Patient-derived induced pluripotent stem cells (iPSCs) represent a novel and powerful tool for in vitro modeling and correction of human diseases.1 In this issue of Blood, Zou et al report on the generation and correction of iPSCs from a patient with X-linked chronic granulomatous disease (X-CGD),2 a severe primary immunodeficiency (PID) because of defects of the NADPH oxidase.3
Mature neutrophils that were differentiated in vitro from the patient's iPSCs showed defective NADPH oxidase activity, thus recapitulating the CGD phenotype. When the patient's iPSCs were transduced with a self-inactivating lentiviral vector (SIN-LV) allowing expression of gp91phox (the NADPH oxidase component that is mutated in X-CGD) and then differentiated into myeloid cells, rapid decrease of gene expression and poor functional rescue were observed, reflecting LV promoter silencing. In contrast, sustained gp91phox expression was observed when zinc-finger nucleases (ZFNs) were used to introduce by homologous recombination (HR) the gp91phox minigene at the AAVS1 locus, a putative safe harbor resistant to silencing.4 Neutrophils that were differentiated from the ZFN-targeted patient-derived iPSCs showed robust NADPH oxidase activity, thus confirming functional rescue (see figure). Successful in vitro correction of the CGD phenotype by targeting iPSCs has also been recently reported in mice by Mukherjee et al.5
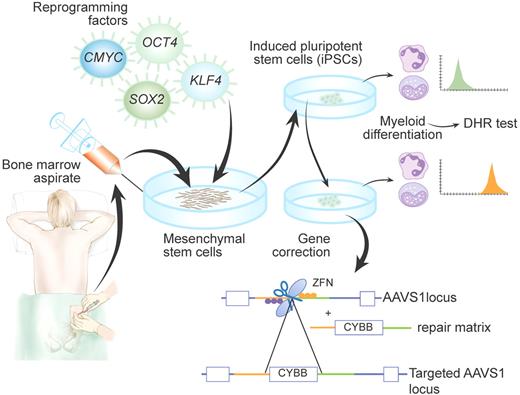
Schematic representation of generation and targeting of induced pluripotent stem cells (iPSCs) derived from a patient with X-linked chronic granulomatous disease (X-CGD), as achieved by Zou et al2 Di-hydrorhodamine (DHR) test was used to confirm that patient-derived iPSCs differentiated into myeloid cells recapitulate the CGD phenotype. Insertion of the CYBB gene into the AAVS1 locus by zinc-finger nucleases (ZFNs) allowed rescue of the cellular phenotype. Professional illustration by Paulette Dennis.
Schematic representation of generation and targeting of induced pluripotent stem cells (iPSCs) derived from a patient with X-linked chronic granulomatous disease (X-CGD), as achieved by Zou et al2 Di-hydrorhodamine (DHR) test was used to confirm that patient-derived iPSCs differentiated into myeloid cells recapitulate the CGD phenotype. Insertion of the CYBB gene into the AAVS1 locus by zinc-finger nucleases (ZFNs) allowed rescue of the cellular phenotype. Professional illustration by Paulette Dennis.
The report by Zou et al is important for several reasons. First, following the recent report of successful generation of iPSCs from patients with various forms of PID,6 this represents the first use of patient-derived iPSCs to model human PIDs in vitro. Furthermore, this study illustrates that disease-specific iPSCs represent a platform to investigate novel approaches to achieve gene correction. Lastly, the authors have performed a thorough analysis of gene-targeted iPSCs to select clones with insertion of gp91phox minigene at only a single AAVS1 allele and no other obvious genomic changes.
The demonstration of stable correction of the disease phenotype by genome editing in iPSCs may have important implications for the development of novel therapeutic approaches to CGD. Hematopoietic cell transplantation (HCT) from HLA-identical donors represents a standard form of treatment for severe cases of CGD, but HCT from alternative donors carries significant risks. Recently, gene therapy for CGD with use of gammaretroviral vectors has proved to be effective for treatment of life-threatening infections, but has been associated with insertional mutagenesis and subsequent myelodysplasia.7 Clonal proliferation because of insertional mutagenesis has been also reported in other trials of gene therapy using retroviral vectors, calling for the development of safer methods. Assessment of the safety profile of newly developed vectors for gene transfer is largely based on in vitro clonogenic assays and serial bone marrow transplantation in mice, but the predictive value of data generated with this approach remains controversial. Generation of patient-derived iPSCs, coupled with robust methods of targeted differentiation and expansion, may represent a significant step forward to assess the safety and efficacy of novel strategies to achieve gene correction.
However, several problems remain to be addressed, as also demonstrated by Zou et al.2 Silencing during reprogramming or differentiation is common after integration of transgene-encoding LV vectors into iPSC genomes. Furthermore, the specific nature of the integration site may affect transgene expression, as confirmed by variable levels of β-globin expression among LV-transduced iPSCs from patients with β-thalassemia.8 Even more importantly, random (or semirandom) integration of viral vectors is associated with risks of insertional mutagenesis.
HR has been used to exchange endogenous DNA sequences with donor DNA templates at specific genomic locations. The efficiency of HR is relatively low, but can be increased on induction of targeted DNA breaks by employing ZFNs9 and homing endonucleases (HEs)10 that can be engineered to recognize specific DNA target sequences. However, these DNA breaks may also be repaired through nonhomologous end-joining pathway, an imprecise repair process that may introduce mutations. Hence the need for careful analysis and selection of targeted cells.
With this important caveat, ZFN- or HE-mediated HR targeting of the disease-associated locus may permit correction of gene mutations while maintaining regulation of gene expression. While this would represent the ideal approach to gene therapy for human genetic diseases, its major limitation is that mutations are often scattered along genes, and correcting each of them would require the development of a large array of customized targeting vectors. Furthermore, this approach is not suitable to correct diseases resulting from gross genomic changes, such as large deletions.
Zou et al have used ZFNs to induce HR at the AAVS1 site, that is, the integration site of adeno-associated virus 2 in the first intron of the ubiquitously expressed PPP1R12C gene. The AAVS1 site is considered a safe harbor for gene insertion. No disease has been associated with mono- or bi-allelic mutations of PPP1R12C. Furthermore, AAVS1-targeted human embryonic stem cells and iPSCs maintain karyotype stability and pluripotency. Targeting of safe harbors is an attractive alternative to true gene correction as it might facilitate correction of a variety of genetic diseases, irrespective of the nature and location of the mutation. However, only limited data are available to confirm that the AAVS1 site is indeed a safe harbor. Recently, Papapetrou et al used a combined bioinformatic and functional approach to identify safe harbor sites after LV-mediated transduction of human iPSCs.8 In that study, definition of safe harbors was based on distance from transcription initiation sites, cancer-related genes, or micro-RNA genes; and location outside transcriptional units or ultraconserved regions of the human genome. As significant progress continues to be made in the functional analysis of the human genome, it is likely that several safe harbors will be identified. Definition of a safe harbor will also likely depend on the nature of the cell type selected for gene targeting and of its progeny of differentiated cells. In this process, patient-derived iPSCs promise to be a great resource to test the safety and efficacy of novel approaches to gene therapy.
Conflict-of-interest disclosure: The author has received research support from Celletics. ■