimer distribution to smaller forms with less hemostatic potential, resulting in continuous bleeding diseases known as VWD.16,17 Most mutations in the A2 domain that are associated with VWD type 2A, the common qualitative defect of VWD, result in enhanced proteolytic susceptibility.18
Recently, it was clearly shown that unfolding of A2 is required for its cleavage by ADAMTS13, and an intermediate state exists with unfolding of the C-terminal region up to and including the β4 strand where the cleavage site is located.19 Further information about the stepwise unfolding of A2 under tensile stress and the consequent exposure of the ADAMTS13 cleavage site were revealed by simulations of force-probe molecular dynamics (MD).20 To further understand the molecular mechanism underlying the shear force–induced cleavage of the VWF A2 domain by ADAMTS13, we carried out structural and functional studies of the A2 domain. We constructed an engineered A2 domain (N1493C/C1670S, ssA2) by introducing an A1/A3-like disulfide bond20 that allowed us to perform further structural studies. While we solved the crystal structure of the engineered A2 domain, Zhang et al reported the crystal structure of a wild-type A2 (wtA2) domain, which provided structural insights into the mechanism of its shear force sensor function.21 Compared with the wtA2 domain, our ssA2 structure reveals a novel Ca2+-binding site on the α3-β4 loop that is absent in the wtA2 structure. To investigate the potential functional role of Ca2+ binding in the shear force–dependent cleavage of the VWF A2 domain by ADAMTS13, we performed MD simulations, mutagenesis studies, and in vitro biochemical assays. Our results show that the binding of Ca2+ stabilizes the A2 domain and impedes its unfolding, thus decreasing the susceptibility of the A2 domain to ADAMTS13 proteolysis. Our structural and biochemical data provide new insights into the regulation of VWF cleavage by ADAMTS13 in hemostasis and thrombosis.
Methods
Cloning, expression, and purification of VWF A2 domain
The gene fragment of the VWF A2 domain (residues Leu1487-Gly1674) was amplified from the cDNA of HEK293T cells and inserted into pET22b (Novagen). An engineered A2 domain (ssA2) that contains the N1493C and C1670S mutations was generated to introduce a disulfide bond that links the N- and C-termini of the A2 domain in a manner analogous to that in the VWF A1 and A3 domains.20
The ssA2 protein and its mutants were expressed in E coli BL21 (DE3) CodonPlus cells. The proteins were purified with nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography followed by gel filtration. The target proteins were concentrated to 15 mg/mL in buffer A (20mM Tris-HCl, pH 8.0, 300mM NaCl, 10% glycerol, and 0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) for crystallization. For inductively coupled plasma atomic emission spectroscopy analysis, the purified ssA2 protein was further dialyzed against buffer B (3mM Tris-HCl, pH 8.0).
For cleavage assays, the wild-type and mutant wtA2 proteins were expressed in HEK293S GNT1− cells, as described previously.21 The target proteins were purified by Ni-NTA affinity chromatography. VWF73 (residues Asp1596-Arg1668) was shown to be the minimal substrate for ADAMTS13.22 The VWF73 gene fragment with a C-terminal hexahistidine tag was inserted into pGEX4T1. The wild-type and mutant glutathione S-transferase (GST)-VWF73 proteins were expressed in E coli BL21 (DE3) CodonPlus cells and purified by Ni-NTA followed by GST affinity chromatography. The purified wtA2 and VWF73 proteins were dialyzed against buffer C (50mM HEPES, pH 7.4, and 150mM NaCl) for cleavage assays.
Crystallization, diffraction data collection, and structure determination and refinement
Crystallization experiments were carried out at 4°C using the hanging drop vapor diffusion method. The crystals of ssA2 and its mutants were grown in drops containing equal volumes of the protein solution and the crystallization solution (0.1M sodium citrate, pH 5.2, and 1.6M [NH4]2SO4). The crystals were cryoprotected with the reservoir solution supplemented with 25% glycerol and then flash-frozen in liquid nitrogen. X-ray diffraction data were collected at −173°C at beam-line 17U of the Shanghai Synchrotron Radiation Facility (Shanghai, China) at a wavelength of 1.0000 Å using a MAR225 detector, and were processed using the HKL2000 (Version 0.98. 698x, HKL Research Inc) software suite.23 Diffraction data statistics are summarized in Table 1.
Statistics of X-ray diffraction data and structure refinement
| . | ssA2 . | ssA2-D1596A . | ssA2-N1602A . | ssA2-D1596A/N1602A . |
|---|---|---|---|---|
| PDB code | 3PPV | 3PPW | 3PPX | 3PPY |
| Diffraction data | ||||
| Space group | P65 | P65 | P65 | P65 |
| Cell parameters | ||||
| a = b/c, Å | 69.8/73.6 | 72.1/76.3 | 72.1/76.3 | 72.5/76.3 |
| Resolution range, Å | 50.0-1.9 | 50.0-1.9 | 50.0-1.9 | 50.0-2.0 |
| (1.97-1.90)* | (1.97-1.90) | (1.97-1.90) | (2.07-2.00) | |
| Observed reflections, no. | 132 324 | 72 418 | 95 704 | 101,783 |
| Unique reflections (I/σ(I) > 0), no. | 15 647 | 17 621 | 17 507 | 15 351 |
| Average redundancy | 8.5 (4.9) | 4.1 (3.1) | 5.5 (4.2) | 6.6 (4.4) |
| I/σ(Ι) | 20.9 (2.5) | 23.1 (6.7) | 16.8 (3.6) | 17.3 (3.4) |
| Completeness, % | 97.0 (87.5) | 99.0 (97.8) | 98.2 (82.5) | 99.3 (96.6) |
| Rmerge, %† | 10.3 (46.9) | 5.8 (27.0) | 12.2 (43.6) | 12.3 (40.9) |
| Refinement and structure model | ||||
| Reflections (Fo > 0σ(Fo)) | 15 645 | 17 597 | 17 506 | 15 335 |
| Working set | 14 864 | 16 707 | 16 615 | 14 552 |
| Free R set | 781 | 890 | 891 | 783 |
| R factor, %‡ | 20.1 (26.5) | 20.4 (29.1) | 20.6 (27.0) | 20.1 (23.6) |
| Free R factor, %‡ | 23.1 (36.9) | 22.6 (33.7) | 23.7 (28.0) | 23.2 (24.6) |
| Subunits/ASU | 1 | 1 | 1 | 1 |
| Average B factor, Å2 | 23.0 | 15.4 | 16.7 | 16.8 |
| Protein main-chain atoms, Å2 | 20.0 | 13.4 | 15.3 | 14.8 |
| Protein side-chain atoms, Å2 | 24.0 | 14.4 | 16.3 | 16.8 |
| RMSD bond lengths, Å | 0.006 | 0.006 | 0.006 | 0.006 |
| RMSD bond angles, degrees | 1.1 | 1.1 | 1.1 | 1.1 |
| Luzzati positional error, Å | 0.228 | 0.237 | 0.227 | 0.222 |
| Ramachandran plot, % | ||||
| Most favored regions, % | 91.1 | 93.0 | 93.0 | 92.5 |
| Allowed regions, % | 8.3 | 6.4 | 6.4 | 6.9 |
| Generously allowed regions, % | 0.6 | 0.6 | 0.6 | 0.6 |
| . | ssA2 . | ssA2-D1596A . | ssA2-N1602A . | ssA2-D1596A/N1602A . |
|---|---|---|---|---|
| PDB code | 3PPV | 3PPW | 3PPX | 3PPY |
| Diffraction data | ||||
| Space group | P65 | P65 | P65 | P65 |
| Cell parameters | ||||
| a = b/c, Å | 69.8/73.6 | 72.1/76.3 | 72.1/76.3 | 72.5/76.3 |
| Resolution range, Å | 50.0-1.9 | 50.0-1.9 | 50.0-1.9 | 50.0-2.0 |
| (1.97-1.90)* | (1.97-1.90) | (1.97-1.90) | (2.07-2.00) | |
| Observed reflections, no. | 132 324 | 72 418 | 95 704 | 101,783 |
| Unique reflections (I/σ(I) > 0), no. | 15 647 | 17 621 | 17 507 | 15 351 |
| Average redundancy | 8.5 (4.9) | 4.1 (3.1) | 5.5 (4.2) | 6.6 (4.4) |
| I/σ(Ι) | 20.9 (2.5) | 23.1 (6.7) | 16.8 (3.6) | 17.3 (3.4) |
| Completeness, % | 97.0 (87.5) | 99.0 (97.8) | 98.2 (82.5) | 99.3 (96.6) |
| Rmerge, %† | 10.3 (46.9) | 5.8 (27.0) | 12.2 (43.6) | 12.3 (40.9) |
| Refinement and structure model | ||||
| Reflections (Fo > 0σ(Fo)) | 15 645 | 17 597 | 17 506 | 15 335 |
| Working set | 14 864 | 16 707 | 16 615 | 14 552 |
| Free R set | 781 | 890 | 891 | 783 |
| R factor, %‡ | 20.1 (26.5) | 20.4 (29.1) | 20.6 (27.0) | 20.1 (23.6) |
| Free R factor, %‡ | 23.1 (36.9) | 22.6 (33.7) | 23.7 (28.0) | 23.2 (24.6) |
| Subunits/ASU | 1 | 1 | 1 | 1 |
| Average B factor, Å2 | 23.0 | 15.4 | 16.7 | 16.8 |
| Protein main-chain atoms, Å2 | 20.0 | 13.4 | 15.3 | 14.8 |
| Protein side-chain atoms, Å2 | 24.0 | 14.4 | 16.3 | 16.8 |
| RMSD bond lengths, Å | 0.006 | 0.006 | 0.006 | 0.006 |
| RMSD bond angles, degrees | 1.1 | 1.1 | 1.1 | 1.1 |
| Luzzati positional error, Å | 0.228 | 0.237 | 0.227 | 0.222 |
| Ramachandran plot, % | ||||
| Most favored regions, % | 91.1 | 93.0 | 93.0 | 92.5 |
| Allowed regions, % | 8.3 | 6.4 | 6.4 | 6.9 |
| Generously allowed regions, % | 0.6 | 0.6 | 0.6 | 0.6 |
RMSD indicates root mean square deviation; and ASU, asymmetric unit.
Numbers in parentheses refer to the highest-resolution shell.
Rmerge = ΣhklΣi∣Ii(hkl)i − 〈I(hkl)〉 ∣ / ΣhklΣiIi(hkl).
R factor = Σ ‖ Fo − Fc ‖ / Σ ∣ Fo ∣.
The ssA2 structure was determined using the molecular replacement method with PHASER (Version 2.1.4; Phaser Scientific Software LLC)24 using the VWF A3 structure (Research Collaboratory for Structural Bioinformatics [RCSB] Protein Data Bank [PDB] code 1AO3)25 as a search model. The initial structure refinement was carried out with CNS (Version 1.2)26 and the manual model building was performed with COOT (Version 8.4.1).27 In the initial difference Fourier maps, it was evident that a disulfide bond formed between Cys1493 and Cys1669, and strong electron density corresponding to a metal ion was detected near the α3-β4 loop (the nomenclature of the secondary structures of ssA2 adopts that of wtA2 by Zhang et al21 ), which was further identified as a Ca2+ ion (see “Results”). The final structure refinement was performed with REFMAC5.28 The structures of the ssA2 mutants were determined using the ssA2 structure as the starting model. The statistics of the structure refinement and the final structure models are listed in Table 1.
Force-probe MD simulations
Molecular modeling and in silico mutagenesis were performed with MOE (2009.10, Chemical Computing Group CCG). A wtA2 model (residues 1495-1671) was constructed based on the ssA2 structure by mutations of Cys1493 back to Asn and Ser1670 back to Cys and subsequent generation of a vicinal disulfide bridge between Cys1669 and Cys1670. The assigned Ca2+ ion was kept or replaced with a water molecule.
All MD simulations and analyses were carried out with the Gromacs suite (version 3.3.3)29 and the OPLS all-atom force field.30 For the equilibrium MD simulations, the protein was solvated in dodecahedral boxes with at least 5500 TIP4p water molecules31 under periodic boundary conditions. The typical protonation states at pH 7 were chosen for ionizable groups of the protein and the necessary amount of counter-ions (Cl− and Na+) were added to ensure a neutral system of physiologic ion strength. A temperature of 27°C and a pressure of 1 bar were assumed. The wtA2 models were simulated each for at least 20 nanoseconds and up to 40 nanoseconds.
For the force-probe MD simulations, the systems were solvated in 19-nm-long rectangular boxes each containing approximately 20 000 water molecules. Harmonic springs were attached to the terminal Cα atoms with spring constants of 500 kJ/(mol/nm2) and then moved away from each other with a velocity of 1.25 nm/ns so that each of the simulations spanned approximately 26 nanoseconds. To keep the system size (ie, the simulation box size) reasonable, the force-probe MD simulations were split into 2 parts. In the beginning, the whole wtA2 domain (residues 1495-1671) was simulated until unfolding of the C-terminal secondary structure elements α6, β6, and α5 was completed. Subsequently, the unfolded region (residues 1634-1671) was removed and the force-probe MD simulations were continued with the truncated fragment.
A2 cleavage assays
The in vitro cleavage assays of the wtA2 and GST-VWF73 proteins included preincubation of the proteins with urea at different concentrations of Ca2+ (step I), followed by administration of ADAMTS13 for the A2 cleavage (step II). In step I, 200 μg/mL of A2 protein in the assay buffer (50mM HEPES, pH 7.5, 150mM NaCl, 50μM ZnCl2, and 0.1% BSA) was incubated at 37°C for 2 hours with different concentrations of urea (1.92M, 2.56M, 3.20M, and 3.84M) and Ca2+ (0mM, 2.5mM, and 5.0mM). For the VWF73 substrates, urea was not added. In step II, for each reaction, 5 μL of the preincubated A2 protein in the assay buffer and 5 μL of 20 μg/mL ADAMTS13 (R&D Systems) in 50mM Tris-HCl (pH 8.5) were mixed and then incubated at 37°C for 1 hour with or without adjustment of the Ca2+ concentration. In some assays, step I was omitted, and 5 μL of the ssA2 protein in the assay buffer containing different concentrations of urea and Ca2+ were incubated with 5 μL of the diluted ADAMTS13 at 37°C for 3 hours. The cleaved C-terminal fragments of the A2 proteins and the intact A2 proteins were detected using Western blotting with the specific antibodies anti-A2C (R&D Systems) and anti-His (Tiangen Biotech), respectively. All experiments were performed at least 3 times.
Results
Crystal structure of VWF A2 domain
The wtA2 protein expressed with the E coli expression system was insoluble, and the refolded wtA2 did not show homogeneity and failed to produce any crystal. Thus, an engineered A2 domain (ssA2) was generated by introducing 2 mutations (N1493C and C1670S) to form a disulfide bond between the N- and C-termini analogous to that in the VWF A1 and A3 domains,20 which allowed further structural studies.
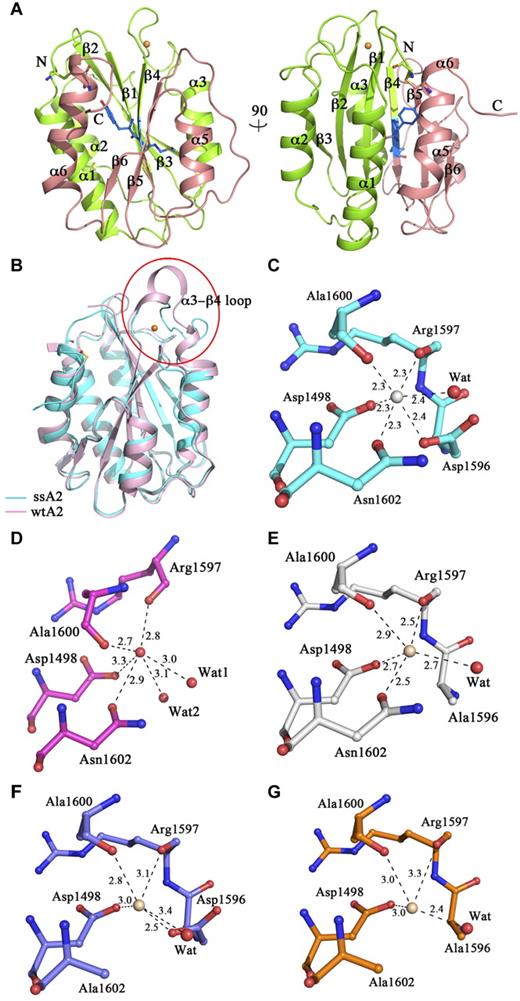
The crystals of ssA2 belong to space group P65 (Table 1), which is different from that of wtA2 (P21).21 The asymmetrical unit contains one ssA2 molecule, corresponding to a Matthews coefficient of 2.3 Å3/Da and a solvent content of 46%. The ssA2 structure was determined at a 1.9 Å resolution, and was refined to an R factor of 20.1% and a free R factor of 23.1% (Table 1). The ssA2 structure assumes a typical Rossmann fold and is very similar to the wtA2 structure with a root mean square deviation of 1.0 Å for all Cα atoms (Figure 1A-B). As observed in the wtA2 structure (PDB code 3GXB),21 Pro1645 takes a cis configuration.
Structure of ssA2. (A) Two views of the overall structure of ssA2. The A2 domain forms a compact structure, with the Tyr1605-Met1606 cleavage site being buried. Residues Tyr1605 and Met1606 are shown as ball-and-stick models and are colored in blue. The region of ssA2 N-terminal to the cleavage site is colored in green and that C-terminal to the site in salmon. The bound Ca2+ ion is shown as a golden sphere. The secondary structure elements of ssA2 are labeled. (B) Structural comparison of ssA2 (cyan) and wtA2 (pink; PDB code 3GXB). The major structural difference occurs in the α3-β4 region. The α3-β4 region is labeled and enclosed in a circle. (C) The Ca2+-binding site of ssA2. The surrounding residues are shown as ball-and-stick models. The coordination bonds between Ca2+ and the ligands are indicated with dashed lines and the bond lengths (Å) are marked. (D-G) The equivalent sites in wtA2 and the ssA2 mutants. The wtA2 protein (D) is colored in magenta, and those of the D1596A (E), N1602A (F), and D1596A/N1602A (G) mutants of ssA2 are colored in white, blue, and orange, respectively. In each of these structures, a molecule (wheat) is bound at this site, although in a geometry different from that of ssA2.
Structure of ssA2. (A) Two views of the overall structure of ssA2. The A2 domain forms a compact structure, with the Tyr1605-Met1606 cleavage site being buried. Residues Tyr1605 and Met1606 are shown as ball-and-stick models and are colored in blue. The region of ssA2 N-terminal to the cleavage site is colored in green and that C-terminal to the site in salmon. The bound Ca2+ ion is shown as a golden sphere. The secondary structure elements of ssA2 are labeled. (B) Structural comparison of ssA2 (cyan) and wtA2 (pink; PDB code 3GXB). The major structural difference occurs in the α3-β4 region. The α3-β4 region is labeled and enclosed in a circle. (C) The Ca2+-binding site of ssA2. The surrounding residues are shown as ball-and-stick models. The coordination bonds between Ca2+ and the ligands are indicated with dashed lines and the bond lengths (Å) are marked. (D-G) The equivalent sites in wtA2 and the ssA2 mutants. The wtA2 protein (D) is colored in magenta, and those of the D1596A (E), N1602A (F), and D1596A/N1602A (G) mutants of ssA2 are colored in white, blue, and orange, respectively. In each of these structures, a molecule (wheat) is bound at this site, although in a geometry different from that of ssA2.
A novel Ca2+-binding site
The only major conformational difference between the ssA2 and wtA2 structures occurs in the region (residues 1591-1602) connecting α3 and β4. This region forms a short 310 helix in wtA2 but a loop in ssA2 (Figure 1B). The disulfide bond between Cys1493 and Cys1669 does not cause any notable conformational change in the nearby regions and therefore is unrelated to the observed conformational change of the α3-β4 loop (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Intriguingly, a metal ion is bound at this loop with evident electron density (supplemental Figure 1B). It is coordinated in a octahedral geometry by 6 ligands: the main-chain carbonyls of Arg1597 (2.3 Å) and Ala1600 (2.3 Å), the side-chain carboxyl of Asp1498 (2.3 Å) and Asp1596 (2.4 Å), the side-chain carbonyl of Asn1602 (2.3 Å), and a water molecule (2.4 Å) (Figure 1C). The metal ion might be Ca2+ because of the typical Ca2+-binding geometry.32 At the equivalent site of wtA2, a molecule was also observed with 6 coordinating ligands (Figure 1D), which was assigned as a water molecule (see “Discussion”).
To identify the type of the metal ion, we performed inductively coupled plasma atomic emission spectroscopy analyses of the protein solution and the buffer. The results showed a higher content of Ca2+ ions (0.0042% vs 0.0011%) and Zn2+ ions (0.0013% vs 0.0002%) in the protein solution than in the buffer alone (Table 2), suggesting that the bound metal ion might have been Ca2+ or Zn2+. For further verification, we collected anomalous dispersion data at the peak wavelengths of Zn2+ (1.28 Å) and Ni2+ (1.49 Å) and the most commonly used wavelength for Ca2+ (1.77 Å), respectively. Evident anomalous signal was only detected at the metal-binding site at the wavelength of 1.77 Å (supplemental Figure 2A), indicating that the bound metal ion was Ca2+ rather than Zn2+ or Ni2+. Because no Ca2+ and Zn2+ ions were added in the crystallization solution, it is most likely that the bound Ca2+ ion was copurified with the protein from the expression system and that the trace of Zn2+ ions in the protein solution were nonspecifically bound to the C-terminal hexahistidine tag.
Inductively coupled plasma atomic emission spectroscopy results
| Metal . | Protein solution, % . | Buffer, % . |
|---|---|---|
| Ca | 0.0042 | 0.0011 |
| Zn | 0.0013 | 0.0002 |
| Cu | 0.0002 | 0.0002 |
| Metal . | Protein solution, % . | Buffer, % . |
|---|---|---|
| Ca | 0.0042 | 0.0011 |
| Zn | 0.0013 | 0.0002 |
| Cu | 0.0002 | 0.0002 |
To investigate whether mutations of residues at the metal-binding site would impede Ca2+ binding, the crystal structures of the D1596A, N1602A, and D1596A/N1602A mutants of ssA2 were determined. In all of these mutant structures, the region encompassing residues 1591-1602 still assumed a loop conformation similar to that in the intact ssA2, and there was evident electron density at the metal-binding site corresponding to a bound metal ion. However, because of the loss of ligand(s), the coordination geometries of the metal ion were distorted with a 5-coordination geometry in the D1596A and N1602A mutants and a 4-coordination geometry in the D1596A/N1602A mutant (Figure 1E-G). The coordination bond lengths were much longer than those in the ssA2 structure but similar to those in the wtA2 structure (Figure 1E-G and supplemental Table 1). In addition, we collected anomalous dispersion data at the wavelength of 1.77 Å for all of these mutants but no anomalous signal was observed, indicating that the bound ions in the mutants were unlikely to be Ca2+. These results suggest that mutations of Asp1596 and Asn1602 impair the ability of ssA2 to bind Ca2+.
Ca2+ binding impedes the unfolding of the A2 domain
To explore the functional role of Ca2+ binding, we conducted simulation studies to examine the effect of Ca2+ binding on the force required for the unfolding of the A2 domain. Based on the ssA2 structure, we constructed a wtA2 model by removing the engineered disulfide bond and then generating a vicinal disulfide bridge between Cys1669 and Cys1670, as observed in the wtA2 structure. The derived wtA2 model was stable in equilibrium MD simulations, the Ca2+-binding site remained intact, and the ion stayed in place, again indicating that the observed loop conformation of the α3-β4 region and Ca2+ binding were unrelated to the engineered disulfide bond. This model was then subjected to force-probe MD simulations with or without Ca2+ being bound.
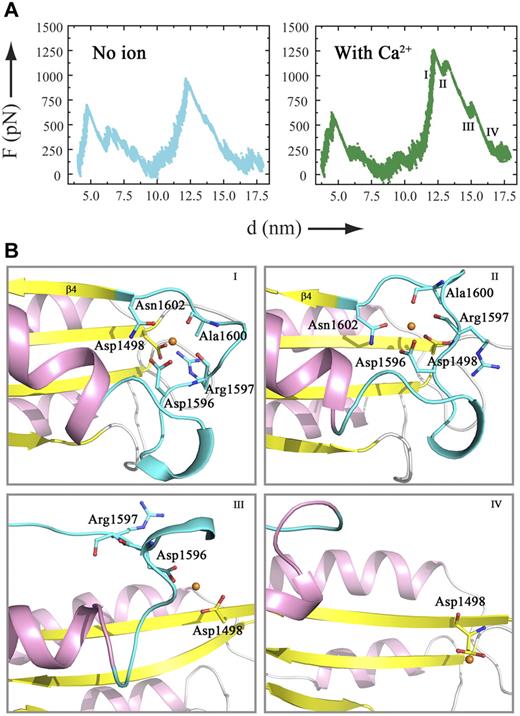
The force-probe MD simulation results of the wtA2 model were compared with those of the wtA2 structure.20 For the initial unfolding of the C-terminal part (α6, β6, and α5), the trajectories of the wtA2 model with and without Ca2+ were similar, resembling those of the wtA2 structure.20 For the unfolding of the central β-sheet (strands β5 and β4), although the force profiles were different for the wtA2 model with a higher force needed to unfold β4, in the absence of Ca2+, the force required for the unfolding of the central β-sheet in the simulations of the wtA2 model was almost the same (∼1000 pN; Figure 2A left panel) in this study as in the previous report.20 Intriguingly, when Ca2+ was bound, the force significantly increased to 1250 pN for the unfolding of strand β4 (Figure 2A right panel), indicating that the interplay between Ca2+ and the involved residues at the metal-binding site impedes the unfolding of the A2 domain, in particular the detachment of β4 from the remaining protein core. Therefore, binding of Ca2+ renders A2 more resistant against pulling forces such as those present in the shear flow of blood.
Force-probe MD simulations of a wtA2 model with or without a Ca2+ bound at the metal-binding site. (A) Force profiles of force-probe MD simulations of the wtA2 model. The α3-β4 region adopts a loop conformation with or without a Ca2+ ion bound at the site equivalent to the Ca2+-binding site of ssA2. The Ca2+ ion is replaced with a water molecule (left panel) or kept at the binding site (right panel). (B) Selected snapshots of the trajectory of force-probe MD simulations of the wtA2 model bound with a Ca2+ ion. For the A2 domain, the α-helices are colored in pink, the β strands in yellow, and the α3-β4 loop in cyan. The β4 strand is marked. The residues that participate in the formation of the Ca2+-binding site are shown as ball-and-stick models and labeled. The bound Ca2+ is shown as a golden sphere.
Force-probe MD simulations of a wtA2 model with or without a Ca2+ bound at the metal-binding site. (A) Force profiles of force-probe MD simulations of the wtA2 model. The α3-β4 region adopts a loop conformation with or without a Ca2+ ion bound at the site equivalent to the Ca2+-binding site of ssA2. The Ca2+ ion is replaced with a water molecule (left panel) or kept at the binding site (right panel). (B) Selected snapshots of the trajectory of force-probe MD simulations of the wtA2 model bound with a Ca2+ ion. For the A2 domain, the α-helices are colored in pink, the β strands in yellow, and the α3-β4 loop in cyan. The β4 strand is marked. The residues that participate in the formation of the Ca2+-binding site are shown as ball-and-stick models and labeled. The bound Ca2+ is shown as a golden sphere.
When Ca2+ was bound, the trajectories of the unfolding of the β4 strand were also different, with the force peak being split into 3 subpeaks representing breakage of interstrand interactions within the central β-sheet, the associated movement of the central β-sheet, and the stepwise disruption of the Ca2+-binding site. At the beginning, Asn1602 was pulled away from the Ca2+-binding site, followed by the movement of β4 (Figure 2B snapshots I and II). After the second subpeak, the Ca2+ ion was only coordinated by Asp1596 and Asp1498 (Figure 2B snapshot III). Finally, the Ca2+-binding site was demolished and β4 was completely unfolded (Figure 2B snapshot IV).
Ca2+ binding protects A2 from cleavage by ADAMTS13
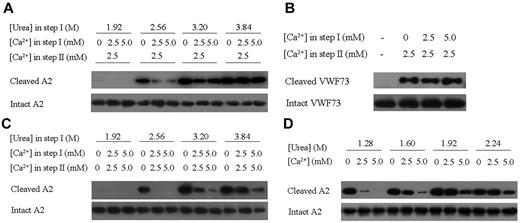
Because unfolding of the A2 domain is essential for the cleavage of VWF by ADAMTS13, we reasoned that the binding of Ca2+ would have an impact on VWF cleavage. This hypothesis was examined using in vitro biochemical assays that included pretreatment of the A2 substrate with urea to facilitate the unfolding process (step I), followed by the proteolysis of A2 by ADAMTS13 (step II) using the wtA2 proteins expressed in HEK293S GNT1− cells with glycosylation (see “Discussion”) but not the engineered ssA2 proteins as substrates. To investigate the effect of Ca2+ on the unfolding process only, various concentrations of Ca2+ (0mM, 2.5mM, and 5.0mM) were added at step I, whereas the Ca2+ concentration was adjusted to the same (2.5mM which is within the normal range of blood Ca2+ concentration) at step II. As shown in Figure 3A, in the presence of high concentration of urea, the wtA2 protein was cleaved by ADAMTS13; however, supplementation of Ca2+ at the concentration of either 2.5mM or 5.0mM during step I significantly suppressed the process, indicating that Ca2+ binding indeed impedes the unfolding of wtA2 and consequently its cleavage by ADAMTS13 (Figure 3A).
Effect of Ca2+ binding on A2 cleavage by ADAMTS13. (A) Examination of the protective effect of Ca2+ with 2-step ADAMTS13 cleavage assays. In step I, the wtA2 protein was denatured under different urea concentrations and Ca2+ was supplemented as indicated. In step II, the Ca2+ concentration was adjusted to 2.5mM. The cleaved and intact A2 proteins were detected with Western blotting using the specific antibodies. (B) Examination of the effect of Ca2+ on the proteolysis of the GST-VWF73 proteins by ADAMTS13. No urea was added in step I. (C) Examination of the effect of Ca2+ on the proteolysis of the wtA2 protein by ADAMTS13 without adjustment of the Ca2+ concentration in step II. (D) Examination of the effect of Ca2+ ion on the proteolysis of wtA2 by ADAMTS13 with 1-step assays.
Effect of Ca2+ binding on A2 cleavage by ADAMTS13. (A) Examination of the protective effect of Ca2+ with 2-step ADAMTS13 cleavage assays. In step I, the wtA2 protein was denatured under different urea concentrations and Ca2+ was supplemented as indicated. In step II, the Ca2+ concentration was adjusted to 2.5mM. The cleaved and intact A2 proteins were detected with Western blotting using the specific antibodies. (B) Examination of the effect of Ca2+ on the proteolysis of the GST-VWF73 proteins by ADAMTS13. No urea was added in step I. (C) Examination of the effect of Ca2+ on the proteolysis of the wtA2 protein by ADAMTS13 without adjustment of the Ca2+ concentration in step II. (D) Examination of the effect of Ca2+ ion on the proteolysis of wtA2 by ADAMTS13 with 1-step assays.
Conversely, if Ca2+ exerts a protective function via the Ca2+-binding site, then the disruption or lack of the Ca2+-binding site would abolish the protective role of Ca2+. VWF73 (corresponding to residues Asp1596-Arg1668 of the A2 domain) is the minimal substrate of ADAMTS13 that is presumably unfolded and susceptible to the cleavage by ADAMTS13 without shear stress or chaotropic agents.22 VWF73 supposedly does not have a Ca2+-binding site because of the absence of both Asp1498 and a defined structure. Consistently, pretreatment of VWF73 with Ca2+ had no effect on the cleavage of this fragment (Figure 3B), indicating the necessity of an intact Ca2+-binding site for the protection of VWF by Ca2+ from ADAMTS13 cleavage.
The metalloprotease ADAMTS13 requires Zn2+33 or Ca2+7,14 for its activity, and it has been reported that the activity of ADAMTS13 is enhanced approximately 160-fold at 5μM Ca2+34 . Therefore, Ca2+ may have 2 opposing effects on VWF cleavage: stabilization of the A2 domain and enhancement of the enzymatic activity of ADAMTS13. Given that the physiologic concentration of Ca2+ remained stable, this combined effect of Ca2+ was examined with the Ca2+ concentration kept unchanged during step I (corresponding to the unfolding process) and step II (corresponding to the proteolytic process). The results shown in Figure 3C show a similar trend as observed in Figure 3A, again demonstrating that the presence of Ca2+ leads to decreased susceptibility of the VWF A2 domain. Considering that the VWF unfolding and ADAMTS13 cleavage occur in the same environment in vivo, the A2 cleavage assay was further modified so that step I and step II were no longer separated, and the A2 unfolding and ADAMTS13 cleavage were carried out under the same conditions (Figure 3D). The results were very similar to those presented in Figure 3C, supporting a protective role of Ca2+ in VWF proteolysis.
Mutations at the Ca2+-binding site impair the protective effect of Ca2+ on A2 cleavage
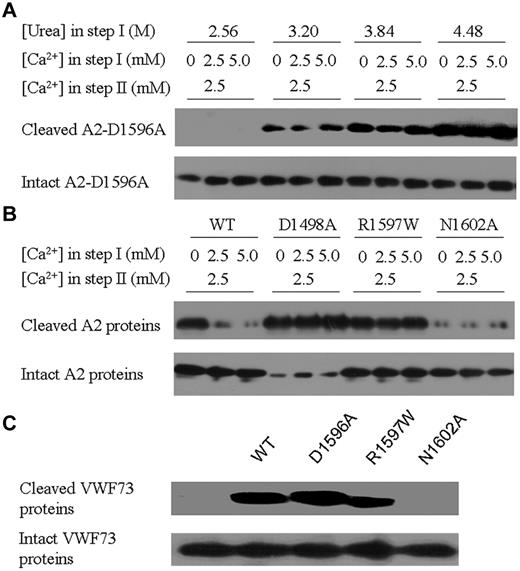
To further investigate the functional role of Ca2+ binding in the protection of the A2 domain, the wtA2 mutants D1596A, D1498A, R1597W, and N1602A, which carry point mutations of the key residues at the Ca2+-binding site, were analyzed with in vitro cleavage assays. At all 3 urea concentrations examined, the cleavage of the D1596A mutant appeared to be independent of the Ca2+ concentration, implying that the side-chain carboxyl is required for exertion of the protective function of Ca2+ binding (Figure 4A). This finding agrees with the critical role of Asp1596 shown by the structural investigations on ssA2 and its D1596A mutant.
Effect of mutations at the Ca2+-binding site on A2 cleavage by ADAMTS13. (A) Examination of the effect of the D1596A mutation on Ca2+ protection of A2. In step I, the D1596A mutant A2 was denatured under different urea concentrations and Ca2+ was supplemented as indicated. In step II, the Ca2+ concentration was adjusted to 2.5mM. The cleaved and intact proteins were detected with Western blotting using the specific antibodies. (B) Examination of the effect of the D1498A, R1597W, and N1602A mutations on Ca2+ protection of A2. In step I, 2.56M urea was added. (C) Examination of the effect of the D1498A, R1597W, and N1602A mutations on the proteolysis of GST-VWF73 by ADAMTS13. No urea was added in step I and the Ca2+ concentration was 2.5mM in both steps.
Effect of mutations at the Ca2+-binding site on A2 cleavage by ADAMTS13. (A) Examination of the effect of the D1596A mutation on Ca2+ protection of A2. In step I, the D1596A mutant A2 was denatured under different urea concentrations and Ca2+ was supplemented as indicated. In step II, the Ca2+ concentration was adjusted to 2.5mM. The cleaved and intact proteins were detected with Western blotting using the specific antibodies. (B) Examination of the effect of the D1498A, R1597W, and N1602A mutations on Ca2+ protection of A2. In step I, 2.56M urea was added. (C) Examination of the effect of the D1498A, R1597W, and N1602A mutations on the proteolysis of GST-VWF73 by ADAMTS13. No urea was added in step I and the Ca2+ concentration was 2.5mM in both steps.
The rest of the mutants were examined with 2-step ADAMTS13 cleavage assays with a fixed urea concentration (2.56M) in step I. The D1498A mutant showed a dramatically increased susceptibility to ADAMTS13, and the presence of a high Ca2+ concentration had no effect on the protection of the A2 unfolding in step I (Figure 4B). In addition to its participation in the formation of the Ca2+-binding site, the side-chain carboxyl group of Asp1498 forms a salt bridge with the side-chain amino group of Arg1597, which stabilizes the position of Arg1597. Therefore, mutation of Asp1498 to Ala might lead to instability of this region, which probably facilitates the unfolding of the A2 domain, partially explaining the substantially increased susceptibility of this mutant. The type 2A VWD-associated R1597W mutant showed similar levels of susceptibility to ADAMTS13 in the presence and absence of Ca2+ in step I (Figure 4B), indicating that Ca2+ protection of A2 cleavage is abolished by the R1597W mutation. Such an effect might have resulted from a change in the configuration of its main-chain carbonyl due to the introduction of a bulky side chain and the loss of the salt-bridging interaction with Asp1498 and the consequent loss or decrease of coordination with Ca2+. Intriguingly, the N1602A mutation led to resistance of A2 to the digestion by ADAMTS13 (Figure 4B).
The mutations D1596A, N1602A, and R1597W were also investigated for the ADAMTS13 minimal substrate VWF73. As expected, mutations D1596A and R1597W exhibited no effect on the susceptibility of VWF73 to ADAMTS13 proteolysis (Figure 4C). Mutation N1602A also led to resistance of the VWF73 fragment to the proteolysis (Figure 4C), although the protective effect of Ca2+ might have been lost due to the disruption of the Ca2+-binding site by the N1602A mutation. It has been shown that the range of the recognized cleavage site for ADAMTS13 encompasses residues Arg1597-Ile1623.35 Our results indicate a critical role of Asn1602 for cleavage of VWF by ADAMTS13, probably because of its proximity to the cleavage site of Tyr1605-Met1606. The detailed functional role(s) of Asn1602 needs to be investigated further.
Discussion
Using structural, computational, and biochemical data, we reveal a novel Ca2+-binding site at the A2 domain of VWF and show that Ca2+ binding at this site protects its unfolding and subsequent proteolysis by ADAMTS13. The VWF A2 domain shares structural similarities with other VWF A domains.25,36,37 Although binding of metal ions has been reported in VWF A domains,37 this is the first time that a metal-binding site has been observed in the A domains of VWF, which is different from the metal-binding site of other VWF A domains.
Comparison of the equivalent sites in the ssA2 and wtA2 structures showed that one major difference lies in the position and configuration of Asp1596 (Figure 1C-D). In the ssA2 structure, Asp1596 is located in the α3-β4 loop and points its side chain inside to coordinate the bound Ca2+ ion, whereas in the wtA2 structure, Asp1596 resides in the 310 helix and the side chain point away from the tentative Ca2+-binding site.38 Despite these differences, a molecule was also detected at the site in the wtA2 structure equivalent to the Ca2+-binding site of ssA2, forming 6 coordination bonds with the surrounding residues and water molecules in a geometry similar to that in our ssA2 structure. However, the bond lengths were much longer than those in the ssA2 structure (2.7-3.3 Å vs 2.3-2.4 Å). Intriguingly, in the D1596A mutant of ssA2, with loss of the coordination of Asp1596, the Ca2+-binding site was partially assembled as in the wtA2 structure, and similarly, a molecule was bound with longer bond lengths (2.5-2.9 Å). Because this molecule is not a Ca2+ ion based on the anomalous dispersion results, by analogy, the molecule bound at the same position of wtA2 was unlikely to be a Ca2+ ion, implying that the partially assembled site is insufficient for Ca2+ binding. Glycosylation of wtA2 occurs at Gln1515 and Gln1574 of the A2 domain of VWF, and mutation of Gln1574, which prevents its glycosylation, leads to increased susceptibility of VWF and VWF A2 to ADAMTS13 proteolysis.39 Indeed, in the wtA2 structure, N-linked glycans were observed at Gln1515 and Gly1574. The glycosylation sites and the Ca2+-binding site were located on opposite sides of the A2 domain, and therefore it is unlikely that glycosylation affected the assembly of the Ca2+-binding site (supplemental Figure 3).
To investigate whether the differences in the conformation of the α3-β4 region and Ca2+ binding between the wtA2 and ssA2 structures were due to the different crystallization conditions, we explored the possibilities of obtaining a wtA2 structure with an ssA2 structure without a Ca2+ ion being bound. Attempts to grow crystals of wtA2 under the crystallization conditions reported here and crystals of ssA2 under the conditions reported by Zhang et al21 were unsuccessful. Instead, we obtained crystals of wtA2 from both the cocrystallization and soaking experiments under the same crystallization conditions used by Zhang et al21 using supplementation of 10mM CaCl2 and crystals of the ssA2 protein treated with 10mM EDTA under the crystallization conditions reported here. No Ca2+ signal was detected in the anomalous dispersion diffraction data for these crystals (data not shown). In the derived wtA2 structures, the α3-β4 region assumed the same 310-helical conformation with a molecule bound at the metal-binding site in a way similar to that in the wtA2 structure (data not shown), indicating that the α-helical conformation of the α3-β4 region and the incapability of binding a Ca2+ are intrinsic properties of the wtA2 structure under the corresponding crystallization conditions. Conversely, in the structure of EDTA-treated ssA2 determined at a 2.0 Å resolution, the Ca2+-binding site was almost identical to that in the structure of ssA2 in complex with Ca2+ (supplemental Figure 2B), although no Ca2+ was bound, implicating that the formation of the binding site in ssA2 is not induced by Ca2+, but rather occurs naturally under the crystallization conditions.
These results indicate that the loop conformation of the α3-β4 region and the capability of binding a Ca2+ are intrinsic properties of the ssA2 structure under the crystallization conditions reported in this study, and that the observed differences between the wtA2 and ssA2 structures might result from the different crystallization conditions. Analysis of the crystal packing of the 2 crystal forms revealed that the α3-β4 region is exposed on the solvent-accessible surface in both structures and is engaged in the interactions with the symmetry-related molecules in the wtA2 (supplemental Figure 4A) but not the ssA2 structure. Moreover, superposition of the ssA2 structure onto the wtA2 structure showed that in the crystal packing of wtA2, the α3-β4 region of the ssA2 would clash with the symmetry-related molecule (supplemental Figure 4B). Therefore, the crystal packing in the wtA2 structure hinders the α3-β4 region from forming a loop structure and therefore prevents complete assembly of the Ca2+-binding site. In solution, it is plausible that the α3-β4 region of VWF A2 might have a high flexibility with the 310-helical conformation seen in the wtA2 structure and the loop conformation seen in the ssA2 structure as 2 quasi-stable conformations, which have been independently captured under different crystallization conditions.
It was reported recently that there is an intermediate unfolded state of A2 that shows unfolding of the C-terminal region up to the β4 strand.19 Of the 5 residues involved in the formation of the Ca2+-binding site identified in this study, 4 (Asp1596, Arg1597, Ala1600, and Asn1602) were located in the C-terminal region of the α3-β4 loop that immediately flanks the β4 strand (supplemental Figure 5). Based on the crystal structure of the catalytic domain of ADAMTS5,40 we constructed a model of ADAMTS13 showing that the active site of the enzyme is a narrow and deep groove (unpublished data), suggesting that the β4 strand needs to be displaced and inserted into the groove for ADAMTS13 cleavage. The displacement of the β4 strand would require a conformational change of the α3-β4 loop and very likely the disruption of the Ca2+-binding site as well, which implies that Ca2+ exerts its protective role through stabilization of the α3-β4 loop and thus restraint of the β4 strand.
PDB accession codes
The structures of ssA2 and its D1596A, N1602A, and D1596A/N1602A mutants have been deposited with the RCSB PDB under accession codes 3PPV, 3PPW, 3PPX, and 3PPY, respectively.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the staff members at the Shanghai Synchrotron Radiation Facility (Shanghai, China) for technical support in diffraction data collection and Professors Xiaohui Zhang (Department of Mechanical Engineering and Mechanics, Lehigh University), Jianfeng Chen (Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China), and Reinhard Schneppenheim (Department of Pediatric Hematology and Oncology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany) for inspiring discussions.
This work was supported by grants from the Ministry of Science and Technology of China (2007CB914302 and 2011CB966301), the National Natural Science Foundation of China (30730028), the Chinese Academy of Sciences (KSCX2-YW-R-107 and SIBS2008002), and the Klaus Tschira foundation. C.B. is grateful for a Feodor Lynen fellowship from the Alexander von Humboldt Foundation.
Authorship
Contribution: M.Z. and X.D. carried out the structural and biochemical studies and drafted the manuscript; C.B. and F.G. conceived the MD simulation studies; C.B. carried out the MD simulation studies and participated in the drafting of the manuscript; H.C. and X.L. participated in the biochemical studies; Y.Z. and T.A.S. provided the initial wtA2 domain sample and crystals; C.Z. participated in the data analysis and drafting of the manuscript; and J.D. conceived of the study, participated in the experimental design and coordination, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation of C.B. is Fritz-Haber-Institut der Max-Planck-Gesellschaft, Theory Department, Berlin-Dahlem, Germany.
Correspondence: Dr Jianping Ding, 320 Yue-Yang Rd, Shanghai 200031, China; e-mail: jpding@sibs.ac.cn.
References
Author notes
M.Z. and X.D. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal