Abstract
We used genome-wide methylation microarrays to analyze differences in CpG methylation patterns in cells relevant to the pathogenesis of myeloma plasma cells (B cells, normal plasma cells, monoclonal gammopathy of undetermined significance [MGUS], presentation myeloma, and plasma cell leukemia). We show that methylation patterns in these cell types are capable of distinguishing nonmalignant from malignant cells and the main reason for this difference is hypomethylation of the genome at the transition from MGUS to presentation myeloma. In addition, gene-specific hypermethylation was evident at the myeloma stage. Differential methylation was also evident at the transition from myeloma to plasma cell leukemia with remethylation of the genome, particularly of genes involved in cell–cell signaling and cell adhesion, which may contribute to independence from the bone marrow microenvironment. There was a high degree of methylation variability within presentation myeloma samples, which was associated with cytogenetic differences between samples. More specifically, we found methylation subgroups were defined by translocations and hyperdiploidy, with t(4;14) myeloma having the greatest impact on DNA methylation. Two groups of hyperdiploid samples were identified, on the basis of unsupervised clustering, which had an impact on overall survival. Overall, DNA methylation changes significantly during disease progression and between cytogenetic subgroups.
Introduction
Upon encounter with antigen, naive B cells undergo somatic hypermutation and class switch recombination in the germinal center, finally differentiating into plasma cells (PCs) residing in the bone marrow.1 Multiple myeloma is a clonal malignancy of these PCs that develops as a consequence of a multistep transformation process. Insight into the molecular mechanisms underlying this transformation process can come from the study of the individual steps leading to myeloma, which is known to evolve from a premalignant state, monoclonal gammopathy of undetermined significance (MGUS), and transforms into myeloma at a rate of 1% per year.2,3 Additional genetic events may transform the myeloma clone further to a more aggressive disease state known as plasma cell leukemia (PCL), in which the clonal cells lose their dependency on the bone marrow microenvironment.
Genomic instability is a characteristic feature of myeloma cells in which translocations involving the IGH locus and MMSET/FGFR3, CCND1, CCND3, MAF, and MAFB occur, as well as numerous structural copy number alterations, including del(1p), del(6q), del(8p), del(13q), del(16q), del(22), and gain of 1q.4-6 However, the mechanisms involved in the progression from MGUS to myeloma are incompletely understood as, although present at decreased frequencies, the genetic markers characteristic of myeloma such as immunoglobulin heavy (IGH) chain rearrangements, hyperdiploidy, and gains and losses of chromosomal regions are also present in MGUS.7,8
Although there has been substantial work performed on the genetics of myeloma, little is known about the epigenetic changes leading to disease progression. Changes in DNA methylation status are one of the key epigenetic features known to regulate gene expression. Methylation changes occur primarily at CpG dinucleotides, which are present at a greater frequency in promoter regions as well as within repeat sequences and transposable elements.9 Hypomethylation in cancer cells mainly occurs within repeat sequences and transposable elements, whereas hypermethylation occurs in promoter regions, particularly of putative tumor suppressor genes.10 Such hypermethylation of DNA is linked with transcriptionally inactive heterochromatin and is associated with methylated histone H3K9 residues.11-13
With the exception of one recent study,14 the epigenetic factors contributing to the pathogenesis of myeloma have been studied on a gene-by-gene basis and, with the use of methylation-specific PCR, several genes have been identified that are hypermethylated, including VHL, XAF1, IRF8, TP53, CDKN2A, CDKN2B, DAPK, SOCS1, CDH1, PTGS2, CCND2, and DCC.15-22 Promoter hypermethylation of cyclin-dependent kinase inhibitor 2A (CDKN2A) and TGFBR2 have been shown to correlate with poor prognosis in myeloma patients, although the prognostic value of CDKN2A hypermethylation remains debatable.16,23,24
In this study we used the Infinium array (Illumina) to analyze CpG island promoter methylation with normal PCs, MGUS, myeloma, and PCL samples to identify methylation changes that may contribute to the pathogenesis of myeloma or that could act as prognostic factors. In addition, we used cytogenetic data available for the myeloma samples to identify methylation changes between known cytogenetic subgroups. These arrays have been used and validated by many groups and favorably correlate with whole-genome methylation sequencing technologies.25-28
Methods
Patient samples and clinical data
The MRC Myeloma IX trial recruited 1970 newly diagnosed patients and comprised 2 arms; the first for older and less-fit patients and the second for younger, fitter patients. The details of the trial have been published elsewhere but, in summary, younger, fitter patients were put on the intensive arm and received autologous transplantation after induction with (1) cyclophosphamide, thalidomide, and dexamethasone or (2) cyclophosphamide, vincristine, doxorubicin, and dexamethasone.29 The nonintensive arm consisted of older patients who were treated with either (1) attenuated cyclophosphamide, thalidomide, and dexamethasone or (2) melphalan and prednisolone. All patients were then randomized to thalidomide maintenance or no thalidomide maintenance. The trial was approved by the MRC Leukemia Data Monitoring and Ethics committee (MREC 02/8/95, ISRCTN68454111).
Bone marrow aspirates were obtained after informed consent. PCs from nonmyeloma patients (normal PC controls, n = 3) and presentation myeloma samples (n = 161) were selected to a purity of > 90% by the use of CD138 microbeads and magnet-assisted cell sorting (Miltenyi Biotech, Bisley, United Kingdom).30 To achieve a sufficient quantity of DNA, some normal PC control samples were pooled. MGUS samples (n = 4) were analyzed by flow cytometry to determine the percentage of PCs within the leukocyte population (range, 0.3%-3.1%) and the percentage of those PCs with an abnormal phenotype (CD19−; CD56+; CD45−; range, 80%-100%). PCs subsequently underwent cell selection with the use of CD138 microbeads as mentioned previously. Samples from PCL patients (n = 7) were not CD138 selected but contained > 90% PC infiltration as determined by microscopy. PCL samples were genetically characterized by fluorescence in situ hybridization (FISH), SNP 6.0 mapping array (Affymetrix), or U133 Plus 2.0 expression array (Affymetrix). The 7 PCL samples consisted of 3 t(4;14), 3 t(11;14), and 1 hyperdiploid sample.
DNA was extracted by the use of commercially available kits (RNA/DNA mini kit or Allprep kit; QIAGEN) according to manufacturer's instructions. DNA quality and quantity were determined on an ND-1000 Spectrophotometer (Nano-Drop Technologies). Interphase FISH analysis was performed on purified PC by use of the micro-FISH technique and probes, which have previously been documented.31,32 In brief, probes to detect t(4;14) (n = 15), t(6;14) (n = 1), t(11;14) (n = 35), t(14;16) (n = 7), t(14;20) (n = 3), del(1p32.3) (n = 21), gain 1q (n = 49), del(17p) (n = 8), and hyperdiploidy (defined by gain of any 2 of chromosomes 5, 9 and 15, n = 73) were used to identify abnormalities. Samples with a split IGH probe but no identified partner were termed unknown translocation.
Methylation arrays
A total of 500 ng of DNA was bisulfite converted by use of the EZ DNA methylation kit (Zymo Research) and subsequently processed for hybridization onto the Infinium humanmethylation27 BeadArray (Illumina) according to manufacturers' protocols. This array interrogates 27 578 CpG dinucleotides encompassing 14 495 genes. In brief, DNA was treated with bisulfite, converting nonmethylated C nucleotides to U (T), whereas methylated C nucleotides remained unaffected. Bisulfite-treated DNA was subsequently amplified, fragmented, and hybridized to locus-specific oligonucleotides on the BeadArray. C or T nucleotides were detected by fluorescence signal from single-nucleotide extension of the DNA fragments. Results were interpreted as a ratio (β value) of methylated signal (C) compared with the sum of methylated and unmethylated signal (C + T) for each locus, where a β-value of 0 represents fully unmethylated DNA and a value of 1 fully methylated DNA. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE GSE21304 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21304).
Bisulfite PCR and sequencing
A total of 100 ng of genomic DNA was bisulfite treated and purified with the Epitect bisulfite kit (QIAGEN) following the manufacturer's instructions. Primers to amplify the regions surrounding methylation array probes were designed and are available in the supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). DNA was amplified by the use of Platinum Taq DNA polymerase (Invitrogen), and reactions were purified and sequenced on a 3500 DNA capillary sequencer (Applied Biosystems). Sequences were analyzed with Sequencher 4.8 (Gene Codes).
Expression array data
Expression array data are available for myeloma samples, have previously been published, and are available under the GEO Series accession number GSE21349 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21349).5
Data analysis
Data were analyzed in GenomeStudio by the use of the methylation module (Illumina). Further analyses were performed with the R and the LIMMA package.33,34 Missing elements in the data were imputed by the use of row means. Differential methylation between samples were identified with an empirical Bayes moderated t test and the resulting P values were adjusted by use of the Benjamini and Hochberg method.35 We considered P < .05 significant. Hierarchical clustering was performed by the Euclidean distance and the Ward method.36 Cluster stability was ascertained with multiscale bootstrap analysis by use of the pvclust R package.37 Approximately unbiased P values were calculated by 100 000 resamplings of the original data. To further investigate the relationships shown with the use of hierarchical clustering, principal component analysis was performed on the methylation data. With the use of a scree plot, the first 3 principal components were deemed to be significant and these data were plotted against one another (supplemental data).
Expression data were normalized by the use of RMA (ie, quantile normalized, median polish).38,39 Correlation between methylation and expression data were investigated with the Pearson correlation between the corresponding probes that mapped to the same gene symbol. Correlations were only considered if they were significantly different (P < .05) from zero.
Results
Methylation profiling of different disease stages in myeloma
Genome-wide methylation profiles were compared between normal B cells (n = 6), normal PCs (n = 3), MGUS samples (n = 4), presentation myeloma (n = 161), PCL (n = 7), and human myeloma cell lines (HMCLs) (n = 9). By the use of XY scatter plots, significant variations in methylation profiles within the B-cell subset of samples that were caused by X-chromosome inactivation in female subjects were noted. To remove these confounding data points, we removed all probes present on the sex chromosomes, leaving 26 486 probes on the array to interrogate.
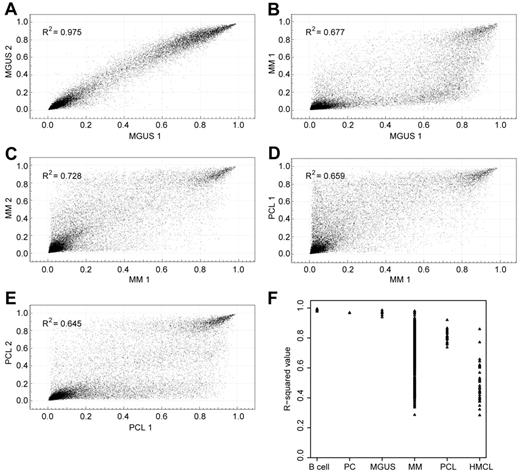
Pairwise comparisons on XY scatter plots were used to determine the variation in methylation within each cell type (Figure 1). Correlation coefficients were determined, and the range and median values are shown in Figure 1 and detailed in Table 1. Median correlation coefficients > 0.9 were observed in premalignant cell types such as B cells, normal PCs, and MGUS PCs, indicating homogeneity of gene methylation between samples. However, in presentation myeloma, as well as in cell lines, the median correlation coefficients were < 0.7, indicating greater heterogeneity of methylation between samples of the same cell type. This heterogeneity may be the result of methylation variation between the different cytogenetic subgroups and is investigated in “Global methylation differences in cytogenetic subgroups.”
XY scatterplots show the overall methylation differences between and within cell types. (A) 2 MGUS samples are very similar. (B) A myeloma sample shows hypomethylation of probes compared with an MGUS sample. (C) Two myeloma samples show heterogeneity of methylation. (D) A PCL sample shows hypermethylation compared with a myeloma sample. (E) 2 PCL samples have equally heterogeneous methylation profiles. (F) Summary of R2 correlation values of samples within each cell type.
XY scatterplots show the overall methylation differences between and within cell types. (A) 2 MGUS samples are very similar. (B) A myeloma sample shows hypomethylation of probes compared with an MGUS sample. (C) Two myeloma samples show heterogeneity of methylation. (D) A PCL sample shows hypermethylation compared with a myeloma sample. (E) 2 PCL samples have equally heterogeneous methylation profiles. (F) Summary of R2 correlation values of samples within each cell type.
Variation in global methylation within cell types
| Cell type (n) . | Range . | Median . |
|---|---|---|
| B cell (6) | 0.9716-0.99 | 0.9833 |
| PC (3) | 0.935-0.9397 | 0.9384 |
| MGUS (4) | 0.8828-0.9730 | 0.9351 |
| Presentation myeloma (161) | 0.0816-0.9611 | 0.6731 |
| PCL (7) | 0.5439-0.8541 | 0.6456 |
| HMCL (9) | 0.2884-0.8834 | 0.472 |
| Cell type (n) . | Range . | Median . |
|---|---|---|
| B cell (6) | 0.9716-0.99 | 0.9833 |
| PC (3) | 0.935-0.9397 | 0.9384 |
| MGUS (4) | 0.8828-0.9730 | 0.9351 |
| Presentation myeloma (161) | 0.0816-0.9611 | 0.6731 |
| PCL (7) | 0.5439-0.8541 | 0.6456 |
| HMCL (9) | 0.2884-0.8834 | 0.472 |
Pairwise XY scatter plots were used to generate Pearson correlation coefficients (r2) between samples of the same cell type.
HMCL indicates human myeloma cell lines; MGUS, monoclonal gammopathy of undetermined significance; PC, plasma cell; and PCL, plasma cell leukemia.
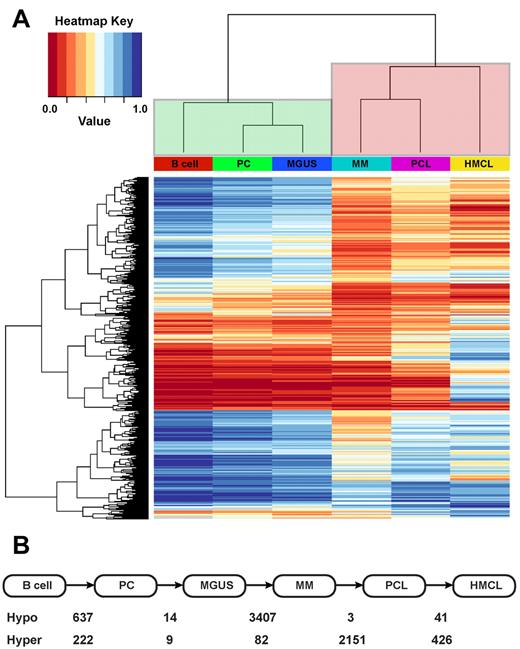
The data for each probe were averaged for each cell type and filtered to remove differences between the cell types characteristic of the multistep pathway seen in the development of myeloma which were not significant (P > .05) or had an average β-value < 0.25 or > 0.75 in both cell types. Overall methylation relationships were analyzed between cell types by cluster analysis by use of the Euclidean method (Figure 2A). The results of this analysis revealed that the overall methylation in these cell types can accurately distinguish between premalignant and malignant cells. MGUS samples cluster closely with normal PCs and B cells, whereas presentation and PCL samples cluster closely with HMCLs. The large branch between nonmalignant and malignant cell types is a consequence of the large difference in global methylation patterns between the 2 cell types, indicating an important role for methylation in the progression of MGUS to myeloma.
DNA methylation distinguishes malignant from nonmalignant phenotypes in myeloma. (A) Hierarchical clustering of overall methylation in cell types can distinguish premalignant (green clade) and malignant phenotypes (red clade). (B) The number of probes differentially methylated in sequential steps of myeloma pathogenesis. Hypo indicates hypomethylated; hyper, hypermethylated at transition.
DNA methylation distinguishes malignant from nonmalignant phenotypes in myeloma. (A) Hierarchical clustering of overall methylation in cell types can distinguish premalignant (green clade) and malignant phenotypes (red clade). (B) The number of probes differentially methylated in sequential steps of myeloma pathogenesis. Hypo indicates hypomethylated; hyper, hypermethylated at transition.
The number of probesets differing between component steps of the pathogenesis to myeloma is shown in Figure 2B. This analysis indicates that there are few methylation changes between normal PCs and the MGUS phenotype. In contrast, there are 3407 probes (1428 genes) that undergo hypomethylation and 82 probes that are hypermethylated from MGUS to presentation myeloma. However, it is the large number of probes that are hypomethylated that are the main cause for the distinct methylation differences between nonmalignant and malignant myelomatous cell types. The probes are designated as being either within or not within a CpG islands, and of the 3407 that underwent hypomethylation, only 655 (19.2%) were within a CpG island, whereas 48 (58.5%) of the 82 that underwent hypermethylation were within a CpG island. Because global hypomethylation of cancers is known to occur outside of CpG islands, this finding is indicative of both global hypomethylation and gene-specific methylation at the transformation of MGUS to myeloma.
At the transition from myeloma to PCL the main changes are hypermethylation of genes (2151 probes, 1802 genes). Of these 2151 probes, 1412 are not within a CpG island, meaning that there is methylation change both within and outside CpG islands. Interestingly, 1168 of these probes (1068 genes) were previously demethylated at the MGUS to myeloma transition, which is consistent with remethylation of previously demethylated genes occurring at the transition to myeloma.
Gene ontology (GO) analysis of the 82 probes (77 genes) hypermethylated at the transition to myeloma indicates that 3 main groups of genes affected are regulation of developmental processes, cell cycle processes, and regulation of transcription. The genes include transcription factors or genes that regulate transcription (ACVR1, ARID3A, BRCA2, C19orf33, CALCA, CBX4, FOXD2, GATA4, HIPK3, HOXB8, HOXD11, ID4, IRF7, LDB1, NCOR2, ONECUT2, RAB37, RUNX2, ZIC1, ZNF385, ZNF560), as well as regulators of cell cycle (ACVR1, AIF1, BCL2, BRCA2, CDKN2B, GAS2L1, ID4, MPHOSPH9, PKMYT1).
Of the 2151 probes (1802 genes) hypermethylated at the myeloma to PCL transition, 739 are annotated as occurring within CpG islands. GO term analysis of these 739 probes reveals that cell-cell signaling (44 genes, P = 5.28 × 10−7), cell development or differentiation (34 genes, P = 3.28 × 10−6), and cell adhesion molecules (45 genes, P = 1.2 × 10−5) are significantly enriched.
Global methylation differences in cytogenetic subgroups
When the dataset is analyzed by the use of an unsupervised clustering approach per sample, rather than per cell type, there are 3 main clusters evident: nonmalignant cells, HMCLs and t(4;14) samples, and other myeloma samples (Figure 3). The clustering was confirmed by principal component analysis (supplemental data). The samples within the main myeloma group can be divided into 5 clades (B-F), and when the samples are annotated according to FISH results, these clades are clearly defined by cytogenetic abnormalities. Clades B and C are mainly hyperdiploid samples, but clade C is more closely related to translocation samples. Clades D through G are predominantly samples with translocations, with clades D and F consisting of t(11;14) samples and clade E of t(14;16) samples. Translocation/cyclin D and University of Arkansas for Medical Sciences expression-based classification40,41 of these subclusters demonstrates that clade D consists of CD-2 samples, whereas clade F can be split into 2 distinct subclusters consisting of those in CD-2 (left branch) and those solely containing CD-1 samples (right branch). Clades G and H are on a separate branch from the majority of myeloma samples and consist of t(4;14) samples and HMCLs, respectively. Other abnormalities such as del(1p), gain 1q, del(13q), del(16q), del(17p), and del(22q) are not associated with nor define specific methylation subgroups nor drive clustering of the samples. Therefore, the main currently known cytogenetic abnormalities that affect methylation in myeloma samples are the translocations [t(4;14), t(11;14), and t(14;16)] and hyperdiploidy. PCL samples did not segregate together but remained within their respective cytogenetic clades, except for one outlying t(4;14) PCL sample, which clustered with the nonmalignant cell types.
Unsupervised hierarchical clustering of samples reveals discrete methylation groups on the basis of cytogenetic abnormalities. Distinct clades are highlighted and labeled A-H. Samples are color coded with cytogenetic data: cell type, translocation, TC, and UAMS classifications and colors are as per the key. Other cytogenetic data are coded as red for present, green for absent, and white for no data. Heatmap key indicates β-value methylation level. UAMS subgroups: CD-1/CD-2, cyclin D1 subgroups; HY, hyperdiploid; LB, low bone disease; MF, MAF; MS, MMSET/FGFR3; PR, proliferation. TC subgroups: D1, cyclin D1; D2, cyclin D2; D3, cyclin D3; MAF, MAF/MAFB; MMSET, MMSET/FGFR3 MAF, v-maf musculoaponeurotic fibrosarcoma oncogene homolog.
Unsupervised hierarchical clustering of samples reveals discrete methylation groups on the basis of cytogenetic abnormalities. Distinct clades are highlighted and labeled A-H. Samples are color coded with cytogenetic data: cell type, translocation, TC, and UAMS classifications and colors are as per the key. Other cytogenetic data are coded as red for present, green for absent, and white for no data. Heatmap key indicates β-value methylation level. UAMS subgroups: CD-1/CD-2, cyclin D1 subgroups; HY, hyperdiploid; LB, low bone disease; MF, MAF; MS, MMSET/FGFR3; PR, proliferation. TC subgroups: D1, cyclin D1; D2, cyclin D2; D3, cyclin D3; MAF, MAF/MAFB; MMSET, MMSET/FGFR3 MAF, v-maf musculoaponeurotic fibrosarcoma oncogene homolog.
On the basis of the aforementioned discussion, it seems that the observed heterogeneity in global methylation within presenting myeloma samples is attributable to the presence of different cytogenetic subgroups within the sample set. To identify the methylation differences driving the clustering, we first split the presentation myeloma samples according to the IgH translocation, comparing each translocation group [t(4;14) n = 15; t(11;14) n = 35; t(14;16) n = 7; t(14;20) n = 3, unknown translocations n = 15] to samples with no split IGH locus (n = 66), as determined by FISH. Methylation β-values were averaged across the samples within each group and analyzed as before. In this analysis the biggest differences were seen in the t(4;14) comparison with 2503 probes (9.4%) with increased methylation in the t(4;14) group compared with those with no split IGH locus and 302 probes with decreased methylation (Table 2). Fewer changes were seen when the t(11;14) (98 hypermethylated and 320 hypomethylated), t(14;16) (26 hypermethylated and 19 hypomethylated), t(14;20) (10 hypermethylated and 1 hypomethylated), and unknown translocations (no differences) were compared against those with no split IGH locus.
Number of probesets significantly altered in methylation status between samples with and without cytogenetic abnormalities
| Cytogenetic abnormality . | Hypomethylated . | Hypermethylated . |
|---|---|---|
| t(4;14) | 302 | 2503 |
| t(14;16) | 19 | 26 |
| t(14;20) | 1 | 10 |
| t(11;14) | 320 | 98 |
| Unknown translocation | 0 | 0 |
| Hyperdiploidy | 134 | 194 |
| Gain 1q | 5 | 13 |
| Del(1p) | 0 | 0 |
| Del(13q) | 21 | 11 |
| Del(16q) | 0 | 0 |
| Del(22q) | 7 | 24 |
| Del(17p) | 1 | 0 |
| Cytogenetic abnormality . | Hypomethylated . | Hypermethylated . |
|---|---|---|
| t(4;14) | 302 | 2503 |
| t(14;16) | 19 | 26 |
| t(14;20) | 1 | 10 |
| t(11;14) | 320 | 98 |
| Unknown translocation | 0 | 0 |
| Hyperdiploidy | 134 | 194 |
| Gain 1q | 5 | 13 |
| Del(1p) | 0 | 0 |
| Del(13q) | 21 | 11 |
| Del(16q) | 0 | 0 |
| Del(22q) | 7 | 24 |
| Del(17p) | 1 | 0 |
Methylation is relative to the control group.
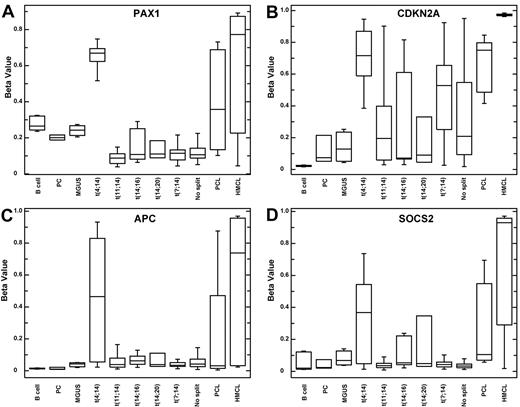
GO term analysis of the 2503 probes (1881 genes) hypermethylated in the t(4;14) samples indicates methylation of genes involved in cell adhesion (147 genes, P = 7.8 × 10−22) and cell-cell signaling (128 genes, P = 1.41 × 10−19). Within the t(4;14) sample group several genes of interest were hypermethylated, some of which were validated by bisulfite-specific PCR (supplemental data). These include adenomatous polyposis coli gene (APC), which is a Wnt signaling pathway antagonist that is involved in cell adhesion, transcriptional activation, and apoptosis. In the t(4;14) subset methylation of APC has a β-value of 0.36-0.41 compared with 0.03-0.07 in samples with no translocation, indicating it is hemimethylated. PAX1, or paired box gene 1, was solely methylated in t(4;14) samples and is a known methylation marker in ovarian cancer. In addition, suppressor of cytokine signaling (SOSC2; 0.45 vs 0.14) and CDKN2A (0.7 vs 0.34) are hypermethylated in t(4;14) samples but were not differentially methylated between MGUS and myeloma. To investigate this difference we separated the data by cell type and, within the myeloma group, by translocation. This analysis revealed that CDKN2A has significantly more methylation in the t(4;14) samples (P = .0003) compared with samples with no split IGH but that myeloma samples as a whole do not have significantly more methylation of CDKN2A than MGUS samples (P = .518; Figure 4). As such, hypermethylation of CDKN2A is significantly prognostic within the myeloma group (comparing β-values < 0.3 vs > 0.3, P = .03), but this is attributable to its association with the poor prognostic t(4;14) subgroup. This observation is different to the situation at CDKN2B, which lies adjacent to CDKN2A in the genome, is fully methylated in all myeloma cytogenetic subgroups and is significantly altered at the transition from MGUS to myeloma.
Boxplots of methylation beta values indicate t(4;14)-specific gene hypermethylation.PAX1 (A), CDKN2A (B), APC (C), and SOCS2 (D).
Boxplots of methylation beta values indicate t(4;14)-specific gene hypermethylation.PAX1 (A), CDKN2A (B), APC (C), and SOCS2 (D).
We investigated the effect of methylation of the genes in the t(4;14) samples further by comparing the data to gene expression data. t(4;14) expression data were compared with samples with no split IGH locus to generate a list of differentially expressed genes. From this analysis, 353 expression probesets were differentially expressed with the corresponding gene differentially methylated, of which 333 had lower expression in t(4;14) samples with increased methylation (supplemental data). The genes with the greatest expression fold changes are shown in Table 3 along with the corresponding methylation changes. C20orf103, which has similarity to LAMP (ie, lysosome-associated membrane protein) domain proteins, was most differentially expressed with a 6.6-fold decrease in expression in t(4;14) samples and a corresponding increase in methylation from 0.231 to 0.472. CD79A was also underexpressed in t(4;14) samples with an increase in methylation. This molecule has been found to have loss of protein expression in a subset of myeloma samples that also have low cyclin D1 expression.42 These are likely to be t(4;14) samples, which are cyclin D2 positive, in which methylation of CD79A has resulted in loss of protein in the cells. Other genes that are underexpressed in t(4;14) samples include glioma tumor suppressor candidate (ie, GLTSCR2) and SOSC2. Conversely, genes hypomethylated in t(4;14) samples with increased expression include DNA methyltransferase 3A, which is responsible for de novo DNA methylation, and insulin receptor 2, which is a tyrosine kinase receptor and mediates phosphoinositide 3-kinase signaling. However, the gene with the largest fold change was collagen triple helix repeat containing-1, which is implicated in promoting cell migration, osteoblastic bone formation, and activation of Wnt signaling pathways.43,44
Because t(4;14) samples most closely resemble HMCLs, with respect to DNA methylation, we investigated whether or not all HMCLs have a t(4;14) methylation profile. We discovered that t(4;14) HMCLs do have hypermethylation of genes that are hypermethylated in t(4;14) samples but additionally at selected loci non-t(4;14). HMCLs also had hypermethylation. For example, C20orf103 hypermethylation is specific to t(4;14) myeloma samples and t(4;14) HMCLs, whereas hypermethylation of CD79A is specific to t(4;14) myeloma samples and all HMCLs, irrespective of translocation (supplemental data). This finding indicates that all HMCLs acquire hypermethylation of genes, in a similar fashion to t(4;14) myeloma samples and PCL samples, but methylation of some genes remain specific to t(4;14).
Samples with cytogenetic abnormalities were also compared with those without the same abnormality. The abnormalities examined were del(1p32.3), gain 1q, del(13q), del(16q), del(17p), del(22q), hyperdiploidy, and any split IGH (Figure 3, Table 2). In comparison with translocation subgroups, these cytogenetic abnormalities are associated with far fewer methylation changes, indicating that these abnormalities are not significantly associated with methylation. The largest methylation changes were noted in the hyperdiploid comparison, in which 134 probes were hypomethylated and 194 were hypermethylated compared with nonhyperdiploid samples. Of interest in the nonhyperdiploid hypomethylated gene list was CCND1, in which 6 probes show a decrease in methylation. However, this difference in methylation did not correlate with a difference in expression of CCND1. When split by translocation group, the decrease in methylation was not limited to the t(11;14) subgroup, which overexpresses CCND1 but was present in all the major translocation groups. In addition, hypermethylation of CCND1 within the hyperdiploid samples did not correlate with translocation/cyclin D (TC) classification status, indicating that methylation of CCND1 at these CpG sites is not linked to expression of the gene.
Genes in t(4;14) samples that are both differentially methylated and differentially expressed compared with samples with no split IGH locus
| Gene . | Chr. . | Expression changes (U133 Plus 2.0) . | Methylation changes (humanmethylation27) . | Description . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Probe . | t(4;14) . | No split . | Fold change . | P . | Probe . | CpG island . | t(4;14) . | No split . | Difference . | P . | |||
| C20orf103 | 20 | 219463_at | 8.038 | 10.772 | −6.656 | .008991 | cg09119967 | False | 0.472 | 0.231 | 0.241 | .001135 | Chromosome 20 open reading frame 103 |
| CD79A | 19 | 1555779_a_at | 7.168 | 8.408 | −2.362 | .006872 | cg04790874 | False | 0.484 | 0.222 | 0.262 | 4.11 × 10−5 | CD79A |
| 205049_s_at | 7.652 | 8.722 | −2.100 | .040305 | |||||||||
| FAM49A | 2 | 208092_s_at | 6.461 | 7.775 | −2.487 | .009265 | cg10106284 | False | 0.287 | 0.167 | 0.12 | .033294 | Family with sequence similarity 49, member A |
| 209683_at | 5.631 | 6.891 | −2.396 | .019702 | |||||||||
| 230276_at | 7.031 | 7.898 | −1.823 | .023463 | |||||||||
| GLTSCR2 | 19 | 234339_s_at | 10.806 | 12.216 | −2.658 | 1.05 × 10−7 | cg16791686 | True | 0.307 | 0.159 | 0.148 | 3.36 × 10−5 | Glioma tumor suppressor candidate region gene 2 |
| 217807_s_at | 13.126 | 14.083 | −1.942 | 1.06 × 10−7 | |||||||||
| GPX1 | 3 | 200736_s_at | 9.846 | 11.311 | −2.760 | .000267 | cg06613840 | True | 0.442 | 0.177 | 0.265 | 2.46 × 10−8 | Glutathione peroxidase 1 |
| cg15900980 | True | 0.386 | 0.082 | 0.304 | 1.16 × 10−6 | ||||||||
| GRM8 | 7 | 216992_s_at | 4.346 | 4.896 | −1.464 | .002383 | cg09868882 | False | 0.532 | 0.267 | 0.265 | 4.34 × 10−8 | Glutamate receptor, metabotropic 8 |
| MAB21L1 | 13 | 206163_at | 3.443 | 4.138 | −1.619 | .037692 | cg05093686 | True | 0.538 | 0.207 | 0.271 | 4.86 × 10−7 | mab-21-like 1 |
| cg12029639 | True | 0.560 | 0.225 | 0.335 | 1.20 × 10−7 | ||||||||
| MBP | 18 | 209072_at | 6.539 | 7.032 | −1.407 | .000475 | cg12555907 | True | 0.841 | 0.411 | 0.43 | 2.61 × 10−8 | Myelin basic protein |
| NME4 | 16 | 212739_s_at | 8.007 | 8.658 | −1.570 | .012902 | cg18676162 | True | 0.321 | 0.178 | 0.143 | .027662 | Nonmetastic cells 4 |
| OSBPL10 | 3 | 231656_x_at | 5.979 | 6.995 | −2.023 | .002011 | cg15840985 | True | 0.329 | 0.170 | 0.159 | .00022 | Oxysterol binding protein-like 10 |
| RPS2 | 16 | 212433_x_at | 13.275 | 14.107 | −1.781 | .000313 | cg18279742 | True | 0.344 | 0.205 | 0.139 | .041014 | Ribosomal protein S2 |
| 203107_x_at | 13.825 | 14.543 | −1.645 | .000254 | |||||||||
| 217466_x_at | 10.499 | 11.145 | −1.564 | .001184 | |||||||||
| SOCS2 | 12 | 203373_at | 7.734 | 8.405 | −1.593 | .041561 | cg04797323 | True | 0.450 | 0.140 | 0.31 | .000257 | Suppressor of cytokine signalling 2 |
| cg06630241 | True | 0.637 | 0.285 | 0.352 | 7.18 × 10−10 | ||||||||
| cg11738543 | True | 0.353 | 0.103 | 0.25 | 5.05 × 10−5 | ||||||||
| cg23412850 | True | 0.330 | 0.059 | 0.271 | 4.24 × 10−6 | ||||||||
| ACADVL | 17 | 200710_at | 10.904 | 10.024 | 1.840 | .000336 | cg24825722 | False | 0.121 | 0.305 | −0.184 | .007423 | Acyl-CoA dehydrogenase, very long chain |
| cg21636577 | False | 0.221 | 0.472 | −0.251 | .000713 | ||||||||
| CTHRC1 | 8 | 225681_at | 12.349 | 8.766 | 11.984 | 4.57 × 10−5 | cg19188612 | True | 0.620 | 0.840 | −0.22 | .00053 | Collagen triple helix containing 1 |
| DNMT3A | 2 | 222640_at | 6.557 | 6.273 | 1.218 | .037539 | cg21629895 | False | 0.339 | 0.491 | −0.152 | .042813 | DNA methyl-transferase 3A |
| GALNT1 | 18 | 201724_s_at | 8.838 | 8.011 | 1.774 | .041426 | cg05714729 | False | 0.212 | 0.383 | −0.171 | .034844 | UDP-N-acetyl-alpha-D-galactosamine |
| GNPTG | 16 | 224887_at | 9.518 | 8.917 | 1.516 | .008847 | cg18146152 | True | 0.534 | 0.748 | −0.214 | .000131 | N-acetyl-glucosamine-1-phosphate transferase, gamma subunit |
| IRS2 | 13 | 209184_s_at | 7.470 | 6.309 | 2.236 | 4.05 × 10−5 | cg14341579 | True | 0.037 | 0.285 | −0.248 | .011963 | Insulin receptor substrate 2 |
| 209185_s_at | 7.711 | 5.849 | 3.636 | 4.90 × 10−6 | cg25802424 | True | 0.037 | 0.265 | −0.228 | .003687 | |||
| LRIG1 | 3 | 211596_s_at | 6.472 | 5.568 | 1.872 | .009466 | cg26131019 | True | 0.057 | 0.310 | −0.253 | .001657 | Leucine-rich repeats and Ig-like domains 1 |
| LRP12 | 8 | 220253_s_at | 4.404 | 3.418 | 1.981 | 1.74 × 10−8 | cg09531892 | True | 0.224 | 0.429 | −0.205 | .03994 | Low-density lipoprotein-related protein 12 |
| 219631_at | 5.758 | 4.230 | 2.883 | 8.52 × 10−7 | |||||||||
| PELI1 | 2 | 218319_at | 12.686 | 11.809 | 1.836 | .033075 | cg15309578 | False | 0.299 | 0.421 | −0.122 | .006695 | Pellino homolog 1 |
| 232213_at | 9.231 | 8.255 | 1.966 | .034372 | |||||||||
| PTPRA | 20 | 213795_s_at | 7.332 | 6.753 | 1.493 | .02016 | cg03115886 | False | 0.443 | 0.601 | −0.158 | .025021 | Protein tyrosine phosphatase receptor type A |
| PTPRCAP | 11 | 204960_at | 9.872 | 8.598 | 2.419 | .004979 | cg05751148 | False | 0.111 | 0.319 | −0.208 | .036243 | Protein tyrosine phosphatase receptor type C associated protein |
| cg20792833 | False | 0.191 | 0.465 | −0.274 | .003583 | ||||||||
| SBNO1 | 12 | 218737_at | 5.164 | 4.624 | 1.454 | .003605 | cg04398275 | False | 0.438 | 0.700 | −0.262 | 2.95 × 10−6 | Strawberry notch homolog 1 |
| SNRK | 3 | 209481_at | 8.785 | 8.049 | 1.666 | .009873 | cg04008913 | False | 0.171 | 0.314 | −0.143 | .031077 | SNF-related kinase |
| ZNF75A | 16 | 227670_at | 5.386 | 4.892 | 1.409 | .02804 | cg02825709 | True | 0.322 | 0.575 | −0.253 | .000649 | Zinc finger protein 75A |
| Gene . | Chr. . | Expression changes (U133 Plus 2.0) . | Methylation changes (humanmethylation27) . | Description . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Probe . | t(4;14) . | No split . | Fold change . | P . | Probe . | CpG island . | t(4;14) . | No split . | Difference . | P . | |||
| C20orf103 | 20 | 219463_at | 8.038 | 10.772 | −6.656 | .008991 | cg09119967 | False | 0.472 | 0.231 | 0.241 | .001135 | Chromosome 20 open reading frame 103 |
| CD79A | 19 | 1555779_a_at | 7.168 | 8.408 | −2.362 | .006872 | cg04790874 | False | 0.484 | 0.222 | 0.262 | 4.11 × 10−5 | CD79A |
| 205049_s_at | 7.652 | 8.722 | −2.100 | .040305 | |||||||||
| FAM49A | 2 | 208092_s_at | 6.461 | 7.775 | −2.487 | .009265 | cg10106284 | False | 0.287 | 0.167 | 0.12 | .033294 | Family with sequence similarity 49, member A |
| 209683_at | 5.631 | 6.891 | −2.396 | .019702 | |||||||||
| 230276_at | 7.031 | 7.898 | −1.823 | .023463 | |||||||||
| GLTSCR2 | 19 | 234339_s_at | 10.806 | 12.216 | −2.658 | 1.05 × 10−7 | cg16791686 | True | 0.307 | 0.159 | 0.148 | 3.36 × 10−5 | Glioma tumor suppressor candidate region gene 2 |
| 217807_s_at | 13.126 | 14.083 | −1.942 | 1.06 × 10−7 | |||||||||
| GPX1 | 3 | 200736_s_at | 9.846 | 11.311 | −2.760 | .000267 | cg06613840 | True | 0.442 | 0.177 | 0.265 | 2.46 × 10−8 | Glutathione peroxidase 1 |
| cg15900980 | True | 0.386 | 0.082 | 0.304 | 1.16 × 10−6 | ||||||||
| GRM8 | 7 | 216992_s_at | 4.346 | 4.896 | −1.464 | .002383 | cg09868882 | False | 0.532 | 0.267 | 0.265 | 4.34 × 10−8 | Glutamate receptor, metabotropic 8 |
| MAB21L1 | 13 | 206163_at | 3.443 | 4.138 | −1.619 | .037692 | cg05093686 | True | 0.538 | 0.207 | 0.271 | 4.86 × 10−7 | mab-21-like 1 |
| cg12029639 | True | 0.560 | 0.225 | 0.335 | 1.20 × 10−7 | ||||||||
| MBP | 18 | 209072_at | 6.539 | 7.032 | −1.407 | .000475 | cg12555907 | True | 0.841 | 0.411 | 0.43 | 2.61 × 10−8 | Myelin basic protein |
| NME4 | 16 | 212739_s_at | 8.007 | 8.658 | −1.570 | .012902 | cg18676162 | True | 0.321 | 0.178 | 0.143 | .027662 | Nonmetastic cells 4 |
| OSBPL10 | 3 | 231656_x_at | 5.979 | 6.995 | −2.023 | .002011 | cg15840985 | True | 0.329 | 0.170 | 0.159 | .00022 | Oxysterol binding protein-like 10 |
| RPS2 | 16 | 212433_x_at | 13.275 | 14.107 | −1.781 | .000313 | cg18279742 | True | 0.344 | 0.205 | 0.139 | .041014 | Ribosomal protein S2 |
| 203107_x_at | 13.825 | 14.543 | −1.645 | .000254 | |||||||||
| 217466_x_at | 10.499 | 11.145 | −1.564 | .001184 | |||||||||
| SOCS2 | 12 | 203373_at | 7.734 | 8.405 | −1.593 | .041561 | cg04797323 | True | 0.450 | 0.140 | 0.31 | .000257 | Suppressor of cytokine signalling 2 |
| cg06630241 | True | 0.637 | 0.285 | 0.352 | 7.18 × 10−10 | ||||||||
| cg11738543 | True | 0.353 | 0.103 | 0.25 | 5.05 × 10−5 | ||||||||
| cg23412850 | True | 0.330 | 0.059 | 0.271 | 4.24 × 10−6 | ||||||||
| ACADVL | 17 | 200710_at | 10.904 | 10.024 | 1.840 | .000336 | cg24825722 | False | 0.121 | 0.305 | −0.184 | .007423 | Acyl-CoA dehydrogenase, very long chain |
| cg21636577 | False | 0.221 | 0.472 | −0.251 | .000713 | ||||||||
| CTHRC1 | 8 | 225681_at | 12.349 | 8.766 | 11.984 | 4.57 × 10−5 | cg19188612 | True | 0.620 | 0.840 | −0.22 | .00053 | Collagen triple helix containing 1 |
| DNMT3A | 2 | 222640_at | 6.557 | 6.273 | 1.218 | .037539 | cg21629895 | False | 0.339 | 0.491 | −0.152 | .042813 | DNA methyl-transferase 3A |
| GALNT1 | 18 | 201724_s_at | 8.838 | 8.011 | 1.774 | .041426 | cg05714729 | False | 0.212 | 0.383 | −0.171 | .034844 | UDP-N-acetyl-alpha-D-galactosamine |
| GNPTG | 16 | 224887_at | 9.518 | 8.917 | 1.516 | .008847 | cg18146152 | True | 0.534 | 0.748 | −0.214 | .000131 | N-acetyl-glucosamine-1-phosphate transferase, gamma subunit |
| IRS2 | 13 | 209184_s_at | 7.470 | 6.309 | 2.236 | 4.05 × 10−5 | cg14341579 | True | 0.037 | 0.285 | −0.248 | .011963 | Insulin receptor substrate 2 |
| 209185_s_at | 7.711 | 5.849 | 3.636 | 4.90 × 10−6 | cg25802424 | True | 0.037 | 0.265 | −0.228 | .003687 | |||
| LRIG1 | 3 | 211596_s_at | 6.472 | 5.568 | 1.872 | .009466 | cg26131019 | True | 0.057 | 0.310 | −0.253 | .001657 | Leucine-rich repeats and Ig-like domains 1 |
| LRP12 | 8 | 220253_s_at | 4.404 | 3.418 | 1.981 | 1.74 × 10−8 | cg09531892 | True | 0.224 | 0.429 | −0.205 | .03994 | Low-density lipoprotein-related protein 12 |
| 219631_at | 5.758 | 4.230 | 2.883 | 8.52 × 10−7 | |||||||||
| PELI1 | 2 | 218319_at | 12.686 | 11.809 | 1.836 | .033075 | cg15309578 | False | 0.299 | 0.421 | −0.122 | .006695 | Pellino homolog 1 |
| 232213_at | 9.231 | 8.255 | 1.966 | .034372 | |||||||||
| PTPRA | 20 | 213795_s_at | 7.332 | 6.753 | 1.493 | .02016 | cg03115886 | False | 0.443 | 0.601 | −0.158 | .025021 | Protein tyrosine phosphatase receptor type A |
| PTPRCAP | 11 | 204960_at | 9.872 | 8.598 | 2.419 | .004979 | cg05751148 | False | 0.111 | 0.319 | −0.208 | .036243 | Protein tyrosine phosphatase receptor type C associated protein |
| cg20792833 | False | 0.191 | 0.465 | −0.274 | .003583 | ||||||||
| SBNO1 | 12 | 218737_at | 5.164 | 4.624 | 1.454 | .003605 | cg04398275 | False | 0.438 | 0.700 | −0.262 | 2.95 × 10−6 | Strawberry notch homolog 1 |
| SNRK | 3 | 209481_at | 8.785 | 8.049 | 1.666 | .009873 | cg04008913 | False | 0.171 | 0.314 | −0.143 | .031077 | SNF-related kinase |
| ZNF75A | 16 | 227670_at | 5.386 | 4.892 | 1.409 | .02804 | cg02825709 | True | 0.322 | 0.575 | −0.253 | .000649 | Zinc finger protein 75A |
Expression values are log-transformed and methylation numbers are β-values.
IGH indicates immunoglobulin heavy; and SNF, sucrose nonfermenting.
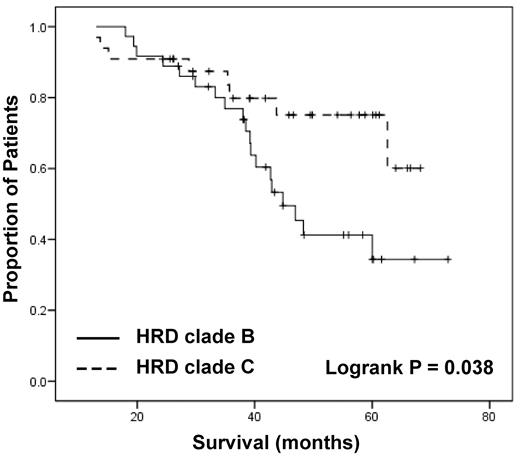
From unsupervised clustering of all samples we found 2 distinct groups of hyperdiploid samples, designated clades B and C (Figure 3). There was no disparity between the 2 groups with respect to the known cytogenetic markers, such as gain 1q or del(13q), which have been used by others to delineate hyperdiploid groups.4 When the overall survival (OS) of samples within clades B and C was analyzed, we found a significant difference (P = .03, median OS 44.8 vs > 70 months; Figure 5), indicating methylation may have both clinical and biologic effects within the hyperdiploid patients. A comparison between these 2 clades on the entire dataset gives 3174 differentially methylated probes, but most of these have a difference < 0.2, leaving 209 probes with a statistically significant difference in methylation. Of these, 11 are more heavily methylated in clade B, which has the poorer OS, and include CDKN2A and CDKN2B (cell-cycle inhibitors) and MAPT (microtubule associated protein).
Two groups of hyperdiploid samples are defined by methylation status, which has an impact on OS.
Two groups of hyperdiploid samples are defined by methylation status, which has an impact on OS.
Changes in methylation from myeloma to PCL
To more clearly delineate the changes in methylation pattern occurring from myeloma to PCL, we compared samples with the same translocation in the 2 disease states. By comparing t(4;14) samples at myeloma (n = 15) and PCL (n = 2) stages, we identified 618 probes (566 genes) as being significantly differentially methylated (supplemental data). These probes were exclusively hypermethylated in t(4;14) PCL compared with t(4;14) myeloma. When the same comparison was performed between t(11;14) myeloma (n = 35) and t(11;14) PCL (n = 3), we identified 566 probes (532 genes) that were differentially methylated, of which 560 were hypermethylated in PCL compared with myeloma. There were 71 genes commonly hypermethylated in both t(4;14) and t(11;14) PCL samples. Although the numbers of PCL samples are limited, the consensus interpretation of these analyses is that there is an increase in methylation of CpG dinucleotides in the promoters of genes at the transition from myeloma to PCL. Pathway analysis of the genes in which methylation levels increase indicates that cytokine-cytokine receptor interaction and Janus kinase/signal transducers and activators of transcription signaling pathways are affected.
Discussion
In this study we used a genome-wide array approach to interrogate the methylation status of more than 27 000 CpG sites. By using a selection of cell types relevant to the multistep pathogenesis of myeloma, we were able to determine the methylation changes that occur from normal PCs, through MGUS to presentation myeloma and PCL. We describe a clear distinction in methylation pattern between nonmalignant cells (B cells, normal PCs, and MGUS cells) compared with malignant PCs (presentation myeloma, PCL, and HMCLs). We also go on to show that the major differences in methylation profile are found at the transition of MGUS to myeloma and myeloma to PCL.
At the transition from MGUS to myeloma, the key feature is an overwhelming loss of methylation. Such global hypomethylation is associated with genome instability in many cancer cell types, including colorectal, gastric, breast, and chronic lymphocytic leukemia.45-48 Genome hypomethylation is frequently linked to altered chromatin structure, changes in DNA methyltransferase activity, loss of imprinting, and increased frequencies of copy number abnormalities. The resulting aberrant transcription and chromosomal instability within clones is likely to contribute to disease progression and is one of the critical differences distinguishing MGUS from myeloma. These results are not unique to the experimental approach used in this experiment and are consistent with a previous analysis of non-CpG element (long interspersed element-1, Alu, and SAT-α) methylation levels, which show decreased methylation levels in myeloma compared with controls.14
We also identified gene-specific hypermethylation at the transition of MGUS to myeloma involving 77 genes. Pathway analysis of the genes affected demonstrates involvement of developmental, cell cycle, and transcriptional regulatory pathways. The genes involved include CALCA, ONECUT2, GATA4, and CDKN2B but not CDKN2A or CDH1, all of which are known to be methylated in other cancer types.49-51 The analysis of differentially methylated genes at the transition from MGUS to myeloma did not identify genes that have been shown to be methylated previously by the use of methylation-specific PCR. However, upon inspection of raw data, we did find hypermethylation of some of these genes, including CDKN2A, CDH1, and DCC in myeloma samples, but the spread of data points across the samples resulted in a P value > .05 (supplemental data).
At the transition from myeloma to PCL, rather than finding further hypomethylation as may have been anticipated, we found further gene-specific hypermethylation, with 1802 genes showing an increase in methylation status. In particular we show remethylation of genes involved in cell signaling and cell adhesion pathways that would be consistent with a mechanism whereby adhesion to the specialized bone marrow niche is impaired, leading to bone marrow–independent growth, allowing the tumor to enter the circulation and proliferate more freely. However, these data were determined by a limited PCL sample cohort and require further investigation.
It is now widely accepted that there are 2 etiologic subgroups of myeloma defined by the presence of either an aberrant class switch recombination event or hyperdiploidy. The relationship between these etiologic subgroups and methylation status is important to understand. From unsupervised clustering of the 161 presentation myeloma samples, it was clear that several independent methylation profiles exist within multiple myeloma: a t(4;14) group, 2 separate t(11;14) groups, and 2 separate hyperdiploid groups. These findings are in contrast to other chromosomal abnormalities, such as del(1p32.1), gain 1q, del(13q), del(16q), del(17p), or del(22), which did not affect clustering of the samples. The most distinct of these methylation profiles belonged to the t(4;14) cytogenetic subgroup, which showed more frequent hypermethylation of genes compared with the other subgroups. Cases with a t(4;14) overexpress 2 potential oncogenes, MMSET and FGFR3, of which MMSET is of particular interest because it encodes a histone methyl transferase, which is known to methylate H3K36 and H4K20 residues and act as a transcriptional repressor.52,53 Pathway analysis of the genes hypermethylated in t(4;14) myeloma indicates a similar phenotype to that of PCL samples. The similarity in methylation profiles between t(4;14) myeloma and PCL suggest that methylation may significantly contribute to the more aggressive clinical phenotype seen in both disease subtypes.
There were a limited number of genes in the t(4;14) subgroup whose methylation status change correlated with gene expression changes. This limited correlation may reflect additional influences from other inactivating methods such as deletions, mutations (through nonsense-mediated decay), and upstream activation or silencing of transcription factors. This is especially true of cell cycle inhibitors such as CDKN2A and CDKN2B, which are also known to be deleted in myeloma samples.5 In particular we have observed deletions of CDKN2C, located at 1p32, which do not correlate completely with loss of gene expression. However, this finding mostly seems to reflect the fact that expression of this gene is lost in > 95% of myeloma samples, presumably through alternate mechanisms.54 We believe that the correlation between methylation and gene expression will be equally complex.
An interesting observation in this study is our description of 2 specific subgroups of hyperdiploid myeloma on the basis of their methylation profile. The TC classification of myeloma, although a significant step forward, arguably does not adequately address the hyperdiploid group. In particular, although the translocation subgroups have a distinct clinical outcome, the hyperdiploid cases, amounting to 50% of the total, are apparently homogeneous in terms of their clinical outcome. Other groups have delineated groups of hyperdiploidy by using mapping or expression array data. These analyses have been able to separate hyperdiploidy into groups on the basis of the presence of 1q+,11+, nuclear factor-κB deregulation, proliferation, or changes in the expression of cancer testis antigens.4,55 Here we show that it is possible to split hyperdiploid samples into 2, on the basis of their methylation profiles, and that each of these groups has a significant difference in OS. These 2 groups are independent of cyclin D expression levels, cytogenetic abnormalities, and of presenting clinical features. Interestingly, we did not find a difference in the methylation status of nuclear factor-κB, proliferation, or cancer testis antigens between the 2 groups, which may have led to the expression changes seen by other groups.
It is important to understand and develop models of how methylation changes may mediate the progressive transformation process from MGUS to myeloma. Although it is clear from this analysis that genome-wide hypomethylation occurs at the transition from MGUS to myeloma, it is not so clear when gene-specific hypermethylation occurs. Hyperdiploidy and the main translocation groups [with the exception of t(4;14)] are present at similar frequencies in MGUS and presentation myeloma. Because these cytogenetic abnormalities are the main defining feature of the methylation subgroups, it may mean that these methylation groups also exist in MGUS cells, but we have not been able to analyze sufficient cases to demonstrate this adequately. At the transition from MGUS to presentation, myeloma secondary hits occur, which result in genome hypomethylation. Whether these hits are mutations of genes controlling DNA methylation, such as DNA methyltransferases, or activation of transposable elements remains to be determined.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the staff at the Haematological Malignancy Diagnostic Service, Leeds; the LRF UK Myeloma Forum Cytogenetics Group, Salisbury; the Clinical Trials Research Unit, Leeds, United Kingdom; and the Cambridge Genomic Services, University of Cambridge, United Kingdom, for processing of methylation arrays.
Research grants and financial support were received from the Leukemia Research Fund, Cancer Research UK, the Bud Flanagan Research Fund, the United Kingdom Department of Health, Myeloma UK, Associazione Italiana Ricerca sul Cancro, and the Biological Research Center of the National Institute for Health Research at the Royal Marsden Hospital.
Authorship
Contribution: B.A.W. designed and performed research, analyzed data, and wrote the paper; C.P.W. analyzed data; L.C. performed research and analyzed data; E.M.S. performed research; K.D.B. performed research; A.N. provided biomaterial; F.E.D. designed research; F.M.R. designed and performed research and analyzed data; and G.J.M. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Brian Walker, Section of Haemato-Oncology, The Institute of Cancer Research, 15 Cotswold Rd, London, SM2 5NG, United Kingdom; e-mail: brian.walker@icr.ac.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal