Abstract
We have developed induced pluripotent stem cells (iPSCs) from a patient with X-linked chronic granulomatous disease (X-CGD), a defect of neutrophil microbicidal reactive oxygen species (ROS) generation resulting from gp91phox deficiency. We demonstrated that mature neutrophils differentiated from X-CGD iPSCs lack ROS production, reproducing the pathognomonic CGD cellular phenotype. Targeted gene transfer into iPSCs, with subsequent selection and full characterization to ensure no off-target changes, holds promise for correction of monogenic diseases without the insertional mutagenesis caused by multisite integration of viral or plasmid vectors. Zinc finger nuclease–mediated gene targeting of a single-copy gp91phox therapeutic minigene into one allele of the “safe harbor” AAVS1 locus in X-CGD iPSCs without off-target inserts resulted in sustained expression of gp91phox and substantially restored neutrophil ROS production. Our findings demonstrate how precise gene targeting may be applied to correction of X-CGD using zinc finger nuclease and patient iPSCs.
Introduction
Patients with X-linked chronic granulomatous disease (X-CGD) have mutations in CYBB encoding the transmembrane gp91phox subunit of phagocyte NADPH oxidase required for microbicidal reactive oxygen species (ROS) production by neutrophils and monocytes.1,2 Patients have life-threatening infections and granulomatous complications. Allogeneic hematopoietic stem cell (HSC) transplantation can cure X-CGD, but most patients lack a suitable donor.3 In addition, graft-versus-host disease remains a significant risk of allogeneic sibling donor or matched unrelated donor transplantation. Gene transfer correction of autologous HSCs lacks these barriers, but current approaches use retrovirus or lentivirus vectors with risk of oncogenic insertional mutagenesis.4-7 More specifically for X-CGD, autologous HSC gene therapy using retrovirus vectors has demonstrated clinical benefit as salvage therapy for life-threatening infection, but long-term gene marking has been low8,9 and insertional mutagenesis-induced life-threatening myelodysplasia has been observed.6,10
Zinc finger nuclease (ZFN)–facilitated homologous recombination (HR)–mediated gene targeting of autologous HSCs could allow precise genomic modification. However, HR rates with HSC are modest,11 and clinical scale-up is problematic. Furthermore, because of limited self-renewal of HSCs in vitro, no applicable method exists by which gene-modified HSCs harboring undesirable insertions, off-target recombination, or other genetic changes can be screened and removed before transplantation.
By contrast, embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) have unlimited proliferative self-renewal potential in vitro,12-14 allowing development of cell therapeutics with precisely defined genetic characteristics. Specific gene targeting can be performed in human ESCs and iPSCs using ZFNs to induce a sequence-specific double-strand DNA break that enhances site-specific HR.15,16 Studies in human ESCs and iPSCs have shown that the AAVS1 locus, which lies within the first intron of the PPP1R12C gene on chromosome 19 and is the common integration site of adeno-associated virus 2, can be used as a nonpathogenic “safe harbor” to target a function-correcting minigene with persistent and strong transgene expression.16-18 The AAVS1 locus has an open chromatin structure that is flanked by insulator elements that shield the integrated cassette from trans-activation or repression.19 Although gene insertion at the AAVS1 locus may affect PPP1R12C gene expression, no haploid insufficiency of PPP1R12C has been reported, and disruption of one AAVS1 allele in transgenic mice and in human ESCs and iPSCs appears to have no adverse effects.18,20
Demonstration that iPSCs can recapitulate disease-specific phenotypes in vitro has been accomplished only with a few monogenic diseases.21-26 In our current study, an important intermediate goal was to demonstrate that we can reproduce the pathognomonic CGD oxidase-negative phenotype in mature neutrophils differentiated from X-CGD iPSCs. As an important proof of principle, we then demonstrate functional correction of the X-CGD defect in these iPSC following ZFN-mediated site-specific HR targeting of a gp91phox minigene to the AAVS1 safe harbor locus, with selection of clones containing an insert at only a single AAVS1 allele and no other genetic changes.
Methods
Approvals for use of human blood and tissues, human iPSCs, and animals
Experimental designs using human samples and iPSCs were approved by the Institutional Review Board (IRB) and Institutional Stem Cell Research Oversight committees at Johns Hopkins University (JHU). Bone marrow or blood was obtained from adult patients with X-CGD and healthy adults at the National Institutes of Health (NIH) Clinical Center, after written informed consent following the Declaration of Helsinki under the auspices of National Institute of Allergy and Infectious Diseases (NIAID) IRB-approved protocols 05-I-0213 and 94-I-0073. Immunodeficient NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJNSG (NSG) mice (The Jackson Laboratory) were used to test pluripotency of human iPSCs before and after gene targeting, based on their ability to form teratomas, as approved by Institutional Animal Care and Use Committees at JHU and NIAID/NIH.
Human iPSC generation and culture
Human X-CGD iPSCs were generated from mesenchymal stem cells (MSCs) derived27 from bone marrow aspirate of an adult male patient with X-CGD. MSCs were reprogrammed with VSV-G pseudotyped, pMX-based retroviral vectors expressing Oct4/Sox2/Klf4/c-Myc28 using procedures described previously.29 Human iPSCs were routinely maintained on CF-1 feeders and passaged as described previously.15 Healthy control human iPS(IMR90) iPSCs were obtained from WiCell as a normal control for iPSC neutrophil differentiation assays.
Pluripotency markers analysis, EB analysis, and teratoma formation assays for human iPSCs
Unfixed or formaldehyde fixed iPSCs were assessed for pluripotency markers as previously described.29 Tri-lineage differentiation potential of iPSCs was tested in vitro by embryoid body (EB) formation assays and in vivo by teratoma formation assays using protocols described previously.29 To test human iPSCs ability to form teratomas in vivo, cells were suspended in hESC-qualified Matrigel (BD Biosciences) and injected into NSG mice. Teratomas formed within 4-8 weeks and were excised for tissue staining and RT-PCR assessment of tissue lineage differentiation. Color and fluorescent images of iPSC markers (in PBS), embryoid bodies (in PBS) and teratomas (embedded sections) were acquired using a Nikon Eclipse TE2000-U inverted microscope with 10×ELWD Plan Fluor/0.3 or 20×ELWD Plan Fluor/0.45 objectives and a Qimaging Micropublisher 5.0 digital camera with QCapture software (v3.1.2). Multi-channel fluorscent images were merged using ImageJ software.
Karyotyping and DNA fingerprinting
Chromosome karyotyping (G-banding) was performed as previously described.29 For each cell line, 20 total metaphases were examined; 5 of the cells were analyzed and karyotyped. For DNA fingerprinting, genomic DNA was isolated using the DNAeasy kit (QIAGEN), and 2 genomic loci D21S2055 and D10S1214 were amplified by PCR using MapPairs human microsatellite marker sets (Invitrogen).
Neutrophil differentiation from human iPSCs
The protocol used for differentiating iPSCs into neutrophils was adapted from a recently published protocol30 developed for neutrophil differentiation from human ESCs, involving EB formation followed by coculture with OP9 stroma cells. The following substitutions were made from the published protocol: StemSpan SFEM (StemCell Technologies) was used for the first 24 hours of EB formation; human interleukin-6 (Peprotech) was used in place of FP6 in subsequent growth media. Following differentiation, cells were collected for analysis of neutrophil surface markers, morphology, and function.
Neutrophil surface marker analyses and morphology
Cells from iPSC neutrophil differentiation cultures were stained with fluorescence-conjugated antibodies to CD13, CD16, and CD45 (BD Biosciences). Flow cytometric analysis of gp91phox expression was performed either by indirect antibody staining with unconjugated 7D5 anti–human flavocytochrome b558 antibody and FITC-conjugated secondary antibody as previously described,31 or with PE-conjugated 7D5 antibody (MBL International) according to manufacturer's protocol. For assessment of neutrophil morphology, cells were deposited onto microscope slides using a Cytospin 3 cytocentrifuge (Thermo Fisher Scientific), then fixed with methanol and stained with modified Giemsa stain (Accustain; Sigma-Aldrich) following manufacturer's protocol.
Zymosan uptake assay of phagocytic capacity
A total of 2 × 105 cells were resuspended in 100 μL of 1× Hanks buffered saline solution (HBSS) containing 1mM CaCl2 and 1mM MgCl2, then incubated with Alexa Fluor 488–conjugated zymosan A BioParticles (Invitrogen) for 30 minutes in a 37°C shaker water bath. Cells were then placed on ice and washed with 4 mL of PBS with centrifugation at 350g for 5 minutes. Cells were resuspended in 1 mL of lyticase (100 U/mL in PBS), incubated 20 minutes at room temperature to remove of extracellular zymosan, washed with PBS containing 1mM EDTA, and analyzed by flow cytometry.
DHR flow cytometry fluorescence assay and nitroblue tetrazolium dye visual microscopic analysis of ROS production
For dihydrorhodamine (DHR) assay, cells were suspended in 400μL of HBSS, 130μM DHR (Invitrogen), and 500 U of catalase (Sigma-Aldrich), and incubated at 37°C for 5 minutes. Samples were then stimulated for ROS production by addition of 100 μL of 400 ng/mL phorbol myristate acetate (PMA; Sigma-Aldrich) in HBSS, followed by incubation at 37°C for 14 minutes, with periodic vortexing; negative control samples were left unstimulated (no PMA addition). Cells were analyzed by flow cytometry for DHR fluorescence as previously described.8
For nitroblue tetrazolium dye (NBT) analysis, cytospin of cells onto microscope slides was performed as described above. Cells were then incubated with 0.25 mL of 1 μg/mL PMA in PBS and 0.25 mL of 0.2% NBT (Sigma-Aldrich) in PBS at 37°C for 30 minutes, fixed with 1.5% paraformaldehyde, then washed 3 times with double-distilled H2O. The formation of blue-black formazan precipitates inside cell vacuoles on ROS reaction with NBT dye was visualized by microscopy. Color images of cytospun neutrophils (both Giemsa stained in Permount and NBT assay in PBS) were acquired using a Ziess ICM405 inverted microscope with 40×LD Plan-achromat/0.6 objective and a Kodak DC290 digital camera with Kodak MSD-290 Acquire software in Adobe Photoshop 6. Contrast or brightness adjustments were performed on whole images using Adobe Photoshop without other processing.
Lentivirus vector transduction of X-CGD iPSCs with Puro-T2A-gp91 vector
The lentivirus vector backbone and packaging plasmids used for this study have been described previously.32,33 The CL20i4rEF1α-Puro-T2A-gp91 self-inactivating lentivirus vector transfer plasmid (see provirus diagram in Figure 3A) contains an internal EF1α (short and intronless) promoter co-expressing a bicistronic puromycin resistance element and (via T2A element) codon-optimized gp91phox. This vector also contains the 400 bp version of chicken insulator cHS4 and was pseudotyped with VSV-G envelope. For lentiviral transduction, iPSCs were passaged onto Matrigel and cultured in mTeSR1 (StemCell Technologies) under feeder-free conditions, to enhance iPSC transduction by preventing preferential virus uptake by mouse embryonic fibroblast (MEF) feeders. The iPSCs were transduced with lentivirus overnight at a multiplicity of infection of 20, in the presence of 5 μg/mL protamine sulfate. After transduction, cells were passaged onto puromycin-resistant MEFs, and starting at least 48 hours after transduction, were treated with 3 μg/mL puromycin for 6 days to select against iPSCs that lack expression of the Puro-T2A-gp91 vector. The resulting polyclonal population of transduced X-CGD iPSCs containing varying sites of vector insertions was then cultured long term or expanded for further characterization and neutrophil differentiation without puromycin selection, unless otherwise specified.
Construction of a p5′AAVS1-SA-puro-pA-CAG-gp91opt-pA-3′AAVS1 minigene donor plasmid for ZFN-mediated HR-targeted expression of gp91phox in X-CGD iPSCs
The codon-optimized gp91phox sequence from the pCL20i4rEF1α-Puro-T2A-gp91 lentivirus was copied by PCR and inserted into the commercially available AAVS1 locus-targeting green fluorescence protein-expressing donor plasmid pAAV-CAGGS-eGFP backbone (Addgene plasmid 22212)16 after excising eGFP. This backbone is flanked at the 5′ and 3′ ends by sequence upstream and downstream of the AAVS1 locus targeting site of the ZFNs. The resultant donor plasmid, p5′AAVS1-SA-puro-pA-CAG-gp91opt-pA-3′AAVS1 (referred to hereafter as the APC-gp91; see upper diagram in Figure 4A) contains a splice acceptor element and 2A linker placed in front of a promoterless puromycin-polyA cassette, which will express the puromycin resistance element only if it is inserted downstream of a constitutively active promoter such as the PPP1R12C promoter. The gp91phox minigene cassette is placed downstream of the puromycin resistance element using the CAG (CMV early enhancer/chicken β-actin) promoter to express a codon-optimized gp91phox cDNA-polyA.
Gene targeting in human X-CGD iPSCs using AAVS1 ZFN mRNAs
A qualified ZFN pair optimized for targeting the AAVS1 locus is commercially available as mRNAs from Sigma-Aldrich (CompoZr AAVS1 targeted integration kit CTI1). ZFNs mRNAs were used instead of DNA plasmids to avoid integration of ZFN DNAs and reduce toxicity by shortening ZFN half-life. Five million human iPSCs were digested for 2-3 minutes using 0.05% trypsin (Invitrogen) and neutralized by trypsin inhibitor (0.5 mg/mL, T6522; Sigma-Aldrich). The cells were centrifuged at 100g for 5 minutes, and resuspended in 100 μL of Amaxa mouse ES nucleofection medium (Amaxa Biosystems) with 20 μg of the APC-gp91 donor plasmid targeting vector and 1 vial (∼ 6 μg of mRNA) of the AAVS1 ZFNs. The cells were transfected using an Amaxa Nucleofector device with program A-23, and then plated immediately onto puromycin-resistant DR4 MEF feeders (GlobalStem) with 10μM ROCK inhibitor Y27632 (Stemgent) and MEF-conditioned medium. Puromycin selection (0.5 μg/mL) was started at 4-5 days after nucleofection. Individual clones were picked a week later and then expanded and maintained without puromycin.
PCR and Southern blot of ZFN-mediated HR-targeted X-CGD iPSC clones
PCR labeling, Southern blotting, and chemiluminescence detection of digoxigenin-labeled DNA probes were performed as previously described.15 After ZFN-mediated HR targeting of the APC-gp91 donor plasmid into X-CGD iPSCs, pooled populations and candidate APC-gp91 targeted clones were screened initially by PCR detection of a 5′ 1033 bp fragment characteristic of the expected targeted integration into the AAVS1 locus. This PCR screen used primers AAVS1U-F2: 5′-CTGCCGTCTCTCTCCTGAGT (within intron 1 of the PPP1R12C gene upstream of the homologous flanking element sequence of APC-gp91) and PuroU-R: 5′-GTGGGCTTGTACTCGGTCAT (within the puromycin resistance element of APC-gp91) with Phusion Hot Start high-fidelity DNA polymerase (New England Biolabs) for 40 cycles of amplification.
The PPP1R12C gene has a Sph1 restriction cleavage site just 5′ of exon 1 with the next downstream Sph1 site in this gene occurring 3′ of exon 3 (see lower diagram in Figure 4A). Because the APC-gp91 donor plasmid contains 2 closely spaced Sph1 restriction sites in the minigene element, ZFN-mediated HR targeting of this plasmid to the AAVS1 site within intron 1 of the PPP1R12C gene changes the genomic Sph1 restriction fragment pattern. Targeted clones passing the first PCR screen were analyzed by Sph1-restriction cleavage of genomic DNA followed by Southern blot analyses with a 3′-probe that corresponds to PPP1R12C exon 3 plus flanking intronic sequence or with a 5′-probe that corresponds to PPP1R12C intron 1 sequence contained in the APC-gp91 5′-flanking homology arm (lower diagram in Figure 4A). The 510 bp 3′-probe was synthesized by PCR using primers: AAV3Pb-F: 5′-GACCCTCAGCCCTGACCAGC and AAV3Pb-R: 5′-TCCCCTCCCAGAAAGACCTGC, and the 705 bp 5′-probe was synthesized by PCR using primers AAV5Pb-F: 5′-GGCCTGGGTCACCTCTACG and AAV5Pb-R: 5′-GAACCAGAGCCACATTAACCG. After Sph1 digest, a targeted insertion that is correctly sited within the AAVS1 locus within intron 1 of the PPP1R12C gene will be detected by the 3′-probe as a 6.0 kb fragment and by the 5′-probe as a 3.8 kb fragment, whereas the wild-type nontargeted AAVS1 locus will appear as a 6.5 kb fragment using either the 3′-probe or the 5′-probe. Absence of the wild-type 6.5 kg fragment signifies bi-allelic targeted insertion into both AAVS1 loci, a characteristic that would disqualify a targeted clone from further consideration. Fragments of any other size detected by the 5′-probe represents random insertions. Further explanation is required to interpret the appearance of fragment bands indicative of random insertions. In addition to the 2 closely spaced Sph1 sites in the APC-gp91 targeting cassette, there is an additional Sph1 restriction site in this donor plasmid outside of the left homology arm at 4.3 kb 5′ upstream. Off-target nonhomologous random insertions will most often include this upstream plasmid Sph1 site, thus generating a plasmid-derived 4.3 kb fragment that can be detected by the 5′-probe, but not by the 3′-probe. This explains why candidate APC-gp91 targeted clones with random insertions will most often have only the same plasmid-derived 4.3 kb fragment as the signal that off-target random insertion has occurred. Occasionally, random insertion fragments of different sizes may be seen.
Sequencing detection of NHEJ mutations in the nontargeted allele of single-allele AAVS1 HR-targeted clones and the predicted top 10 other off-target sites in the genome
The DNA double-strand cleavage induced by ZFNs that does not result in homologous recombination with donor APC-gp91 plasmid is subject to annealing by cellular DNA repair machinery. This repair can maintain the wild-type sequence, or nonhomologous end joining (NHEJ) may introduce loss or gain in base pairs. Genomic DNA from clones selected for sequencing analysis of the remaining putative wild-type AAVS1 locus were PCR-amplified using the primers spanning the AAVS1 ZFN target site that would generate an ∼ 500 bp fragment from the unaltered WT site (AAVSCEL-F: 5′-TTCGGGTCACCTCTCACTCC and AAVSCEL-R: 5′-GGCTCCATCGTAAGCAAACC), but an ∼ 5000 bp fragment from the APC-gp91 targeted site. The short elongation time with Taq DNA polymerase (72°C, 30s) selectively amplified the nontargeted integration, putative wild-type AAVS1 allele producing only the ∼ 500 bp band, which was subsequently purified and sequenced.
The top 10 putative off-target sites for the AAVS1 ZFNs used in the current study have been previously identified by SELEX (Systematic Evolution of Ligands by Exponential) enrichment.16 We amplified all top 10 putative off-targets using Phusion Hot Start high-fidelity DNA polymerase for 40 cycles of amplification, using the primer sets previously published,16 except for the reverse primer for off-target site 2, which was changed to 5′-CAGGAAGAGCAGGAGATGAGGAGTT for increased PCR product specificity.
Flow cytometric screening of ZFN-mediated HR-targeted clones
For flow cytometric analyses of gp91phox expression in targeted clones, trypsin-digested single cells were first stained with TRA-1-60 primary and secondary antibodies, followed by PE-conjugated 7D5 antibody (described in “Neutrophil surface marker analyses and morphology) for 30 minutes. Flow cytometry gating on TRA-1-60+ cells allowed specific assessment of gp91phox expression in the undifferentiated pluripotent human iPSC population.
Results
Production and characterization of X-CGD iPSC lines
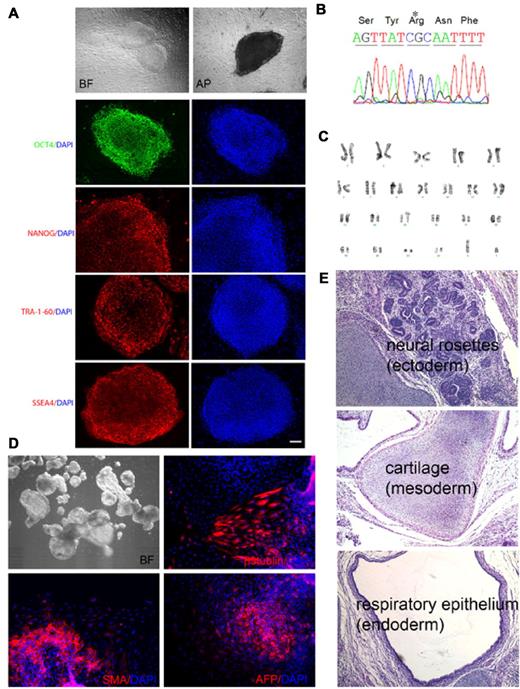
We established iPSCs reprogrammed from bone marrow mesenchymal stem cells (MSCs) of an adult male patient with X-CGD with 458T→G mutation in CYBB exon 5 (Leu153Arg). This missense mutation adversely affects gp91phox protein stability, abolishing neutrophil oxidase activity. After reprogramming, colonies exhibiting human ESC-like morphology and expressing TRA-1-60 pluripotency marker were expanded. Four lines were subjected to full characterization as outlined in Table 1. As shown in Figure 1 for the X-CGD iPSC line iXC1 as an example, all were positive for additional pluripotency markers (Figure 1A), retained the expected mutation in exon 5 of the CYBB gene (Figure 1B), exhibited normal karyotype (Figure 1C), and were capable of differentiation into 3 germ layers by either in vitro EB formation (Figure 1D) or in vivo teratoma formation (Figure 1E). Additionally, DNA fingerprinting of all X-CGD iPSC lines matched the original patient MSCs, but not other iPSCs or ESCs cultured in the laboratory during reprogramming (not shown).
Summary of 4 human X-CGD iPSC lines that are fully characterized
| X-CGD iPSC line . | Pluripotency markers . | DNA fingerprinting . | Karyotype . | EB formation . | Teratoma formation . | T→G CYBB mutation . |
|---|---|---|---|---|---|---|
| iXC1 | All expressed | Identity + | 46,XY | + | + | Present |
| iXC6 | All expressed | Identity + | 46,XY | + | + | Present |
| iXC9 | All expressed | Identity + | 46,XY | + | + | Present |
| iXC17 | All expressed | Identity + | 46,XY | + | + | Present |
| X-CGD iPSC line . | Pluripotency markers . | DNA fingerprinting . | Karyotype . | EB formation . | Teratoma formation . | T→G CYBB mutation . |
|---|---|---|---|---|---|---|
| iXC1 | All expressed | Identity + | 46,XY | + | + | Present |
| iXC6 | All expressed | Identity + | 46,XY | + | + | Present |
| iXC9 | All expressed | Identity + | 46,XY | + | + | Present |
| iXC17 | All expressed | Identity + | 46,XY | + | + | Present |
The 4 X-CGD iPSC lines were characterized in detail by all of the listed criteria. All 4 lines were subjected to the same analysis (shown in detail in Figure 1 for line iXC1) including expression of pluripotency markers (alkaline phosphatase activity, TRA-1-60, SSEA-4, OCT4, and NANOG), presence of normal male karyotype, presence of the patient's 458T→G mutation in exon 5 of CYBB, and ability to form EBs ex vivo and teratomas in NSG mice in vivo containing all 3 embryonic germ layer cells and tissues. DNA fingerprinting for all lines confirmed derivation from the patient primary MSCs.
Characterization of X-CGD iPSC line iXC1. Line iXC1 (A) exhibits characteristic human ESC-like morphology in bright field (BF) microscopy (top left) and expresses pluripotency markers that include alkaline phosphatase (AP) activity (top right) and fluorescence antibody staining for OCT4, NANOG, TRA-1-60, and SSEA-4 (left panels in green and red). Blue staining (right panels) are 4′-6-Diamidino-2-phenylindole (DAPI) staining of nuclei (bar = 100 μm); (B) has the CYBB 458T→G mutation; (C) has normal male karyotype; (D) forms embryoid bodies in vitro shown by BF (top left) microscopic appearance, and by expression of β-tubulin class III neuroectoderm marker (top right), smooth muscle actin (SMA) mesoderm marker (bottom left), and α-fetoprotein (AFP) endoderm marker (bottom right); (E) forms teratomas in vivo in NSG mice that include all 3 germ layers, identified here as neural rosette ectoderm (top), cartilage mesoderm (middle), and respiratory epithelial endoderm (bottom). Not shown is that RT-PCR analysis of cDNA generated from the iXC1 teratoma confirmed presence of mRNA specific for human PAX6 (ectoderm), CD34 (mesoderm), and α-feto-protein (endoderm).
Characterization of X-CGD iPSC line iXC1. Line iXC1 (A) exhibits characteristic human ESC-like morphology in bright field (BF) microscopy (top left) and expresses pluripotency markers that include alkaline phosphatase (AP) activity (top right) and fluorescence antibody staining for OCT4, NANOG, TRA-1-60, and SSEA-4 (left panels in green and red). Blue staining (right panels) are 4′-6-Diamidino-2-phenylindole (DAPI) staining of nuclei (bar = 100 μm); (B) has the CYBB 458T→G mutation; (C) has normal male karyotype; (D) forms embryoid bodies in vitro shown by BF (top left) microscopic appearance, and by expression of β-tubulin class III neuroectoderm marker (top right), smooth muscle actin (SMA) mesoderm marker (bottom left), and α-fetoprotein (AFP) endoderm marker (bottom right); (E) forms teratomas in vivo in NSG mice that include all 3 germ layers, identified here as neural rosette ectoderm (top), cartilage mesoderm (middle), and respiratory epithelial endoderm (bottom). Not shown is that RT-PCR analysis of cDNA generated from the iXC1 teratoma confirmed presence of mRNA specific for human PAX6 (ectoderm), CD34 (mesoderm), and α-feto-protein (endoderm).
Differentiation of mature neutrophils from X-CGD iPSC and control iPSC
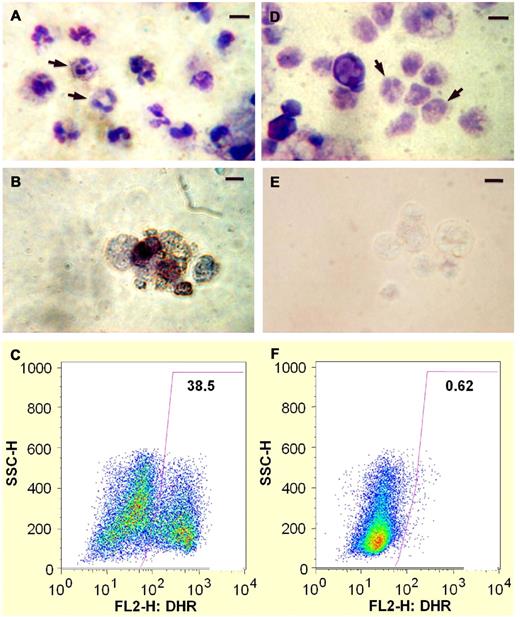
We differentiated human iPSC lines derived either from healthy control or from the X-CGD patient into a population containing approximately 30%-40% of cells with mature neutrophil morphology (Figure 2A and D). By flow cytometry, > 97% of cells expressed CD45 pan-leukocyte marker, 27.9%-34.7% expressed CD13 neutrophil marker, 73.7%-78.9% expressed CD16 neutrophil/myeloid marker, and > 70% were capable of zymosan particle phagocytosis (not shown). ROS production was measured by nitro NBT dye reduction and by DHR 123 fluorescence flow cytometry assay.8 Neutrophils derived from healthy control iPSCs were strongly oxidase-positive by NBT and DHR assays (Figure 2B-C), whereas neutrophils derived from X-CGD iPSC lines were devoid of oxidase activity by NBT and DHR (Figure 2E-F). Thus, neutrophils differentiated in vitro from X-CGD patient-derived iPSCs model the pathognomonic ROS-negative phenotype of CGD.
Neutrophil differentiation of X-CGD iPSC line iXC9 and normal control human iPSC line iPS(IMR90). Panels A-C show cytospin Giemsa stain, cytospin NBT assay, and DHR flow cytometric analysis, respectively, of neutrophils arising from differentiation of normal control iPSC line iPS(IMR90). Panels D-F are cytospin Giemsa stain, cytospin NBT assay, and DHR flow cytometric analysis, respectively, of neutrophils arising from differentiation of X-CGD iPSC line iXC9. Arrows in the cytospin Giemsa stain images (A,D) indicate some of the cells with mature neutrophil morphology (bar = 10 μm). Cytospin NBT assay shows blue-black formazan precipitates resulting from robust ROS production by PMA-activated neutrophils derived from normal control iPSCs (B), but lack of formazan precipitates indicating no ROS production by neutrophils derived from X-CGD iPSCs (E). The DHR dot plot images (y-axis side scatter; x-axis DHR fluorescence) demonstrate robust ROS production (high fluorescence) by 38.5% of PMA-activated cells representing mature neutrophils derived from normal control iPSCs (C), but no evidence of any significant ROS production by PMA-activated neutrophils derived from X-CGD iPSCs (F).
Neutrophil differentiation of X-CGD iPSC line iXC9 and normal control human iPSC line iPS(IMR90). Panels A-C show cytospin Giemsa stain, cytospin NBT assay, and DHR flow cytometric analysis, respectively, of neutrophils arising from differentiation of normal control iPSC line iPS(IMR90). Panels D-F are cytospin Giemsa stain, cytospin NBT assay, and DHR flow cytometric analysis, respectively, of neutrophils arising from differentiation of X-CGD iPSC line iXC9. Arrows in the cytospin Giemsa stain images (A,D) indicate some of the cells with mature neutrophil morphology (bar = 10 μm). Cytospin NBT assay shows blue-black formazan precipitates resulting from robust ROS production by PMA-activated neutrophils derived from normal control iPSCs (B), but lack of formazan precipitates indicating no ROS production by neutrophils derived from X-CGD iPSCs (E). The DHR dot plot images (y-axis side scatter; x-axis DHR fluorescence) demonstrate robust ROS production (high fluorescence) by 38.5% of PMA-activated cells representing mature neutrophils derived from normal control iPSCs (C), but no evidence of any significant ROS production by PMA-activated neutrophils derived from X-CGD iPSCs (F).
Transduction of X-CGD iPSCs with VSV-G pseudotyped CL20i4rEF1α-Puro-T2A-gp91 self-inactivating lentivector
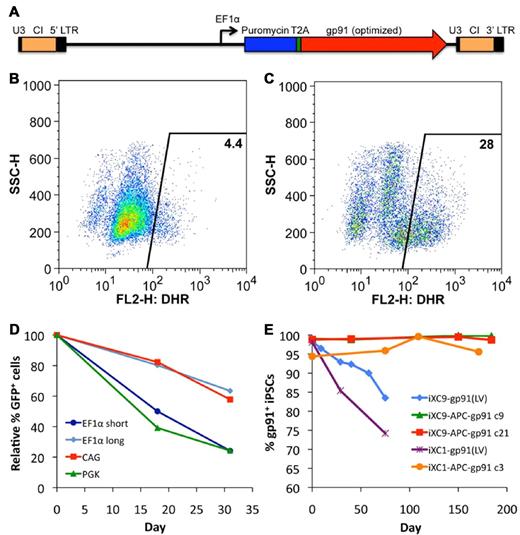
Transduction of X-CGD iPSCs with CL20i4rEF1α-Puro-T2A-gp91 (Figure 3A) lentivector encoding puromycin resistance and gp91phox followed by 6 days of puromycin selection resulted in expression of gp91phox transgene by almost all of the cells in the puromycin-selected population. However, when lentivector-transduced X-CGD iPSCs were subjected to culture conditions inducing differentiation to mature neutrophils in the absence of continued puromycin selection, there was a rapid decrease in the number of cells expressing gp91phox such that only ∼ 4% of the resultant mature neutrophils were weakly oxidase-positive in the DHR assay (Figure 3B). Continued puromycin selection during neutrophil differentiation of lentivector-transduced X-CGD iPSCs maintained expression of gp91phox (not shown) and 28% of the resultant cells demonstrated significant correction of ROS generation by DHR assay (Figure 3C), a percentage similar to that seen with populations of neutrophils differentiating from normal control iPSCs (see Figures 2C and 6B). These observations suggest that a significant proportion, although not all, of lentivector genomic inserts are subject to accelerated silencing of gp91phox expression during differentiation of iPSCs to neutrophils.
Lentivirus vector gene transfer mediated expression of gp91phox or GFP in X-CGD iPSCs. (A) Schematic of CL20i4rEF1α-Puro-T2A-gp91 self-inactivating bicistronic lentivirus vector (LV) insert with internal EF1α (short intronless) promoter co-expressing puromycin resistance gene and (via T2A element) codon-optimized gp91phox. This vector also contains the 400 bp version of chicken insulator cHS4 (CI). Not shown are the series of closely related CL20 lentivectors constructs with the same backbone used for the experiment in panel D of this figure, but expressing GFP instead of gp91phox, using EF1α short, EF1α long, CAG, or PGK promoters. (B) Flow cytometric DHR analysis of ROS production by neutrophils differentiated from LV-transduced line iXC9 X-CGD iPSC shows only 4% of cells are weakly DHR positive (compare with Figure 2C and F), even though almost all of the transduced and puromycin selected iXC9 cells strongly expressed gp91phox just before differentiation (as shown in panel E). (C) Flow cytometric DHR analysis of ROS production by neutrophils differentiated from the same LV-transduced iXC9 line, but maintained in puromycin selection during the differentiation, results in 28% of cells DHR-positive, a value similar to that in Figures 2C and 6B for the normal control. (D) Promoter-based differences in GFP lentivirus expression in unselected polyclonal populations of transduced iXC9 iPSCs during neutrophil differentiation. GFP expression during in vitro neutrophil differentiation of iXC9 iPSCs transduced with lentivirus containing GFP expressed from EF1α short, EF1α long, CAG, or PGK promoter. The proportion of cells remaining GFP positive was calculated relative to the initial level (range 73%-90%) observed in the undifferentiated iPSCs at the start of the observation period, which was set to 100% at baseline to allow direct comparison of the proportional change. (E) Shown is the decrease over time in culture of gp91phox transgene expression measured by flow cytometry in iXC9 or iXC1 transduced with CL20i4rEF1α-Puro-T2A-gp91 lentivirus (blue diamonds and purple asterisks), compared with the very long-term stability of expression of gp91phox from the AAVS1 locus targeted APC-gp91 minigene in iXC9 and iXC1 (green triangles, red squares, and orange circles). For all of these transduced or gene-targeted X-CGD iPS lines, day 0 on this graph corresponds to the time at which lines completed puromycin selection and were then grown thereafter for the period of time shown in the absence of puromycin.
Lentivirus vector gene transfer mediated expression of gp91phox or GFP in X-CGD iPSCs. (A) Schematic of CL20i4rEF1α-Puro-T2A-gp91 self-inactivating bicistronic lentivirus vector (LV) insert with internal EF1α (short intronless) promoter co-expressing puromycin resistance gene and (via T2A element) codon-optimized gp91phox. This vector also contains the 400 bp version of chicken insulator cHS4 (CI). Not shown are the series of closely related CL20 lentivectors constructs with the same backbone used for the experiment in panel D of this figure, but expressing GFP instead of gp91phox, using EF1α short, EF1α long, CAG, or PGK promoters. (B) Flow cytometric DHR analysis of ROS production by neutrophils differentiated from LV-transduced line iXC9 X-CGD iPSC shows only 4% of cells are weakly DHR positive (compare with Figure 2C and F), even though almost all of the transduced and puromycin selected iXC9 cells strongly expressed gp91phox just before differentiation (as shown in panel E). (C) Flow cytometric DHR analysis of ROS production by neutrophils differentiated from the same LV-transduced iXC9 line, but maintained in puromycin selection during the differentiation, results in 28% of cells DHR-positive, a value similar to that in Figures 2C and 6B for the normal control. (D) Promoter-based differences in GFP lentivirus expression in unselected polyclonal populations of transduced iXC9 iPSCs during neutrophil differentiation. GFP expression during in vitro neutrophil differentiation of iXC9 iPSCs transduced with lentivirus containing GFP expressed from EF1α short, EF1α long, CAG, or PGK promoter. The proportion of cells remaining GFP positive was calculated relative to the initial level (range 73%-90%) observed in the undifferentiated iPSCs at the start of the observation period, which was set to 100% at baseline to allow direct comparison of the proportional change. (E) Shown is the decrease over time in culture of gp91phox transgene expression measured by flow cytometry in iXC9 or iXC1 transduced with CL20i4rEF1α-Puro-T2A-gp91 lentivirus (blue diamonds and purple asterisks), compared with the very long-term stability of expression of gp91phox from the AAVS1 locus targeted APC-gp91 minigene in iXC9 and iXC1 (green triangles, red squares, and orange circles). For all of these transduced or gene-targeted X-CGD iPS lines, day 0 on this graph corresponds to the time at which lines completed puromycin selection and were then grown thereafter for the period of time shown in the absence of puromycin.
These observations led us to examine differences in lentivector internal promoter susceptibility to silencing during differentiation of iPSCs to neutrophils. We used our self-inactivating insulated lentivector backbone to construct vectors expressing eGFP from each of 4 candidate internal promoters: EF1α short, EF1α long (containing splice sites), CAG, and PGK. Using these eGFP lentivectors, EF1α short and PGK promoters were particularly susceptible to silencing during neutrophil differentiation from iPSCs, but EF1α long and CAG promoters facilitated more stable expression of eGFP during differentiation (Figure 3D) and no change in the mean fluorescence index of eGFP expression (not shown). These experiments supported the use of CAG promoter in the next phase of our studies to express gp91phox from APC-gp91 minigene donor plasmid used for ZFN AAVS1 safe harbor targeting. Even without differentiation to neutrophils, long-term culture of CL20i4rEF1α-Puro-T2A-gp91–transduced X-CGD iPSC lines in the absence of puromycin resulted in progressive loss of gp91phox expression (Figure 3E), a problem not observed to occur with targeted APC-gp91 minigene, as will be discussed.
ZFN-mediated HR targeting of APC-gp91 minigene to the AAVS1 safe harbor site in X-CGD iPSC
The unlimited expansion potential of human iPSCs allows for clonal selection and genetic screening of individual gene-modified clones. A very recent publication demonstrated that it is possible to screen lentivector-transduced iPSCs to select clones with vector inserts only at sites in the genome that allow stable corrective transgene expression and minimal effect on nearby genes.26 More direct approaches include placing the corrective minigene into a predefined safe harbor site or directly repairing the CYBB mutation. Targeted repair of specific mutations within CYBB would be the ideal, but requires designing a different ZFN gene-targeting strategy for each of the many known X-CGD mutations throughout CYBB. Additionally, almost 5% of X-CGD cases involve nonrepairable large deletions of CYBB. Therefore, we employed ZFNs designed to precisely target our puromycin selectable gp91phox minigene DNA plasmid with AAVS1 homology arms (APC-gp91) to the AAVS1 locus within intron 1 of the PPP1R12C gene, a well-characterized safe harbor site (Figure 4A). The availability of a pair of previously qualified high-affinity and -specificity ZFNs, each recognizing 12bp of adjacent DNA sequences (separated by 6bp) at the AAVS1 locus,16 further made this site a reasonable first choice as our proof-of-principle demonstration of precisely targeted correction of X-CGD.
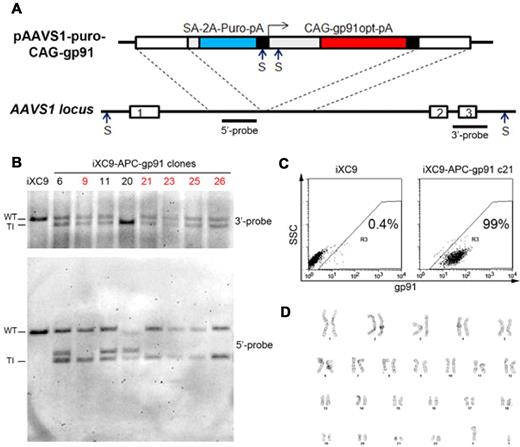
Analysis of APC-gp91 minigene plasmid ZFN-mediated HR targeting to the AAVS1 locus in X-CGD iPSCs. (A) Upper schematic of APC-gp91 minigene donor plasmid (pAAVS1-puro-CAG-gp91) and lower schematic of the PPP1R12C gene structure that includes exons 1, 2, and 3 flanking the AAVS1 locus that is within intron 1. Dotted lines indicate AAVS1 homology arms within the targeting donor plasmid. The puromycin-polyA promotorless cassette (SA-2A-Puro-pA) relies on a splice accepter plus 2A linker to use the PPP1R12C promoter for expression. The gp91phox minigene cassette uses CAG (CMV early enhancer/chicken β-actin) promoter to express a codon-optimized gp91phox cDNA-polyA. Restriction enzyme Sph1 (S) sites and 3′- and 5′-probes used for Southern blot are shown in both schematics. Not shown for the donor plasmid is a Sph1 restriction site 4.3 kb upstream of the 2 closely spaced Sph1 restriction sites within the schematic of pAAVS1-puro-CAG-gp91 plasmid. This upstream Sph1 site would not be present with a correctly targeted insert, but is often included with off-target random insertion of the donor plasmid. (B) Sph1 restriction Southern blot analysis of some of the APC-gp91 minigene targeted iXC9 (iXC9-APC-gp91) clones. 3′-probe (upper blot) detects wild-type allele (WT) 6.5 kb fragment in all clones except no. 20 (in which both alleles are targeted) and the 3′-targeted integration (TI) 6.0 kb fragment. 5′-probe (lower blot) detects WT 6.5 kb fragment in all but clone 20, the 5′ TI 3.8 kb fragment in all clones, and additional fragment(s) generated by random insertion in some clones (note the commonly seen 4.3 kb fragment derived entirely from donor plasmid sequence that is indicative of random insertion, and the 5.8 kb random insertion fragment seen in clone 20). From this particular analysis, clones indicated in red containing no random insert and a single-copy APC-gp91 insert in AAVS1 (nos. 9, 21, 23, 25, 26) were chosen for NHEJ screening (Table 3). (C) Flow cytometry gp91phox transgene expression by iXC9-APC-gp91 c21 (right panel) and lack of gp91phox expression by parental X-CGD iPSC clone iXC9 (left panel) (y-axis side scatter; x-axis fluorescent anti-gp91phox). (D) iXC9-APC-gp91 c21 normal karyotype.
Analysis of APC-gp91 minigene plasmid ZFN-mediated HR targeting to the AAVS1 locus in X-CGD iPSCs. (A) Upper schematic of APC-gp91 minigene donor plasmid (pAAVS1-puro-CAG-gp91) and lower schematic of the PPP1R12C gene structure that includes exons 1, 2, and 3 flanking the AAVS1 locus that is within intron 1. Dotted lines indicate AAVS1 homology arms within the targeting donor plasmid. The puromycin-polyA promotorless cassette (SA-2A-Puro-pA) relies on a splice accepter plus 2A linker to use the PPP1R12C promoter for expression. The gp91phox minigene cassette uses CAG (CMV early enhancer/chicken β-actin) promoter to express a codon-optimized gp91phox cDNA-polyA. Restriction enzyme Sph1 (S) sites and 3′- and 5′-probes used for Southern blot are shown in both schematics. Not shown for the donor plasmid is a Sph1 restriction site 4.3 kb upstream of the 2 closely spaced Sph1 restriction sites within the schematic of pAAVS1-puro-CAG-gp91 plasmid. This upstream Sph1 site would not be present with a correctly targeted insert, but is often included with off-target random insertion of the donor plasmid. (B) Sph1 restriction Southern blot analysis of some of the APC-gp91 minigene targeted iXC9 (iXC9-APC-gp91) clones. 3′-probe (upper blot) detects wild-type allele (WT) 6.5 kb fragment in all clones except no. 20 (in which both alleles are targeted) and the 3′-targeted integration (TI) 6.0 kb fragment. 5′-probe (lower blot) detects WT 6.5 kb fragment in all but clone 20, the 5′ TI 3.8 kb fragment in all clones, and additional fragment(s) generated by random insertion in some clones (note the commonly seen 4.3 kb fragment derived entirely from donor plasmid sequence that is indicative of random insertion, and the 5.8 kb random insertion fragment seen in clone 20). From this particular analysis, clones indicated in red containing no random insert and a single-copy APC-gp91 insert in AAVS1 (nos. 9, 21, 23, 25, 26) were chosen for NHEJ screening (Table 3). (C) Flow cytometry gp91phox transgene expression by iXC9-APC-gp91 c21 (right panel) and lack of gp91phox expression by parental X-CGD iPSC clone iXC9 (left panel) (y-axis side scatter; x-axis fluorescent anti-gp91phox). (D) iXC9-APC-gp91 c21 normal karyotype.
Using the APC-gp91 donor plasmid that we designed for this study and the previously described15 ZFN mRNA pair optimized to target the AAVS1 locus, we reproducibly achieved high-efficiency targeted integrations in each of 2 of our X-CGD iPSC lines after nucleofection and puromycin selection (Table 2). As described in “PCR and Southern blot of ZFN-mediated HR-targeted X-CGD iPSC clones” and indicated in the third column of Table 2, our initial screen of clones by PCR for detection of the 1033 bp band characteristic of the expected targeted integration into the AAVS1 locus demonstrated that 75% of clones had an integrant in at least one AAVS1 allele.
Summary of the outcomes of ZFN-mediated HR targeting of APC-gp91 minigene to the AAVS1 locus in X-CGD iPSCs
| X-CGD iPSC line . | Passage number . | No. with insert present by PCR . | No. with AAVS1 targeting confirmed by Southern blot . | No. with inserts in both AAVS1 alleles . | No. with random insertion . | No. with NHEJ of WT AAVS1 allele . | No. passing all criteria . |
|---|---|---|---|---|---|---|---|
| iXC1 | p26 | 6/6 | 5/5 tested | 2/5 | 1/5 | 1/3 tested | 2 |
| iXC9 | p43-46 | 21/30 | 15/15 tested | 1/15 | 4/15 | 3/5 tested | 2 |
| X-CGD iPSC line . | Passage number . | No. with insert present by PCR . | No. with AAVS1 targeting confirmed by Southern blot . | No. with inserts in both AAVS1 alleles . | No. with random insertion . | No. with NHEJ of WT AAVS1 allele . | No. passing all criteria . |
|---|---|---|---|---|---|---|---|
| iXC1 | p26 | 6/6 | 5/5 tested | 2/5 | 1/5 | 1/3 tested | 2 |
| iXC9 | p43-46 | 21/30 | 15/15 tested | 1/15 | 4/15 | 3/5 tested | 2 |
X-CGD iPSC lines iXC1 and iXC9, at passage numbers indicated, were used for APC-gp91 minigene ZFN targeting to the AAVS1 locus to yield 6 and 30 resultant clones after puromycin selection, respectively. By initial PCR screen, all 6 iXC1-APC-gp91 clones and 21 of 30 iXC9-APC-gp91 clones yielded the 5′ 1033 bp product expected from targeted integration into the AAVS1 locus. Of these, 5 from iXC1 and 15 from iXC9 were selected for additional characterization by Southern blot analysis (examples of this analysis for some iXC9-APC-gp91 clones are shown in Figure 4B) to determine which clones had targeted inserts at one versus both AAVS1 alleles, and which had random insert(s). Clones having targeted integration at only one AAVS1 allele and no random integration were further tested by sequencing to determine which had NHEJ additions or deletions at the putative wild-type (WT) AAVS1 allele (Table 3). This rigorous screen left 2 iXC1-APC-gp91 clones (c1, c3) and 2 iXC9-APC-gp91 clones (c9, c21) that passed all genetic analysis criteria.
Detailed genetic characterization of candidate APC-gp91 X-CGD iPSC clones
Clones screening positive by PCR for APC-gp91 minigene targeting to the AAVS1 safe harbor were further analyzed to select those with only a single allele AAVS1 locus insertion, with no random inserts, and with no NHEJ mutations at either the nontargeted AAVS1 allele or at the top 10 off-target sites. Southern blot analyses performed to confirm APC-gp91 targeting into one or both AAVS1 alleles and to detect off-target integrations showed that ∼ 75% (15/20) of clones examined from the initial screen were correctly targeted without additional random integrations (Figure 4B and Table 2), the majority of which inserted APC-gp91phox into only one of the 2 AAVS1 alleles. However, DNA sequencing did reveal NHEJ-mediated small deletions or insertions at the nontargeted wild-type AAVS1 allele in ∼ 50% of these clones (Table 3). Further examination of the top 10 possible off-target sites of AAVS1 ZFNs identified previously by sequence similarities16 revealed no evidence of off-target NHEJ DNA mutations in the AAVS1-targeted iPSC clones of interest (Figure 5).
Analysis of NHEJ by sequencing of the remaining putative wild-type AAVS1 locus
| Reference sequence | TGTCCCCTCCACCCCACAGTGGGGCCACTAGGGACAGGATTGGTGACAGA |
| iXC9-APC-gp91 c9 | TGTCCCCTCCACCCCACAGTGGGGCCACTAGGGACAGGATTGGTGACAGA |
| iXC9-APC-gp91 c21 | TGTCCCCTCCACCCCACAGTGGGGCCACTAGGGACAGGATTGGTGACAGA |
| iXC9-APC-gp91 c23 | TGTCCCCTCCACCCCACAGTGGGGCCA––––––––AGGATTGGTGACAGA |
| iXC9-APC-gp91 c25 | TGTCCCCTCCACCCCACAGTGGGGccagCCACTAGGGACAGGATTGGTGACAGA |
| iXC9-APC-gp91 c26 | TGTCCCCTCCACCCCACAGTGGGGCCA–––-GGACAGGATTGGTGACAGA |
| iXC1-APC-gp91 c1 | TGTCCCCTCCACCCCACAGTGGGGCCACTAGGGACAGGATTGGTGACAGA |
| iXC1-APC-gp91 c3 | TGTCCCCTCCACCCCACAGTGGGGCCACTAGGGACAGGATTGGTGACAGA |
| iXC1-APC-gp91 c5 | TGTCCCCTCCACCCCACAGTGGGGcaCCACTAGGGACAGGATTGGTGACAGA |
| Reference sequence | TGTCCCCTCCACCCCACAGTGGGGCCACTAGGGACAGGATTGGTGACAGA |
| iXC9-APC-gp91 c9 | TGTCCCCTCCACCCCACAGTGGGGCCACTAGGGACAGGATTGGTGACAGA |
| iXC9-APC-gp91 c21 | TGTCCCCTCCACCCCACAGTGGGGCCACTAGGGACAGGATTGGTGACAGA |
| iXC9-APC-gp91 c23 | TGTCCCCTCCACCCCACAGTGGGGCCA––––––––AGGATTGGTGACAGA |
| iXC9-APC-gp91 c25 | TGTCCCCTCCACCCCACAGTGGGGccagCCACTAGGGACAGGATTGGTGACAGA |
| iXC9-APC-gp91 c26 | TGTCCCCTCCACCCCACAGTGGGGCCA–––-GGACAGGATTGGTGACAGA |
| iXC1-APC-gp91 c1 | TGTCCCCTCCACCCCACAGTGGGGCCACTAGGGACAGGATTGGTGACAGA |
| iXC1-APC-gp91 c3 | TGTCCCCTCCACCCCACAGTGGGGCCACTAGGGACAGGATTGGTGACAGA |
| iXC1-APC-gp91 c5 | TGTCCCCTCCACCCCACAGTGGGGcaCCACTAGGGACAGGATTGGTGACAGA |
Shown are the results from sequencing the AASV1 ZFN target site in 5 iXC9-APC-gp91 clones and 3 iXC1-APC-gp91 clones that contained one allele of targeted integration and no random integrations. The putative wild-type allele was PCR-amplified (as described in “Sequencing detection of NHEJ mutations”) and products were sequenced to look for NHEJ-mediated mutations. The upper case letters indicate matches to reference AAVS1 sequence, where the bolded underlined upper case letters indicate target sequences to which the ZFN pair was designed to bind. Bolded underlined lower case letters indicate base pair inserts, and bolded dashed lines indicate base pair deletions relative to the reference. Four of the 8 clones fully match the reference sequence with no inserts or deletions.
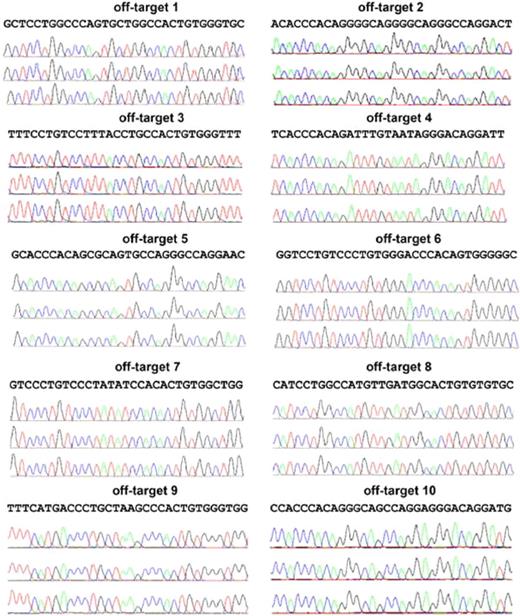
Analysis of the top 10 putative off-targets for AAVS1 ZFNs in targeted clones to rule out NHEJ mutations at those sites. Ten SELEX-predicted genomic off-targets were PCR-amplified and products sequenced for iXC1-APC-gp91 c3 (top chromatograph), iXC9-APC-gp91 c9 (middle chromatograph), and iXC9-APC-gp91 c21 (bottom chromatograph) for each of the 10 sites as indicated. Because the PCR products are from a homogenous population in each clone, mutations in the off-target sequence will show as single/double peaks/nucleotides that are different from reference sequence. None of these 3 clones showed any mutations in the off-target sequences.
Analysis of the top 10 putative off-targets for AAVS1 ZFNs in targeted clones to rule out NHEJ mutations at those sites. Ten SELEX-predicted genomic off-targets were PCR-amplified and products sequenced for iXC1-APC-gp91 c3 (top chromatograph), iXC9-APC-gp91 c9 (middle chromatograph), and iXC9-APC-gp91 c21 (bottom chromatograph) for each of the 10 sites as indicated. Because the PCR products are from a homogenous population in each clone, mutations in the off-target sequence will show as single/double peaks/nucleotides that are different from reference sequence. None of these 3 clones showed any mutations in the off-target sequences.
After applying these extensive genetic screening criteria for choosing clones with the desired targeting outcome characteristics, we identified 4 clones satisfying all genetic screening characteristics (Tables 2–3 and Figure 4). Three of these clones designated iXC9-APC-gp91 c9, iXC9-APC-gp91 c21, and iXC1-APC-gp91 c3 were subjected to more detailed functional testing.
Function of iXC9-APC-gp91 c9, iXC9-APC-gp91 c21, and iXC1-APC-gp91 c3
These 3 clones maintained high-level gp91phox expression (Figures 3E and 4C) in > 95% of cells for > 4 months of culture without puromycin selection, exhibited normal karyotypes (Figure 4D), maintained human ESC/iPSC-like morphology, expressed pluripotency markers, retained the exon 5 mutation in CYBB, and differentiated into 3 germ layers by EB or teratoma formation assays (not shown).
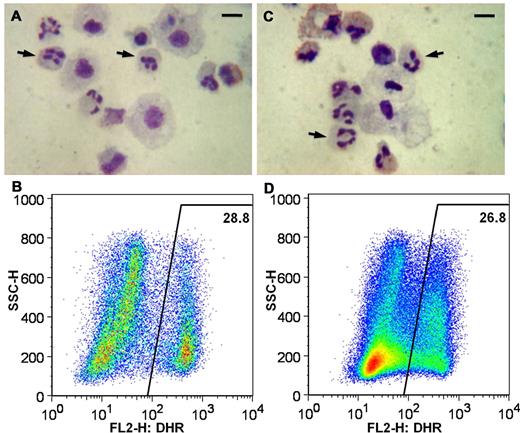
All 3 of these fully characterized APC-gp91phox targeted X-CGD iPSC clones could be differentiated to mature neutrophils with restored oxidase activity. An example of this is shown in Figure 6, where mature neutrophils differentiated in the same experiment from the normal control human iPSC line, iPS(IMR90), (Figure 6A-B) are compared with neutrophils differentiated from the gp91phox minigene-corrected X-CGD iPSC line iXC1-APC-gp91 c3 (Figure 6C-D). In this experiment, neutrophils derived from healthy control iPSCs were strongly oxidase-positive in the DHR assay (Figure 6B). Neutrophils derived from the minigene-corrected X-CGD iPSC clone are equally oxidase-positive in the DHR assay (Figure 6D), in marked contrast to the parental X-CGD iPSC clone–derived oxidase-negative neutrophils shown in Figure 2F. This demonstrates full functional correction of the pathognomonic neutrophil oxidase defect by ZFN-mediated HR gene targeting of X-CGD patient-derived iPSCs.
Functional restoration of ROS production in neutrophils differentiated from iXC1-APC-gp91 c3. Panels A and B are cytospin Giemsa stain and DHR flow cytometric analysis, respectively, of neutrophils arising from differentiation of normal control iPSC line iPS(IMR90); and panels C and D are cytospin Giemsa stain and DHR flow cytometric analysis, respectively, of neutrophils arising from differentiation of ZFN-targeted minigene-corrected X-CGD iPSC clone iXC1-APC-gp91 c3. Arrows in the cytospin Giemsa-stain images (A,C) indicate some of the cells with mature neutrophil morphology (bar = 10 μm). The DHR dot plot images (y-axis side scatter; x-axis DHR fluorescence) demonstrate robust ROS production (high fluorescence) by 28.8% and from 26.8% of cells, respectively, representing PMA-activated neutrophils derived from normal control iPSCs (B) and from ZFN-targeted minigene-corrected X-CGD iPSC clone iXC1-APC-gp91 c3 (D).
Functional restoration of ROS production in neutrophils differentiated from iXC1-APC-gp91 c3. Panels A and B are cytospin Giemsa stain and DHR flow cytometric analysis, respectively, of neutrophils arising from differentiation of normal control iPSC line iPS(IMR90); and panels C and D are cytospin Giemsa stain and DHR flow cytometric analysis, respectively, of neutrophils arising from differentiation of ZFN-targeted minigene-corrected X-CGD iPSC clone iXC1-APC-gp91 c3. Arrows in the cytospin Giemsa-stain images (A,C) indicate some of the cells with mature neutrophil morphology (bar = 10 μm). The DHR dot plot images (y-axis side scatter; x-axis DHR fluorescence) demonstrate robust ROS production (high fluorescence) by 28.8% and from 26.8% of cells, respectively, representing PMA-activated neutrophils derived from normal control iPSCs (B) and from ZFN-targeted minigene-corrected X-CGD iPSC clone iXC1-APC-gp91 c3 (D).
Discussion
Our work is the first demonstration that neutrophils differentiated from X-CGD patient-specific iPSCs fully model the oxidase-negative disease phenotype. We also demonstrate ZFN-mediated specific targeting of an oxidase function-correcting minigene to the AAVS1 safe harbor site in X-CGD iPSCs, resulting in full functional correction of the CGD phenotype. We outline the extensive genetic screening that must be performed to ensure that corrected clones have only a single allele AAVS1 locus insert, retain wild-type sequence at the nontargeted AAVS1 allele, have no random insertions of the donor plasmid, and have no off-target NHEJ mutations at the top off-target sites potentially targetable with this ZFN set. This provides a detailed roadmap for more universal application of this ZFN targeting approach in iPSCs for correction of other monogenic disorders. For clinical application, deep sequencing would be required to more fully characterize genetic changes in iPSCs that differ from patient primary cells.
In our initial studies of gene correction of the X-CGD iPSCs using lentivirus vector encoding gp91phox, we were surprised to encounter a significant level of silencing of insulated lentivector during differentiation of neutrophils from a polyclonal population of transduced X-CGD iPSCs. Long-term expression of gp91phox from the lentivirus transduction in the same polyclonal population was relatively stable in the undifferentiated iPSCs. In addition, we also previously had found that a version of this vector without the puromycin and T2A element, CL20i4rEF1α-gp91, could mediate long-term high-level gp91phox expression and functional correction in neutrophils arising in vivo from X-CGD patient CD34+ hematopoietic stem cells engrafted for 7 weeks in a NOD/SCID mouse xenograft model.33 This myeloid differentiation-associated gene silencing of lentivector transgene in iPSCs was significantly improved when the CAG promoter was substituted for the EF1α (short intronless) promoter in the lentivector, but was not fully abrogated. Even with the EF1α short promoter, if puromycin selection was used during neutrophil differentiation, then oxidase correction in the DHR assay was similar to neutrophils differentiating from normal iPSCs (Figure 3C). This suggests that there are sites of lentivector insertion that are protected from the myeloid differentiation-associated silencing. Thus, in theory it could have been possible to screen for lentivector-transduced clones of X-CGD iPSCs that maintained expression of the transgene during myeloid differentiation.
The extensive screening of stable-expressing lentivector-transduced iPSC clones is the approach that was successfully taken in a recent report of the correction of human thalassemia iPSCs using a lentivirus vector previously developed specifically for high expression of β-globin.26 In that study, one transduced thalassemia iPSC clone meeting all their criteria was able to be selected for stable high expression of β-globin from insert sites that by additional analysis had minimal effects on nearby genes. Because the silencing we encountered with lentivectors transducing iPSCs was increased during differentiation, the requirement to differentiate each clone to myeloid cells and to determine gene expression or silencing made this approach less practical. For our studies, we took the alternative approach of targeting a corrective minigene specifically to a predesigned safe harbor site such as the AAVS1 locus, which has previously been shown to permit stable expression of transgenes, and which is also a location that has been shown to have minimal effects on nearby genes.18,20 It may be that one or the other of these gene correction approaches in iPSCs will be the more practical, depending on the disease entity that is being corrected and particular promoter/transgene combination that is required for functional correction.
Management of life-threatening fungal infections in CGD remains a particularly difficult challenge, in that some fungal infections cannot be eradicated with standard management and slowly progress to fatal outcome. Irradiated granulocytes collected from G-CSF–stimulated donors are used as salvage therapy in such settings. This may help because small numbers of oxidase-normal neutrophils work in synergy with CGD neutrophils to kill fungi.34 It is not practical to find HLA-matched granulocyte donors, but patients treated with random donor granulocytes rapidly develop anti-HLA antibodies that limit effectiveness of granulocyte transfusions and have risk of severe transfusion reactions. A source of even small numbers of HLA-autologous oxidase-normal neutrophils to treat selected CGD patients with incurable fungus infection would circumvent the need for random donor granulocyte transfusions. Gene-corrected autologous X-CGD iPSCs such as those described in our report could be used to generate small numbers of mature functional neutrophils ex vivo for infusion to help manage severe fungus infection unresponsive to conventional therapy. Mature neutrophils derived from differentiating iPSCs are end-stage differentiated cells that have no capacity for permanent engraftment nor ability to proliferate, thus circumventing some of the safety concerns for the potential of iPSCs to form teratomas or generate other oncogenic events in subjects.
Our studies demonstrate that ZFN targeting of a disease-corrective donor plasmid minigene to the AAVS1 safe harbor site is a broadly applicable approach to correction of monogenic disorders in iPSCs. Clinical application would require rederivation of X-CGD iPSCs under GMP conditions using methods that are virus-free,35,36 leaving no genetic traces from the reprogramming process. Furthermore, for some applications, methods of deriving transplantable numbers of purified HSCs retaining marrow reconstitution potential from genetically corrected iPSCs still must be developed.37 Despite the current technical barriers to clinical scale-up of iPSC culture and to efficient generation of transplantable HSCs from iPSC, there are compelling reasons4-7 to develop the precise gene-targeting functional correction of iPSCs that we have demonstrated to replace current virus vectors for gene therapy of monogenic inherited disorders.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr S. Rosenzweig, Dr M. Sadat, and other members of the Malech and Cheng Laboratories for technical advice and support. We thank the Johns Hopkins DNA synthesis and sequencing facility for sequencing of PCR fragments derived from iPSC clones.
Research conducted at JHU was supported by NIH grant RO1 HL073781 and Maryland Stem Cell Research Fund Exploratory Grant 2010-MSCRFE-0044-00. Research conducted at NIH was supported by NIAID intramural program projects Z01-AI-000644 and Z01-AI-000988. This work was supported by Grant Award J4G/09/10 from the Chronic Granulomatous Disorder Research Trust (United Kingdom) to L.C., H.L.M., and J.Z.
National Institutes of Health
Authorship
Contribution: J.Z. established and characterized iPSCs and performed targeted gene correction and analyses; C.L.S. established MSCs, conducted promoter silencing studies, and performed neutrophil differentiation and analyses; B.-K.C. assisted with iPSC derivation and culture; U.C. performed ROS and neutrophil morphology analyses; J.P. constructed and generated lentiviral vectors; H.W. assisted with neutrophil marker analyses; S.N.D. performed karyotype analyses; L.C. and H.L.M. supervised the project; and J.Z., C.L.S., L.C., and H.L.M. wrote and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Linzhao Cheng, Institute for Cell Engineering, Johns Hopkins University School of Medicine, 733 N Broadway, BRB780, Baltimore, MD 21205; e-mail: lcheng2@jhmi.edu; or Harry L. Malech, MD, LHD/NIAID/NIH, Bldg 10, Rm 5-3750, 10 Center Dr, MSC-1456, Bethesda, MD 20892-1456; e-mail: hmalech@nih.gov.
References
Author notes
J.Z. and C.L.S. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal