Abstract
T cell–mediated heterologous immunity to different pathogens is promising for the development of immunotherapeutic strategies. Aspergillus fumigatus and Candida albicans, the 2 most common fungal pathogens causing severe infections in immunocompromised patients, are controlled by CD4+ type 1 helper T (TH1) cells in humans and mice, making induction of fungus-specific CD4+ TH1 immunity an appealing strategy for antifungal therapy. We identified an immunogenic epitope of the A fumigatus cell wall glucanase Crf1 that can be presented by 3 common major histocompatibility complex class II alleles and that induces memory CD4+ TH1 cells with a diverse T-cell receptor repertoire that is cross-reactive to C albicans. In BALB/c mice, the Crf1 protein also elicits cross-protection against lethal infection with C albicans that is mediated by the same epitope as in humans. These data illustrate the existence of T cell–based cross-protection for the 2 distantly related clinically relevant fungal pathogens that may foster the development of immunotherapeutic strategies.
Introduction
The saprophytic mold Aspergillus fumigatus and Candida albicans, a commensal of the mucocutaneous membranes, are the most common fungal pathogens among immunocompromised patients. Both fungi cause severe invasive infections and are responsible for substantial morbidity and mortality despite effective antifungal treatment.1 Immunotherapeutic approaches are promising to enhance pathogen-specific immunity and thereby limit infectious complications. Previously, adoptively transferred A fumigatus-specific type 1 helper T (TH1) cell clones generated using heat-inactivated fungus as antigen were capable of ameliorating invasive aspergillosis without causing adverse effects in patients after hematopoietic stem cell transplantation (HSCT).2 The beneficial effect of adoptive T-cell therapy could be increased if the transferred T cells were reactive not only to A fumigatus but also to other fungal pathogens. Despite genetic and morphologic disparities and distinct clinical diseases, the host immunity to different fungi is quite similar. In invasive aspergillosis as well as in invasive candidiasis TH1-biased immune responses correlate with protective immunity and resistance, whereas type 2 helper T (TH2)–biased responses generally lead to an exacerbation of disease.3-5 TH1 cell activation is instrumental to clear the infection by improving the effector function of innate immune cells through the release of proinflammatory cytokines.4
Previous data in mice have suggested the existence of cross-protective immunity between different fungi. For example, vaccination with Saccharomyces yeast protected mice against subsequent lethal infection with other fungi.6,7 The mechanism of this cross-protection is not yet fully elucidated and could be based either on unspecific immunostimulatory effects indiscriminately enhancing the effector function of innate immune cells or on adaptive immunity triggering antigen-specific antibody– or T cell–mediated responses. Because different fungi share conserved gene products and T-cell receptor (TCR) binding degeneracy allows recognition of several similar peptides by a single TCR,8 the presence of memory TH1 cells responding to immunodominant major histocompatibility complex (MHC) class II peptide antigens common to several fungi is not unlikely. Therefore, we sought to determine whether A fumigatus antigens can induce TH1 cells with cross-reactivity to other fungal pathogens in humans and whether these antigens would be similarly cross-protective in mice.
Methods
Cloning, expression, and purification of recombinant proteins
Aspergillus fumigatus superoxide dismutase,9 catalase,10 peptidase,11 and 1,3-β-glucanosyl-transferase Gel112 were produced in the yeast Pichia pastoris as described previously.13 Extracellular cell wall glucanase Crf113 (cDNA provided by Utz Reichard, University of Göttingen, Germany), the major allergen, and cytotoxin AspF114 were expressed in the human cell line phoenix GALV.15 We introduced the mutation AspF1 His136Leu to diminish toxicity in the eukaryotic producer cells.16 Transgene expression of the pcDNA3.1-TOPO vector (Invitrogen) was enhanced with 1mM sodium butyrate (Sigma-Aldrich). Producer cells were lysed with 50mM Tris-HCl, pH 7.4, 150mM NaCl, 1mM EDTA (ethylenediaminetetraacetic acid), and 1% Triton X-100, and the recombinant proteins were purified under native conditions using M2-agarose according to the manufacturer's instructions (Sigma-Aldrich).
Peptides
A peptide library covering the complete A fumigatus Crf1 sequence consisting of 94 15-mer peptides overlapping by 4 amino acids (aa) was divided into subpools of 10 peptides and 94 single peptides (Crf1/p1-Crf1/p94; PANATecs). Three 11-mer peptides of Crf1/p40/41 overlapping by 2 aa (QETFHTYTIDW, TFHTYTIDWTK, and HTYTIDWTKDA) were obtained from JPT Peptide Technologies. Cytomegalovirus (CMV) peptide pp65283-297 (KPGKISHIMLDVAFT) was obtained from ProImmune. The 15-mer peptides A flavus Crf1162-176 (HTYTIDWNKDQTTWS), A terreus ATEG_05185159-173 (HTYTVNWSKDQIQWL), A nidulans Crf1162-176 (HTYTVTWSPDAISWI), A niger Crf1160-174 (HTYTVEWTESATTWS), and C albicans Crh1161-175 (HTYVIDWTKDAVTWS) were provided by S.S.
Fungal strains, generation of fungal extracts, and inactivated fungi
Fungal strains A fumigatus D141, A nidulans FGSCA4, and C albicans SC5314 and clinical isolates A fumigatus, A terreus, A flavus, A niger, C albicans, C glabrata, and C krusei were provided by Sven Krappmann and Prof Joachim Morschhäuser (both University of Würzburg, Germany), and Reno Frei (University Hospital Basel, Switzerland).
Aspergillus conidia were cultured for 0, 4, 8, or 12 hours and Candida species for 24 hours. Fungi were then inactivated with 70% ethanol for 30 minutes or by heating to 96°C for 60 minutes. Fungal extracts of A fumigatus were generated as described previously.17 Protein content was measured by Bradford protein assay (Perbio Science) according to the manufacturer's instructions. Inactivated fungi and extracts were tested for sterility on Sabouraud dextrose agar for 3 days.
Isolation of T cells and generation of DCs
Transfection of DCs with Crf1 mRNA
Aspergillus fumigatus Crf1 cDNA was cloned into the pGEM4Z vector (provided by Kris Thielemans, Vrije University of Brussels, Belgium) and in vitro–transcribed using an mMESSAGE mMACHINE T Ultra kit (Ambion) according to the manufacturer's instructions. Because the Crf1 cDNA lacked the endogenous signal sequence, it was fused to the N-terminal domain of the invariant chain to enhance antigen presentation by MHC class II.20 Maturated DCs were electroporated with 15 μg of mRNA at 300 V and 150 μF using the EasyjecT Plus apparatus (Equibio) in serum-free medium (Opti-MEM; Gibco).19
T-cell expansion, T-cell cloning, restimulation of T-cell clones, and quantification of cytokines by IFN-γ, ELISA, and enzyme-linked immunosorbent spot assays and cytometric bead array
For analysis of recombinant proteins and epitope mapping, 1 × 107 PBMCs were incubated with 5 μg/mL protein or peptide for 7 days in addition of 5 U/mL IL-2 (Proleukin; Chiron). Antigen specificity was analyzed by IFN-γ enzyme-linked immunosorbent assay (ELISA) after restimulation with autologous monocytes incubated overnight with 5 μg/mL protein in a responder-to-stimulator (R:S) ratio of 5:1. Epitope mapping was performed by IFN-γ enzyme-linked immunosorbent spot assay after stimulation with peptide-pulsed autologous DCs at an R:S ratio of 10:1.
For T-cell expansion, 1 × 107 PBMCs were incubated with 5 μg/mL peptide, and 5 U/mL IL-2 was added. On day 7, T cells were restimulated with autologous peptide-pulsed monocytes at an R:S ratio of 5:1, and 10 ng/mL IL-7 and IL-15 (R&D Systems) were added.
For T-cell cloning, 6 × 107 PBMCs were stimulated overnight with 5 μg/mL peptide and 1 μg/mL CD40 antibody (HB14; Miltenyi Biotec). CD154+ cells were isolated by magnetic separation (Miltenyi Biotec). Selected cells were cultured with γ-irradiated autologous PBMCs (ratio, 1:50), and 5 U/mL IL-2 was added until day 7. After day 7 the culture was supplemented with 10 ng/mL IL-7 and IL-15. On day 14, T-cell clones were generated as described previously.21 T-cell clones were stimulated either with mature peptide-pulsed (2.5 μg/mL) or Crf1 mRNA-transfected DCs, immature DCs incubated for 24 hours with A fumigatus cell extract (20 μg/mL) in the presence of cytokine maturation cocktail, or immature DCs incubated for 24 hours with inactivated A fumigatus or C albicans fungus (multiplicity of infection 3) in an R:S ratio of 5:1. Levels of IFN-γ, granulocyte macrophage–colony-stimulating factor (GM-CSF), IL-10, and IL-5 in culture supernatant of human T-cell clones were quantified using ELISA or cytometric bead array (BD Biosciences) according to the manufacturer's instructions.
TCR binding affinity, MHC class II restriction, and Vβ repertoire analysis
TCR binding affinity was determined by peptide titration. Autologous DCs pulsed with decreasing amounts of peptide (3 to 3 × 10−4 μg/mL) were used for T-cell stimulation in a ratio of 1:10.
Human leukocyte antigen (HLA) restriction was analyzed with partially matched allogeneic Epstein-Barr virus-lymphoblastoid cell lines (LCLs; provided by Patricia Comoli [University Pavia, Italy], and Inge Jedema [Leiden University Medical Center, The Netherlands]) pulsed with 10−3 to 1 μg/mL peptide using IFN-γ ELISA.
The TCR Vβ repertoire was analyzed by flow cytometry (IOTest Beta Mark kit; Beckman Coulter), polymerase chain reaction (PCR),22 and sequence analysis.23 A phycoerythrin-conjugated Crf1/p41-specific HLA-DRB1*0401 MHC class II tetramer was used according to the manufacturer's instructions (Beckman Coulter).
Animals
Female C57BL/6 and BALB/c mice, 8 to 10 weeks old, were purchased from Charles River Laboratories (Calco). Homozygous IFN-γ–deficient mice on BALB/c were bred under specific pathogen-free conditions at the Animal Facility of Perugia University, Perugia, Italy. Experiments were performed according to the Italian Approved Animal Welfare Assurance A-3143-01.
Experimental models of coinfection and vaccination
Mice were infected with 2 × 107 resting A fumigatus conidia intranasally,6 with 108C albicans intragastrically,24 or with both fungi. For coinfection, mice were infected a week before the subsequent infection. In the vaccination model, mice were injected intranasally with 2 × 107Aspergillus resting conidia/20 μL saline once, 14 days before infection, or with 5 μg of purified Crf1p antigen6 and 10nM murine cytosine guanine dinucleotide (CpG) oligodeoxynucleotide 1862, administered 14, 7, and 3 days before intragastric C albicans infection.6 Mice were immunosuppressed intraperitoneally with 150 mg/kg cyclophosphamide a day before infection. Mice were monitored for fungal growth (colony-forming units [CFUs]/organ expressed as mean ± SE) and inflammatory pathology. Mice dying of fungal challenge routinely underwent necropsy for histopathologic confirmation of infection.6 Sections (3-4 μm) of paraffin-embedded tissues were stained using the periodic acid-Schiff procedure (Bio-Optika). For differential counts, cytospin preparations from stomachs or lungs were made and stained with May-Grunwald Giemsa reagents (Carlo Erba Reagents). At least 200 cells/cytospin preparation were counted, and the absolute number of each cell type was calculated. Photographs were taken using a high-resolution Olympus BX51 microscope, with an Olympus DP71 camera using Olympus Cell P 3.3 software. The objective lenses were Plan 20×/0.40 Ph1 for histology and Plan N 100×/1.25 Oil Ph3 for BAL. The image medium was EVKITT mounting medium (Bio-Optika) for histology and Olympus Immersion Oil for BAL. Adobe Photoshop CS3 was used for image processing.
Real-time PCR
Real-time reverse transcription-PCR was performed from purified CD4+ T cells from thoracic (pulmonary aspergillosis) or mesenteric (gastrointestinal candidiasis) lymph nodes as described previously.6
Proliferation assay
For mice, DCs (1 × 105) and CD4+ T cells (5 × 105) were purified from spleens untreated or vaccinated with A fumigatus or C albicans, by magnetic separation of MicroBeads (Miltenyi Biotec), and then they were cocultured with and without anti-CD3ϵ antibody (clone 145-2C11; BD Pharmingen), heat-inactivated Aspergillus or Candida (at a cell:fungi ratio of 2:1), or 5 μg/mL Crf1/p22 or Crf1/p41 peptide. Human T-cell lines (1 × 105) were restimulated on day 20 with peptide-pulsed DCs (1 × 104). Proliferation was measured by thymidine labeling.6 The data are expressed as mean counts per minute ± SE of triplicate cultures.
Statistical analysis
Data were analyzed by Prism 5.0 (GraphPad Software). Student t test or analysis of variance and Bonferroni test were used to determine the statistical significance (P) of differences in organ clearance and in vitro assays. The data reported are either from one representative experiment of 3 independent experiments (for in vivo data) or are pooled from 3 experiments (for in vitro assays). The in vivo groups consisted of 6 mice/group.
Results
The p41 epitope of the A fumigatus cell wall glucanase Crf1 is highly immunogenic in healthy individuals
Given that the identification of potential protective fungal antigens is rather intricate because of the extensiveness of fungal genomes, we analyzed 6 different A fumigatus recombinant proteins based on their previously reported immunogeneic reactivity in mice and humans.6,14 Most of the proteins, such as superoxide dismutase, catalase, the major allergen and cytotoxin AspF1 and 1,3-β-glucanosyl-transferase Gel1, elicited only weak IFN-γ responses in some donors. By contrast, glycosylphosphatidylinositol-anchored extracellular cell wall glucanase Crf1 and secreted protein peptidase induced high IFN-γ in 6 of 7 and 5 of 10 tested donors, respectively (data not shown). Because of the broad response and the sequence similarity to the C albicans ortholog Crh1, in subsequent experiments we focused on the protein Crf1. The response to Crf1 was mediated by CD4+ TH1 cells, because MHC class II blocking antibody nearly abrogated IFN-γ release (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). To identify Crf1-specific MHC class II epitopes, we mapped PBMCs of 7 healthy individuals with different HLA-DRB1* alleles with a custom-made peptide library consisting of 94 overlapping 15-mer peptides. After overnight stimulation, no IFN-γ response by enzyme-linked immunosorbent spot assay was detected, indicating that the peripheral memory T-cell repertoire of Crf1 is low. By prestimulation with subpools for 7 days and restimulation with the respective single peptides, we identified 10 potential candidate epitopes (p1, p9, p41, p48, p49, p58, p59, p69, p84, and p89; supplemental Table 1). Based on the results obtained by epitope mapping, the probability of antigen presentation by certain MHC class II alleles assessed by epitope prediction,25 previously reported immunogenic reactivity,26 and sequence similarity to the C albicans ortholog Crh1, we generated CD4+ TH1 T-cell clones specific for the peptides p1, p22, p27, p41, p48, p59, p69, and p84 (Figure 1A; supplemental Table 1).
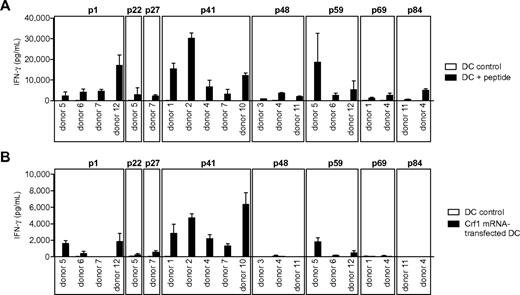
Response of peptide-specific T-cell clones to endogenously processed antigen presented by DCs. (A) T-cell clones from different donors specific for the peptides p1, p22, p27, p41, p48, p59, p69, and p84 were stimulated with peptide-pulsed DCs or (B) DCs transfected with Crf1 mRNA (n = 3). Number of T-cell clones reacting to Crf1 mRNA-transfected DCs/number of peptide-specific T-cell clones: p1 donor 5, 3/3; p1 donor 6, 1/5; p1 donor 7, 0/6; p1 donor 12, 10/10; p22 donor 5, 1/2; p27 donor 7, 1/3; p41 donor 1, 11/11; p41 donor 2, 16/16; p41 donor 4, 2/2; p41 donor 7, 4/4; p41 donor 10, 3/3; p48 donor 3, 0/7; p48 donor 4, 0/4; p48 donor 11, 0/5; p59 donor 5, 1/3; p59 donor 6, 2/2; p59 donor 12, 1/7; p69 donor 1, 0/3; p69 donor 4, 0/2; p84 donor 11, 0/5; p84 donor 4, 0/4. Shown is 1 clone per antigen and donor.
Response of peptide-specific T-cell clones to endogenously processed antigen presented by DCs. (A) T-cell clones from different donors specific for the peptides p1, p22, p27, p41, p48, p59, p69, and p84 were stimulated with peptide-pulsed DCs or (B) DCs transfected with Crf1 mRNA (n = 3). Number of T-cell clones reacting to Crf1 mRNA-transfected DCs/number of peptide-specific T-cell clones: p1 donor 5, 3/3; p1 donor 6, 1/5; p1 donor 7, 0/6; p1 donor 12, 10/10; p22 donor 5, 1/2; p27 donor 7, 1/3; p41 donor 1, 11/11; p41 donor 2, 16/16; p41 donor 4, 2/2; p41 donor 7, 4/4; p41 donor 10, 3/3; p48 donor 3, 0/7; p48 donor 4, 0/4; p48 donor 11, 0/5; p59 donor 5, 1/3; p59 donor 6, 2/2; p59 donor 12, 1/7; p69 donor 1, 0/3; p69 donor 4, 0/2; p84 donor 11, 0/5; p84 donor 4, 0/4. Shown is 1 clone per antigen and donor.
To determine whether the different Crf1 peptide epitopes can be actually processed and presented by DCs, the T-cell clones were stimulated with DCs transfected with Crf1 mRNA. All generated p41-specific CD4+ T-cell clones were activated readily, whereas only some of the p1-, p22-, p27-, and p59-specific CD4+ T-cell clones responded to endogenously processed antigen. None of the CD4+ T-cell clones specific for the peptides p48, p69, and p84 responded to Crf1 mRNA-transfected DCs (Figure 1B).
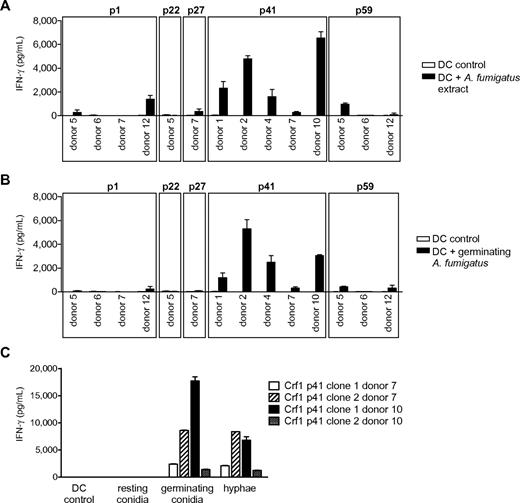
We next assessed the efficiency of T-cell activation by naturally occurring fungal antigen. Because fungal surface structures can influence DC function via engagement of Toll-like and other pattern recognition receptors,24,27,28 we first stimulated the T-cell clones with DCs incubated with fungal extract to minimize the risk of unspecific T-cell responses. All Crf1/p41-specific T-cell clones of all 5 donors and most Crf1/p1-specific T-cell clones of 2 of 4 donors responded to A fumigatus extract, whereas only very few T-cell clones specific for Crf1/p27 and Crf1/p59 and none of the Crf1/p22-specific T-cell clones could be activated (Figure 2A). All T-cell clones responding to fungal extract could likewise be activated by inactivated fungus (Figure 2B). Because A fumigatus changes its morphology and concurrently also its antigenic properties during conversion from inert fungal spores to invasive pathogenic hyphae,29 we wanted to ascertain which morphotypes of A fumigatus are recognized by the Crf1/p41-specific T-cell clones. Although inactivated resting conidia elicited no or only weak responses, germinating conidia and outgrown hyphae induced high T-cell responses (Figure 2C).
Response of Crf1 peptide-specific T-cell clones to stimulation with native A fumigatus antigen. (A) T-cell clones from different donors specific for p1, p22, p27, p41, and p59 that responded to stimulation with Crf1 mRNA-transfected DCs were stimulated with A fumigatus cell extract or (B) inactivated germinating A fumigatus conidia (n = 3). Shown is 1 clone per antigen and donor. (C) Crf1/p41-specific T-cell clones respond to stimulation with DCs incubated with germinating A fumigatus conidia (6 hours) or hyphae (12 hours) but not or only weakly to resting conidia (0 hours; n = 3).
Response of Crf1 peptide-specific T-cell clones to stimulation with native A fumigatus antigen. (A) T-cell clones from different donors specific for p1, p22, p27, p41, and p59 that responded to stimulation with Crf1 mRNA-transfected DCs were stimulated with A fumigatus cell extract or (B) inactivated germinating A fumigatus conidia (n = 3). Shown is 1 clone per antigen and donor. (C) Crf1/p41-specific T-cell clones respond to stimulation with DCs incubated with germinating A fumigatus conidia (6 hours) or hyphae (12 hours) but not or only weakly to resting conidia (0 hours; n = 3).
TH1 cells contribute to antifungal immunity by augmenting the effector functions of innate immune cells, particularly monocytes and neutrophils, via secretion of cytokines such as IFN-γ and GM-CSF.30-32 Culture supernatant of activated p41-specific T-cell clones contained high concentrations of both cytokines (supplemental Figure 2) and in addition showed a tendency to improve killing of A fumigatus conidia by monocytes, although differences in conidial uptake by the phagocytes cannot be excluded (supplemental Figure 3).
These findings suggest that most healthy donors harbor highly reactive TH1 cells specific for the A fumigatus Crf1/p41 epitope that are capable of improving fungal killing by innate immune cells, whereas immunity to all other analyzed peptides seems to be less widespread.
Aspergillus fumigatus Crf1/p41-specific T-cell clones are cross-reactive to C albicans
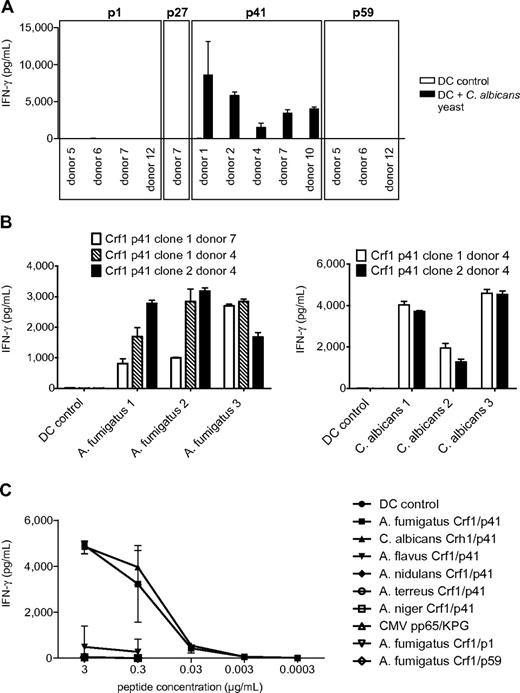
Candida albicans is the second most prevalent fungal pathogen in HSCT recipients, and the aa sequence of the A fumigatus protein Crf1 shows some sequence similarity to its ortholog Crh1 in C albicans. Therefore, we ascertained whether the T-cell clones that recognized endogenously processed A fumigatus are also cross-reactive to C albicans. All Crf1/p41-specific T-cell clones of all donors recognized and responded to C albicans, whereas none of the Crf1/p1-, Crf1/p27-, and Crf1/p59-specific T-cell clones could be activated (Figure 3A). The A fumigatus Crf1/p41 epitope and the corresponding C albicans epitope are nearly identical differing only in 1 aa, where valine is substituted for threonine at position 4 (supplemental Figure 4A). Because these observations were made in established strains, we also stimulated T-cell clones with different clinical isolates of A fumigatus and C albicans and could further confirm this cross-reactivity of the Crf1/p41 epitope (Figure 3B).
Aspergillus fumigatus Crf1/p41-specific T-cell clones show highly specific cross-reactivity to C albicans and can be activated by different clinical isolates of both fungi. (A) All A fumigatus Crf1/p41-specific T-cell clones from different donors respond to stimulation with inactivated C albicans yeast (n = 3). Shown is 1 clone per antigen and donor. (B) Aspergillus fumigatus Crf1/p41-specific T-cell clones respond to different inactivated clinical isolates of germinating A fumigatus conidia and C albicans yeast (n = 3). (C) Aspergillus fumigatus Crf1/p41-specific T-cell clones from donors with different MHC class II restriction were stimulated with A fumigatus Crf1/p41 peptide, the homologous peptides of C albicans, A flavus, A nidulans, A terreus, and A niger (supplemental Figure 4B) or the unrelated peptides CMV pp65 KPG (KPGKISHIMLDVAFT) and A fumigatus Crf1/p1 and p59 (supplemental Table 1; representative experiments).
Aspergillus fumigatus Crf1/p41-specific T-cell clones show highly specific cross-reactivity to C albicans and can be activated by different clinical isolates of both fungi. (A) All A fumigatus Crf1/p41-specific T-cell clones from different donors respond to stimulation with inactivated C albicans yeast (n = 3). Shown is 1 clone per antigen and donor. (B) Aspergillus fumigatus Crf1/p41-specific T-cell clones respond to different inactivated clinical isolates of germinating A fumigatus conidia and C albicans yeast (n = 3). (C) Aspergillus fumigatus Crf1/p41-specific T-cell clones from donors with different MHC class II restriction were stimulated with A fumigatus Crf1/p41 peptide, the homologous peptides of C albicans, A flavus, A nidulans, A terreus, and A niger (supplemental Figure 4B) or the unrelated peptides CMV pp65 KPG (KPGKISHIMLDVAFT) and A fumigatus Crf1/p1 and p59 (supplemental Table 1; representative experiments).
We then analyzed whether our T-cell clones might cross-react to other clinically relevant Aspergillus and Candida species, such as A flavus, A nidulans, A niger, A terreus, C glabrata, and C krusei because they also express proteins with sequence similarity to A fumigatus Crf1. With the exception of the Crf1/p1-specific T-cell clones of 1 of 4 donors that showed weak cross-reactivity to A flavus, A nidulans, and A terreus, none of the Crf1/p27-, Crf1/p41-, and Crf1/p59-specific T-cell clones showed any significant cross-reactivity to these fungi (data not shown). To further confirm the specificity of TCR engagement and T-cell activation, we stimulated Crf1/p41-specific T-cell clones from donors with different MHC class II restriction with the homologous 15-mer peptides of C albicans Crh1 and of other clinically relevant Aspergillus species, including A flavus, A nidulans, A niger, and A terreus, and the unrelated peptides Crf1/p1 and Crf1/p59 and of CMV pp65, and we could show that T-cell activation is highly specific (Figure 3C). The weak cross-reactivity to the Crf1/p41 homolog of A flavus is possibly explained by its higher homology to the A fumigatus sequence than of the other Aspergillus species (supplemental Figure 4B).
Thus, Crf1/p41-specific TH1 cells represent a core repertoire in antifungal immunity, being highly specific and reactive to A fumigatus and also to C albicans, the second most important pathogenic fungus in HSCT patients.
Cross-reactive Crf1/p41-specific CD4+ TH1 cells are diverse in their Vβ repertoire and HLA II restriction
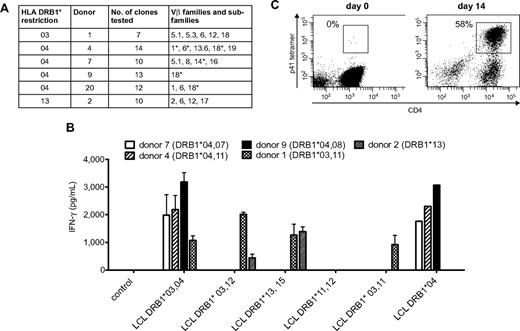
Because Crf1/p41-specific T cells can be activated by A fumigatus as well as C albicans and all humans are in permanent contact with both pathogens via inhalation of fungal spores or commensalism, we reasoned that the T-cell repertoire of memory cells recognizing this epitope should be broad and polyclonal. We therefore determined the TCR Vβ chains of 7 to 14 Crf1/p41-specific CD4+ T-cell clones of each donor by flow cytometry, PCR, and nucleotide sequence analysis. The TCRs of each donor belonged to 1 to 5 different Vβ families and showed additional variations on the level of nucleotide sequence in Vβ chains of the same family, indicating a diverse repertoire of Crf1/p41-specific T cells (Figure 4A).
Cross-reactive Crf1/p41-specific T cells show a diverse TCR Vβ repertoire, multiple MHC class II restriction, and high proliferative capacity. (A) The TCR Vβ repertoire of p41-specific CD4+T-cell clones is diverse in different donors. TCR Vβ chains were determined by flow cytometry, PCR, nucleotide sequence analysis, or a combination of these techniques. *TCRs differ on the level of nucleotide sequence. Nomenclature is according to Wei et al.50 (B) The Crf1/p41 epitope can be presented by HLA-DRB1*03, 04, and 13 and allows cross-recognition between HLA-DRB1*03 and 13. The HLA restriction of Crf1/p41-specific T-cell clones from 5 different donors was determined by stimulation with peptide-pulsed unmatched or partially matched LCLs (n = 2; for LCL DRB1*04, n = 1). (C) Crf1/p41-specific T cells are not terminally differentiated and can be rapidly expanded in vitro. 107 PBMCs could be expanded to a median of 3.5 × 107 cells (range, 3-4.9 × 107 cells) within 14 days, and the frequency of antigen-specific cells as determined by an HLA-DRB1*04 restricted tetramer reached levels of 42% to 63% (median, 50.6%). T cells were restimulated with autologous peptide-pulsed monocytes on day 7, and culture medium was supplemented with IL-2, IL-7, and IL-15 (representative experiment, n = 8).
Cross-reactive Crf1/p41-specific T cells show a diverse TCR Vβ repertoire, multiple MHC class II restriction, and high proliferative capacity. (A) The TCR Vβ repertoire of p41-specific CD4+T-cell clones is diverse in different donors. TCR Vβ chains were determined by flow cytometry, PCR, nucleotide sequence analysis, or a combination of these techniques. *TCRs differ on the level of nucleotide sequence. Nomenclature is according to Wei et al.50 (B) The Crf1/p41 epitope can be presented by HLA-DRB1*03, 04, and 13 and allows cross-recognition between HLA-DRB1*03 and 13. The HLA restriction of Crf1/p41-specific T-cell clones from 5 different donors was determined by stimulation with peptide-pulsed unmatched or partially matched LCLs (n = 2; for LCL DRB1*04, n = 1). (C) Crf1/p41-specific T cells are not terminally differentiated and can be rapidly expanded in vitro. 107 PBMCs could be expanded to a median of 3.5 × 107 cells (range, 3-4.9 × 107 cells) within 14 days, and the frequency of antigen-specific cells as determined by an HLA-DRB1*04 restricted tetramer reached levels of 42% to 63% (median, 50.6%). T cells were restimulated with autologous peptide-pulsed monocytes on day 7, and culture medium was supplemented with IL-2, IL-7, and IL-15 (representative experiment, n = 8).
To evaluate the utility of the Crf1/p41 epitope for potential clinical application, we defined the HLA restriction of Crf1/p41-specific CD4+ T-cell clones from different donors by stimulation with peptide-pulsed partially matched LCLs. We found indications that presentation of Crf1/p41 is mediated by HLA-DRB1*03, 04, and 13 and that in some instances DRB1*03-restricted TCRs might be able to recognize Crf1/p41 peptide presented by DRB1*13 and vice versa (Figure 4B). Because presentation of Crf1/p41 also could be mediated by other MHC class II alleles, we verified the HLA restriction by stimulation of PBMCs from additional donors. All HLA-DRB1*03, 04, and 13 donors reacted to Crf1/p41, whereas all 5 donors lacking HLA-DRB1*03, 04, and 13 did not (data not shown). Further analysis elucidated that the 11-mer peptide HTYTIDWTKDA is the minimal epitope of Crf1/p41 for all 3 HLA alleles (data not shown).
Together, these results demonstrate that the Crf1/p41 epitope induces an oligoclonal immune response in the majority of individuals and can be presented by multiple MHC class II alleles.
Cross-reactive Crf1/p41-specific CD4+ TH1 cells are not terminally differentiated and show high proliferative capacity in response to antigen
The feasibility of immunotherapeutic strategies such as vaccination or adoptive T-cell transfer is intimately linked to the capacity of antigens to efficiently activate and expand pathogen-specific T cells. Using an HLA-DRB1*0401–restricted Crf1/p41 tetramer, we identified within the CD4+ T-cell fraction 1 to 2 tetramer-positive cells/105 CD4+ T cells. Despite this extremely low precursor frequency, we were able to generate Crf1/p41-specific T-cell lines within 14 days, with a median percentage of antigen-specific T cells of 50.6% (Figure 4C). These Crf1/p41-specific CD4+ T-cell lines produced high amounts of the TH1 cytokine IFN-γ (2 705-35 927 pg/mL) and specifically proliferated on peptide stimulation (supplemental Figure 5).
Infection with A fumigatus conidia or vaccination with Crf1 protein protects mice against subsequent infection with C albicans
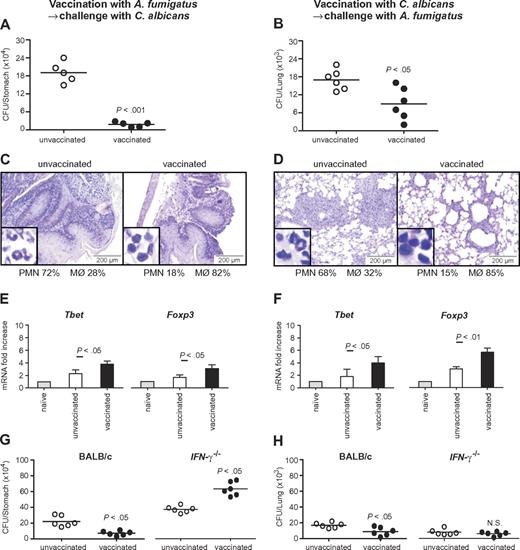
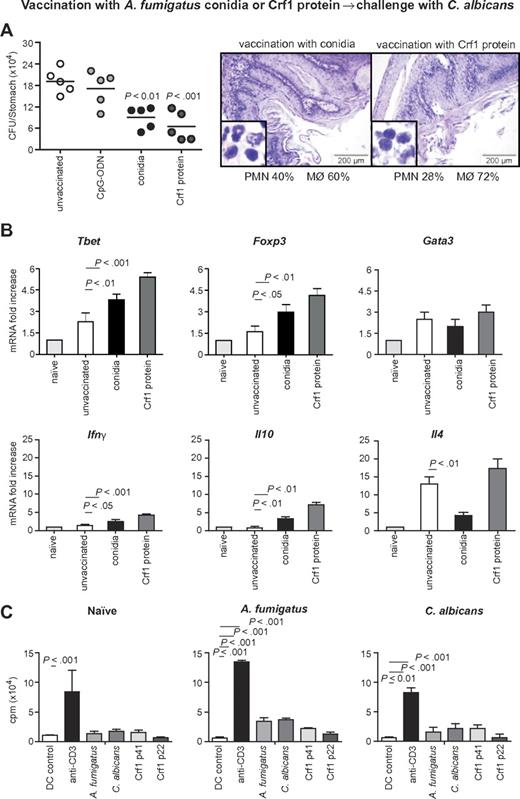
To assess the relevance of the cross-reactive T-cell repertoire in vivo, we infected immunosuppressed C57BL/6 mice either intrapulmonarily with A fumigatus or intragastrically with C albicans a week before rechallenge with C albicans or A fumigatus, respectively. The results show that preinfection with either fungus significantly restricted the fungal growth after rechallenge (Figure 5A-B), and ameliorated inflammatory pathology in the respective target organs, the lung and stomach, as judged by the reduced numbers of abscesses, acanthosis, and local inflammatory cell recruitment (Figure 5C-D). Furthermore, preinfection with A fumigatus or C albicans activated Tbet+/Foxp3+ CD4+ T cells in the respective draining lymph nodes (Figure 5E-F). Cross-protection by either fungus was greatly reduced in IFN-γ–deficient mice, further suggesting that protection is mediated by cross-protective TH1 cells (Figure 5G-H). To verify whether the Aspergillus versus Candida cross-protection can be mediated by the Aspergillus Crf1 protein, we vaccinated mice with Aspergillus conidia or the Crf1 protein and CpG as adjuvant before C albicans infection. We have already shown that the Crf1 protein confers vaccine-induced resistance to Aspergillus.6 We found that both vaccination strategies reduced the fungal burden and inflammatory pathology in the stomach of mice with candidiasis (Figure 6A). This correlated with the activation of Tbet+/Foxp3+ CD4+ T cells, and their respective cytokines, IFN-γ and IL-10, in mesenteric lymph nodes. As expected, IL-4–producing Gata3+ CD4+ T cells were not activated (Figure 6B). Antigen-specific proliferation of CD4+ spleen cells in untreated mice or animals vaccinated with A fumigatus or C albicans suggests that the observed cross-protection could be mediated by T cells recognizing the same epitope p41 of the Crf1 protein that mediates cross-reactivity in humans. In contrast, the previously described epitope p2226 did not induce significant proliferation in vaccinated animals (Figure 6C).
Cross-protection between A fumigatus and C albicans in mice is mediated by TH1 immunity. (A,C,E,G) Cross-protection by A fumigatus against gastrointestinal candidiasis. Mice treated with resting A fumigatus conidia intranasally a week before intragastric injection with Calbicans show reduced fungal growth (A), inflammatory cell recruitment (insets), and acanthosis compared with untreated animals (C). (B,D,F,H) Cross-protection by C albicans against invasive pulmonary aspergillosis. Mice treated with C albicans intragastrically a week before intranasal injection of resting A fumigatus conidia show reduced fungal growth (B) and inflammatory cell recruitment and amelioration of lung inflammatory pathology compared with untreated animals (D). (E-F) Infection with A fumigatus (E) or with C albicans (F) showed a significant increase in protective TH1 and regulatory T cells. (G-H) Cross-protection between A fumigatus and C albicans is dependent on IFN-γ. IFN-γ–deficient mice treated with A fumigatus (G) or C albicans (H) show no reduced fungal growth in the lungs or the stomach, respectively. The reduced fungal burden in IFN-γ–deficient mice is because of compensatory cytokines. Shown are fungal growth (CFUs/organ, mean ± SE), stomach (C) and lung histology (D), differential cell counts in cytospin preparations from stomachs (C) and lungs (D) and transcription factor mRNA expression in CD4+ T cells from mesenteric (candidiasis, E) or thoracic (aspergillosis, F) lymph nodes a week after infection. Shown are the results of one representative experiment of 3 independent experiments (for in vivo data) or pooled from 3 experiments (for in vitro assays). Bars represent SE. Photographs were taken using a high-resolution Olympus BX51 microscope, using Olympus Cell P 3.3 software. P, vaccinated versus unvaccinated animals.
Cross-protection between A fumigatus and C albicans in mice is mediated by TH1 immunity. (A,C,E,G) Cross-protection by A fumigatus against gastrointestinal candidiasis. Mice treated with resting A fumigatus conidia intranasally a week before intragastric injection with Calbicans show reduced fungal growth (A), inflammatory cell recruitment (insets), and acanthosis compared with untreated animals (C). (B,D,F,H) Cross-protection by C albicans against invasive pulmonary aspergillosis. Mice treated with C albicans intragastrically a week before intranasal injection of resting A fumigatus conidia show reduced fungal growth (B) and inflammatory cell recruitment and amelioration of lung inflammatory pathology compared with untreated animals (D). (E-F) Infection with A fumigatus (E) or with C albicans (F) showed a significant increase in protective TH1 and regulatory T cells. (G-H) Cross-protection between A fumigatus and C albicans is dependent on IFN-γ. IFN-γ–deficient mice treated with A fumigatus (G) or C albicans (H) show no reduced fungal growth in the lungs or the stomach, respectively. The reduced fungal burden in IFN-γ–deficient mice is because of compensatory cytokines. Shown are fungal growth (CFUs/organ, mean ± SE), stomach (C) and lung histology (D), differential cell counts in cytospin preparations from stomachs (C) and lungs (D) and transcription factor mRNA expression in CD4+ T cells from mesenteric (candidiasis, E) or thoracic (aspergillosis, F) lymph nodes a week after infection. Shown are the results of one representative experiment of 3 independent experiments (for in vivo data) or pooled from 3 experiments (for in vitro assays). Bars represent SE. Photographs were taken using a high-resolution Olympus BX51 microscope, using Olympus Cell P 3.3 software. P, vaccinated versus unvaccinated animals.
Aspergillus fumigatus Crf1 protein mediates cross-protection to C albicans. Mice were vaccinated intranasally with resting A fumigatus conidia 14 days before infection or with purified Crf1 protein and murine CpG oligodeoxynucleotide 1862 14, 7, and 3 days before intragastric infection with C albicans. Note the reduced fungal growth, inflammatory cell recruitment (A, insets), and acanthosis in mice vaccinated with A fumigatus conidia or Crf1 protein as opposed to control mice shown in Figure 5C. Panel B shows the increase in protective TH1 and regulatory T-cell transcription factors and cytokine levels in vaccinated mice with A fumigatus conidia or the Crf1 protein, whereas the levels of TH2 transcription factors and cytokines remain unchanged. Shown are fungal growth (CFUs/organ, mean ± SE), stomach histology, differential cell counts in cytospin preparations from stomachs, and transcription factor and cytokine mRNA expression in CD4+ T cells from mesenteric lymph nodes a week after infection with C albicans. Shown are the results of one representative experiment of 3 independent experiments (for in vivo data) or pooled from 3 experiments (for in vitro assays). Bars represent SE. Photographs were taken using a high-resolution Olympus BX51 microscope, using Olympus Cell P 3.3 software. P, vaccinated vs unvaccinated animals. (C) Proliferation of purified CD4+ spleen cells from untreated mice or animals vaccinated with A fumigatus or C albicans in response to control DCs, anti-CD3ϵ antibody, heat-inactivated A fumigatus or C albicans, or A fumigatus Crf1 p41 and p22 peptides. The data are expressed as mean counts per minute ± SE of triplicate cultures. P, stimulated vs control DCs.
Aspergillus fumigatus Crf1 protein mediates cross-protection to C albicans. Mice were vaccinated intranasally with resting A fumigatus conidia 14 days before infection or with purified Crf1 protein and murine CpG oligodeoxynucleotide 1862 14, 7, and 3 days before intragastric infection with C albicans. Note the reduced fungal growth, inflammatory cell recruitment (A, insets), and acanthosis in mice vaccinated with A fumigatus conidia or Crf1 protein as opposed to control mice shown in Figure 5C. Panel B shows the increase in protective TH1 and regulatory T-cell transcription factors and cytokine levels in vaccinated mice with A fumigatus conidia or the Crf1 protein, whereas the levels of TH2 transcription factors and cytokines remain unchanged. Shown are fungal growth (CFUs/organ, mean ± SE), stomach histology, differential cell counts in cytospin preparations from stomachs, and transcription factor and cytokine mRNA expression in CD4+ T cells from mesenteric lymph nodes a week after infection with C albicans. Shown are the results of one representative experiment of 3 independent experiments (for in vivo data) or pooled from 3 experiments (for in vitro assays). Bars represent SE. Photographs were taken using a high-resolution Olympus BX51 microscope, using Olympus Cell P 3.3 software. P, vaccinated vs unvaccinated animals. (C) Proliferation of purified CD4+ spleen cells from untreated mice or animals vaccinated with A fumigatus or C albicans in response to control DCs, anti-CD3ϵ antibody, heat-inactivated A fumigatus or C albicans, or A fumigatus Crf1 p41 and p22 peptides. The data are expressed as mean counts per minute ± SE of triplicate cultures. P, stimulated vs control DCs.
Discussion
Aspergillus fumigatus and C albicans are the 2 most common fungal pathogens causing severe invasive infections among immunocompromised patients. Although new antifungal drugs for prophylaxis and treatment have reduced incidence and overall mortality of invasive fungal infections, breakthrough infections as well as toxicity counterbalance this development. In this study, we identified the epitope p41 of the A fumigatus extracellular cell wall glucanase Crf1 as a promising new target for antifungal immunotherapeutic strategies because it induces TH1 cells that are cross-reactive to C albicans in mice and humans. Moreover, the Crf1/p41 epitope can be presented by at least 3 different MHC class II alleles, covering 59.9% of the white population,33 and it activates an oligoclonal T-cell response with a diverse repertoire of TCR Vβ chains. This cross-reactive T-cell epitope is especially interesting for clinical application because Crf1/p41-specific memory CD4+ T cells can be rapidly expanded ex vivo despite low precursor frequencies. Particularly, T cell–mediated cross-reactivity between A fumigatus and C albicans seems to be not only restricted to humans but also similarly observed in an animal model, because mice vaccinated with A fumigatus conidia or Crf1 protein are protected against lethal infection with C albicans and vice versa, suggesting that T cell–mediated cross-protection could be of significant relevance in antifungal immunity.
Antifungal CD4+ TH1 immunity plays a pivotal role in the clearance of most fungal infections, including invasive aspergillosis and candidiasis. Immunotherapeutic strategies activating antifungal TH1 CD4+ immunity, such as adoptive transfer of A fumigatus-specific CD4+ TH1 cells and vaccination with diverse fungal antigens, have already shown therapeutic efficacy in mouse models34 and HSCT recipients.2 However, potent and safe antigens for broad clinical application are hampered by lack of identification of target epitopes. Identification of immunodominant proteins is complicated because of potential unspecific stimulation of the PBMCs depending on the expression system such as Escherichia coli, Pichia pastoris, or human cell lines used for the generation of the recombinant proteins and the low precursor frequencies of Aspergillus-specific T cells detectable in peripheral blood. Different approaches have been used so far. Of all proteins tested, the A fumigatus glycosylphosphatidylinositol-anchored extracellular cell wall glucanase Crf1 described in our study and by other groups seems to be the most potent candidate antigen.26,34,35 We could substantiate this observation by identifying T cells specific for 4 Crf1 peptide epitopes that can be activated by DCs presenting endogenously processed fungal antigen. Of these 4 epitopes, Crf1/p41 was by far the most robust epitope because all CD4+ T-cell clones from multiple donors responded to this peptide as well as to fungal antigen. In contrast, we have discovered several other peptides that activated CD4+ T cells that were not specific for A fumigatus and did not respond to fungal antigen. This unspecific activation of T cells is often observed when unphysiologically high doses of peptides are used for T-cell stimulation, but it is not indicative of physiologic T-cell activation.8,36 In summary, we characterized for the first time CD4+ TH1 T-cell clones generated by stimulation with peptides that can be activated by whole A fumigatus processed by DCs.
Crf1/p41 seems to be an exceptionally potent A fumigatus antigen, because all T-cell clones showed strong reactivity toward the profoundly different fungal pathogens A fumigatus and C albicans. The discovery of a defined peptide epitope activating TH1 cells with cross-reactivity to the 2 most important fungal pathogens in humans is to our knowledge a novel finding and only little is known in mouse models.37 The concept of heterologous immunity to different pathogens mediated by cross-reactive T cells has until now only been described in detail for viral infections,38 for example, for influenza virus39,40 and flavivirus.41,42 The discovery of this mechanism has initiated the development of new vaccines that will target conserved protein domains of different flaviviral strains for induction of cross-protection. In our confirmatory murine experiments, TH1-associated transcription factors and cytokine mRNA levels as well as experiments with IFN-γ knockout mice suggest that TH1 cells indeed play a crucial role in mediating cross-protection between the 2 fungi A fumigatus and C albicans. Furthermore, the same epitope mediating cross-reactivity in humans is also cross-reactive in mice. Together, our data illustrate for the first time the existence of T cell–based cross-protection between distantly related fungal pathogens in humans as well as mice that could therefore be considered in general as a strategy for induction and boosting of a broad specific cellular immunity.
Mathematic models have suggested that far more immunogenic T-cell epitopes are existent than TCRs, being randomly generated and selected via the thymus in a single individual.43 Nevertheless, the adaptive cellular immune response is capable of protecting higher evolutionary organisms from pathogens. This is facilitated on the molecular level between TCRs and MHC peptide complexes by several mechanisms that include induced fitting of the TCRs, differential TCR docking capabilities, structural degeneracy in the interaction between TCRs and MHC complexes, molecular mimicry of the epitope, and antigen-dependent tuning of peptide-MHC flexibility. In line with this theory, the cross-reactivity observed in our study is probably because of molecular mimicry. Aspergillus fumigatus Crf1/p41 and the corresponding epitope of the C albicans homolog Crh1 differ only in 1 aa, and both epitopes are recognized by the same TCR. This aa substitution is probably located at the MHC nonanchor site P2 because Tyr163 and Asp166 represent optimal P1 and P4 anchor residues, respectively, in the epitope binding to HLA-DRB1*03 or -DRB1*04. Interestingly, cross-reactivity is observed in spite of a nonanchor residue substitution.40 This phenomenon was universally observed in different T-cell clones derived from various donors. In contrast, the p41 epitopes of other Aspergillus species differing in 3 to 7 aa from the A fumigatus sequence are no longer cross-reactive, with the exception of A flavus that shows very weak cross-reactivity in some of the Crf1/p41-specific T-cell clones, which is possibly explained by its higher homology to the A fumigatus sequence than that of the other Aspergillus species. This high specificity raises the concern that adoptive transfer of Crf1/p41-specific T cells could lead to mutations in the Crf1 glucanase of the fungi because of increased pressure. Because this concern cannot be excluded, a combination of multiple different T-cell epitopes would be advantageous in clinical applications. We further observed that CD4+ p41-specific HLA-DRB1*03–restricted T-cell clones could be activated with peptide presented by HLA-DRB1*13 and vice versa, which is in line with the recently described structural degeneracy of epitope presentation by different MHC class II molecules.
The cross-reactivity of Crf1/p41-specific T cells to 2 of the most important fungal pathogens in immunocompromised hosts makes this epitope conceptually an interesting target for immunotherapeutic approaches for prevention and treatment of common fungal infections. Cross-reactivity could be observed in several different fungal isolates, implying that the epitope is generally conserved. Notably, because A fumigatus changes its morphology during germination of inert conidia to invasive hyphae, Crf1/p41 is probably expressed only after germination and during hyphal growth. Therefore, expression of Crf1 occurs in the pathogenic form of the fungus when angioinvasion and organ destruction take place and cellular immunity is activated. The unresponsiveness to resting conidia could either result from selective expression of Crf1 during fungal growth in accordance with its function in cell wall construction12,44 or from insufficient T-cell activation because of inadequate DC costimulation.44,45
So far, antifungal cross-protection has been only described in the context of antibody-mediated immunity. Antibodies targeting cell wall structures such as β-glucan conferred protection against a variety of fungal infections,46 and the protective potential of vaccination with Saccharomyces yeast against intravenous challenge with A fumigatus and Coccidioides also is probably mediated by antibodies.7,47 Similarly as our in vivo model, these studies were not able to fully elucidate the immunologic mechanism whereby Aspergillus and Candida infections are controlled. Our in vitro experiments and previous murine studies48 indicate that the stimulatory effect of TH1 cells on antifungal activity of innate immune cells is probably based on the synergistic action of secreted cytokines, including IFN-γ and colony-stimulating factors such as GM-CSF, which in our human experiments could both be detected in the culture supernatant of activated T-cell clones in high amounts. There was also evidence that supernatant from p41-specific T-cell clones activated with A fumigatus is able to enhance the antifungal activity of human monocytes. Moreover, the in vivo model substantiates that IFN-γ knockout mice are not protected against infection, suggesting the crucial role of TH1 cells in mediating cross-protection between the 2 fungi A fumigatus and C albicans.
Basically, 2 immunotherapeutic strategies—adoptive T-cell transfer and vaccination—could enhance the pathogen-specific immune system and may thereby limit fungal infections. Because immunosuppressed patients, such as recipients of HSCT, are predisposed for fungal infections that are unresponsive to immunization because of impaired B-cell reconstitution, transfer of either specific T cells or antibodies seem to be more beneficial. Adoptive T-cell transfer seems even more favorable because it is able to induce antigen-specific immune reconstitution with persistent long-term antigen-specific activity.49
The clinical utility of this epitope is further strengthened because Crf1/p41–specific T cells have a broad MHC restriction, covering nearly 60% of the white population, and it activates an oligoclonal TCR Vβ repertoire in healthy individuals. The feasibility of immunotherapeutic strategies is highly dependent on the capacity of antigens to efficiently expand pathogen-specific memory T cells. In fact, we were able to demonstrate that the cross-reactive fungal CD4+ T-cell population can be readily expanded in vitro, implying that they do not represent terminally differentiated T cells. This high proliferative capacity is mandatory for immunotherapeutic intervention such as vaccination and adoptive T-cell therapies in immunocompromised hosts for prevention and treatment of common fungal infections.
In summary, we describe how the pathogen-specific T-cell response is selectively shaped to respond to common fungal pathogens in humans. These data may have implications for the development of vaccination strategies and T-cell therapy for various infectious diseases.
The online version of the article contains a data supplement.
Presented in abstract form at the 51st annual meeting of the American Society of Hematology, New Orleans, LA, December 5-8, 2009,51 and at the 52nd annual meeting of the American Society of Hematology, Orlando, FL, December 4-7, 2010.52
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Prof Utz Reichard for the provision of cDNA, Prof Joachim Morschhäuser and Dr Reno Frei for provision of fungal strains, and Drs Patricia Comoli and Inge Jedema for provision of LCLs. We also thank Michaela Dümig for excellent technical assistance and Profs Manuel Battegay, Hans Stauss, and Hans-Georg Rammensee for helpful discussions.
This study was supported by BayImmuNet (M.S.T.); Specific Targeted Research Project MANASP (LSHE-CT-2006), contract 037899 (FP6; L.R., J.P.L., J.L., M.S.T., H.E.); and the Swiss National Foundation (grant PBBS P3 123164; N.K.).
National Institutes of Health
Authorship
Contribution: C.S. and N.K. designed, performed, and analyzed experiments with human lymphocytes and wrote the manuscript in cooperation with M.S.T.; S.B., T.Z., and S.M. performed and analyzed the animal experiments; M.K., S.L., B.C., and E.W. performed experiments with human lymphocytes; S.S. contributed to the writing of the manuscript; S.K. provided vital reagents and contributed to the writing of the manuscript; H.E., J.-P.L., and J.L. provided vital reagents; L.R. designed the animal research and contributed to the writing of the manuscript; M.S.T. designed the research with human lymphocytes and wrote the paper in cooperation with C.S. and N.K.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Max S. Topp, Medizinische Klinik und Poliklinik II, Universitaetsklinikum Wuerzburg, Oberduerrbacher Str 6, 97080 Würzburg, Germany; e-mail: topp_m@klinik.uni-wuerzburg.de.
References
Author notes
C.S. and N.K. are joint first authors of this article.
L.R. and M.S.T. contributed equally to this work.