Abstract
Fetal liver and adult bone marrow hematopoietic stem cells (HSCs) renew or differentiate into committed progenitors to generate all blood cells. PRDM16 is involved in human leukemic translocations and is expressed highly in some karyotypically normal acute myeloblastic leukemias. As many genes involved in leukemogenic fusions play a role in normal hematopoiesis, we analyzed the role of Prdm16 in the biology of HSCs using Prdm16-deficient mice. We show here that, within the hematopoietic system, Prdm16 is expressed very selectively in the earliest stem and progenitor compartments, and, consistent with this expression pattern, is critical for the establishment and maintenance of the HSC pool during development and after transplantation. Prdm16 deletion enhances apoptosis and cycling of HSCs. Expression analysis revealed that Prdm16 regulates a remarkable number of genes that, based on knockout models, both enhance and suppress HSC function, and affect quiescence, cell cycling, renewal, differentiation, and apoptosis to various extents. These data suggest that Prdm16 may be a critical node in a network that contains negative and positive feedback loops and integrates HSC renewal, quiescence, apoptosis, and differentiation.
Introduction
Hematopoietic stem cells (HSCs) can self-renew and differentiate into all cell types of the hematopoietic system and are regulated by interacting intrinsic and extrinsic mechanisms.1 Among intrinsic mechanisms, several transcriptional regulators involved as partners of leukemogenic fusion proteins, such as Mll2-4 and Evi1,5 are required for normal HSC function, whereas others, such as Runx16,7 and Scl,8,9 are essential for the establishment of HSCs during development. PR domain-containing 16 (PRDM16), a 140-kDa zinc finger protein, was originally discovered as a fusion partner in t(1:3)(p36;q21) translocations in acute myeloblastic leukemia (AML)10,11 and later in t(1;21)(p36;q22) translocations fused to RUNX1.12,13 In addition, elevated PRDM16 expression, because of promoter hypomethylation, is frequently observed in karyotypically normal AML.14 Deletion of the PR domain, which shows homology with a SET chromatin remodeling domain and is also present in EVI1,10 appears important for the leukemogenic properties of human PRDM16. Translocations involving PRDM16 invariably delete the PR domain,10-13 whereas PR-deleted Prdm16 causes AML in p53−/− mice.14 Furthermore, both Prdm16 and Evi1 are frequent targets of insertional mutagenesis in mice, causing deletion of the PR domain.15 Overexpression of Prdm16 expands HSCs in vitro. However, these expanded HSCs cause a myeloproliferative disease after transplantation.16 Prdm16 has also been shown to be critical for the development of brown adipose tissue in the mouse. PRDM16 is a transcriptional cofactor and interacts with the ligand-activated transcription factor peroxisome proliferator-activated receptor-γ and with CCAAT/enhancer-binding protein-β.17,18
Although its involvement in leukemic translocations and high expression in karyotypically normal AML suggest a physiologic role for Prdm16 in hematopoiesis, this role has not been established yet. Therefore, we analyzed the role of Prdm16 in hematopoiesis.
Methods
Mice
C57BL/6J mice (CD45.2+ B6) were purchased from The Jackson Laboratory and C57BL/6.SJL-PtprcaPep3b/BoyJ (CD45.1+ B6 mice) from the National Cancer Institute. Sperm from Prdm16Gt(OST67423)Lex mice (Lexicon Genetics)19 was reconstituted by in vitro fertilization in the Mouse Genetics Shared Resource of the Mount Sinai School of Medicine. Animals were housed in a specific pathogen-free facility. Experiments and animal care were performed in accordance with the Mount Sinai Institutional Animal Care and Use Committee.
Mouse genotyping
Genotyping for Prdm16 was done using a forward primer in gene-trap vector (5′-AAATGGCGTTACTTAAGCTAGCTTGC-3′) and in intron 1 (5′-AAATGGCGTTACTTAAGCTAGCTTGC-3′) and a reverse primer in exon 2 (5′-CCATCTGAGGTCGTCTGAAACTGG-3′), yielding a 231-bp band from a wt allele and a 122-bp band from a deleted allele.
Antibodies and cytokines
Fluorescein isothiocyanate-conjugated anti-CD2, anti-CD3ϵ, anti-CD8α, anti-CD4, anti-CD19, anti-B220, anti-Gr1, anti-Mac1, anti-CD48, phycoerythrin-conjugated anti-Flt3, PECy7-conjugated streptavidin, and allophycocyanin-AlexaFluor-750-conjugated anti-c-kit were purchased from eBioscience. Fluorescein isothiocyanate-conjugated anti-CD41, phycoerythrin-conjugated anti-Sca1, anti-CD34, peridinin chlorophyll protein-Cy5.5-conjugated anti-Mac1, streptavidin, allophycocyanin-conjugated anti-c-kit, anti-IgM, goat anti-rat antibody, PerCP-conjugated streptavidin, PECy7-conjugated anti-CD19, anti-hCD4, allophycocyanin-Cy7–conjugated streptavidin, anti-CD19, anti-CD8, and Pacific blue-conjugated anti-B220 were purchased from BD Biosciences PharMingen. Phycoerythrin-, allophycocyanin-, and PECy7-conjugated anti-CD150 and Pacific blue-conjugated anti-Sca1 were purchased from BioLegend. Lineage cocktail included CD2, CD3ϵ, CD8α, CD4, CD19, B220, Gr1, Mac1, and Ter119, as well as CD41 and CD48 when noted,
Cell sorting and flow cytometry
Bone marrow (BM) and fetal liver (FL) cells were isolated by cell sorting as described previously.20 Flow cytometric analysis was performed on a 5-laser LSRII with DiVa software (BD Biosciences) and analyzed using FlowJo software. For analysis of β-galactosidase activity in Prdm16+/− BM cells, 5 × 106Prdm16+/− or wt BM cells were resuspended in 100 μL of phosphate-buffered saline/2% fetal calf serum before loading with 100 μL of 2mM fluorescein di-D-β-galactopyranoside (Invitrogen) in dH2O and followed by incubation at 37°C for 60 seconds. Uptake was stopped by addition of 10 volumes of ice-cold phosphate-buffered saline/2% fetal calf serum, and the reaction was allowed to proceed for 1 to 3 hours on ice in the dark. 4,6-diamidino-2-phenylindole (1 μg/mL, Sigma-Aldrich) was added before analysis, for dead cell exclusion. For measurement of apoptosis, FL cells were stained with the HSC cocktail, washed and resuspended at 0.5 × 106 cells/200 μL in binding buffer, and stained with annexin V-phycoerythrin (BD Biosciences, PharMingen) as recommended by the manufacturer.
Colony assays
A total of 2.5 × 104 E15 to E18 FL cells/mL were plated in triplicate in 35-mm nontissue culture plates in methylcellulose M3434 medium supplemented with recombinant cytokines according to the manufacturer's instructions (StemCell Technologies). Myeloid and erythroid colony formation was scored after 7 days.
Quantitative RT-PCR
Total RNA, isolated using RNeasy Mini Kit (QIAGEN), was reverse-transcribed with SuperScript III (Invitrogen), according to the manufacturer's instructions, and cDNA was amplified with a specific primer and probe mix for Prdm16 (TaqMan Gene Expression Assay, Applied Biosystems), using 18S RNA as an internal control. Thermal cycling conditions were 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute on a StepOnePlus Real-Time PCR System. Analysis was done using the comparative threshold cycle (Ct) method (ΔΔCt).
Gene expression analysis
Five LSKCD150+ FL cells from E15 Prdm16−/− and wt littermate mice were sorted directly into the mixture of CellsDirect 2x Reaction Mix (CellsDirect One-Step qRT PCR Kit, Invitrogen), 0.2x TaqMan Assay Mix (Applied Biosystems), and SuperScript III RT/Platinum Taq Mix (Invitrogen). Reverse transcription and specific target amplification were serially performed as follows: 15 minutes at 50°C, 2 minutes at 95°C, 22 cycles of 15 seconds at 95°C, and 4 minutes at 60°C. Preamplified cDNA was diluted with TE buffer (1:5) and was used for the real-time polymerase chain reaction (PCR). Gene expression was analyzed using BioMark 96–96 Dynamic Array (Fluidigm) using inventoried TaqMan Gene Expression Assay (Applied Biosystems). The PCR profile was 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Data were analyzed using BioMark Real-Time PCR Analysis Software, Version 2.0 (Fluidigm), and the geometric mean of the ΔΔCt values for the different sorted samples was represented.
Transplantation assays
A total of 0.5 to 2 × 106 FL or BM cells (CD45.2+, B6.129 background, the donor) were mixed with an equal number of T cell-depleted wt C57BL/6.SJL-PtprcaPep3b/BoyJ (CD45.1+B6) or CD45.1+CD45.2+ C57BL/6.SJL-PtprcaPep3b/BoyJ.C57BL/6F1 (CD45.1+CD45.2+B6) competitor BM cells, and injected into lethally irradiated (500 cGy followed by 700 cGy 3 hours later) B6.129F1 hosts (CD45.2+). Wt controls were always littermates. After 12 to 16 weeks, peripheral blood (PB) and BM were analyzed. A total of 2 × 106 BM cells were then transplanted into lethally irradiated secondary B6.129F1 hosts. In transplantation experiments with purified cells, 300 BM LT-HSC (LSKCD150+) or 500 LSK FL or adult BM LSK cells were mixed with 0.2 to 0.5 × 106 T cell-depleted wt CD45.1+B6 or CD45.1+CD45.2+B6 BM cells. Reconstitution ratios were calculated by dividing the fraction of donor (CD45.2+) to competitor (CD45.1+ or CD45.1+CD45.2+) cells within the B (CD19+) and myeloid (Mac1+Gr1+) lineages. Reconstitution ratios are a direct representation of the relative potency of the tested HSC populations. T cells were not analyzed, as donor and recipient could not be distinguished, and as remaining host-derived hematopoietic cells are mostly T cells after lethal irradiation.20 To measure the effect of secondary transplantation, the logarithm of the reconstitution ratio was compared in primary and secondary hosts. An advantage of log transformation is that a ratio less than 1 will give a negative value, and negative ratios will extend over the same numerical range as positive ones (eg, a ratio of 0.01 gives a log ratio of −2, a ratio of 100 gives a log ratio of +2), thus normalizing the data.21 Accordingly, data are presented as the difference between input log donor/competitor (ie, the log ratio in the primary recipient) and the output log donor/competitor (ie, the log ratio 3 months after reconstitution of the secondary recipients) and are referred to as Δ(log ratio).
Homing assay
E18 FL cells were labeled with carboxyfluorescein diacetate succinimidyl diester (CFSE) according to manufacturer's instructions (Invitrogen). More than 99% of cells were stained, and fluorescence intensity ranged between 104 and 105. CFSE-stained cells from Prdm16−/−, Prdm16+/−, or wt mice were intravenously injected into B6.129F1 lethally irradiated mice (12 Gy, one dose). After 3 hours, the BM was harvested and homing efficiency was calculated as previously described.22
Statistical analysis
For statistical analysis, paired or unpaired Student t test were used. When more than 2 groups were compared, one-way ANOVA was used. Results are expressed as mean ± SEM. Bonferroni correction was applied to determine statistically significant differences in Fluidigm expression analysis.
Results
Selective expression of Prdm16 in the earliest stem and progenitor cells
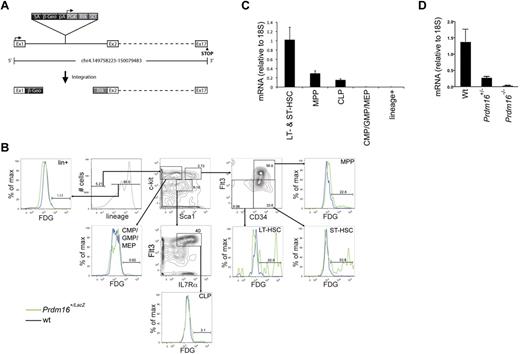
We examined the expression pattern of Prdm16 in the BM of Prdm16-deficient mice generated in a gene-trap screen using a LacZ-expressing vector. In these mice, LacZ insertion into the first intron of Prdm16 leads to β-galactosidase expression under the control of endogenous Prdm16 regulatory elements but termination of PRDM16 translation after the first exon (Figure 1A).19 Flow cytometric LacZ staining using fluorescein di-D-β-galactopyranoside in adult heterozygous mice revealed exclusive expression in lineage−Sca1+kit+ (LSK) hematopoietic stem and progenitor cells (HSPCs). Within the LSK population, expression was highest in short-term (ST, LSKCD34+Flt3−)23 and long-term (LT, LSKCD34−Flt3−)23 HSCs, and was lower in multipotential progenitors (MPPs, LSKCD34+Flt3+).23 No LacZ was detected in common lymphoid progenitors (CLPs, lin−Sca1lokit−IL7Rα+Flt3+)23 and lin−Sca1−kit+ (LS−K)23 cells, which contain common myeloid, granulocyte/macrophage, and megakaryocyte/erythroid progenitors (Figure 1B). Similarly, quantitative PCR showed predominant expression of Prdm16 mRNA in HSCs, although some expression of Prdm16 mRNA was also observed in CLPs (Figure 1C). Next, we verified that Prdm16 mRNA was undetectable in Prdm16−/− embryos. Although Prdm16 mRNA was nearly undetectable in brains of Prdm16−/− embryos, expression was reduced to 20.2% ± 0.06% of that in wt controls in Prdm16+/− embryos, significantly different from the expected 50% (P = .02, Figure 1D) and indicating that Prdm16 expression is haploinsufficient.
Expression of Prdm16 in adult BM. (A) Schematic representation of the genomic Prdm16 region in Prdm16Gt(OST67423)Lex (Prdm16−/−) mice. Adapted from Zambrowicz et al19 with permission. (B) Expression of LacZ, measured by flow cytometry after staining with fluorescein di-D-β-galactopyranoside, in the BM of Prdm16Gt(OST67423)Lex/+ mice. CMP indicates common myeloid progenitor; GMP, granulocyte/macrophage progenitor; MEP, megakaryocyte erythroid progenitor; LT-HSC, long-term hematopoietic stem cell; and ST-HSC, short-term hematopoietic stem cell. (C) Expression of Prdm16 mRNA in HSCs (LSKFlt3−, composed of ST- and LT-HSCs), MPPs, CLPs, and lineage+ cells (n = 3). (D) Expression of Prdm16 mRNA in brain from Prdm16−/−, Prdm16+/−, and wt embryos.
Expression of Prdm16 in adult BM. (A) Schematic representation of the genomic Prdm16 region in Prdm16Gt(OST67423)Lex (Prdm16−/−) mice. Adapted from Zambrowicz et al19 with permission. (B) Expression of LacZ, measured by flow cytometry after staining with fluorescein di-D-β-galactopyranoside, in the BM of Prdm16Gt(OST67423)Lex/+ mice. CMP indicates common myeloid progenitor; GMP, granulocyte/macrophage progenitor; MEP, megakaryocyte erythroid progenitor; LT-HSC, long-term hematopoietic stem cell; and ST-HSC, short-term hematopoietic stem cell. (C) Expression of Prdm16 mRNA in HSCs (LSKFlt3−, composed of ST- and LT-HSCs), MPPs, CLPs, and lineage+ cells (n = 3). (D) Expression of Prdm16 mRNA in brain from Prdm16−/−, Prdm16+/−, and wt embryos.
We conclude that Prdm16 mRNA is expressed specifically in the most immature HSPCs and that Prdm16+/− mice are haploinsufficient with respect to Prdm16 mRNA expression.
Hematopoietic profile of Prdm16–deficient mice
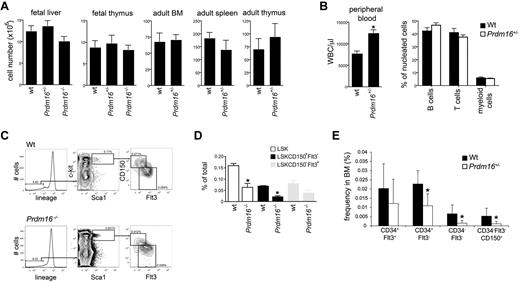
When Prdm16+/− mice were mated, the distribution of genotypes among embryos at all stages of development followed Mendelian ratios (wt: 22; Prdm16+/−: 47; Prdm16−/−: 22). However, no Prdm16−/− live-born pups were ever observed, indicating that Prdm16−/− mice die during or very shortly after birth. Cellularity of FL and thymus of Prdm16−/− and Prdm16+/− embryos at E16 to E18 was similar to that of wt embryos (Figure 2A). In adult mice, cellularity of BM, spleen, and thymus was also similar in wt and Prdm16+/− mice (Figure 2A). However, total PB white blood cell counts in adult Prdm16+/− mice were higher than in wt mice (Figure 2B). As the distribution of B, T, and myeloid cells was similar, this increase involved all these lineages (Figure 2B). The fraction of developing B cells (AA4.1+B220+CD19− pre-pro-B cells, AA4.1+B220+CD19+ pre- and pro-B cells, AA4.1+IgM+ immature B cells)24 as well as the fraction of developing myeloid (Mac1+Gr1+) and erythroid (Ter119+) cells were similar in Prdm16−/−, Prdm16+/−, and wt FL. T-cell development in the thymus (as measured by the fraction of lin−CD25−c-kit+ early thymic precursors, CD4−CD8− double-negative, CD4+CD8+ double-positive, and CD4 and CD8 single-positive cells)25,26 of Prdm16−/−, Prdm16+/−, and wt embryos did not differ (not shown). Similarly, in adult Prdm16+/− mice, B, T, myeloid, and erythroid development was similar to that in wt mice (not shown).
Hematologic profile of Prdm16-deficient mice. (A) Cellularity of FL and thymus, and of adult BM, thymus, and spleen in Prdm16-deficient mice (n = 3–5). (B) WBC count (left panel) and lineage distribution (measured by flow cytometric analysis of CD19, Thy1, and Mac1/Gr1) in the PB of adult Prdm16+/− and wt mice (n = 5). *P = .0006. (C) Representative example of flow cytometric analysis of the fraction of LSK HSPCs, LSK CD150−Flt3+ MPPs, and LSK CD150+Flt3− HSCs in E15 Prdm16−/− and wt FL cells. (D) Frequency of the populations in panel C in E13.5 to E15.5 FL from Prdm16−/− and wt embryos. Frequencies obtained as the percentage of cells in doublet discriminated scatter (n = 4-6). *P < .05, one-way analysis of variance. (E) Frequencies of subpopulations of LSK cells in the BM of Prdm16+/− mice or wt littermates. Lineage cocktail included anti–CD41 and CD48 (n = 8). *P < .03.
Hematologic profile of Prdm16-deficient mice. (A) Cellularity of FL and thymus, and of adult BM, thymus, and spleen in Prdm16-deficient mice (n = 3–5). (B) WBC count (left panel) and lineage distribution (measured by flow cytometric analysis of CD19, Thy1, and Mac1/Gr1) in the PB of adult Prdm16+/− and wt mice (n = 5). *P = .0006. (C) Representative example of flow cytometric analysis of the fraction of LSK HSPCs, LSK CD150−Flt3+ MPPs, and LSK CD150+Flt3− HSCs in E15 Prdm16−/− and wt FL cells. (D) Frequency of the populations in panel C in E13.5 to E15.5 FL from Prdm16−/− and wt embryos. Frequencies obtained as the percentage of cells in doublet discriminated scatter (n = 4-6). *P < .05, one-way analysis of variance. (E) Frequencies of subpopulations of LSK cells in the BM of Prdm16+/− mice or wt littermates. Lineage cocktail included anti–CD41 and CD48 (n = 8). *P < .03.
Next, we phenotypically analyzed the stem and progenitor cell compartment of Prdm16-deficient mice. In Prdm16−/− E13.5 to 15.5 embryos, the frequency of LSK HSPCs, LSKCD150−Flt3+ MPPs, and in particular LSKCD150+Flt3− HSCs27 (FL HSCs, in contrast to adult HSCs, express CD34),28,29 was decreased compared with wt littermates (Figure 2C-D). In Prdm16+/− FL, only the LSKCD150+Flt3− fraction was decreased significantly (not shown). Similarly, adult Prdm16+/− mice showed reduced frequency of LT- and ST-HSCs, whereas MPPs were less profoundly affected (Figure 2E). As BM and FL cellularity were similar in wt and Prdm16-deficient mice (Figure 2A), these differences in HSPC frequencies are reflective of differences in absolute number.
These phenotypic data indicate that, consistent with predominant expression of Prdm16 in the most primitive HSPC compartments, deletion of Prdm16 decreases their frequency in BM and FL. Prdm16 is furthermore haploinsufficient in this respect.
Prdm16 deficiency causes a defect in HSCs
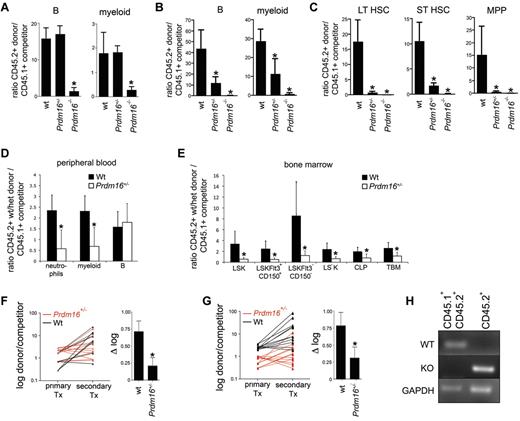
To assess progenitor function, we performed colony assays. Myeloid and erythroid colony formation was mildly affected compared with wt in Prdm16+/− (82% ± 3.5%, P < .0001) and Prdm16−/− (77% ± 3.5%, P = .0003) FL cells and adult BM cells (52% ± 4.1%, P < .0001) (n = 4 triplicate experiments for each genotype, not shown), indicating a subtle defect at the level of progenitor cells consistent with a limited decrease in phenotypically defined progenitors, such as MPPs. To assess HSC function, we performed competitive repopulation studies. We injected 0.5 × 106 wt littermate, Prdm16+/−, or Prdm16−/− FL cells (CD45.2+, B6.129 background) together with an equal number of T cell-depleted wt CD45.1+ or CD45.1+CD45.2+ B6 BM cells into lethally irradiated B6.129F1 (CD45.2+) hosts. T-cell depletion of competitors was performed to avoid both graft-versus-host disease and rejection of donor cells by competitor cells. T-cell reconstitution was not analyzed, as donor and recipient could not be distinguished, and as remaining host-derived hematopoietic cells are mostly T cells after lethal irradiation.20 Although immunophenotypically defined HSCs were decreased 3-fold in Prdm16−/− FL, the contribution from Prdm16−/− cells to the myeloid and B lineages was 10-fold lower compared with that of wt FL and Prdm16+/− cells after 12 weeks (Figure 3A), and was undetectable after 5 months (Figure 3B). The contribution of Prdm16+/− FL cells was similar to that of wt cells after 12 weeks but was also severely reduced after 5 months (Figure 3A-B). Furthermore, in the BM, contribution of Prdm16+/− and in particular of Prdm16−/− FL cells to the LT-HSC, ST-HSC, and MPP compartments was extremely low compared with wt cells 5 months after transplantation, indicative of a severe defect in self-renewal capacity (Figure 3C). In competitive repopulation studies using adult BM, Prdm16+/− cells showed an initial repopulation defect in the myeloid lineage at 12 weeks (Figure 3D), probably a reflection of the fact that, because of their faster turnover, myeloid cells are a more sensitive indicator of HSC function. In the recipient BM, Prdm16+/− HSC contribution to all BM HSPC subfractions was decreased, however (Figure 3E). After transfer into secondary recipients, Prdm16+/− BM cells lost repopulation capacity compared with wt cells in the B lineage (P = .02, Figure 3F), whereas the loss in the myeloid lineage was near statistical significance (P = .07, Figure 3G).
Function of Prdm16–deficient FL and BM cells. (A) Ratio between donor (Prdm16−/−, Prdm16+/−, or wt FL cells) and competitor (T cell–depleted CD45.1+ or CD45.1+CD45.2+ B6 BM cells) cells among PB myeloid and B cells 12 weeks after transplantation of 0.5 × 106 cells of each population into lethally irradiated B6.129F1 (CD45.2+) mice (n = 3 for wt, n = 12 for Prdm16+/−, n = 14 for Prdm16−/−). *P < .01, one-way analysis of variance. (B-C) Ratio between donor (Prdm16−/−, Prdm16+/−, or wt FL cells) and competitor (T cell–depleted CD45.1+ B6 BM cells) cells among PB myeloid and B cells (B) and among BM LT-HSCs, ST-HSCs, and MPPs (C) 20 weeks after transplantation as in panel A. (D-E) Competitive repopulation of 2 × 106 wt or Prdm16+/− BM cells (CD45.2+) with 2 × 106 C57BL/6 (CD45.1+) BM cells in B6.129F1 recipients after 12 to 16 weeks. Data presented as donor/competitor ratio within a specific compartment in PB (D) and BM (E) (n = 6). *P < .02. TBM indicates total BM. (F-G) Shift in donor/competitor ratio within B cells (F) and myeloid cells (G) after serial transplantation of 2 × 106 BM cells from primary recipients into lethally irradiated secondary B6.129F1 CD45.2+ recipients 12 to 14 weeks after primary transplantation. The right hand panels of panels F and G show the difference in log(donor/competitor) between primary and secondary recipients (n = 12 secondary recipients for each genotype). *P = .07 for myeloid cells. *P = .01 for B cells. Tx indicates transplantation. (H) Genomic PCR for the wt and mutant alleles in the myeloid and B cells of the spleens of recipients of CD45.2+Prdm16−/− cells and CD45.1+CD45.2+ B6 competitor cells.
Function of Prdm16–deficient FL and BM cells. (A) Ratio between donor (Prdm16−/−, Prdm16+/−, or wt FL cells) and competitor (T cell–depleted CD45.1+ or CD45.1+CD45.2+ B6 BM cells) cells among PB myeloid and B cells 12 weeks after transplantation of 0.5 × 106 cells of each population into lethally irradiated B6.129F1 (CD45.2+) mice (n = 3 for wt, n = 12 for Prdm16+/−, n = 14 for Prdm16−/−). *P < .01, one-way analysis of variance. (B-C) Ratio between donor (Prdm16−/−, Prdm16+/−, or wt FL cells) and competitor (T cell–depleted CD45.1+ B6 BM cells) cells among PB myeloid and B cells (B) and among BM LT-HSCs, ST-HSCs, and MPPs (C) 20 weeks after transplantation as in panel A. (D-E) Competitive repopulation of 2 × 106 wt or Prdm16+/− BM cells (CD45.2+) with 2 × 106 C57BL/6 (CD45.1+) BM cells in B6.129F1 recipients after 12 to 16 weeks. Data presented as donor/competitor ratio within a specific compartment in PB (D) and BM (E) (n = 6). *P < .02. TBM indicates total BM. (F-G) Shift in donor/competitor ratio within B cells (F) and myeloid cells (G) after serial transplantation of 2 × 106 BM cells from primary recipients into lethally irradiated secondary B6.129F1 CD45.2+ recipients 12 to 14 weeks after primary transplantation. The right hand panels of panels F and G show the difference in log(donor/competitor) between primary and secondary recipients (n = 12 secondary recipients for each genotype). *P = .07 for myeloid cells. *P = .01 for B cells. Tx indicates transplantation. (H) Genomic PCR for the wt and mutant alleles in the myeloid and B cells of the spleens of recipients of CD45.2+Prdm16−/− cells and CD45.1+CD45.2+ B6 competitor cells.
To assure that the CD45.2+ B and myeloid cells in the reconstituted recipients were from donor and not of host origin, we isolated CD45.1+CD45.2+ and CD45.2+ cells that expressed either Mac1 (myeloid cells) or CD19 (B cells) from spleen of recipients of Prdm16−/− FL cells, extracted the genomic DNA, and amplified using primers specific for the wt or mutant Prdm16 allele. In the CD45.1+CD45.2+ competitor cells, only the wt allele was detected, whereas in the CD45.2+ donor population, only the mutant allele was observed (Figure 3H). These data confirm that the host did not contribute to the CD45.2+ B and myeloid populations.
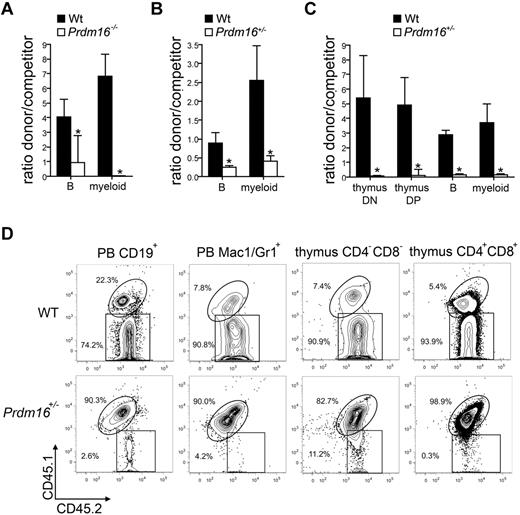
To confirm that the defect in competitive repopulation capacity was intrinsic to the HSCs, 300 FL LSK cells (Figure 4A), 500 adult BM LSK cells (Figure 4B), or 300 adult BM LT-HSCs (LSKCD34−Flt3−, Figure 4C) were transplanted together with 2 × 105 T cell–depleted CD45.1+ or CD45.1+CD45.2+ B6 competitor cells into lethally irradiated B6.129F1 (CD45.2+) mice. A representative example of the flow cytometric analysis of recipients of adult Prdm16+/− and wt LT-HSCs is shown in Figure 4B. For all cell populations tested, a defect in competitive repopulation of Prdm16-deficient cells was observed 12 to 16 weeks after transplantation. In recipients of adult BM LT-HSCs, we also analyzed contribution to developing T cells in the thymus (CD4−CD8− double-negative and CD4+CD8+ double-positive cells), as in contrast to mature T cells, which are long-lived and relatively radioresistant, these cell populations turn over rapidly and can therefore not be derived from the host. The T-cell potential of Prdm16+/− cells was decreased to the same extent as their B and myeloid potential (Figure 4C-D).
Function of Prdm16-deficient HSPCs. (A-C) Ratio between donor (300 Prdm16−/− or wt FL LSK cells (n = 4) (A), 500 Prdm16+/− or wt adult BM LSK cells (n = 9) (B), or 300 Prdm6+/− or wt adult BM LSKCD34−Flt3− cells (n = 4) (C) and competitor (0.5 × 106 T cell-depleted CD45.1+ or CD45.1+CD45.2+ B6 BM cells) cells among PB myeloid and B cells, and (C) thymic CD4+CD8+ double-positive and CD4−CD8− double-negative developing T cells, 10 to 15 weeks after transplantation into lethally irradiated B6.129F1 (CD45.2+) mice. *P < .01. (D) Representative example of donor (CD45.2+) and competitor (CD45.1+CD45.2+) reconstitution in doublet discriminated PB CD19+ B and Mac1+Gr1+ myeloid cells, and in thymic CD4−CD8− double-negative and CD4+CD8+ double-positive cells after competitive transplantation with wt or Prdm16+/− adult BM LT-HSCs.
Function of Prdm16-deficient HSPCs. (A-C) Ratio between donor (300 Prdm16−/− or wt FL LSK cells (n = 4) (A), 500 Prdm16+/− or wt adult BM LSK cells (n = 9) (B), or 300 Prdm6+/− or wt adult BM LSKCD34−Flt3− cells (n = 4) (C) and competitor (0.5 × 106 T cell-depleted CD45.1+ or CD45.1+CD45.2+ B6 BM cells) cells among PB myeloid and B cells, and (C) thymic CD4+CD8+ double-positive and CD4−CD8− double-negative developing T cells, 10 to 15 weeks after transplantation into lethally irradiated B6.129F1 (CD45.2+) mice. *P < .01. (D) Representative example of donor (CD45.2+) and competitor (CD45.1+CD45.2+) reconstitution in doublet discriminated PB CD19+ B and Mac1+Gr1+ myeloid cells, and in thymic CD4−CD8− double-negative and CD4+CD8+ double-positive cells after competitive transplantation with wt or Prdm16+/− adult BM LT-HSCs.
We conclude that Prdm16 is critical for HSC function, maintenance, and renewal and that this effect is intrinsic to HSCs and not dependent on the microenvironment.
Enhanced apoptosis in Prdm16−/− HSCs
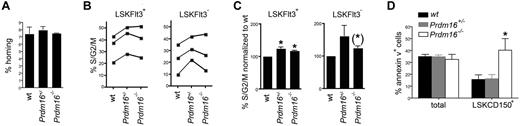
Next, we attempted to define a mechanism underlying the loss of HSCs in Prdm16-deficient mice. Homing of Prdm16−/−, Prdm16+/−, and wt FL cells was measured by injecting CFSE-labeled cells in lethally irradiated hosts, and determining the absolute number of CFSE-labeled cells that homed to the BM 3 hours after injection.22 No difference in homing was observed (Figure 5A). These data argue against a homing defect in Prdm16-deficient HSPCs. Next, we analyzed cell cycle activity in FL HSPCs by staining using 4,6-diamidino-2-phenylindole. As cycling activity of HSPCs decreased with advancing fetal development (Figure 5B), data were normalized to those obtained from wt littermates (Figure 5C). Although a limited increase in cycling activity was observed in both Prdm16+/− and Prdm16−/− LSKFlt3+ MPPs, this increase was just short of statistical significance in LSKFlt3− HSCs. Next, we examined apoptosis by staining fetal liver cells for annexin V. Compared with wt HSCs, the annexin V+ fraction was increased in Prdm16−/− HSCs, but not in Prdm16+/− HSCs (Figure 5D). No difference in apoptosis was observed in MPPs (not shown) or in total FL cells (Figure 5D).
Effects of Prdm16 on homing, apoptosis, and cell cycling. (A) Homing of Prdm16−/−, Prdm16+/−, and wt E16 FL cells after transplantation into lethally irradiated CD45.1+ recipients (n = 3). (B) Percentage of FL LSKFlt3+ and LSKFlt3− cells from Prdm16−/−, Prdm16+/−, and wt mice in S/G2/M phase of the cell cycle as measured by 4,6-diamidino-2-phenylindole staining. Each experiment contained 2 to 4 embryos per genotype. (C) Fraction of FL LSKFlt3+ and LSKFlt3− cells from Prdm16−/−, Prdm16+/−, and wt mice in S/G2/M phase normalized to the values of wt littermate embryos (n = 3 litters). *P < .05. (*)P = .08. (D) Percentage of apoptotic total FL and LSKCD150+ FL cells in Prdm16−/−, Prdm16+/−, and wt embryos (n = 3 for each genotype). *P = .05, one-way ANOVA.
Effects of Prdm16 on homing, apoptosis, and cell cycling. (A) Homing of Prdm16−/−, Prdm16+/−, and wt E16 FL cells after transplantation into lethally irradiated CD45.1+ recipients (n = 3). (B) Percentage of FL LSKFlt3+ and LSKFlt3− cells from Prdm16−/−, Prdm16+/−, and wt mice in S/G2/M phase of the cell cycle as measured by 4,6-diamidino-2-phenylindole staining. Each experiment contained 2 to 4 embryos per genotype. (C) Fraction of FL LSKFlt3+ and LSKFlt3− cells from Prdm16−/−, Prdm16+/−, and wt mice in S/G2/M phase normalized to the values of wt littermate embryos (n = 3 litters). *P < .05. (*)P = .08. (D) Percentage of apoptotic total FL and LSKCD150+ FL cells in Prdm16−/−, Prdm16+/−, and wt embryos (n = 3 for each genotype). *P = .05, one-way ANOVA.
Thus, whereas cycling activity was mildly increased in the HSPC compartment, apoptosis was specifically increased in Prdm16−/− HSCs.
Expression analysis of Prdm16−/− and wt HSCs
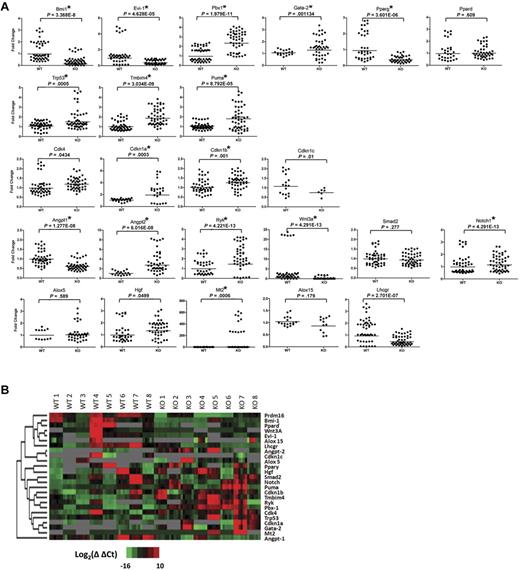
We used a microfluidic quantitative PCR array (Fluidigm)30 to compare expression of a select set of genes in purified wt and Prdm16−/− FL HSCs. Five LSKCD150+ FL cells were sorted per well. Eight triplicate groups of 5 Prdm16−/− and wt littermate HSCs were analyzed for the expression of 29 genes (Figure 6). Genes known to be involved in HSC function, renewal, and maintenance (Bmi1, Pbx1, Evi1, and Gata2),5,31-33 apoptosis (Trp53, Puma, and Tmbim4),34-38 cell cycle regulation (Cdk4, Cdkn1a, Cdk1nb, and Ckdn1c),39,40 as well as cytokines and genes involved in cytokine signaling (Notch1, Angpt1, Angpt2, Ryk, Wnt3a, and Smad2)41-45 were selected. Prdm16−/− neural stem cells (NSCs) have been reported to display increased oxidative stress.46 Therefore, several genes previously reported to be decreased in Prdm16−/− ventricular zone NSCs (Hgf, Lhcgr, Mt2, Pdk4, Crym, Ogn, Pparss2, and Hmgcs2)46 as well as 2 genes involved in oxidative stress and reported to be up-regulated in Bmi1−/− BM cells (Alox5 and Alox15)47 were examined. Finally, as Prdm16 interacts with the ligand-activated transcription factor peroxisome proliferator-activated receptor-γ and increases its ligand-induced transcriptional activity18 in brown fat adipogenesis, we also measured expression of Pparg and its homolog, Ppard. Results are summarized in Figure 6.
Expression analysis of Prdm16−/− and wt FL HSCs. (A) Scatter dot plots of the expression of the genes, as measured using Fluidigm multiplex quantitative PCR, indicated on top of each panel in 8 groups of 5 Prdm16−/− or wt FL LSKCD150+ HSCs. *P < .002, threshold required after Bonferroni correction. All experimental replicates of the amplification step and the microfluidic quantitative PCR chip are shown. Undetectable expression is defined by a Ct value of 35 or greater. (B) Heat map of the expression levels for Prdm16 and 24 genes shown in panel A.
Expression analysis of Prdm16−/− and wt FL HSCs. (A) Scatter dot plots of the expression of the genes, as measured using Fluidigm multiplex quantitative PCR, indicated on top of each panel in 8 groups of 5 Prdm16−/− or wt FL LSKCD150+ HSCs. *P < .002, threshold required after Bonferroni correction. All experimental replicates of the amplification step and the microfluidic quantitative PCR chip are shown. Undetectable expression is defined by a Ct value of 35 or greater. (B) Heat map of the expression levels for Prdm16 and 24 genes shown in panel A.
No detectable signal was obtained for Pdk4, Crym, Ogn, Pparss2, and Hmgcs2 (not shown). Other oxidative stress-responsive genes were expressed at similar levels in wt and Prdm16−/− HSCs, except for Mt2, expression of which was increased in Prdm16−/− HSCs and Lhcgr, which was lower (Figure 6). These data indicate that the mechanisms underlying the defects in HSC maintenance in Prdm16−/− mice probably differ from those involved in the reported defects in NSC function.46
The expression of the negative cell cycle regulators Cdkn1a and Cdk1nb was mildly higher in Prdm16−/− than in wt HSCs. Expression of Trp53 (p53) and one of its prime transcriptional targets, Puma,34 as well as that of the potential Bax inhibitor Tmbim4 was higher in Prdm16−/− than in wt HSCs. These observations may explain the increased apoptosis observed in Prdm16−/− HSCs. Several transcriptional regulators that are known to be critical for HSC maintenance5,31-33 were differentially expressed in wt and Prdm16−/− HSCs. Expression of Bmi1 and Evi1 was lower, whereas expression of Pbx1 and Gata2 was higher in Prdm16−/− than in wt HSCs. A profound decrease in Pparg expression was observed in Prdm16−/− HSCs. Expression of the Ppar family member Ppard was similar in wt and Prdm16−/− HSCs, however. Finally, the expression of the number of cytokines and cytokine receptors was significantly different in wt and Prdm16−/− cells. Expression of Angpt1 and Wnt3a was suppressed, whereas expression of the Wnt receptor Ryk48 (which has been reported to be expressed specifically in LT-HSCs)45 and of Angpt2 was increased. Taken together, these data suggest that Prdm16 regulates HSCs through multiple, and sometimes opposing, mechanisms.
Discussion
We have shown here that, within the hematopoietic system, Prdm16 is expressed very selectively in the earliest stem and progenitor compartments, and, consistent with this expression pattern, is required for the maintenance of the HSC pool during development and after transplantation.
A HSC defect in Prdm16−/− mice was also very recently described by Chuikov et al.46 We noted some differences in our studies. Although many of their experiments were performed using cells isolated from newborn mice, we never observed live-born Prdm16−/− pups. As the origin of the mice used in our studies was the same as that of the mice used in the studies of Chuikov et al,46 the cause of this difference is unclear but may be related to the backcrossing of Prdm16−/− mice onto C57Bl/6 background in their study. We also observed leukocytosis in adult Prdm16+/− mice. Given the normal cellularity of hematopoietic organs and the profound HSC defect observed after transplantation of Prdm16+/− BM or purified HSPCs, this finding is probably explained by an organismal effect of Prdm16 that is independent of its cell autonomous role in HSCs. Furthermore, Chuikov et al46 did not observe a defect in HSC number and function in heterozygous Prdm16+/− mice, although they did report an NSC defect in Prdm16+/− mice. In contrast, we found that Prdm16 displays both expression and functional haploinsufficiency with respect to HSC function. This difference, too, may be the result of the different genetic background of the mice used in our studies (B6.126) compared with the studies of Chuikov et al,46 where the mice were backcrossed onto the C57BL/6 background. Haploinsufficiency has previously been described in Evi1-deleted mice.5 The phenotype of Evi1−/− mice is qualitatively remarkably similar, although quantitatively more severe, compared with that of Prdm16−/− mice.5 As Evi1 expression was down-regulated in Prdm16−/− HSCs (Figure 6), Prdm16 may act, at least in part, through regulation of Evi1 expression.
The mechanism of action of Prdm16 is probably more complex, however. Maintenance of the HSC function in the long-term requires a critical balance between renewing and differentiating proliferation, quiescence, and survival, as well as appropriate homeostatic responses to damage and stress.1,34 Both in mouse models where HSC quiescence is increased, such as Mef−/− and Bmi1−/− mice,31,49 and in models where HSC cell cycle activity is increased, such as Gfi−/−, Evi1−/−, and Pbx1−/− mice,5,33,50 long-term HSC function is severely compromised. Prdm16−/− NSCs have been reported to display increased oxidative stress and to express less hepatocyte growth factor (Hgf) mRNA. Addition of HGF could partially reverse the NSC defect in Prdm16−/− mice.46 We examined the expression of genes, including Hgf, which were down-regulated in Prdm16−/− NSCs.46 Only one of those, luteinizing hormone/choriogonadotropin receptor (Lhcgr) was also decreased in Prdm16−/− HSCs, whereas another gene, Mt2 (metallothionein 2), a reported indirect target of Prdm16,46 was actually increased in HSCs, rather than decreased, as was reported for NSCs.46 Furthermore, the expression of 2 genes involved in oxidative stress responses and up-regulated in the BM of Bmi1−/− mice, Alox5 and Alox15, was also similar in Prdm16−/− and wt HSCs.47 These observations suggest that the mechanisms underlying the HSC defect in Prdm16-deficient mice may be partially overlapping with, but are nevertheless substantially different from, those causing the NSC defect.
At least one of the mechanisms underlying the HSC defect in Prdm16−/− mice may be increased apoptosis. Consistent with this finding, expression of Trp53 (p53) and its prime target, Puma,34 as well as Tmbim4, a potential Bax inhibitor,38 was increased. Trp53−/− mice have an expanded HSPC compartment,34,35 whereas Puma−/− mice are resistant to hematopoietic failure caused by irradiation and cytotoxic drugs.36,37 It is interesting to note, however, that Prdm16 deletion affects transcription or stability of p53 mRNA, as p53 is typically regulated post-transcriptionally.34 An interesting observation made by Chuikov et al is that, in contrast to NSCs, Prdm16−/− HSCs appeared to contain lower levels of reactive oxygen species than wt HSCs.46 This is surprising as we found decreased Bmi1 expression in Prdm16−/− HSCs and as Bmi1 deletion causes increased oxidative stress.47 As p53 has been shown to decrease oxidative stress in some conditions,51 it is possible that this observation is, at least in part, explained by increased p53 expression in Prdm16−/− HSCs. Somewhat puzzling was that no significant increase in the fraction of apoptotic cells was observed in Prdm16+/− HSCs. It is possible that the difference between wt and Prdm16+/− HSCs with respect to apoptosis as determined by annexin V staining was too subtle to detect. However, given the severity of the HSC defect in Prdm16+/− mice, which quantitatively approached that of Prdm16−/− mice, it is more likely that Prdm16 affects HSC function through multiple mechanisms and that the threshold level of Prdm16 expression required to affect these mechanisms varies.
This possibility is strongly suggested by the subtle but consistent increase in cell cycle activity in the HSPC compartment that was, in contrast to the fraction of apoptotic cells, similar in Prdm16+/− and Prdm16−/− mice. It is unclear whether this increase in cycling activity is a result of selective depletion of quiescent HSCs, or of more generally enhanced cycling activity of the HSC compartment, or both. Interestingly, expression analysis revealed increased expression of several genes that are negative regulators of cell cycling in HSPCs.34,35,39,40 These include, in addition to p53, Cdkn1a (p21) and Cdkn1b (p27). Furthermore, one of the most striking results of our expression analysis was decreased Bmi1 expression in Prdm16−/− HSCs. Bmi1 is a polycomb gene that suppresses the expression of the tumor suppressors p16ink4a and p19arf and is critical for HSC renewal.31,52 Prdm16 must therefore affect cell cycling through multiple opposing mechanisms.
Prdm16 directly or indirectly regulates a remarkable number of transcriptional regulators that are critical for HSC function and act through overlapping but probably distinct mechanisms. Some of the genes that are critical for HSC function, such as Evi1 and Bmi1, are down-regulated,5,31 but others, such as Gata2 and Pbx1,32,33 are up-regulated. Furthermore, p53, a negative regulator of HSC function,34,35 is up-regulated. Thus, Prdm16 induces changes in HSCs that, based on well-characterized knockout models, both enhance and suppress HSC function, and affect quiescence, cell cycling, renewal, differentiation, and apoptosis to various extents. These expression data suggest that Prdm16 may be a critical node in a network that contains negative and positive feedback loops and regulates and integrates HSC renewal, quiescence, differentiation, and apoptosis. The role of PPAR family members in HSCs is unclear. Pparg and Ppard, but not Ppara, are expressed in HSCs (not shown). Prdm16 deletion decreases expression of Pparg mRNA. The transcriptional activity of Pparg is enhanced by Prdm16 during brown fat adipogenesis.18 The fact that Prdm16 deletion decreases expression of a transcription factor of which it increases the activity suggests a positive feedback loop. It will therefore be of interest to examine the effect of deletion of Pparg and to study any compensatory effects of both Pparg and Ppard on HSC function.
Our expression data also suggest a role for Prdm16 in the regulation of autocrine and paracrine signaling in and among HSCs and suggest that cell autonomously produced cytokines and their receptors may be part of this HSC regulatory network. Expression of Angpt1 (Angiopoietin-1 or Ang-1) was suppressed in Prdm16−/− HSCs. Ang-1 enhances HSC quiescence by signaling through Tie-2.41 On the other hand, expression of Angpt2 (Ang-2), which antagonizes Ang-1-induced Tie-2 signaling,42 was increased. Furthermore, the decrease of Wnt3a expression in Prdm16−/− HSCs is of interest and may explain part of the HSC defect. FL Wnt3a−/− HSCs show a repopulation defect after transplantation into adult recipients.43 Whereas one interpretation of these findings is that HSCs generated in a Wnt3a-deficient environment are functionally abnormal, another, possibly more plausible explanation may be that cell autonomous production of Wnt3a is critical for sustained HSC function. It is also interesting to note that Ryk, a noncanonical Wnt receptor that has been shown to be relatively specifically expressed in LT-HSCs,45,48 is also regulated directly or indirectly by Prdm16.
Taken together, we have shown here that Prdm16 plays a critical role in long-term HSC maintenance and renewal. Prdm16 is probably a major and integrative node in a network that involves at the very least Evi1, Pbx1, Gata2, Bmi1, the downstream Bmi1 targets p16inka and p19arf, p53 and its targets, as well as cytokine receptors and cell autonomously produced cytokines, and regulates HSC renewal, cycling, and apoptosis.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health (grants RO1 HL073760 and RO1 AG 016327; H.-W.S.).
National Institutes of Health
Authorship
Contribution: F.A. performed hematopoietic profile analysis, studies using purified progenitor and stem cells, expression studies, homing, and apoptosis; S.A. performed studies in adult mice; A.L. and P.K. assisted in most experiments; A.S., D.-F.L., and I.R.L. assisted and provided advice in microfluidics array expression analysis; B.Y.Z. performed part of the studies on fetal mice; and H.-W.S. designed experiments, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hans-Willem Snoeck, Department of Gene and Cell Medicine, Mount Sinai of School of Medicine, Gustave L. Levy Place, Box 1496 New York, NY 10029; e-mail: hans.snoeck@mssm.edu.
References
Author notes
F.A. and S.A. contributed equally to this study.