Abstract
To identify new markers for minimal residual disease (MRD) detection in acute lymphoblastic leukemia (ALL), we compared genome-wide gene expression of lymphoblasts from 270 patients with newly diagnosed childhood ALL to that of normal CD19+CD10+ B-cell progenitors (n = 4). Expression of 30 genes differentially expressed by ≥ 3-fold in at least 25% of cases of ALL (or 40% of ALL subtypes) was tested by flow cytometry in 200 B-lineage ALL and 61 nonleukemic BM samples, including samples containing hematogones. Of the 30 markers, 22 (CD44, BCL2, HSPB1, CD73, CD24, CD123, CD72, CD86, CD200, CD79b, CD164, CD304, CD97, CD102, CD99, CD300a, CD130, PBX1, CTNNA1, ITGB7, CD69, CD49f) were differentially expressed in up to 81.4% of ALL cases; expression of some markers was associated with the presence of genetic abnormalities. Results of MRD detection by flow cytometry with these markers correlated well with those of molecular testing (52 follow-up samples from 18 patients); sequential studies during treatment and diagnosis-relapse comparisons documented their stability. When incorporated in 6-marker combinations, the new markers afforded the detection of 1 leukemic cell among 105 BM cells. These new markers should allow MRD studies in all B-lineage ALL patients, and substantially improve their sensitivity.
Introduction
Leukemia relapse is the major cause of treatment failure for patients with acute lymphoblastic leukemia (ALL).1-4 Relapse originates from leukemic cells that are resistant to chemotherapy but become undetectable after initial treatment in most cases. Nevertheless, methods more sensitive than microscopic examination can demonstrate leukemic cells in a proportion of samples with no morphologic evidence of leukemia, a finding termed “minimal residual disease (MRD).”5 MRD is currently the most powerful prognostic indicator in childhood ALL,6-16 and there is strong evidence supporting its prognostic significance in adult ALL.17-21 Thus, MRD monitoring has been introduced into many contemporary treatment protocols for risk assignment and selection of therapeutic regimens.1-4 MRD measurements are also clinically useful in patients with relapsed ALL who achieve a second remission,22-24 can help optimize the timing of hematopoietic stem cell transplantation,25 and guide decisions about donor lymphocyte infusion posttransplantation.26
Among methods for detecting MRD in ALL, PCR amplification of Ag-receptor genes has proven to be valuable and has been extensively standardized,27 but the technical expertise and instrumentation required limit its application to specialized centers. PCR amplification of fusion transcripts may also provide useful clinical information but its applicability in ALL is restricted by the fact that molecular targets currently adaptable to routine MRD studies are present in only a minority of patients.27 Flow cytometric detection of leukemia-specific markers has been shown to predict outcome in numerous clinical correlative studies.7,9,10,13,14,17,18,20 The method holds potential for wider applicability than molecular techniques because flow cytometric methods for leukemia diagnosis are already established at most cancer centers worldwide.5
MRD studies by flow cytometry rely on panels of Abs to define unique immunophenotypic signatures of leukemic cells which must distinguish leukemic blasts from their normal counterparts, the CD19+CD10+ lymphoid progenitors of the BM (“hematogones”).5,5,27-29 Standard 4-color flow cytometry can detect 1 leukemic cell in up to 10 000 normal BM or peripheral blood cells but this task typically requires considerable interpretative expertise. Therefore, the identification of new leukemia markers that are easily detectable and are stably expressed in a large proportion of ALL cases should simplify the application of MRD studies, help extend their benefit to all patients and possibly enhance the sensitivity of MRD detection.
In this study, we first screened for genes expressed at different levels in ALL cells and their normal counterparts by comparing genome-wide gene expression profiles of 270 cases of newly diagnosed B-lineage ALL to those of highly purified normal CD19+CD10+ cells. Genes that had a substantially abnormal expression in leukemic cells were then tested by flow cytometry to assess levels of protein expression. The most promising molecules were examined in detail for their usefulness as MRD markers.
Methods
Patients
BM samples were collected at presentation from 470 patients, ages < 1-18 years, with newly diagnosed B-lineage ALL; 270 samples were included in genome-wide gene expression studies, and 200 were tested to validate the gene expression findings by flow cytometry. We also studied BM samples obtained during therapy in 51 patients and samples obtained at relapse in 9. The diagnosis of B-lineage ALL was unequivocal and based on morphology, cytochemistry, and cell marker expression. To determine gene expression and immunophenotype of normal lymphoid progenitors, we studied BM samples collected from 22 healthy donors aged 2-25 years (median, 10 years) during the harvest of BM for stem cell transplantation, and BM samples obtained from 27 patients with B-lineage ALL, 7 with T-lineage ALL and 5 with acute myeloid leukemia (AML) during therapy. These studies were approved by the Institutional Review Board, with informed consent obtained in accordance with the Declaration of Helsinki from donors, patients, their parents or their guardians, and assent from the patients, as appropriate.
Leukemic and normal mononuclear cells were collected after centrifugation on a density gradient (AccuPrep; Nycomed) and washed 3 times in PBS. Leukemic cells used in gene expression studies were cryopreserved. Normal BM CD19+ cells used for gene expression analysis were first enriched using a MACS separation system (Miltenyi Biotec) from fresh samples, yielding > 98% CD19+ cell purity. Cells were then stained with anti-CD19 conjugated to PE and anti-CD10 conjugated to FITC (both from Becton Dickinson) and CD19+CD10+ cells were sorted using a MoFlo high-speed FACS (Cytomation). All samples were processed or cryopreserved within 5 hours of collection.
Gene expression array studies
Gene expression profiling studies were performed as previously described.30 Briefly, we isolated total RNA from freshly thawed B-lineage ALL cells and flow sorted normal CD19+CD10+ cells using the TRIzol reagent (Invitrogen). After generating cDNA, we prepared biotin-labeled cRNA hybridization solutions according to the protocols of Affymetrix. The solutions were hybridized to HG-U133A oligonucleotide microarrays (Affymetrix). After staining with PE-conjugated streptavidin, the arrays were read with a laser confocal scanner (Agilent). Signal values were computed from the image files using Affymetrix GeneChip Operating Software. Intensity values for a total of 22 283 probe sets on the U133A microarray were obtained. All microarray data are available on the Gene Expression Omnibus (GEO) under accession number GSE28497.
Flow cytometric analysis and MRD studies
To verify the differential expression of gene products in ALL cells, we used flow cytometry with the Abs listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In most tests, the Abs were used in combination with anti-CD19 conjugated to allophycocyanin, anti-CD10 conjugated to either FITC or PE, and anti-CD34 conjugated to peridinin chlorophyll protein (PerCP). Isotype-matched nonreactive Abs were used as controls. For cell staining, monunucleated cells were washed in PBS containing 0.5% BSA and 0.5% sodium azide (PBSA), mixed with rabbit serum to block surface Fc receptors, incubated with the Abs for 10 minutes at 20°C in the dark, washed twice in PBSA and fixed with 0.5% paraformaldehyde. For intracellular markers, cells were permeabilized and fixed before exposure to Abs using 8E, a reagent prepared in our laboratory from a proprietary formula. Measurements of Ab labeling were performed by multiparameter flow cytometry, using a FACSCalibur flow cytometer (Becton Dickinson).
Studies of MRD by flow cytometry were performed using combinations of mAbs that identified leukemia-associated immunophenotypes determined at diagnosis. Cell marking was performed using a combination of 4 markers simultaneously and, where indicated, with 6-marker combinations. Data acquisition and analysis was done as previously described, using FACSCalibur or LSRII (Becton Dickinson) flow cytometers, and CellQuest, DiVa (both from Becton Dickinson), FlowJo (TreeStar) and/or Kaluza (Beckman Coulter) software.5 Determination of MRD by PCR amplification of immunoglobulin and T-cell receptor genes was done as previously described.16 The results of MRD studies by the 2 methods were recorded independently.
Results
Identification of genes differentially expressed in leukemic and normal immature B cells
To identify genes over or underexpressed in ALL cells, we compared genome-wide expression data from 270 B-lineage ALL samples and CD19+CD10+ lymphoid progenitors obtained from the BM of 4 healthy donors. A total of 133 probe sets (corresponding to 112 genes) had signals higher than 2-fold of the highest value obtained among normal CD19+CD10+ cells in 75% or more of ALL cases studied; conversely, 192 probe sets (corresponding to 165 genes) had signals at least 50% lower than the lowest signal among the normal CD19+CD10+ cells in 75% or more of ALL cases. When the criteria for inclusion was extended to genes that were differentially expressed in at least 25% of ALL cases, 1405 probe sets were overexpressed and 1474 were underexpressed (supplemental Tables 2-3).
Among the genes that were differentially expressed, there were some previously shown to be abnormally expressed in ALL cells. Thus, CD58 was overexpressed in 81.9% of cases,31 and WT1 in 32.6% of cases,32 whereas PAX5 was underexpressed in 86.7%33 and CD38 in 73.7% of cases.34 Among the 23 ALL specimens with MLL gene rearrangements, 100% overexpressed galectin-1 and 91.3% underexpressed CD10, characteristic features of this subset of ALL35,36 ; most of these cases also overexpressed a cohort of genes (eg, FLT3, LMO2, ADAM10, MEIS1) previously reported to be associated with MLL-rearranged leukemia.37 As expected, all 26 cases with TCF3-PBX1 overexpressed PBX1,38 and, among the 62 cases with ETV6-RUNX1, 46.8% overexpressed CD13 and 29.0% CD33.39
Validation of gene expression array results by flow cytometry
Among the genes differentially expressed by gene array analysis, there were some already widely used for MRD studies by flow cytometry, such as, CD58, CD38, CD13, and CD34,14,29,31,34 suggesting the possibility that other useful markers could be present among the remaining genes. To prioritize genes for validation by flow cytometry, we applied the following inclusion criteria: (1) differential expression in at least 25% of cases of ALL, or 40% of cases of a genetic subtype of ALL; (2) overexpression in leukemic cells by at least 3-fold of the maximum value in normal cells, or underexpression by 3-fold of the minimum value in normal cells; and (3) commercial availability of specific Abs conjugated to fluorochromes suitable for flow cytometry. Guided by these criteria, we selected the 30 genes (25 overexpressed in ALL and 5 underexpressed) listed in Table 1 and determined whether the differential expression measured by microarray analysis at mRNA level corresponded to differential expression of the encoded proteins.
Expression level in ALL cells and normal CD19+ CD10+ cells of the genes selected for study
| Probe set . | Gene symbol/CD number . | Range expression in normal CD19+ CD10+ (n = 4)* . | Range expression in ALL (n = 270)* . | % of ALL cases with higher/lower expression . | ||
|---|---|---|---|---|---|---|
| Min . | Max . | Min . | Max . | |||
| Overexpressed in ALL | ||||||
| 203685_at | BCL2 | 149.1 | 229 | 150.1 | 5203.8 | 90 |
| 201841_s_at | HSPB1 | 15.6 | 302 | 36.4 | 8938.9 | 87 |
| 207643_s_at | TNFRSF1A/CD120a | 94.4 | 234.7 | 28 | 3029.5 | 84 |
| 209933_s_at | CD300A | 59.1 | 126.1 | 29.3 | 1481.7 | 81 |
| 202910_s_at | CD97 | 149.2 | 303.4 | 31.5 | 6537 | 80 |
| 212298_at | NRP1/CD304 | 5.6 | 10.6 | 3.3 | 2155.1 | 80 |
| 204563_at | SELL/CD62L | 288.1 | 415.7 | 205 | 16 680.8 | 79 |
| 210916_s_at | CD44 | 40.8 | 109.2 | 18.8 | 4864.2 | 78 |
| 215177_s_at | ITGA6/CD49f | 31.5 | 145.9 | 14.6 | 12 400.9 | 75 |
| 209583_s_at | CD200 | 313.2 | 423.9 | 42.3 | 3955.4 | 72 |
| 203939_at | NT5E/CD73 | 54.3 | 121.1 | 5 | 5133.9 | 67 |
| 213620_s_at | ICAM2/CD102 | 871.1 | 983 | 233.8 | 7321.1 | 65 |
| 210895_s_at | CD86 | 31.9 | 106.9 | 13.6 | 1392.7 | 60 |
| 204116_at | IL2RG/CD132 | 604 | 1213.2 | 574.9 | 8123.4 | 56 |
| 201028_s_at | CD99 | 1148.8 | 2047.8 | 325.4 | 19 384.2 | 50 |
| 201887_at | IL13RA1 | 81.2 | 159.2 | 30.4 | 2829.5 | 41 (59 of BCR-ABL1) |
| 208654_s_at | CD164 | 754.9 | 1274.4 | 667.6 | 15 234.8 | 39 (71 of hyperdiploid; 59 of BCR-ABL1) |
| 209963_s_at | EPOR | 49.4 | 127.9 | 17.8 | 1950.4 | 36 |
| 212151_at | PBX1 | 30.4 | 114.4 | 16.5 | 10 825.3 | 36 (100 of TCF3-PBX1) |
| 205987_at | CD1C | 16 | 249.7 | 9.7 | 7020.6 | 27 (71 of hyperdiploid) |
| 204863_s_at | IL6ST/CD130 | 119.4 | 160.9 | 8.5 | 837.1 | 27 |
| 205718_at | ITGB7 | 12.1 | 208.5 | 10.9 | 3223.5 | 25 (50 of BCR-ABL1) |
| 206148_at | IL3RA/CD123 | 53.6 | 260.6 | 18.7 | 3497.2 | 22 (71 of hyperdiploid) |
| 208651_x_at | CD24 | 3125.7 | 4310.4 | 258.6 | 14 231.4 | 17 (54 of TCF3-PBX1) |
| 209617_s_at | CTNNA1 | 23.3 | 68.7 | 6 | 424.5 | 9 (40.3 of ETV6-RUNX1) |
| Underexpressed in ALL | ||||||
| 205297_s_at | CD79B | 11 257.6 | 17 637 | 19.1 | 8701 | 96 |
| 204440_at | CD83 | 1759.5 | 4557.9 | 47.8 | 9729.2 | 91 |
| 209795_at | CD69 | 6458.6 | 7502.8 | 31.6 | 16 301.1 | 77 |
| 204192_at | CD37 | 5624.7 | 13 251.7 | 225.6 | 10 302.9 | 67 (81 of ETV6-RUNX1) |
| 215925_s_at | CD72 | 1888.1 | 4238.1 | 9.1 | 4968.5 | 57 |
| Probe set . | Gene symbol/CD number . | Range expression in normal CD19+ CD10+ (n = 4)* . | Range expression in ALL (n = 270)* . | % of ALL cases with higher/lower expression . | ||
|---|---|---|---|---|---|---|
| Min . | Max . | Min . | Max . | |||
| Overexpressed in ALL | ||||||
| 203685_at | BCL2 | 149.1 | 229 | 150.1 | 5203.8 | 90 |
| 201841_s_at | HSPB1 | 15.6 | 302 | 36.4 | 8938.9 | 87 |
| 207643_s_at | TNFRSF1A/CD120a | 94.4 | 234.7 | 28 | 3029.5 | 84 |
| 209933_s_at | CD300A | 59.1 | 126.1 | 29.3 | 1481.7 | 81 |
| 202910_s_at | CD97 | 149.2 | 303.4 | 31.5 | 6537 | 80 |
| 212298_at | NRP1/CD304 | 5.6 | 10.6 | 3.3 | 2155.1 | 80 |
| 204563_at | SELL/CD62L | 288.1 | 415.7 | 205 | 16 680.8 | 79 |
| 210916_s_at | CD44 | 40.8 | 109.2 | 18.8 | 4864.2 | 78 |
| 215177_s_at | ITGA6/CD49f | 31.5 | 145.9 | 14.6 | 12 400.9 | 75 |
| 209583_s_at | CD200 | 313.2 | 423.9 | 42.3 | 3955.4 | 72 |
| 203939_at | NT5E/CD73 | 54.3 | 121.1 | 5 | 5133.9 | 67 |
| 213620_s_at | ICAM2/CD102 | 871.1 | 983 | 233.8 | 7321.1 | 65 |
| 210895_s_at | CD86 | 31.9 | 106.9 | 13.6 | 1392.7 | 60 |
| 204116_at | IL2RG/CD132 | 604 | 1213.2 | 574.9 | 8123.4 | 56 |
| 201028_s_at | CD99 | 1148.8 | 2047.8 | 325.4 | 19 384.2 | 50 |
| 201887_at | IL13RA1 | 81.2 | 159.2 | 30.4 | 2829.5 | 41 (59 of BCR-ABL1) |
| 208654_s_at | CD164 | 754.9 | 1274.4 | 667.6 | 15 234.8 | 39 (71 of hyperdiploid; 59 of BCR-ABL1) |
| 209963_s_at | EPOR | 49.4 | 127.9 | 17.8 | 1950.4 | 36 |
| 212151_at | PBX1 | 30.4 | 114.4 | 16.5 | 10 825.3 | 36 (100 of TCF3-PBX1) |
| 205987_at | CD1C | 16 | 249.7 | 9.7 | 7020.6 | 27 (71 of hyperdiploid) |
| 204863_s_at | IL6ST/CD130 | 119.4 | 160.9 | 8.5 | 837.1 | 27 |
| 205718_at | ITGB7 | 12.1 | 208.5 | 10.9 | 3223.5 | 25 (50 of BCR-ABL1) |
| 206148_at | IL3RA/CD123 | 53.6 | 260.6 | 18.7 | 3497.2 | 22 (71 of hyperdiploid) |
| 208651_x_at | CD24 | 3125.7 | 4310.4 | 258.6 | 14 231.4 | 17 (54 of TCF3-PBX1) |
| 209617_s_at | CTNNA1 | 23.3 | 68.7 | 6 | 424.5 | 9 (40.3 of ETV6-RUNX1) |
| Underexpressed in ALL | ||||||
| 205297_s_at | CD79B | 11 257.6 | 17 637 | 19.1 | 8701 | 96 |
| 204440_at | CD83 | 1759.5 | 4557.9 | 47.8 | 9729.2 | 91 |
| 209795_at | CD69 | 6458.6 | 7502.8 | 31.6 | 16 301.1 | 77 |
| 204192_at | CD37 | 5624.7 | 13 251.7 | 225.6 | 10 302.9 | 67 (81 of ETV6-RUNX1) |
| 215925_s_at | CD72 | 1888.1 | 4238.1 | 9.1 | 4968.5 | 57 |
ALL indicates acute lymphoblastic leukemia; Min, minimum; and Max, maximum.
Data expressed as units calculated by MAS5.0 to a median target intensity of 500.
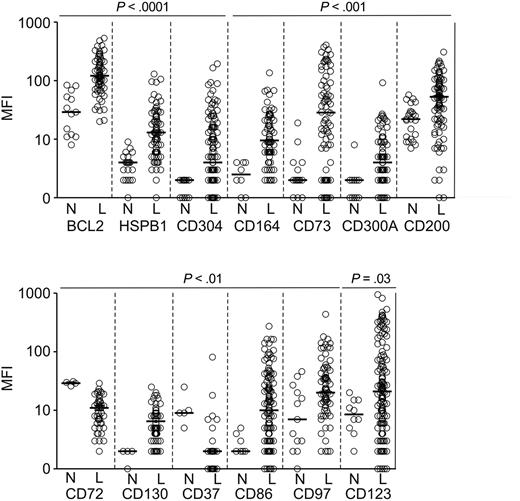
After confirming the specificity of the Abs with known positive and negative target cells (listed in supplemental Table 1), we tested their reactivity using ALL samples obtained at diagnosis (n = 200) and leukemia-free BM samples (n = 61). Importantly, we included in these comparisons not only BM specimens from healthy donors (n = 22) but also BM from children with ALL (MRD-negative according to PCR amplification of Ag-receptor genes) and AML (MRD negative by flow cytometry) during chemotherapy (n = 39), some with a high proportion of hematogones. We first analyzed overall expression in the ALL versus normal CD19+CD10+ B-cell progenitor groups and found that for 13 of the 30 markers tested, the difference in overall expression was statistically significant (P < .05): 11 of the 25 proteins encoded by the overexpressed genes were expressed at a significantly higher level in B-lineage ALL cells, while 2 of the 5 proteins encoded by underexpressed genes had a significantly lower expression (Figure 1).
Markers expressed at significantly different levels in ALL cells and in CD19+CD10+ B-cell progenitors as determined by flow cytometry. Shown are mean fluorescence intensity (MFI) values obtained in CD19+ leukemic lymphoblasts from BM samples of patients with newly diagnosed ALL (“L”) and BM CD19+CD10+ cells from healthy donors or from patients with leukemia in remission and no evidence of MRD (“N”). Each symbol indicates results of one sample; horizontal bars correspond to median values within each group.
Markers expressed at significantly different levels in ALL cells and in CD19+CD10+ B-cell progenitors as determined by flow cytometry. Shown are mean fluorescence intensity (MFI) values obtained in CD19+ leukemic lymphoblasts from BM samples of patients with newly diagnosed ALL (“L”) and BM CD19+CD10+ cells from healthy donors or from patients with leukemia in remission and no evidence of MRD (“N”). Each symbol indicates results of one sample; horizontal bars correspond to median values within each group.
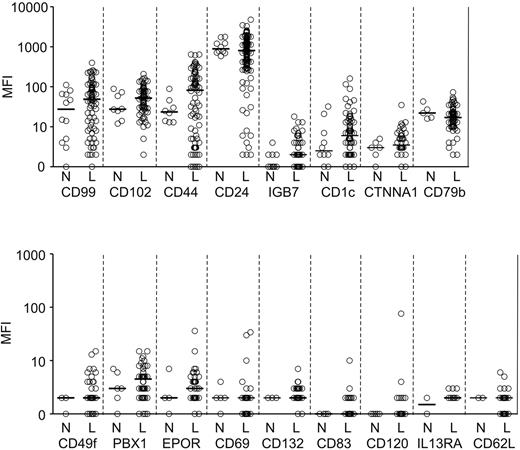
For the remaining 17 markers, the difference in overall expression between ALL cells and normal B-cell progenitors was not statistically significant (Figure 2). However, for some of these markers, a substantial proportion of ALL cases showed overexpression or underexpression. Therefore, we determined the number of ALL cases that expressed a given marker at levels higher than the highest mean fluorescence intensity (MFI) value recorded among normal B-cell progenitors and had an MFI > 10 (a typical threshold for positive staining with our flow cytometer settings). For markers that were underexpressed in ALL cells, we calculated the number of ALL cases with expression lower than the lowest MFI measured in normal B-cell progenitors, but excluded markers where normal B-cell progenitors had an MFI < 10. By using these criteria, 22 of the 30 markers were differentially expressed in 5%-81.4% (median, 35%) of cases (Table 2). Thus, CD44 was differentially expressed in 81.4% cases studied, BCL2 was overexpressed in 76.6%, HSPB1 (heat shock protein 27) in 63.4% and CD73 in 54.5%. Notably, CD44 and CD24 were overexpressed in some cases and underexpressed in others. Importantly, some of the markers, such as CD97, CD99, and CD102, appeared to be overexpressed in a much larger proportion of cases when the comparisons included only resting BM samples. However, when regenerating specimens rich in hematogones were included, it became clear that expression in leukemic cells for many of these cases was not outside the range of normality. Of the 30 markers studied, only 8 (EPOR, CD1c, CD120a, CD37, CD62L, CD132, CD83, and IL13RA1) showed no clear differential expression by the above criteria and were excluded from further studies.
Markers expressed at levels not significantly different (P > .05) in ALL cells and CD19+CD10+ B-cell progenitors as determined by flow cytometry. Shown are mean fluorescence intensity (MFI) values obtained in CD19+ leukemic lymphoblasts from BM samples of patients with newly diagnosed ALL (“L”) and BM CD19+CD10+ cells from healthy donors or from patients with leukemia in remission and no evidence of MRD (“N”). Each symbol indicates results of one sample; horizontal bars correspond to median values within each group.
Markers expressed at levels not significantly different (P > .05) in ALL cells and CD19+CD10+ B-cell progenitors as determined by flow cytometry. Shown are mean fluorescence intensity (MFI) values obtained in CD19+ leukemic lymphoblasts from BM samples of patients with newly diagnosed ALL (“L”) and BM CD19+CD10+ cells from healthy donors or from patients with leukemia in remission and no evidence of MRD (“N”). Each symbol indicates results of one sample; horizontal bars correspond to median values within each group.
Expression of the new markers in ALL cells relative to their expression in bone marrow CD19+CD10+ cells from healthy individuals as determined by flow cytometry
| Marker . | No. of ALL cases studied . | No. of cases with overexpression (%)* . | No. of cases with underexpression (%)† . |
|---|---|---|---|
| CD44 | 86 | 46 (53.5) | 24 (27.9) |
| BCL2 | 77 | 59 (76.6) | |
| HSPB1 | 93 | 59 (63.4) | |
| CD73 | 77 | 42 (54.5) | |
| CD24 | 139 | 16 (11.5) | 55 (39.6) |
| CD123 | 142 | 72 (50.7) | |
| CD72 | 45 | 22 (48.9) | |
| CD86 | 135 | 63 (46.7) | |
| CD200 | 95 | 41 (43.2) | |
| CD79b | 74 | 31 (41.9) | |
| CD164 | 75 | 31 (41.3) | |
| CD304 | 129 | 37 (28.7) | |
| CD97 | 81 | 22 (27.2) | |
| CD102 | 76 | 15 (19.7) | 2 (2.6) |
| CD99 | 77 | 17 (22.1) | |
| CD300A | 92 | 18 (19.6) | |
| CD130 | 71 | 12 (16.9) | |
| PBX1 | 64 | 4 (11.8) | |
| CTNNA1 | 44 | 4 (9.1) | |
| ITGB7 | 90 | 7 (7.8) | |
| CD69 | 33 | 2 (6.1) | |
| CD49f | 38 | 2 (5.3) | |
| EPOR | 45 | 2 (4.4) | |
| CD1C | 86 | 3 (3.5) | |
| CD120a | 35 | 1 (2.9) | |
| CD37 | 46 | 1 (2.2) | |
| CD62L | 34 | 0 | |
| CD132 | 33 | 0 | |
| CD83 | 33 | 0 | |
| IL13RA1 | 10 | 0 |
| Marker . | No. of ALL cases studied . | No. of cases with overexpression (%)* . | No. of cases with underexpression (%)† . |
|---|---|---|---|
| CD44 | 86 | 46 (53.5) | 24 (27.9) |
| BCL2 | 77 | 59 (76.6) | |
| HSPB1 | 93 | 59 (63.4) | |
| CD73 | 77 | 42 (54.5) | |
| CD24 | 139 | 16 (11.5) | 55 (39.6) |
| CD123 | 142 | 72 (50.7) | |
| CD72 | 45 | 22 (48.9) | |
| CD86 | 135 | 63 (46.7) | |
| CD200 | 95 | 41 (43.2) | |
| CD79b | 74 | 31 (41.9) | |
| CD164 | 75 | 31 (41.3) | |
| CD304 | 129 | 37 (28.7) | |
| CD97 | 81 | 22 (27.2) | |
| CD102 | 76 | 15 (19.7) | 2 (2.6) |
| CD99 | 77 | 17 (22.1) | |
| CD300A | 92 | 18 (19.6) | |
| CD130 | 71 | 12 (16.9) | |
| PBX1 | 64 | 4 (11.8) | |
| CTNNA1 | 44 | 4 (9.1) | |
| ITGB7 | 90 | 7 (7.8) | |
| CD69 | 33 | 2 (6.1) | |
| CD49f | 38 | 2 (5.3) | |
| EPOR | 45 | 2 (4.4) | |
| CD1C | 86 | 3 (3.5) | |
| CD120a | 35 | 1 (2.9) | |
| CD37 | 46 | 1 (2.2) | |
| CD62L | 34 | 0 | |
| CD132 | 33 | 0 | |
| CD83 | 33 | 0 | |
| IL13RA1 | 10 | 0 |
ALL indicates acute lymphoblastic leukemia; and MFI, mean fluorescence intensity.
Number of ALL cases that expressed the indicated marker at levels higher than the highest MFI value recorded among normal B-cell progenitors, and had an MFI higher than 10.
Number of ALL cases that expressed the indicated marker at levels lower than the lowest MFI measured in normal B-cell progenitors, excluding cases in which a marker had an MFI lower than 10 on normal B-cell progenitors.
Validation of the new markers for MRD detection
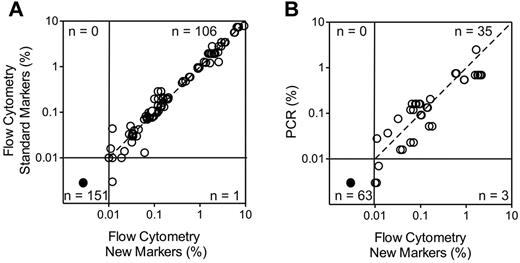
To determine the reliability of the new markers to identify leukemic cells in clinical samples, we studied 128 BM samples collected during treatment (46 during or at the end of remission induction therapy and 82 during postremission therapy) from 51 patients with B-lineage ALL in whom expression of the markers on the leukemic cells had been measured at diagnosis. The markers included in these studies were the top 16 differentially expressed markers listed in Table 2, for a total of 258 tests. The markers were incorporated in 4 color combinations including, in addition to the new marker, CD19, CD10 and CD34, and were compared with standard marker combinations used in our laboratory (supplemental Table 4). Using a threshold of 0.01% ALL cells to define MRD positivity, no discordant results were observed except for one comparison in which MRD was negative with the standard markers and 0.012% with the new markers. Overall, there was an excellent correlation in MRD estimates between new and standard markers (r = 0.9816, P < .0001 by Spearman regression analysis of the MRD-positive tests; Figure 3A).
Relation between results of MRD monitoring with the new markers and those of standard MRD assays. (A) Comparison with flow cytometry using standard markers. The top 16 differentially expressed markers listed in Table 2 were included for a total of 258 tests (number of tests for each marker: CD44, 15; BCL2, 18; HSPB1, 32; CD73, 15; CD24, 7; CD123, 34; CD72, 10; CD86, 21; CD200, 29; CD79b, 5; CD164, 7; CD304, 12; CD97, 20; CD102, 10; CD99, 8; CD300A, 15). Spearman regression analysis of positive MRD results by both methods: r = 0.9816, P < .0001). (B) Comparison with PCR amplification of Ag-receptor genes performed in a subset of the samples (r = 0.8178, P < .0001). Each symbol represents results obtained by 4-color flow cytometry including Abs against CD19, CD10, CD34 and one of the new markers. The dashed line is the line of identity.
Relation between results of MRD monitoring with the new markers and those of standard MRD assays. (A) Comparison with flow cytometry using standard markers. The top 16 differentially expressed markers listed in Table 2 were included for a total of 258 tests (number of tests for each marker: CD44, 15; BCL2, 18; HSPB1, 32; CD73, 15; CD24, 7; CD123, 34; CD72, 10; CD86, 21; CD200, 29; CD79b, 5; CD164, 7; CD304, 12; CD97, 20; CD102, 10; CD99, 8; CD300A, 15). Spearman regression analysis of positive MRD results by both methods: r = 0.9816, P < .0001). (B) Comparison with PCR amplification of Ag-receptor genes performed in a subset of the samples (r = 0.8178, P < .0001). Each symbol represents results obtained by 4-color flow cytometry including Abs against CD19, CD10, CD34 and one of the new markers. The dashed line is the line of identity.
In a subset of 52 samples from 18 patients studied with some of the markers (CD44, BCL2, HSPB1, CD73, CD24, CD123, CD86, CD200, CD304, CD97, CD99, CD102, and CD300a), MRD estimates by PCR amplification of clonally rearranged Ig and TCR genes were also available. Using the threshold of 0.01% ALL cells to define MRD positivity, MRD was negative (< 0.01%) by flow cytometry and PCR analysis in 35 of the 52 samples studied. By contrast, MRD was ≥ 0.01% according to both methods in 15 samples. Two additional samples had MRD ≥ 0.01% by flow cytometry while PCR showed detectable signals but below the 0.01% threshold: 0.007% and 0.003%. Figure 3B shows the relation between flow cytometric and PCR measurements for each marker. Among the 101 tests performed, MRD was < 0.01% by flow cytometry in 63 and ≥ 0.01% in 38, with a good correlation in the MRD-positive estimates by the 2 techniques (r = 0.8178, P < .0001).
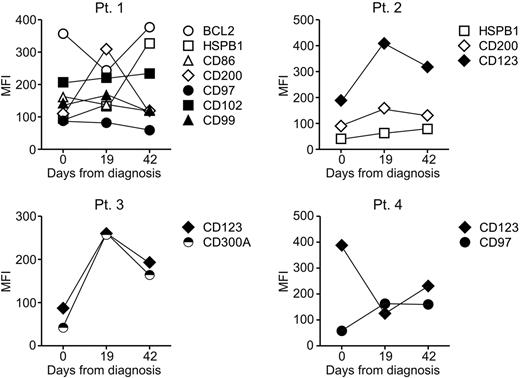
To determine stability of the new markers during the course of the disease, a prerequisite for reliable MRD tracking,40 we compared their level of expression to that recorded during MRD monitoring in 10 patients who had persistent MRD during remission induction therapy (day 19 and 42 from diagnosis). Figure 4 illustrates the results obtained in 4 representative cases: although levels of expression fluctuated during therapy, they consistently remained outside the range of normality in all cases studied. The long-term stability of the markers was assessed by comparing the immunophenotype of paired samples collected at diagnosis and relapse from 9 patients whose leukemic cells at diagnosis expressed some of the newly identified markers identified. As shown in Table 3, abnormal marker expression at diagnosis reverted to expression within the normal range at relapse in 7 of 55 (12.7%) of comparisons. In the remaining cases, marker expression remained abnormal. Importantly, in all 9 patients studied at least one marker remained abnormally expressed at relapse. In 58 additional comparisons where the markers at diagnosis were within the normal range, there were 10 instances (17.2%) in which expression became abnormal at relapse. These results suggest that false-negative results because of phenotypic shifts affecting these markers would be unlikely.
Expression of the new markers before, during and at the end of remission induction therapy. Symbols indicate mean fluorescence intensity (MFI) of each marker as measured on ALL cells at diagnosis and on residual leukemic lymphoblasts detected on days 19 and 42 of remission induction therapy. At all time points, all markers were expressed at levels that exceeded those measured in normal CD19+CD10+ cells.
Expression of the new markers before, during and at the end of remission induction therapy. Symbols indicate mean fluorescence intensity (MFI) of each marker as measured on ALL cells at diagnosis and on residual leukemic lymphoblasts detected on days 19 and 42 of remission induction therapy. At all time points, all markers were expressed at levels that exceeded those measured in normal CD19+CD10+ cells.
Expression of new markers in paired samples of B-lineage ALL collected at diagnosis and at relapse
| Marker . | Patient 1 . | Patient 2 . | Patient 3 . | Patient 4 . | Patient 5 . | Patient 6 . | Patient 7 . | Patient 8 . | Patient 9 . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D . | R . | D . | R . | D . | R . | D . | R . | D . | R . | D . | R . | D . | R . | D . | R . | D . | R . | |
| CD44 | — | — | 233 | 235 | 372 | —* | 325 | —* | — | — | — | — | 3 | 4 | 208 | 93 | 292 | 307 |
| BCL2 | 254 | 249 | 240 | 262 | — | — | 286 | 326 | 144 | 137 | 332 | 562 | 150 | 151 | 87 | 137 | 273 | 188 |
| HSPB1 | 18 | 47 | 44 | 94 | — | 74† | 169 | 143 | 141 | 165 | 64 | 351 | 23 | 267 | 15 | 158 | 80 | 36 |
| CD73 | 30 | 39 | 37 | 64 | 56 | 149 | 29 | 105 | — | — | 95 | 30 | 33 | —* | 168 | 33 | 103 | 63 |
| CD24 | — | — | 338 | 504 | — | 333† | 204 | 146 | 104 | 2 | — | 277† | — | — | — | — | — | 77† |
| CD123 | — | — | 62 | 31 | — | — | 45 | 133 | — | — | — | — | — | — | — | — | — | 25† |
| CD72 | 6 | 7 | NT | NT | NT | NT | — | — | — | — | 5 | 3 | 8 | 13 | 6 | 6 | 4 | 4 |
| CD86 | — | — | 26 | 23 | — | — | 33 | —* | — | — | — | — | — | — | — | — | — | — |
| CD200 | 86 | 65 | 107 | 88 | — | — | 80 | 201 | 260 | 154 | — | — | 102 | 126 | — | 72† | 83 | —* |
| CD164 | — | — | 18 | 49 | — | — | — | — | — | — | — | — | — | 29† | NT | NT | NT | NT |
| CD97 | — | — | 119 | 53 | — | — | 84 | 200 | — | — | — | 54† | — | — | — | — | — | — |
| CD102 | — | — | — | — | — | — | 96 | 92 | — | — | — | — | — | 100† | — | — | — | 136† |
| CD99 | — | — | — | — | — | — | — | 329† | 184 | —* | 124 | —* | — | — | — | — | 122 | 488 |
| Marker . | Patient 1 . | Patient 2 . | Patient 3 . | Patient 4 . | Patient 5 . | Patient 6 . | Patient 7 . | Patient 8 . | Patient 9 . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D . | R . | D . | R . | D . | R . | D . | R . | D . | R . | D . | R . | D . | R . | D . | R . | D . | R . | |
| CD44 | — | — | 233 | 235 | 372 | —* | 325 | —* | — | — | — | — | 3 | 4 | 208 | 93 | 292 | 307 |
| BCL2 | 254 | 249 | 240 | 262 | — | — | 286 | 326 | 144 | 137 | 332 | 562 | 150 | 151 | 87 | 137 | 273 | 188 |
| HSPB1 | 18 | 47 | 44 | 94 | — | 74† | 169 | 143 | 141 | 165 | 64 | 351 | 23 | 267 | 15 | 158 | 80 | 36 |
| CD73 | 30 | 39 | 37 | 64 | 56 | 149 | 29 | 105 | — | — | 95 | 30 | 33 | —* | 168 | 33 | 103 | 63 |
| CD24 | — | — | 338 | 504 | — | 333† | 204 | 146 | 104 | 2 | — | 277† | — | — | — | — | — | 77† |
| CD123 | — | — | 62 | 31 | — | — | 45 | 133 | — | — | — | — | — | — | — | — | — | 25† |
| CD72 | 6 | 7 | NT | NT | NT | NT | — | — | — | — | 5 | 3 | 8 | 13 | 6 | 6 | 4 | 4 |
| CD86 | — | — | 26 | 23 | — | — | 33 | —* | — | — | — | — | — | — | — | — | — | — |
| CD200 | 86 | 65 | 107 | 88 | — | — | 80 | 201 | 260 | 154 | — | — | 102 | 126 | — | 72† | 83 | —* |
| CD164 | — | — | 18 | 49 | — | — | — | — | — | — | — | — | — | 29† | NT | NT | NT | NT |
| CD97 | — | — | 119 | 53 | — | — | 84 | 200 | — | — | — | 54† | — | — | — | — | — | — |
| CD102 | — | — | — | — | — | — | 96 | 92 | — | — | — | — | — | 100† | — | — | — | 136† |
| CD99 | — | — | — | — | — | — | — | 329† | 184 | —* | 124 | —* | — | — | — | — | 122 | 488 |
Values indicate MFI as determined by flow cytometry.
ALL indicates acute lymphoblastic leukemia; D, diagnosis; R, relapse; MFI, mean fluorescence intensity, NT, not tested; and —, MFI within the normal range.
Significant change in expression: from abnormal expression at diagnosis to expression within the normal range at relapse.
Significant change in expression: from expression within the normal range at diagnosis to abnormal expression at relapse.
Association of the new markers with genetic subtypes of ALL
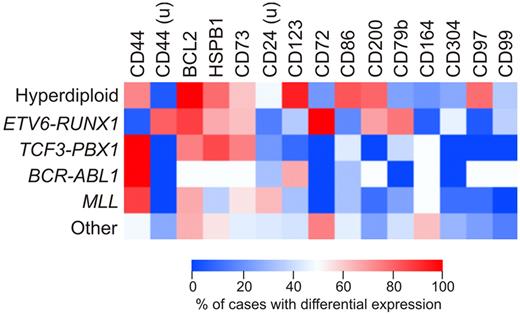
We determined whether expression of the new markers identified in this study was associated with known genetic subtypes of ALL, such as hyperdiploidy (51-65 chromosomes), ETV6-RUNX1, TCF3-PBX1, BCR-ABL1, or MLL gene rearrangements. Figure 5 shows the results of this analysis as a heat map including markers that were differentially expressed in at least 20% of cases. Expression of some markers was clearly related to ALL genetic subtype. Thus, among hyperdiploid (51-65 chromosomes) ALL cases, there was a significantly higher prevalence of CD123 (P < .0001 by Fisher exact test), CD86 (P < .0001), CD200 (P = .0003), and CD97 (P < .0001) overexpression compared with the other cases without this genetic abnormality. Among cases with ETV6-RUNX1, there was a higher prevalence of CD200 overexpression (P < .0001), and of CD44 (P < .0001), CD72 (P = .0073), and CD79b (P = .0109) underexpression. Finally, abnormal expression of CD164 was most prevalent among cases lacking all the genetic abnormalities analyzed (P = .002).
Differential expression of the new markers according to the genetic subtype of ALL. Heatmap shows the percentage of cases among the main genetic subtypes of childhood ALL in which the markers studied were differentially expressed by flow cytometry. Percentages refer to cases in which each marker was overexpressed in ALL cases compared with CD19+CD10+ cells from nonleukemic BM samples; markers underexpressed in ALL cells are indicated by “u.” All markers differentially expressed in at least 20% of cases were included.
Differential expression of the new markers according to the genetic subtype of ALL. Heatmap shows the percentage of cases among the main genetic subtypes of childhood ALL in which the markers studied were differentially expressed by flow cytometry. Percentages refer to cases in which each marker was overexpressed in ALL cases compared with CD19+CD10+ cells from nonleukemic BM samples; markers underexpressed in ALL cells are indicated by “u.” All markers differentially expressed in at least 20% of cases were included.
Comparisons between new markers and established leukemia-associated immunophenotypes
In a proportion of patients with ALL, MRD cannot currently be monitored because of the lack of suitable immunophenotypes5,27 ; in other patients, leukemic cells express only one set of markers, and have an increased risk of false-negative MRD results because of immunophenotypic shifts.7 The availability of additional markers should allow MRD studies in patients whose ALL cells currently lack suitable leukemia-associated immunophenotypes and minimize the risk of false-negative results. To test this prediction, we assessed the expression of the markers discovered in this study in 171 ALL diagnostic samples: 11 lacked detectable leukemia-associated immunophenotypes with the standard panel of Abs used in our laboratory (see supplemental Table 4 for list of standard markers), and another 37 had only one immunophenotypic abnormality detectable with the standard panel. The new markers detected a phenotypic abnormality suitable for MRD study in all of the 11 cases previously lacking a leukemia-associated immunophenotype, and identified additional abnormalities in 36 of the 37 cases with one standard abnormality. Thus, the new markers substantially improved the potential for flow cytometric monitoring of MRD in 47 of 48 cases who could not be adequately studied previously. The number of new markers differentially expressed ranged from 2 to 12 (median, 6). In 45 of the 47 cases, 1 or more markers were expressed at levels that exceeded by at least 2-fold the normal range, allowing a particularly clear identification of ALL cells.
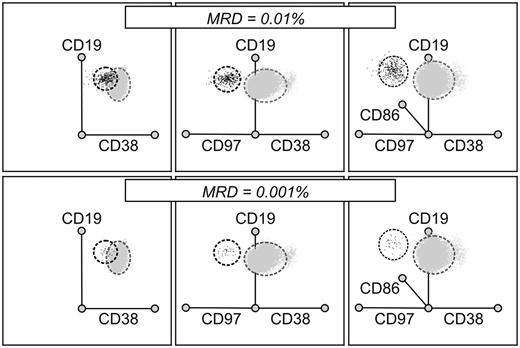
The availability of additional markers of leukemia is likely to improve the resolution of leukemic and normal cells during flow cytometric analysis. This notion was clearly demonstrated in experiments in which mixtures of leukemia and normal cells were analyzed using either one standard phenotypic abnormality (eg, underexpression of CD38), or additional abnormalities revealed by the new markers. An example of such an experiment is shown in Figure 6: by examining 3 abnormalities simultaneously (underexpression of CD38, overexpression of CD97 and CD86), the discrimination between normal and leukemic CD19+, CD10+, CD34+ cells is much improved, allowing the unequivocal detection of ALL cells at a level of 0.001%.
Additional leukemia-associated markers improve resolution of MRD. Files containing flow cytometric data from 5 nonleukemic BM samples cells (gray dots) and one diagnostic ALL sample (black dots) were merged and analyzed as radial plots using the Kaluza software after selecting all CD19+CD10+CD34+ cells. Mixtures containing 0.01% ALL cells (top row) and 0.001% ALL cells (bottom row) are shown. Underexpression of CD38 alone, a standard MRD marker, could not discriminate well between ALL cells and normal CD19+ CD10+ CD34+ cells (left panels) in this case; the discrimination was improved by analyzing expression of CD97 (middle panels), and further improved by the inclusion of CD86 and the use of a 3-dimensional space (right panels).
Additional leukemia-associated markers improve resolution of MRD. Files containing flow cytometric data from 5 nonleukemic BM samples cells (gray dots) and one diagnostic ALL sample (black dots) were merged and analyzed as radial plots using the Kaluza software after selecting all CD19+CD10+CD34+ cells. Mixtures containing 0.01% ALL cells (top row) and 0.001% ALL cells (bottom row) are shown. Underexpression of CD38 alone, a standard MRD marker, could not discriminate well between ALL cells and normal CD19+ CD10+ CD34+ cells (left panels) in this case; the discrimination was improved by analyzing expression of CD97 (middle panels), and further improved by the inclusion of CD86 and the use of a 3-dimensional space (right panels).
Discussion
Progress in the identification of new leukemia-specific markers has historically relied either on serendipitous observations or on painstaking comparisons of the expression of individual markers in normal and leukemic cells.5,29,41 In this study, we searched for new markers of MRD in B-lineage ALL by defining differences in genome-wide gene expression between leukemic and normal cells. We focused our analysis on normal BM cells with immunophenotypic features similar to those of leukemic lymphoblasts, ie BM CD19+CD10+ cells, which are the most difficult to distinguish from ALL lymphoblasts during MRD studies by flow cytometry.5,29 Among genes differentially expressed by genome-wide expression array analysis, we selected 30 for further studies based on their highly abnormal expression at the mRNA level in a substantial proportion of ALL cases and on the availability of specific Abs suitable for routine flow cytometric studies. We found that 22 of the 30 markers were also differentially expressed by flow cytometry. When those that were differentially expressed in at least 20% of cases (n = 16) were applied to study MRD in clinical samples, they yielded results that correlated well with those obtained by standard flow cytometric methods and PCR-based analyses, and were generally stably expressed during the course of the disease. The addition of the new markers to the current panels allowed the identification of unique leukemia profiles in all patients and afforded the detection of 1 leukemic cell in 100 000 normal BM cells, thus significantly enhancing the power of flow cytometric monitoring of MRD in ALL.
The conceptual underpinning for the current study was provided by the results of a pilot study in which we used first-generation gene arrays containing probes for approximately 4000 genes to compare the gene expression profiles of diagnostic ALL samples from 4 patients and CD19+CD10+ BM immature B cells from 2 healthy donors.31 Despite its technical limitations, this early proof-of-principle study allowed the identification of CD58 as a useful MRD marker.14,31 In the present study, we used a much more powerful tool (ie, gene expression arrays probing approximately 23 000 genes) and a much larger set of samples (270 cases of B-lineage ALL) encompassing the spectrum of genetic abnormalities occurring in ALL to identify other markers. CD58 was one of the genes that were differentially expressed in this analysis. This, together with the identification of other genes previously shown to be differentially expressed in ALL cells confirmed the dependability of the approach. More importantly, numerous differentially expressed genes were discovered.
One potential limitation of the array-based screening is that transcript expression might not always predict levels of protein expression. Nevertheless, for 13 of the 30 markers tested by flow cytometry, there was a good concordance between results of microarray measurements and those by flow cytometry, with statistically significant differences in protein expression between the leukemia and control groups. Expression of 9 additional markers was not significantly different between the 2 groups, but we identified several ALL cases with expression at levels clearly distinct from those found during normal B-cell development. Finally, 8 of the 30 markers tested were generally undetectable by flow cytometry despite a relative abundance of mRNA transcripts measured by microarray analysis. Whether any of these genes, or other genes found to be differentially expressed in ALL cells but not tested by flow cytometry, could be useful targets for PCR-based studies of MRD remains to be determined. One precedent for using differentially expressed genes for MRD studies by quantitative PCR is represented by WT1,32 which was found to be overexpressed in 36% of cases of ALL in our study (supplemental Table 2). The database of differentially expressed genes that we generated can now be mined in search for other targets for molecular MRD monitoring.
Expression of some of the genes was associated with ALL subtypes defined by genetic abnormalities. Among the most striking was the overexpression of several markers in hyperdiploid (51-65 chromosomes) ALL cases, including CD86, CD97 and CD123 (the latter confirming an earlier observation by Djokic et al),42 underexpression of CD44 and CD72 in cases with ETV6-RUNX1, and overexpression of CD200 in both genetic subtypes. In contrast to the consistent overexpression of PBX1 at the mRNA level in TCF3-PBX1 cases, PBX1 expression by flow cytometry in these cases was unremarkable. Because the purpose of our study was to improve MRD monitoring in ALL, we did not address the functional relevance of the molecules found to be differentially expressed. Our findings, however, should spur correlative studies to determine the prognostic significance of the differentially expressed genes, and experiments to ascertain whether they play a role in leukemogenesis and could be targeted for therapy. To this end, overexpression of BCL2 in ALL is associated with prolonged survival of leukemic cells43 and this molecule can be targeted by specific inhibitors44 ; HSBP1 (heat shock protein 27), CD86 (B7.2), and CD304 (neuropilin-1) overexpression have been associated with drug resistance and disease progression in AML cells45-47 ;expression of CD200 carries an adverse prognosis in patients with multiple myeloma48 ; and Abs against CD99 and CD123 have been shown to have effective antitumor activity in murine models of Ewing sarcoma49 and AML,50 respectively.
Morphologic studies of treatment response are being supplanted by MRD monitoring in contemporary ALL protocols, thus changing the criteria that define complete remission and leukemia relapse.1-4 Although clinically useful, MRD assays are currently limited by their technical complexity and, in the case of flow cytometry, by the requirement for expert interpretative skills. Both molecular and flow cytometric methods also require identification of the leukemia-associated targets at diagnosis, a task that it is difficult to accomplish in referral centers where diagnostic (or relapse) samples may not be available. The discovery of new markers of ALL should help widening the applicability of MRD testing and possibly allow reliable MRD studies without prior knowledge of the leukemic cells' immunophenotype, a concept that remains to be conclusively proven. By incorporating the top 10 differentially expressed markers discovered in this study in our MRD Ab panels (Table 4) we have been able to identify leukemia-associated immunophenotypes to monitor MRD in the last 155 consecutive B-lineage ALL patients enrolled at our institution. The sensitivity of this approach should surpass the standard 0.01% threshold. Ongoing studies suggest that the approach reported here can also identify new markers for universal and sensitive MRD monitoring in AML.
Marker combinations currently used in our laboratory for MRD studies in B-lineage ALL incorporating the top 10 differentially expressed markers discovered in this study
| FITC . | PE . | PerCP . | Allophycocyanin . | PE-Cy7 . | APC-H7 . | Horizon v450 . |
|---|---|---|---|---|---|---|
| CD38 | CD24* | CD34 | CD19 | CD10 | CD45 | CD44* |
| CD38 | CD58 | CD34 | CD19 | CD10 | CD45 | CD44* |
| CD38 | CD73* | CD34 | CD19 | CD10 | CD45 | CD15 |
| CD38 | CD200* | CD34 | CD19 | CD10 | CD45 | CD44* |
| CD66c | CD123* | CD34 | CD19 | CD10 | CD45 | CD86* |
| CD72* | CD13 | CD34 | CD19 | CD10 | CD45 | CD33 |
| CD79b* | HSPB1* | CD34 | CD19 | CD10 | CD45 | Bcl2* |
| FITC . | PE . | PerCP . | Allophycocyanin . | PE-Cy7 . | APC-H7 . | Horizon v450 . |
|---|---|---|---|---|---|---|
| CD38 | CD24* | CD34 | CD19 | CD10 | CD45 | CD44* |
| CD38 | CD58 | CD34 | CD19 | CD10 | CD45 | CD44* |
| CD38 | CD73* | CD34 | CD19 | CD10 | CD45 | CD15 |
| CD38 | CD200* | CD34 | CD19 | CD10 | CD45 | CD44* |
| CD66c | CD123* | CD34 | CD19 | CD10 | CD45 | CD86* |
| CD72* | CD13 | CD34 | CD19 | CD10 | CD45 | CD33 |
| CD79b* | HSPB1* | CD34 | CD19 | CD10 | CD45 | Bcl2* |
MRD indicates minimal residual disease; ALL, acute lymphoblastic leukemia; PerCP, peridin chlorophyll protein; and PE-Cy7, PE–Cyanine 7.
Marker from this study.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants CA60419 and CA21765 from the National Cancer Institute and by the American Lebanese Syrian Associated Charities (ALSAC).
National Institutes of Health
Authorship
Contribution: E.C.-S. performed cell sorting and flow cytometric studies, and analyzed data; G.S., X.S., S.S., and J.R.D. performed gene expression array studies and analyzed data; C.C., L.K., P.L., and M.M. performed flow cytometric studies; P.S. performed molecular studies of MRD; C.-H.P. led the clinical protocols under which the samples studied were collected and annotated; and D.C. initiated the study with E.C.S., analyzed data, and wrote the manuscript with the contribution of all other authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dario Campana, MD, PhD, Department of Oncology, St Jude Children's Research Hospital, 262 Danny Thomas Pl, Memphis TN 38105; e-mail: dario.campana@stjude.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal