Abstract
A recent clinical trial for adrenoleukodystrophy (ALD) showed the efficacy and safety of lentiviral vector (LV) gene transfer in hematopoietic stem progenitor cells. However, several common insertion sites (CIS) were found in patients' cells, suggesting that LV integrations conferred a selective advantage. We performed high-throughput LV integration site analysis on human hematopoietic stem progenitor cells engrafted in immunodeficient mice and found the same CISs reported in patients with ALD. Strikingly, most CISs in our experimental model and in patients with ALD cluster in megabase-wide chromosomal regions of high LV integration density. Conversely, cancer-triggering integrations at CISs found in tumor cells from γretroviral vector–based clinical trials and oncogene-tagging screenings in mice always target a single gene and are contained in narrow genomic intervals. These findings imply that LV CISs are produced by an integration bias toward specific genomic regions rather than by oncogenic selection.
Introduction
Stable genetic modification of hematopoietic stem/progenitor cells (HSPCs) is achieved with retroviral vectors (RVs) that integrate into the cell genome and express a therapeutic transgene.1 Transplantation of genetically modified autologous HSPCs provides a therapeutic option for patients with genetic disorders.1-3 However, in clinical trials for X-linked severe combined immunodeficiency (X-SCID) and chronic granulomatous disease (CGD) oncogenesis triggered by γRV-mediated insertional mutagenesis has occurred. Leukemic or myelodysplastic cell clones in patients from these trials harbored RV integrations at common insertion sites (CISs) targeting recurrently LMO2 or MDS1-EVI1, PRDM16, SETBP1, and other genes.4-7
Alternative to γRVs, HIV-derived self-inactivating lentiviral vectors (LVs) transduce human HSPCs efficiently and display a superior safety profile with respect to γRVs as shown in in vitro and in vivo preclinical mouse models.8-11 Moreover, good efficacy and safety of LVs has also been documented in a recent HSPC-based clinical trial for X-linked adrenoleukodystrophy (ALD).3 However, a careful LV integration site analysis in derived cells from patients with ALD showed that relevant numbers of CISs were present.3 This observation raises concerns12 because the detection of CISs is a well-established hallmark of insertional mutagenesis in mice13,14 and clinical trials.5,7,15 Thus, it is possible that the occurrence of CISs in the ALD clinical trial is a still silent effect of genotoxicity. To understand whether CISs generated by LV integrations are the product of genotoxicity we generated our own dataset of LV integrations in human HSPCs and their progeny after engraftment in immunodeficient mice and studied the integration pattern and the clonal repertoire of vector-marked cells in in vitro culture and in vivo. Moreover, we performed an extensive comparison between our dataset and the integrations found in the ALD clinical trial and in other gene therapy trials that reported insertional leukemogenesis, as well as in mice subjected to RV-mediated oncogene tagging. From our own integration data and the meta-analysis of the other integration datasets we provide evidence that the driving force leading to the appearance of CISs in LV-transduced HSPCs from the ALD clinical trial reflects a previously unappreciated bias of LVs in integration site selection rather oncogenic selection.
Methods
LV production and isolation and transduction of human HSPCs
LV.ARSA (arylsulfatase A) and LV.GFP (green fluorescent protein) were produced with the use of the pCCLsin.cPPT.hPGK.hARSA.WPREmut6 and the pCCLsin.cPPT.hPGK.GFP.Wpre transfer plasmids.16 Vesicular stomatitis virus–pseudotyped LV-concentrated stocks were produced and titered as described.17 Human HSPCs were obtained by positive selection of CD34-expressing cells (CD34 progenitor cell isolation kit, MACS; Miltenyi Biotec) from BM aspirates, mobilized peripheral blood (MPB), or CB of healthy donors on collection with informed consent in accordance with the Declaration of Helsinki (TIGET01 protocol, approved by San Raffaele Scientific Institute Ethical Committee). Alternatively, purified CD34+ cells from BM of healthy donors were provided by Lonza (Human Bone Marrow CD34+ Progenitors 2M-101; Lonza). Soon after purification or thawing, cells were placed in culture on retronectin-coated wells (T100A Takara) in CellGro SCGM medium (2001 CellGenix) at a concentration of 1-1.5 × 106 cells/mL in the presence of a standard cocktail of cytokines18 for 24-48 hours of prestimulation. Cells were then transduced with LV.ARSA or LV.GFP (at a MOI of 100-200) for 12 hours. One or 2 hits of transduction were performed. At the end of transduction, cells were counted and collected for clonogenic assays, flow cytometry, and in vivo studies. Remaining cells were plated in IMDM/10% FBS with cytokines (IL-3, 60 ng/μL; IL-6, 60 ng/μL; SCF, 300 ng/μL) and cultured for a total of 14 days. Thereafter, cells were collected for molecular and flow cytometric studies.
Rag2−/−Il2rg−/− mice transplantation and engraftment evaluation
Rag2−/−Il2rg−/− mice were obtained from the Central Institute for Experimental Animals, Nogawa, Japan, and maintained in our animal facility according to approved protocols. Three-day-old mice were sublethally irradiated (450 cGy) 24 hours before intravenous injection of untransduced and unmanipulated or transduced CD34+ cells. HSPC transduction was performed as described in “LV production and isolation and transduction of human HSPCs.” Ten to 12 weeks after transplantation, mice were killed; BM, spleen, and thymus were collected; and multicolor cytofluorimetric analysis to assess human cell engraftment and differentiation was performed as previously described.16 All procedures were performed according to protocols approved by the Animal Care and Use Committee of the San Raffaele Scientific Institute (IACUC no. 325 and no. 353) and communicated to the Ministry of Health and local authorities according to Italian law.
Quantitative PCR
Genomic DNA was extracted from CD34+ liquid culture samples with QIAamp DNA Blood Mini Kit (QIAGEN), and from murine tissues with the Blood & Cells DNA Midi Kit (QIAGEN) after o/n digestion with proteinase K (Roche). LV sequences were detected by quantitative PCR on 50 ng of total genomic DNA (primers and probe: forward, 5′-TAC TGA CGC TCT CGC ACC-3′; reverse, 5′-TCT CGA CGC AGG ACT CG-3′; probe, 5′-FAM-ATC TCT CTC CTT CTA GCC TC-MGB) and normalized on the hTert gene (primers and probe: forward, 5′-GGA CAC GTT ATT TAC CCT GTT TCG-3′; reverse, 5′-GGT GAA CCT CGT AAG TTT ATG CAA-3′; probe, 5′-VIC-TCA GGA CGT CGA GTG GAC ACG GTG-TAMRA-3′). Absolute quantifications were plotted on standard curves prepared with serial dilutions of genomic DNA from the CEMA301 clone.19 Vector copy number (VCN) was then calculated as described.20
Linear amplification mediated–PCR and genomic integration site analysis
The sequences of linear amplification mediated (LAM)–PCR primers and procedures for LV integration site retrieval have been previously described.10,21 Briefly, 10-100 ng of genomic DNA was used as template for LAM-PCR and initiated with a 50-cycle linear PCR and restriction digest with the use of Tsp509I or HpyCH4IV and ligation of a restriction site–complementary linker cassette. The first exponential biotinylated PCR product was captured by magnetic beads and reamplified by a nested PCR. LAM-PCR products were separated by Spreadex gel electrophoresis (Elchrom Scientific) to verify the presence and number of bands. LAM-PCR was shotgun cloned into the TOPO TA vector (Invitrogen) and sequenced by Sanger sequencing (GATC Biotech) or directly sequenced by 454 pyrosequencing after a PCR reamplification with the use of oligonucleotides with specific 4-8 nucleotide sequence tags for sample identification. Sequences were validated and classified with specific PERL scripts and aligned to the human genome (freeze March 2006; University of California Santa Cruz; UCSC) or with the use of the UCSC BLAT genome browser.22 Genes targeted by vector integrations were considered those nearest to the integration site.
Statistical analysis
Statistical analyses were made by one-way ANOVA for repeated measurements with the use of Bonferroni test for post hoc analysis after significant main effect of the treatment (95% confidence interval) or with Student t test (95% confidence interval). Overrepresented on gene ontology classes were identified by the DAVID-EASE software 2.0 (http://david.abcc.ncifcrf.gov/home.jsp) with the use of the stringency setting “high.“ Differences in targeting frequency at gene classes between in vitro and in vivo datasets or between datasets originated by BM-, MPB-, and CB-derived CD34+ cells were scored by the Fisher exact test with the use of the GraphPad Prism Software Version 5.03. Automated generation and statistical evaluation of the chromosomal frequency distributions and the distributions around CIS centers of the different integration datasets were performed with ad hoc scripts with the use of R statistical software Version 2.10.1.
Grubbs test for outlier analysis was performed with the online “Outlier Calculator” tool (http://www.graphpad.com/quickcalcs/Grubbs1.cfm) with the use of the values of gene integration frequency corrected by the gene size to which was added a constant value of 100 Kb (calculated with the UCSC Hg18 RefSeq genomic coordinates for transcript start and end ± 50 Kb).
For all statistical comparisons unless otherwise specified, P < .05 was considered significant.
Results
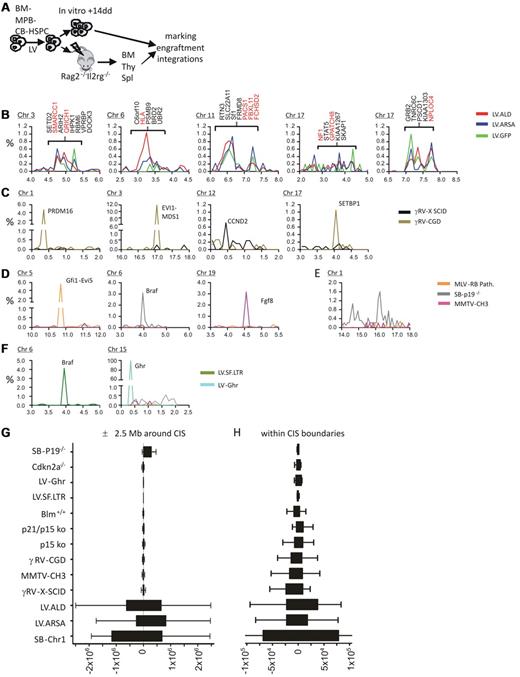
We analyzed the integration profile of different LVs expressing therapeutic20 or GFP transgenes in human CD34+ HSPCs in vitro and in vivo after engraftment in Rag2−/−Il2rg−/− mice that received a transplant (Figure 1A; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). More than 2300 unique genomic integration sites were mapped by LAM-PCR21 and high-throughput 454 pyrosequencing from 5 distinct in vitro samples and 13 mice that received a transplant (supplemental Table 1). LVs displayed the expected tendency to integrate within genes (data not shown). No enrichment in preferentially targeted gene classes was found from in vitro to in vivo (supplemental Figure 2). Adopting previously described statistical criteria,23 31 CISs were identified in our datasets (supplemental Table 2), accounting for 5.2%-8.6% of all integrations. No enrichment in CISs was found from the in vitro to the in vivo condition (supplemental Table 2). Interestingly, 85% (11 of 13) of the CISs hit by ≥ 4 integrations found in our dataset from human mouse hematochimeras matched the CISs identified in the ALD clinical trial (Table 1). Moreover, five 10-20 Mb–wide genomic regions containing ≥ 5 CISs, among which ≥ 1 overlapped with the integration dataset from the ALD clinical trial. Strikingly, ≤ 76% of all LV integrations at CISs were clustered in these 5 genomic regions (accounting for 2.5% of the human genome) located in chromosomes 3, 6, 11, and 17 (2 regions) (Figure 1B; supplemental Figure 2A; supplemental Table 3). This LV CIS distribution is clearly different from those obtained with analyzing well-characterized oncogenic CISs from γRV-based clinical trials for X-SCID7 and CGD5 (674 and 760 integrations analyzed, respectively) and oncogene-tagging screenings in mice14,24,25 with γ-retroviruses and Sleeping Beauty (SB) transposon (21 511 integrations analyzed from 17 studies; Figure 1C-D; supplemental Figure 3B-C). Indeed, oncogenic CISs always appeared as isolated and sharp peaks in the frequency distribution of integrations along the chromosomes. The only exception was found in SB transgenic mice that showed on chromosome 1 a higher integration frequency over a wide genomic region near the transposon concatemer (Figure 1E); this enrichment, however, is caused by a “local hopping” effect and not by oncogenic selection. Considering the distribution of integrations mapping in a 5 Mb-wide genomic region centered on each CIS, we found that, in the case of genotoxic CISs, 90% of integrations were located, at most, in a central 100-Kb region (n = 50 CISs analyzed), whereas LV integrations were evenly spread over the whole region (n = 81 LV CISs; Figure 1G; supplemental Figure 4; P value ranging from 2 × 10−16 to 7.7 × 10−5, according to bin size and comparison group; Fisher exact test). These findings indicate that LV CISs are embedded in wide regions of high-integration density. Interestingly, the distribution of LV CIS integrations within the boundaries of the CIS itself was significantly more spread than for oncogenic CISs (supplemental Figure 4C). The tight clustering of integrations at genotoxic CISs suggests that selectable oncogene activation events, independently from the type of vector used, may occur preferentially with integrations targeting specific genomic regions. Probably, integrations in close proximity of regulatory regions or within specific introns favor oncogene overexpression or the formation of aberrantly spliced oncogenic proteins. To test the validity of this rule also for LVs, we induced hematopoietic tumors by injecting a previously described genotoxic LV10 in Cdkn2a−/− mice and analyzed the integrations in tumors. The CISs generated in this LV-based insertional mutagenesis model, as well as in another in vitro study,26 displayed the same narrow clustering pattern described above for genotoxic CISs (as shown for the top-ranking CIS, the Braf oncogene; Figure 1F). Of note, cell clones harboring LV CIS integrations were not preferentially enriched in vivo and did not predominate over other repopulating cells, both in our study here (Figure 2) and in the ALD clinical trial.3
Identification of LV CISs in human HSPCs from hematochimeric mice and ALD clinical trial and comparative analysis of integration distribution within the CISs and in the surrounding chromosomal regions in datasets with documented insertional mutagenesis events. (A) Experimental strategy for LV integration site profiling in human CD34+ HSPCs derived from BM, MPB, and CB. On ex vivo transduction with LVs expressing a therapeutic (arylsulfatase A; LV.ARSA) or marker (LV.GFP) gene, cells were transplanted into immunodeficient mice (Rag2−/−Il2rg−/−), and a portion was cultured in vitro for 14 days. BM, thymus (Thy), and spleen (Spl) from mice that received a transplant were harvested 12 weeks after transplantation. Vector copy number, engraftment, and integration site analysis were then performed on the available samples. (B) Frequency distributions of LV integrations at 5 chromosomal regions targeted at high frequency. The bin size used for the chromosomal distributions is 1 Mb. The y-axis is the percentage of the total integrations of each dataset; the x-axis is chromosomal coordinates in megabase ×10. Genes at CIS locations are indicated for the ALD dataset and in red when common between the ALD and our datasets from panel A. (C) Frequency distributions of γRV integrations surrounding validated genotoxic CISs found in X-SCID and CGD clinical trials. (D) Frequency distributions of γ-retroviruses or SB transposon integrations surrounding validated genotoxic CISs found in tumors generated in different insertional mutagenesis studies. (E) Frequency distribution of SB transposon integrations at chromosome 1 near the transposon concatemer locus in transgenic mice. (F) Frequency distribution of genotoxic LV integrations targeting (left) Braf in hematopoietic tumors from Cdkn2a−/− mice and (right) the Ghr gene in IL-3–independent cell clones from Bokhoven et al.26 (G-H) Distribution of vector integrations around CIS centers. (G) Tukey box-and-whisker graph representing the distance of vector integrations from the center of CISs found in each dataset in a ± 2.5-Mb region (x-axis, units in base pair). (H) Tukey box-and-whisker graph representing the distance of vector integrations from the center of each CIS within the CIS interval. The center of each CIS was calculated as the position closest to the highest number integrations within the CIS interval. The tighter clustering of genotoxic integrations within CIS boundaries, although suggestive of positional constrains for cancer gene–activating integrations, it does not test if the integration frequency at the CIS is significantly different with respect to other regions and therefore cannot be used to discriminate between different CIS types.
Identification of LV CISs in human HSPCs from hematochimeric mice and ALD clinical trial and comparative analysis of integration distribution within the CISs and in the surrounding chromosomal regions in datasets with documented insertional mutagenesis events. (A) Experimental strategy for LV integration site profiling in human CD34+ HSPCs derived from BM, MPB, and CB. On ex vivo transduction with LVs expressing a therapeutic (arylsulfatase A; LV.ARSA) or marker (LV.GFP) gene, cells were transplanted into immunodeficient mice (Rag2−/−Il2rg−/−), and a portion was cultured in vitro for 14 days. BM, thymus (Thy), and spleen (Spl) from mice that received a transplant were harvested 12 weeks after transplantation. Vector copy number, engraftment, and integration site analysis were then performed on the available samples. (B) Frequency distributions of LV integrations at 5 chromosomal regions targeted at high frequency. The bin size used for the chromosomal distributions is 1 Mb. The y-axis is the percentage of the total integrations of each dataset; the x-axis is chromosomal coordinates in megabase ×10. Genes at CIS locations are indicated for the ALD dataset and in red when common between the ALD and our datasets from panel A. (C) Frequency distributions of γRV integrations surrounding validated genotoxic CISs found in X-SCID and CGD clinical trials. (D) Frequency distributions of γ-retroviruses or SB transposon integrations surrounding validated genotoxic CISs found in tumors generated in different insertional mutagenesis studies. (E) Frequency distribution of SB transposon integrations at chromosome 1 near the transposon concatemer locus in transgenic mice. (F) Frequency distribution of genotoxic LV integrations targeting (left) Braf in hematopoietic tumors from Cdkn2a−/− mice and (right) the Ghr gene in IL-3–independent cell clones from Bokhoven et al.26 (G-H) Distribution of vector integrations around CIS centers. (G) Tukey box-and-whisker graph representing the distance of vector integrations from the center of CISs found in each dataset in a ± 2.5-Mb region (x-axis, units in base pair). (H) Tukey box-and-whisker graph representing the distance of vector integrations from the center of each CIS within the CIS interval. The center of each CIS was calculated as the position closest to the highest number integrations within the CIS interval. The tighter clustering of genotoxic integrations within CIS boundaries, although suggestive of positional constrains for cancer gene–activating integrations, it does not test if the integration frequency at the CIS is significantly different with respect to other regions and therefore cannot be used to discriminate between different CIS types.
CIS genes targeted multiple times (N Hits) within the integration datasets
| No. of hits . | Genes (interval in Kb) . |
|---|---|
| 4 | CARD8 (23), NSD1 (90), QRICH1 (31), SAPS2 (47), USP48 (48) |
| 5 | GPATCH8 (53) |
| 6 | FCHSD2 (272) |
| 7 | NPLOC4 (76), SMARCC1 (95), NF1 (140) |
| 8 | PACS1 (110), HLA (542) |
| 9 | FBXL11 (107) |
| No. of hits . | Genes (interval in Kb) . |
|---|---|
| 4 | CARD8 (23), NSD1 (90), QRICH1 (31), SAPS2 (47), USP48 (48) |
| 5 | GPATCH8 (53) |
| 6 | FCHSD2 (272) |
| 7 | NPLOC4 (76), SMARCC1 (95), NF1 (140) |
| 8 | PACS1 (110), HLA (542) |
| 9 | FBXL11 (107) |
The maximum distance between integrations targeting the same CIS is indicated. Note that only CIS genes targeted by ≥ 4 integrations are shown. With the exception of SAPS2 and USP48, all other CIS genes found in our experimental dataset matched the CISs of the ALD clinical trial. See supplemental Table 2 for further details on the identified CISs.
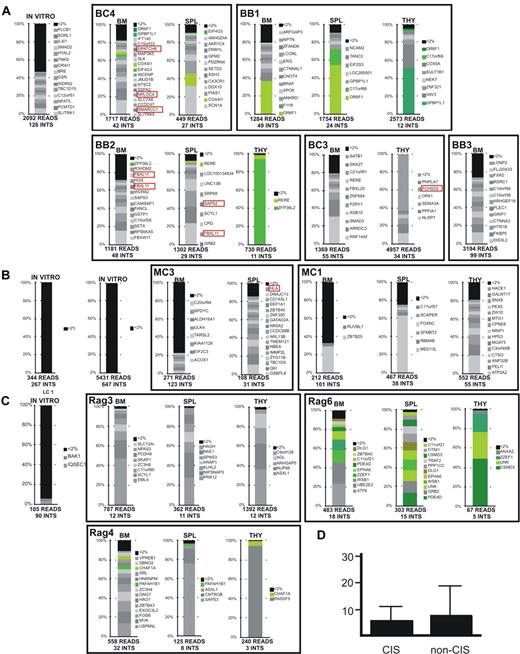
Relative retrieval frequency of sequencing reads of integration sites in in vitro and in vivo samples. Retrieval frequency of sequencing reads corresponding to a unique LV integration site from the in vitro culture and the indicated organs of mice that received a transplant with CD34+ HSPCs derived from BM (A), MPB (B), and CB (C) cells. Within a red box are represented the integrations at CISs (considered only CISs constituted by ≥ 4 integrations). LAM-PCR products were sequenced by 454-pyrosequencing or Sanger chemistry. Each bar shows the percentage of reads for each integration site in the sample dataset. The total number of reads and of unique integration sites (INTS) in each sample dataset is given. Integrations represented by < 2% of the total reads in the dataset were pooled and shown in black at the top of each bar (< 2%). Integrations represented by > 2% of the total sequencing reads are shown individually with the symbol of the targeted gene. Identical integrations found in different organs of the same mouse are shown in green. (D) The averaged percentage of sequencing reads representing CIS integrations and non-CIS integrations was not statistically different.
Relative retrieval frequency of sequencing reads of integration sites in in vitro and in vivo samples. Retrieval frequency of sequencing reads corresponding to a unique LV integration site from the in vitro culture and the indicated organs of mice that received a transplant with CD34+ HSPCs derived from BM (A), MPB (B), and CB (C) cells. Within a red box are represented the integrations at CISs (considered only CISs constituted by ≥ 4 integrations). LAM-PCR products were sequenced by 454-pyrosequencing or Sanger chemistry. Each bar shows the percentage of reads for each integration site in the sample dataset. The total number of reads and of unique integration sites (INTS) in each sample dataset is given. Integrations represented by < 2% of the total reads in the dataset were pooled and shown in black at the top of each bar (< 2%). Integrations represented by > 2% of the total sequencing reads are shown individually with the symbol of the targeted gene. Identical integrations found in different organs of the same mouse are shown in green. (D) The averaged percentage of sequencing reads representing CIS integrations and non-CIS integrations was not statistically different.
The canonical statistical approaches for the identification of biologically significant CISs contained in a given genomic interval assume that integrations are randomly distributed along the genome.23,27 To correct the significance of CISs for biases of vector genomic integration, we devised an additional CIS validation step that takes into account the relative frequency of integration at the genomic region surrounding the CIS interval. We decided that the best approach was to measure and compare the integration frequency within the genomic intervals defined by transcription units rather than the entire flanking genomic intervals that may contain large intergenic regions without integrations. Therefore, our analysis is focused on the comparison of integration frequencies at genes, some of which are the culprits of oncogenesis and the preferred targets of different vector platforms. In our rationale, for a CIS to be considered the result of genetic selection (genotoxic), the integration frequency at the CIS target-gene interval must be high enough to be considered a significant outlier with respect to the integration frequency at other genes contained in the flanking genomic regions (genes targeted at least by 1 integration). However, if the integration frequency at the CIS target-gene is not statistically different from the integration frequency of other flanking genes, it will imply that the CIS is embedded in a wider region of similar integration frequency and thus probably the product of a vector-specific integration bias. The gene integration frequency is defined as the ratio between the number of integrations targeting a given gene and its size. To determine whether the integration frequency at a CIS target-gene is high enough to be considered a significant outlier with respect to other genes contained in the surrounding regions, we performed the Grubbs test for outliers (supplemental Statistical Material). We applied the Grubbs test for outliers to the gene integration frequencies of 9 γRV CISs reported in the X-SCID and CGD clinical trials, the CISs targeting Braf both in mouse Cdkn2a−/− LV.SF.LTR-marked histiocytic sarcomas and in SB transposon–marked Arf−/− sarcomas, and finally on the several LV CISs identified in the 5 genomic regions in the ALD clinical trial and in the human/mouse hematochimeras in this study (Figure 3; supplemental Statistical Material).
Graphical representation of the Grubbs test for outliers results on gene integration frequencies in ∼ 10-20 Mb genomic regions around CIS. The y-axis is the ratio Z that measures how distant is the integration frequency of a given gene with respect to the average of all genes analyzed (genes targeted by ≥ 1 integration contained within the specified genomic interval). The x-axis is the chromosomal position of the gene (coordinates in base pairs). A negative or positive ratio Z value implies that the gene is targeted at a frequency below or above the average, respectively. The red lines indicate the threshold beyond which the values can be considered significantly different. The red triangles indicate genes considered as CISs in previous publications with the use of the classic statistical approach (in parentheses the number of integrations targeting each gene). (A) Two examples of ratio Z of gene integration frequency at genomic regions around CISs from γRV-based X-SCID and CGD clinical trials. LMO2 (targeted by 7 integrations) and CCND2 (7 integrations) appear to be targeted at a significantly higher frequency by this test (see supplemental Statistical Material for analyses of other CISs from the same clinical trials). (B) Ratio Z of gene integration frequency at the genomic region around Braf (68 integrations) and the neighboring genes in histiocytic sarcomas form Cdkn2a−/− mice injected with LV.SF.LTR. (C) A very similar integration profile is found at Braf (24 integrations) in sarcomas from Arf−/− SB transposon/transposase transgenic mice. (D) Ratio Z representation of 2 genomic regions at LV CISs in common between the ALD clinical trial and the hematochimeric model show that none of the identified CISs is a significant outlier.
Graphical representation of the Grubbs test for outliers results on gene integration frequencies in ∼ 10-20 Mb genomic regions around CIS. The y-axis is the ratio Z that measures how distant is the integration frequency of a given gene with respect to the average of all genes analyzed (genes targeted by ≥ 1 integration contained within the specified genomic interval). The x-axis is the chromosomal position of the gene (coordinates in base pairs). A negative or positive ratio Z value implies that the gene is targeted at a frequency below or above the average, respectively. The red lines indicate the threshold beyond which the values can be considered significantly different. The red triangles indicate genes considered as CISs in previous publications with the use of the classic statistical approach (in parentheses the number of integrations targeting each gene). (A) Two examples of ratio Z of gene integration frequency at genomic regions around CISs from γRV-based X-SCID and CGD clinical trials. LMO2 (targeted by 7 integrations) and CCND2 (7 integrations) appear to be targeted at a significantly higher frequency by this test (see supplemental Statistical Material for analyses of other CISs from the same clinical trials). (B) Ratio Z of gene integration frequency at the genomic region around Braf (68 integrations) and the neighboring genes in histiocytic sarcomas form Cdkn2a−/− mice injected with LV.SF.LTR. (C) A very similar integration profile is found at Braf (24 integrations) in sarcomas from Arf−/− SB transposon/transposase transgenic mice. (D) Ratio Z representation of 2 genomic regions at LV CISs in common between the ALD clinical trial and the hematochimeric model show that none of the identified CISs is a significant outlier.
The approach does find significant ratio Z outliers for γRV CISs at LMO2, CCND2, RUNX1, EVI1-MDS1, SETBP1, and PRDM16 (targeted, respectively by 7, 9, 5, 94, 9, and 37 integrations) from the X-SCID and CGD clinical trials (Figure 3A; supplemental Statistical Material). Other γRV CISs at EGR (4 integrations), BCL2 (4 integrations), and BACH2 (5 integrations) did not appear to be targeted at a significantly higher frequency with respect to flanking genes. This approach is not influenced by the integration site selection of the different vector platforms as identifies the genotoxic murine CISs at Braf targeted both in LV.SF.LTR-induced Cdkn2a−/− histiocytic sarcomas and in SB transposon–induced Arf−/− sarcomas (Figure 3B-C).
However, LV CISs from the ALD trials and our human/mouse hematochimeras, even if targeted by high numbers of integrations (eg, 29, 27, and 19 integrations, respectively, targeting PACS1, FBXL11, and TNRC6C) did not show a significantly higher targeting frequency with respect to flanking genes that were also identified as CISs by the canonical statistical analysis (Figure 3D; supplemental Statistical Material).
Discussion
We showed that the genomic integration profile of LVs expressing therapeutic and marker transgenes in human hematopoietic cells engrafted in immunodeficient mice is remarkably similar to the integration profile observed in LV-treated patients with ALD. These data indicate that xenotransplantation models are a valid surrogate for the study of integration profiles of LVs in human HSPCs in vivo. Moreover, the LV CISs found in our datasets overlapped for the 85% with those reported in the ALD clinical trial.
Although the well-established role of CISs in genotoxicity has fueled concerns about the safety of LV CISs identified in the ALD clinical trial,12 our analysis highlights important differences with respect to the known genotoxic CISs identified in malignant cell clones from mouse oncogene-tagging screenings or γRV-based clinical trials. Indeed, the LV CISs in the ALD clinical trial and in our human/mouse hematochimeras clustered in the megabase-wide genomic regions with an overall higher integration frequency with respect to other chromosomal regions. Differently, genotoxic CISs are distributed along chromosomes as isolated sharp peaks and always targeting a single gene, the culprit of oncogenesis. Because the features characterizing genotoxic CISs are consistent across different vector platforms (retroviruses, γRV, transposons, and genotoxic LV.SF.LTR) and tumor types (hematopoietic and mammary), it suggest that a different mechanism, other than genetic selection, may drive the formation of LV CISs in our preclinical and ALD studies. In support to this notion, it is unlikely that genetic selection would preferentially favor integrations deregulating cancer genes clustered in specific genomic intervals and not the many other well-known oncogenes spread along the genome.
Moreover, differently than the known genotoxic CIS integrations marking leukemic or dominant cell clones,5,28,29 the LV CIS integrations in the ALD clinical trial and our human/mouse hematochimeras are not enriched from in vitro to in vivo conditions, or during time after transplantation, and are not overrepresented (dominant) with respect to other integrations. Note, however, that without further experimental evidence, it is not possible to formally exclude that any of the CIS integrations or even any integration of the dataset, regardless the CIS status or the type of targeted gene, could be the result of selection.
Our findings highlight also the need of more stringent statistical tools for interpreting the presence of CISs identified in future clinical trials. Canonical CIS statistics assume that integrations are distributed randomly across the genome and do not take in account the integration biases intrinsic to a given vector. Therefore, CISs in genomic regions targeted at high frequency will be considered identical to those in which only one gene is targeted at high frequency. Alternative statistical methods for CIS validation should consider the size of the datasets analyzed and the local genomic integration biases. We developed a new approach for the validation of CIS significance on the basis of the comparison of the integration frequency at the CIS gene with respect to other genes contained in the surrounding genomic regions. With the use of the Grubbs test for outliers we were able to distinguish well-validated genotoxic CISs generated with the use of 3 different vector platforms (genotoxic LV, SB transposon, and γ-retrovirus/RV). Some CISs from the γRV-based clinical trials were not found to be outliers by this test. Whether the lack of significance is because of a low sensitivity of this specific test or to a true lack of genotoxicity is unclear. However, the LV CISs in the ALD clinical trial and our human/mouse hematochimeras mapped in 10-20 Mb–wide chromosomal regions together with other genes also targeted by CISs at a similar integration frequency and were not found to be outliers by this test. On the basis of our rationale, a CIS cannot be considered a significant outlier when in the same 10-20 Mb–wide chromosomal region multiple CISs are present with a similar integration frequency. The reasons why CIS target genes display a higher integration frequency with respect to others genes within the same interval remain obscure. Possibly, cellular protein-mediated tethering of the lentiviral preintegration complex at gene-dense genomic regions with high transcriptional activity could be responsible for the observed LV integration preferences and CIS formation in our pr-clinical and ALD studies.30-44 More refined statistical methods capable of detecting multiple outliers within a population of values may be required to pinpoint multiple genes targeted at a significantly higher frequency with respect to the average gene integration frequency (eg, using the Chauvenet criterion, Peirce criterion, Bayesian models, and others).45,46
One of the strengths of our outlier-detection approach is that it takes advantage of the integration pattern originating from the same vector-specific dataset and in similar experimental conditions to perform statistical comparisons with respect to the flanking genomic regions, without the need of random- or neutral-control integration profiles. This is important because vector-specific integration profiles from in vitro or nonleukemic cells from patients who received a transplant cannot be formally assumed as neutral, because genotoxic integrations may be selected in vitro or may be present in “normal” hematopoietic cells in vivo before full-blown neoplastic transformation occurs. Moreover, in future studies it will be useful to study larger γRV integration datasets28,47,48 to possibly improve the strength of the analyses and to perform comparisons with CISs identified by Kernel deconvolution-based and k-mean clustering analysis methods.49,50
Overall, our findings highlight a previously unappreciated feature of LV integration that invalidates the predictive value on genotoxicity of standard CIS statistics for this class of vectors. Moreover, our meta-analysis provides a way to distinguish alarming CISs originating from gain-of-function mutations from those probably originating from biases in integration site selection. The lack of evident signs of genotoxicity during the 2-year follow-up of the ALD clinical trial and in our human/mouse hematochimeras, the widespread distribution of LV integrations at and around CISs, and the integration preference for specific genomic regions altogether suggest that the LV CIS integrations found in the ALD clinical trial are probably the result of an intrinsic integration bias toward selected megabase-wide genomic regions in HSPCs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Lucia Sergi Sergi for LV production and Laura Lorioli, Rossano Cesari, and Anna Zingale for help with quantitative PCR analysis.
This work was supported by grants from the Association for International Cancer Research (AICR 09-0784; E.M.), EU Clinigene NoE (LSHB-CT-2006-018933; E.M.), Italian Telethon (TIGET grants; A.B., L.N., and E.M.), EU (HEALTH-2009-222878, PERSIST; L.N.), the Italian Ministries of University and Research and of Health (L.N.), and the European Leukodystrophy Association (ELA; A.B. and L.N.).
Authorship
Contribution: A.B. designed the research, analyzed data, and wrote the manuscript; C.C.B., D.C., M.R., M.C., F.B., T.P., A.C., and J.S. performed research and analyzed data; E.R., G.Z., and S.M. performed the bioinformatics and statistical analyses; P.A. and N.C. provided original integration data; and C.v.K., M.S., L.N., and E.M. designed the research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Eugenio Montini, San Raffaele Telethon Institute for Gene Therapy (HSR-TIGET), via Olgettina 58, Milan 20132, Italy; e-mail: montini.eugenio@hsr.it.
References
Author notes
L.N. and E.M. share senior authorship.