Abstract
Considerable effort has been spent identifying prognostic biomarkers in classic Hodgkin lymphoma (cHL). The aim of our study was to search for possible prognostic parameters in advanced-stage cHL using a proteomics-based strategy. A total of 14 cHL pretreatment tissue samples from younger, advanced-stage patients were included. Patients were grouped according to treatment response. Proteins that were differentially expressed between the groups were analyzed using 2D-PAGE and identified by liquid chromatography mass spectrometry. Selected proteins were validated using Western blot analysis. One of the differentially expressed proteins, the carbohydrate-binding protein galectin-1 (Gal-1), was further analyzed using immunohistochemistry HC and its expression was correlated with clinicopathologic and outcome parameters in 143 advanced-stage cHL cases. At the univariate level, high Gal-1 expression in the tumor microenvironment was correlated with poor event-free survival (P = .02). Among younger (≤ 61 years) patients, high Gal-1 was correlated with poorer overall and event-free survival (both P = .007). In this patient group and at the multivariate level, high Gal-1 expression retained a significant predictive impact on event-free survival. Therefore, in addition to its functional role in cHL-induced immunosuppression, Gal-1 is also associated with an adverse clinical outcome in this disease.
Introduction
The tumor lesion in classic Hodgkin lymphoma (cHL) is characterized by the presence of the typical Hodgkin and Reed-Sternberg (HRS) tumor cells, largely outnumbered by nonneoplastic inflammatory bystanders that include B and T lymphocytes (predominantly Th2-type and FoxP3+ regulatory T cells), macrophages, eosinophils, basophils, and plasma cells.1
Over the past decades, the treatment of cHL has improved considerably, and current therapeutic strategies succeed in curing > 80% of patients. However, 20%-30% still experience relapse or refractory disease, many subsequently succumbing to death as a result of either treatment complications or disease progression.2 Prompted by observations in other lymphoma subtypes such as follicular lymphoma,3 increased attention has been drawn to the possible biologic role and prognostic significance of the tumor-infiltrating inflammatory cells in cHL. Several recent studies have focused on this area, using gene-expression profiling to identify new prognostic markers in cHL.4,5 Although these studies have led to the identification of several candidate genes, no specific biologic marker has yet been introduced as a reliable tool for pretherapeutic risk assessment.
Several factors may influence the proteomic machinery and affect the final properties of the encoded protein at several levels: translational inhibition/activation, posttranslational modifications, inhibition/activation of the proteins, specific degradations, etc. Consequently, there can be considerable discrepancy between the expression level of RNA and that of the corresponding protein product.6 Therefore, proteomics-based approaches have recently been used in HL to identify protein patterns that may provide pathogenetic and prognostic clues.7,8 However, these studies were only performed in cell lines and still await confirmation in an in vivo setting.
Galectin-1 (Gal-1), a member of a growing family of animal lectins with affinity to poly-N-acetyllactosamine–enriched glycoconjugates, has recently emerged as a regulator of inflammatory responses, angiogenesis, and tumor progression.9,10 This glycan-binding protein has been identified as a critical mediator of the immunosuppressive activity of cHL because it is responsible for creating the Th2/regulatory T cell–skewed microenvironment typical of the disease.11,12 Moreover, the combination of Gal-1 and c-Jun has been proposed as a diagnostic biomarker for distinguishing cHL from other lymphomas with shared morphologic and molecular features.13
The aim of the present study was to provide data from an “in vivo” setting by identifying differentially expressed proteins in primary diagnostic tissue samples from treatment-sensitive versus treatment-refractory cHL patients. This study constitutes the first approach seeking to correlate protein expression, as assessed by proteomics, with treatment response and clinical outcome in advanced-stage cHL. Protein expression was evaluated in both the neoplastic and nonneoplastic compartments of tumor biopsies.
Methods
Patients
A total of 14 cHL pretreatment tissue samples were selected for 2D-PAGE and liquid chromatography–tandem mass spectrometry (LC-MS/MS). The tissue samples were embedded in optimal cutting temperature medium (Tissue-Tek; Sakura) compound, snap-frozen in isopentane in liquid nitrogen at the time of diagnosis, and stored at −80°C.
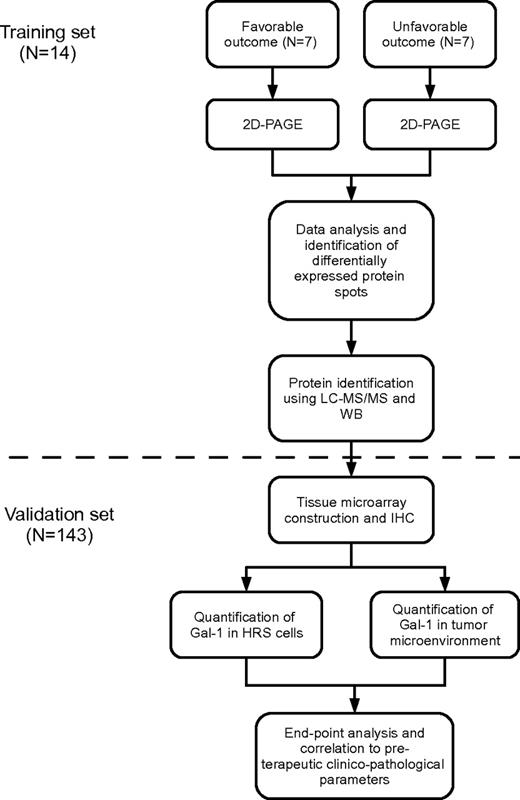
On the basis of responses to initial treatment, 2 patient cohorts were identified. The clinical features of these patients are detailed in Table 1. One cohort was characterized by a favorable treatment response, defined as a sustained complete remission for a minimum of 36 months. The second was characterized by an unfavorable response to treatment, defined as either progressive/refractory disease or relapse within a year after the end of first-line therapy. To maximize comparability between the 2 cohorts in terms of protein expression, patients in the 2 groups were matched with regard to age (range 20-35 vs 14-51, median 24 vs 27 years; P = .52), clinical stage (IB, IIB, III, or IV), and cHL histology (nodular sclerosis or mixed cellularity). The workflow is outlined in Figure 1.
Clinical characteristics of patients selected for proteome comparison
| No. . | Age, y . | Histology . | Ann Arbor stage . | B symptoms . | Treatment . | Outcome . |
|---|---|---|---|---|---|---|
| 1 | 21 | MC | III | absent | ABVD | FR |
| 2 | 24 | NS | II | present | ABVD/COPP + RT | FR |
| 3 | 24 | NS | IV | present | ABVD/COPP | FR |
| 4 | 26 | NS | II | present | ABVD/COPP + RT | FR |
| 5 | 35 | NS | III | absent | ABVD + RT | FR |
| 6 | 20 | NS | III | present | ABVD/COPP + RT | FR |
| 7 | 25 | NS | III | present | ABVD/COPP + RT | FR |
| 8 | 51 | cHL-NOS | I | present | ABVD/COPP + RT | UR |
| 9 | 41 | NS | IV | present | ABVD/COPP + RT | UR |
| 10 | 14 | NS | III | present | ABVD/COPP | UR |
| 11 | 21 | NS | II | present | ABVD/COPP | UR |
| 12 | 27 | NS | II | present | ABVD/COPP | UR |
| 13 | 33 | NS | IV | present | ABVD | UR |
| 14 | 23 | NS | III | present | ABVD/COPP + RT | UR |
| No. . | Age, y . | Histology . | Ann Arbor stage . | B symptoms . | Treatment . | Outcome . |
|---|---|---|---|---|---|---|
| 1 | 21 | MC | III | absent | ABVD | FR |
| 2 | 24 | NS | II | present | ABVD/COPP + RT | FR |
| 3 | 24 | NS | IV | present | ABVD/COPP | FR |
| 4 | 26 | NS | II | present | ABVD/COPP + RT | FR |
| 5 | 35 | NS | III | absent | ABVD + RT | FR |
| 6 | 20 | NS | III | present | ABVD/COPP + RT | FR |
| 7 | 25 | NS | III | present | ABVD/COPP + RT | FR |
| 8 | 51 | cHL-NOS | I | present | ABVD/COPP + RT | UR |
| 9 | 41 | NS | IV | present | ABVD/COPP + RT | UR |
| 10 | 14 | NS | III | present | ABVD/COPP | UR |
| 11 | 21 | NS | II | present | ABVD/COPP | UR |
| 12 | 27 | NS | II | present | ABVD/COPP | UR |
| 13 | 33 | NS | IV | present | ABVD | UR |
| 14 | 23 | NS | III | present | ABVD/COPP + RT | UR |
FR indicates favorable response; UR, unfavorable response; MC, mixed cellularity; NS, Nodular sclerosis; NOS, not otherwise specified; and RT, radiotherapy.
Flowchart describing the 2-step study strategy. Shown is the training-set analysis followed by the validation-set analysis. IHC indicates immunohistochemistry.
Flowchart describing the 2-step study strategy. Shown is the training-set analysis followed by the validation-set analysis. IHC indicates immunohistochemistry.
Where possible, data generated from the proteomic analysis were validated in a cohort of 143 cHL patients using immunohistochemistry (IHC) on tissue microarrays (TMAs) constructed from formalin-fixed, paraffin-embedded primary pretreatment tumor biopsies (Table 3). Clinical and follow-up data were obtained from clinical records. For deceased patients, the precise date of death was obtained from the Danish Civil Registration System. All primary diagnostic tumor biopsies were reviewed and reclassified according to the World Health Organization classification of tumors of the hematopoietic and lymphoid tissues.14 The patients were treated at Aarhus University Hospital (Denmark) between 1990 and 2007 according to standard guidelines. Patients were uniformly treated with “ABVD/COPP” (Adriamycin, bleomycin, vinblastine, dacarbazine/cyclophosphamide, vincristine, procarbazine, and prednisone) chemotherapy. Additional radiotherapy was given in case of pretherapeutic bulk or localized residual masses. Treatment response was assessed using standardized guidelines.15 The study was approved by the Regional Ethical Committee and by the Danish Data Protection Agency.
Sample collection and preparation
From each cHL sample, a total of 70 sections of 20 μm were cut and transferred into empty Eppendorf tubes. H&E stains were made before and after this process to ensure the presence of HRS cells throughout the samples. Subsequently, 350 μL of 3-10 NL lysis buffer containing 9M urea, 2% (wt/vol) Triton X-100, 2% (wt/vol) DTT, and 2% (vol/wt) IPG-buffer (Amersham) were added to the cut sections.
Determination of total protein content
To ensure loading of equal protein amounts for both 2D-PAGE and 1D Western blotting (WB), the protein concentration was determined using a noninterfering assay (488250; Calbiochem).
2D-PAGE
The 2D-PAGE procedure was performed essentially as described previously16,17 with a few modifications. In summary, 100 μg of protein was used from each sample. Isoelectric focusing was performed using 18-cm pH 3-10 or pH 4-7 nonlinear IPG strips. The IPG strips were rehydrated overnight (∼ 20 hours) in 150 μL of rehydration solution containing 8M urea, 2% (wt/vol) CHAPS, 0.3% (wt/vol) DTT, 2% (vol/wt) IPG-buffer, and bromphenol blue together with the 100-μg protein sample in 200 μL of 3-10 NL lysis buffer in a Immobiline DryStrip Reswelling Tray. After rehydration, the samples were run at 500 V for 1 minute, 500 V for 5 hours, and 3500 V for 14.5 hours in a Multiphor II system (PerkinElmer) in a gradient mode at 17°C (MultiTemp III Thermostatic Circulator; GE Healthcare Life Sciences). The IPG strips were equilibrated twice: first in 20 mL of equilibration buffer consisting of 0.6% (wt/vol) Tris-HCl (pH 6.8), 6M urea, 30% (vol/vol) glycerol, 1% (wt/vol) SDS, and 0.05% (wt/vol) DTT, and then in 4.5% (wt/vol) iodoacetamide and bromophenol blue. For the second dimension, 12% SDS-PAGE gels were run at 50 V overnight (∼ 20 hours), and transferred to fixation solution containing 50% ethanol, 12% acetic acid, and 0.05% formaldehyde for a minimum of 1 hour.
Silver staining
To visualize the proteins separated by 2D-PAGE, we used the Vorum protocol as described previously.18 Gels were washed 3 times for 20 minutes each in 35% ethanol, then pretreated for 2 minutes in 0.02% Na2S2O3;5H2O and washed in water 3 times for 3 minutes each. The gels were stained with 0.2% AgNO3 and 0.028% formaldehyde for 20 minutes. Gels were then rinsed twice in water for 1 minute, developed in 6% Na2CO3, 0.0185% formaldehyde, 0.0004% Na2S2O3;5H2O, and the reaction was stopped using 40% (vol/vol) ethanol and 12% (vol/vol) acetic acid. This solution was changed once and the gels were dried between cellophane sheets and sealed.
Data analysis
Two analyses were performed with first-dimension separation using isoelectric focusing at either pH 3-10 or pH 4-7. Thirteen patients were included in each analysis. (pH 3-10: patients 27, 35, 69, 79, 138, 174, 182, 201, 243, 333, 415, 463, and 464; pH 4-7: patients 35, 69, 79, 138, 174, 182, 201, 243, 333, 415, 462, 463, and 464). The gels were scanned with a GS-710 Calibrated Imaging Densitometer (Bio-Rad) using the Quantity One software package Version 4.0.2. Gels were then imported into the PDQuest Advanced 2D Gel Analysis Software Version 3.0 (Bio-Rad) that assigned a volume to each spot proportional to the amount of protein. In brief, the gels were normalized against the total density and, after background subtraction, the spots were automatically defined and quantified. One gel in each study (pH 3-10: patient 79; pH 4-7: patient 464) was chosen as a reference gel (master gel). In the reference gel, each spot was assigned a number and all other gels were aligned to the reference gel and all matched spots were numbered accordingly. All matches were critically evaluated and the necessary editions and corrections were performed manually. Spots that showed a significant (Mann-Whitney U test, P < .05) and at least 2-fold differential expression were cut out of the gel and subjected to in-gel tryptic digestion.
Protein identification by LC-MS/MS
Sample preparation and MS were performed essentially as described previously.19 In brief, proteins were in-gel trypsinized overnight, pretreated with acetonitrile, reduced with DTT, blocked with iodoacetamide, cleaved by trypsin, and the peptides were extracted by 1 change of NH4HCO3 and 3 changes of formic acid in acetonitrile. Finally, the extracted peptides were dried. The dried fragmented protein samples were dissolved in 6 μL of buffer A (97.7% H2O, 2% acetonitrile, and 0.3% formic acid). Peptide separation before MS/MS analysis was performed using an inert nano-liquid chromatography system (LC Packings). The samples were then analyzed on a Q-TOF Premier mass spectrometer (Waters). The processed data were used to search the Swiss-Prot Database (version 57.7, 57.8, or 2010_12 containing 497.293, 509.019, or 523.151 sequences, respectively) or in a few cases the mammalian part (64 853 sequences) or the human part (20 405 sequences) using the online version of the Mascot MS/MS Ion Search facility (Matrix Science).20 Searching was performed with doubly and triply charged ions with 2 missed cleavages, a peptide mass tolerance of 20 or 50 ppm, and an MS/MS tolerance of 0.05 Da.
Antibodies
The primary rabbit polyclonal anti–Gal-1 antibody generated in the laboratory of one of the authors (G.A.R.) was described previously and found to be specific for Gal-1 detection in cHL13 (WB dilution 1:100; IHC dilution 1:5000). Other antibodies used were purchased from commercial suppliers: anti-PSME1 (HPA006632; WB dilution 1:100, IHC dilution 1:400; ATLAS Antibodies), anti-GNAI3 (XW-7212; WB dilution 1:200 ProSci Incorporated), anti-Prdx1 (HPA007730; WB dilution 1:100, IHC dilution 1:75; Atlas Antibodies), anti-FOXP3 (clone 236A/E7; dilution 1:50, Biocare Medical), anti–granzyme-B (clone GrB-7; dilution 1:20, Dako), anti-CD68 (clone KP-1; dilution 1:800; Dako), anti-CD163 (clone 10D6; dilution 1:50, Novocastra), and anti-latent membrane protein-1 (anti–LMP-1; clones CS 1-4; dilution 1:100, Dako).
WB
WB was carried out in 11 or 12 of 14 patients from the cohort. One patient per cohort could not be analyzed because of the limited amount of protein left after the 2D-PAGE experiments. The same amount of protein was added to each lane of the WBs used for one comparison, either 5 μg per lane in the case of Gal-1 or 10 μg per lane in the case of guanine nucleotide-binding protein G(k) subunit α (GNAI3), peroxiredoxin-1 (PRDX1), proteasome activator complex subunit 1 (PSME2), and desmoplakin (DESP). The samples were loaded on Novex 10%-20% Tris-glycine gels, 1.0-mm, 10-well (Invitrogen) and run for 2 hours at 125 V. After transfer by electroblotting (200 mA for 2 hours) to Hybond-H Extra, nitrocellulose membranes, 0.45 mm (Amersham Biosciences/GE Healthcare), the blots were blocked with 5% skim milk in PBS-T (0.05% Tween-20 in 80mM Na2HPO4, 20mM NaH2PO4, and 100mM NaCl, pH 7.5) and incubated overnight at 4°C. The blots were washed 3 times in PBS-T, incubated for 1 hour with the primary antibody, washed again 3 times in PBS-T, and incubated with HRP-conjugated secondary antibodies (P217, P260; Dako or ab16349; Abcam; diluted 1:5000). Development of blots was done using the enhanced chemiluminescence system (Amersham International) after 4 PBS-T washes. Digital photos were taken and analyzed by Fujifilm LAS-3000 and Image Reader LAS-3000 (Fuji). Quantification of the bands in the WBs was done with the Multi Gauge V3.0 analysis program (Fuji). In brief, boxes were created around each band and a background box was made in a reference area. The pixel intensities were measured and the mean of the pixel intensity from the reference area was subtracted from each pixel in the bands. The intensities of the bands were then analyzed with a Mann-Whitney U test to find significant differences (P < .05).
“Housekeeping” proteins are often used as references to check for proper analyses of the WBs because the expression of these proteins is assumed to be at a constant level under varying conditions. However, there is increasing evidence from proteomic studies that “housekeeping” proteins such as actin, tubulin, and GAPDH may be inadequate as internal standards because their expression can be affected by a large number of factors.19,21-23 Our present observations are in agreement with these findings, because we found 2 often used “housekeeping” proteins, actin and GAPDH, to be differentially expressed in tumor biopsies. Therefore, they were not considered adequate as reference proteins in the present study. Instead, we chose to normalize the WBs to the total protein concentration present in the tumor sample. For this purpose, the protein concentration was measured using a noninterfering protein assay (see “Determination of total protein content”).
IHC and in situ hybridization
IHC staining was performed using 4-μm paraffin-embedded tissue sections. Gal-1, FoxP3, granzyme-B, CD68, CD163, and LMP-1 stainings were performed on the Benchmark automated staining system (Ventana Medical Systems). For PRDX1, the Xmatrx automated staining system (BioGenex Laboratories) was used. The staining for PSME1 was performed manually. Slides were incubated with the primary antibodies for 32 minutes (Gal-1, PSME1, FoxP3, granzyme-B, CD68, CD163, and LMP-1) or 60 minutes (PRDX1) at 37°C. For primary antibody detection, the Ventana Ultraview DAB detection kit and the Dako Envision Flex+ visualization system were used. Tissues from malignant melanoma (Gal-1), tonsil tissue (FoxP3 and granzyme-B), colon (PSME1), lung (PRDX1), liver (CD68 and CD163), and Epstein-Barr virus–positive (EBV+) cHL specimens (LMP-1 and EBER) were used as positive controls. The primary antibodies were omitted in negative controls. For LMP-1 staining, the signal was enhanced with a Ventana amplification kit. Slides were counterstained with hematoxylin. Nonisotopic in situ hybridization for EBV-encoded RNAs 1 and 2 (EBER) was performed and scored as described previously.24
TMA construction
TMA blocks containing triplicate 1-mm cores from each patient paraffin-embedded sample were constructed with a manual tissue microarrayer (Beecher Instruments) according to standard guidelines.25 Each block also contained samples of normal lymph node, placenta, kidney, and liver tissue as internal controls and guides.
Quantification of Gal-1 expression in HRS cells
Case in which > 50% of the HRS cells reacted with the antibody were considered positive. Intra-observer variation was estimated in 30 patients and the match between readings was found to be excellent with an overall agreement of 89%.
Quantification of Gal-1 expression in nonneoplastic bystander cells
Quantification of immunohistochemical stained sections was performed by stereologic analysis using a light microscope equipped with a computer-assisted stereology system (CAST; Visiopharm). The microscope had a motorized stage controlled by a computer for systematic uniform random sampling of fields of view. A Gal-1–positive cell profile in the tumor microenvironment was defined as nuclear staining of any non-HRS cell in the tumor microenvironment (eg, macrophages, plasma cells, lymphocytes, or endothelial cells). A minimum of ∼ 100 Gal-1–positive cell profiles was counted from each patient sample (median for the entire cohort: 232). Cell profiles were counted with a 40× lens at a final screen magnification of 1423× using a 2D unbiased counting frame.26 Areas containing either necrosis or artifacts were ignored. To evaluate the intra-observer variation, the Pearson correlation coefficients were calculated in a subset of patients (n = 30). The correlation coefficient was found to be 0.91 and, in accordance with a cut-point between the third and fourth quartile, the percentage of correctly classified samples was 93%. The numbers of FoxP3- and granzyme-B–positive cell profiles per square millimeter were quantified in a similar manner. The CD68+ or CD163+ volume fraction was determined by point counting with a 20× lens at a total magnification of 739×.
Statistical analysis
Patient characteristics were compared using the Wilcoxon rank-sum test, Kruskal-Wallis equality-of-populations rank test, and χ2 when appropriate. The follow-up time was defined as the time from diagnosis to either last follow-up or a given event. Event-free survival (EFS) was measured from the date of diagnosis to either disease progression or relapse, discontinuation of treatment for any reason, or censoring date. Overall survival (OS) was measured from the date of diagnosis to the date of death for any reason or censoring date. For correlation of patient survival and Gal-1 expression, patients were grouped into quartiles. The quartile of patients with the highest Gal-1 expression was also analyzed against the lower 3 quartiles taken as one group. This corresponded to a cut point of 1825 cell profiles/mm2, which was used throughout the data analysis. Survival was estimated by the Kaplan-Meier method and compared using a log-rank test. A Cox proportional multivariate model was used to assess the possible association between Gal-1 and OS and EFS, while adjusting for those factors that had a significance level < .10 at the univariate level. Before the Cox regression analysis, the assumption of proportional hazards was assessed graphically. Correlation between continuous variables was determined using the Kendall correlation. Two-sided P < .05 was considered statistically significant and P < .10 was considered as borderline significant. All statistical analyses were done using STATA software Version 10.1.
Results
Identification of differentially expressed proteins using 2D-PAGE and LC-MS/MS
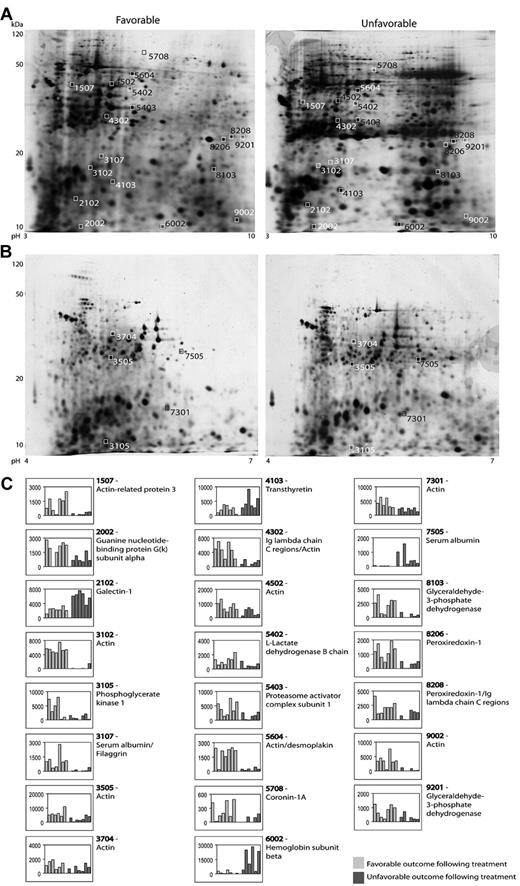
Figure 2 shows representative 2D gels from the pH 3-10 IPG and pH 4-7 IPG experiments. In total, 37 protein spots were found to be differentially expressed in patients with advanced-stage Hodgkin lymphoma. All of these were excised and analyzed for protein identification. Using LC-MS/MS and a subsequent search in the Swiss-Prot Database, 23 spots resulted in clear protein identification. Four of these spots contained more than one protein; for example, spot 4302 contained both the Ig λ chain C region and actin. Moreover, some proteins were identified in more than one spot (eg, PRDX1 in 2 spots: 8206 and 8208). Table 2 summarizes the identification information, including protein name, accession number, molecular weight, Mascot score, and function. Five of the listed proteins were further validated by WB analysis.
2D-gel analysis. (A-B) Representative gels (pH 3-10 IPG and pH 4-7 IPG) from 2D-PAGE. The 4-digit numbers are the specific spot numbers of the proteins listed in Table 2. (C) Histograms of all identified proteins displaying the spot intensities. Each patient is represented by 1 column and columns are stacked according to treatment response (favorable vs unfavorable outcome).
2D-gel analysis. (A-B) Representative gels (pH 3-10 IPG and pH 4-7 IPG) from 2D-PAGE. The 4-digit numbers are the specific spot numbers of the proteins listed in Table 2. (C) Histograms of all identified proteins displaying the spot intensities. Each patient is represented by 1 column and columns are stacked according to treatment response (favorable vs unfavorable outcome).
List of identified proteins
| Spot no. . | Identified peptides . | Protein name . | SwissProt accession . | Size, kDa . | Mascot score . | Biological processes† . |
|---|---|---|---|---|---|---|
| pH 3–10 | ||||||
| 1507 | DREVGIPPEQSLETAK EFNKYDTDGSK | Actin-related protein 3 | ARP3_HUMAN | 47.4 | 77 | Cellular component movement, regulation of actin filament polymerization, activation of dendrite morphogenesis, regulation of myosin II filament assembly or disassembly, response to antibiotic |
| 2002 | DGGVQACFSRSR | Guanine nucleotide-binding protein G(k) subunit alpha | GNAI3_HUMAN | 40.5 | 24‡ | G-protein–coupled receptor protein signaling pathway, negative regulation of adenylate cyclase activity, negative regulation of synaptic transmission, signal transduction, vesicle function |
| 2102 | VRGEVAPDAK FNAHGDANTIVCNSK | Galectin-1 | LEG1_HUMAN | 14.7 | 154 | Activation of I-kB kinase/NF-kB cascade, apoptotic programmed cell, death, cellular response to glucose stimulus, multicellular organismal response to stress, myoblast cell differentiation, negative regulation of cell-substrate adhesion |
| 3102 | QEYDESGPSIVHR | Actin§ | ACTB_HUMAN | 42 | 86 | Blood coagulation, platelet activation, platelet degranulation |
| 3107 | TCVADESAENCDK | Serum albumin | ALBU_HUMAN | 69.4 | 23‡ | Transport, negative regulation of programmed cell death and apoptosis, maintenance of mitochondrial location, cellular response to starvation |
| 3107 | HHEASSRADSSR | Filaggrin | FILA_HUMAN | 435 | 32 | Keratinocyte differentiation, multicellular organismal development |
| 4103 | GSPAINVAVHVFR | Transthyretin | TTHY_HUMAN | 16 | 96 | Transport |
| 4302* | AGVETTTPSK | Ig lambda chain C regions | LAC_HUMAN | 11.3 | 65 | Involved in immune response |
| 4302* | QEYDESGPSIVHR | Actin§ | ACTB_HUMAN | 42 | 87 | Blood coagulation, platelet activation, platelet degranulation |
| 4502 | DSYVGDEAQSK DSYVGDEAQSKR VAPEEHPVLLTEAPLNPK LCYVALDFEQEMATAASSSSLEK SYELPDGQVITIGNER DLYANTVLSGGTTMYPGIADR QEYDESGPSIVHR | Actin§ | ACTG_HUMAN | 42 | 601 | Adherens junctions organization, axon guidance, cell junction assembly, cell-cell junction organization, cellular component movement, response to calcium ion |
| 5402 | LIAPVAEEEATVPNNK SLADELALVDVLEDK VIGSGCNLDSAR LKDDEVAQLKK | L-lactate dehydrogenase B chain | LDHB_HUMAN | 37 | 298 | Cellular carbohydrate metabolic process, glycolysis, lactate metabolic process NAD metabolic process, oxidation-reduction process, pyruvate metabolic process |
| 5403 | IEDGNNFGVAVQEK QPHVGDYR | Proteasome activator complex subunit 1 | PSME1_HUMAN | 28.7 | 136 | Regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle, proteasomal ubiquitin-dependent protein catabolism |
| 5604* | DSYVGDEAQSK DSYVGDEAQSKR | Actin§ | ACTA_HUMAN | 42 | 85 | Muscle contraction, regulation of blood pressure, response to virus, vascular smooth muscle contraction |
| 5604* | ALLQAILQTEDMLK ETQSQLETER | Desmoplakin | DESP_HUMAN | 331.8 | 37 | Adherens junction organization, cell-cell adhesion, development of epidermis, keratinocyte differentiation, peptide cross-linking, wound healing |
| 5708 | AAPEASGTPSSDAVSR | Coronin-1A | COR1A_HUMAN | 51.0 | 52 | Actin modulating activity, calcium ion transport, cell-substrate adhesion, cell movement, involved in immune response (T cell response, T cell homeostasis, leukocyte chemotaxis, phagocytosis), regulation of cell shape |
| 6002 | VHLTPEEK VNVDEVGGEALGR | Hemoglobin subunit beta | HBB_HUMAN | 16.0 | 105 | Nitric oxide transport, oxygen transport, regulation of nitric oxide biosynthesis, regulation of blood pressure, regulation of blood vessel size |
| 8103 | AGAHLQGGAK | Glyceraldehyde-3-phosphate dehydrogenase | G3P_HUMAN | 36 | 39 | Glucose metabolism, glycolysis, oxidation, and reduction processes |
| 8206 | ATAVMPDGQFK | Peroxiredoxin 1 | PRDX1_HUMAN | 22.1 | 30 | Cell, proliferation, cell redox homeostasis, erythrocyte homeostasis, hydrogen peroxide catabolic process, NK cell mediated cytotoxicity, oxidation and reduction processes, regulation of NF-kappaB import into nucleus, regulation of stress-activated MAPK cascade, removal of superoxide radicals, skeletal system development |
| 8208* | IGHPAPNFK ATAVMPDGQFK QITVNDLPVGR | Peroxiredoxin 1 | PRDX1_HUMAN | 22.1 | 25‡ | Cell, proliferation, cell redox homeostasis, erythrocyte homeostasis, hydrogen peroxide catabolic process, NK cell mediated cytotoxicity, oxidation and reduction processes, regulation of NF-kB import into nucleus, regulation of stress-activated MAPK cascade, removal of superoxide radicals, skeletal system development |
| 8208* | AGVETTTPSK | Ig lambda chain C regions | LAC_HUMAN | 11.3 | 32‡ | Involved in immune response |
| 9002 | DLYANTVLSGGTTMYPGIADR | Actin§ | ACTB_HUMAN | 42 | 99 | Blood coagulation, platelet activation, platelet degranulation |
| 9201 | LVINGNPITIFQER AGAHLQGGAK IISNASCTTNCLAPLAK | Glyceraldehyde-3-phosphate dehydrogenase | G3P_HUMAN | 36 | 186 | Glucose metabolism, glycolysis, oxidation and reduction processes |
| pH 4–7 | ||||||
| 7505 | TCVADESAENCDK ETYGEMADCCAK AAFTECCQAADK | Serum albumin | ALBU_HUMAN | 69.4 | 182 | Transport, negative regulation of programmed cell death and apoptosis, maintenance of mitochondrial location, cellular response to starvation |
| 3105 | GCITIIGGGDTATCCAK | Phosphoglycerate kinase 1 | PGK1_HUMAN | 45 | 55 | Carbohydrate metabolic process, gluconeogenesis, glucose metabolic process, glycolysis, phosphorylation |
| 3505 | VAPEEHPVLLTEAPLNPK | Actin§ | ACTB_HUMAN | 42 | 60 | Blood coagulation, platelet activation, platelet degranulation |
| 3704 | SYELPDGQVITIGNER | Actin§ | ACTA_HUMAN | 42 | 56 | Muscle contraction, regulation of blood pressure, response to virus, vascular smooth muscle contraction |
| 7301 | VAPEEHPVLLTEAPLNPK | Actin§ | ACTB_HUMAN | 42 | 73 | Blood coagulation, platelet activation, platelet degranulation |
| Spot no. . | Identified peptides . | Protein name . | SwissProt accession . | Size, kDa . | Mascot score . | Biological processes† . |
|---|---|---|---|---|---|---|
| pH 3–10 | ||||||
| 1507 | DREVGIPPEQSLETAK EFNKYDTDGSK | Actin-related protein 3 | ARP3_HUMAN | 47.4 | 77 | Cellular component movement, regulation of actin filament polymerization, activation of dendrite morphogenesis, regulation of myosin II filament assembly or disassembly, response to antibiotic |
| 2002 | DGGVQACFSRSR | Guanine nucleotide-binding protein G(k) subunit alpha | GNAI3_HUMAN | 40.5 | 24‡ | G-protein–coupled receptor protein signaling pathway, negative regulation of adenylate cyclase activity, negative regulation of synaptic transmission, signal transduction, vesicle function |
| 2102 | VRGEVAPDAK FNAHGDANTIVCNSK | Galectin-1 | LEG1_HUMAN | 14.7 | 154 | Activation of I-kB kinase/NF-kB cascade, apoptotic programmed cell, death, cellular response to glucose stimulus, multicellular organismal response to stress, myoblast cell differentiation, negative regulation of cell-substrate adhesion |
| 3102 | QEYDESGPSIVHR | Actin§ | ACTB_HUMAN | 42 | 86 | Blood coagulation, platelet activation, platelet degranulation |
| 3107 | TCVADESAENCDK | Serum albumin | ALBU_HUMAN | 69.4 | 23‡ | Transport, negative regulation of programmed cell death and apoptosis, maintenance of mitochondrial location, cellular response to starvation |
| 3107 | HHEASSRADSSR | Filaggrin | FILA_HUMAN | 435 | 32 | Keratinocyte differentiation, multicellular organismal development |
| 4103 | GSPAINVAVHVFR | Transthyretin | TTHY_HUMAN | 16 | 96 | Transport |
| 4302* | AGVETTTPSK | Ig lambda chain C regions | LAC_HUMAN | 11.3 | 65 | Involved in immune response |
| 4302* | QEYDESGPSIVHR | Actin§ | ACTB_HUMAN | 42 | 87 | Blood coagulation, platelet activation, platelet degranulation |
| 4502 | DSYVGDEAQSK DSYVGDEAQSKR VAPEEHPVLLTEAPLNPK LCYVALDFEQEMATAASSSSLEK SYELPDGQVITIGNER DLYANTVLSGGTTMYPGIADR QEYDESGPSIVHR | Actin§ | ACTG_HUMAN | 42 | 601 | Adherens junctions organization, axon guidance, cell junction assembly, cell-cell junction organization, cellular component movement, response to calcium ion |
| 5402 | LIAPVAEEEATVPNNK SLADELALVDVLEDK VIGSGCNLDSAR LKDDEVAQLKK | L-lactate dehydrogenase B chain | LDHB_HUMAN | 37 | 298 | Cellular carbohydrate metabolic process, glycolysis, lactate metabolic process NAD metabolic process, oxidation-reduction process, pyruvate metabolic process |
| 5403 | IEDGNNFGVAVQEK QPHVGDYR | Proteasome activator complex subunit 1 | PSME1_HUMAN | 28.7 | 136 | Regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle, proteasomal ubiquitin-dependent protein catabolism |
| 5604* | DSYVGDEAQSK DSYVGDEAQSKR | Actin§ | ACTA_HUMAN | 42 | 85 | Muscle contraction, regulation of blood pressure, response to virus, vascular smooth muscle contraction |
| 5604* | ALLQAILQTEDMLK ETQSQLETER | Desmoplakin | DESP_HUMAN | 331.8 | 37 | Adherens junction organization, cell-cell adhesion, development of epidermis, keratinocyte differentiation, peptide cross-linking, wound healing |
| 5708 | AAPEASGTPSSDAVSR | Coronin-1A | COR1A_HUMAN | 51.0 | 52 | Actin modulating activity, calcium ion transport, cell-substrate adhesion, cell movement, involved in immune response (T cell response, T cell homeostasis, leukocyte chemotaxis, phagocytosis), regulation of cell shape |
| 6002 | VHLTPEEK VNVDEVGGEALGR | Hemoglobin subunit beta | HBB_HUMAN | 16.0 | 105 | Nitric oxide transport, oxygen transport, regulation of nitric oxide biosynthesis, regulation of blood pressure, regulation of blood vessel size |
| 8103 | AGAHLQGGAK | Glyceraldehyde-3-phosphate dehydrogenase | G3P_HUMAN | 36 | 39 | Glucose metabolism, glycolysis, oxidation, and reduction processes |
| 8206 | ATAVMPDGQFK | Peroxiredoxin 1 | PRDX1_HUMAN | 22.1 | 30 | Cell, proliferation, cell redox homeostasis, erythrocyte homeostasis, hydrogen peroxide catabolic process, NK cell mediated cytotoxicity, oxidation and reduction processes, regulation of NF-kappaB import into nucleus, regulation of stress-activated MAPK cascade, removal of superoxide radicals, skeletal system development |
| 8208* | IGHPAPNFK ATAVMPDGQFK QITVNDLPVGR | Peroxiredoxin 1 | PRDX1_HUMAN | 22.1 | 25‡ | Cell, proliferation, cell redox homeostasis, erythrocyte homeostasis, hydrogen peroxide catabolic process, NK cell mediated cytotoxicity, oxidation and reduction processes, regulation of NF-kB import into nucleus, regulation of stress-activated MAPK cascade, removal of superoxide radicals, skeletal system development |
| 8208* | AGVETTTPSK | Ig lambda chain C regions | LAC_HUMAN | 11.3 | 32‡ | Involved in immune response |
| 9002 | DLYANTVLSGGTTMYPGIADR | Actin§ | ACTB_HUMAN | 42 | 99 | Blood coagulation, platelet activation, platelet degranulation |
| 9201 | LVINGNPITIFQER AGAHLQGGAK IISNASCTTNCLAPLAK | Glyceraldehyde-3-phosphate dehydrogenase | G3P_HUMAN | 36 | 186 | Glucose metabolism, glycolysis, oxidation and reduction processes |
| pH 4–7 | ||||||
| 7505 | TCVADESAENCDK ETYGEMADCCAK AAFTECCQAADK | Serum albumin | ALBU_HUMAN | 69.4 | 182 | Transport, negative regulation of programmed cell death and apoptosis, maintenance of mitochondrial location, cellular response to starvation |
| 3105 | GCITIIGGGDTATCCAK | Phosphoglycerate kinase 1 | PGK1_HUMAN | 45 | 55 | Carbohydrate metabolic process, gluconeogenesis, glucose metabolic process, glycolysis, phosphorylation |
| 3505 | VAPEEHPVLLTEAPLNPK | Actin§ | ACTB_HUMAN | 42 | 60 | Blood coagulation, platelet activation, platelet degranulation |
| 3704 | SYELPDGQVITIGNER | Actin§ | ACTA_HUMAN | 42 | 56 | Muscle contraction, regulation of blood pressure, response to virus, vascular smooth muscle contraction |
| 7301 | VAPEEHPVLLTEAPLNPK | Actin§ | ACTB_HUMAN | 42 | 73 | Blood coagulation, platelet activation, platelet degranulation |
LC-MS/MS and subsequent search in the Swiss-Prot database resulted in more than one possible protein identification.
Biological processes are taken from the Gene Ontology (GO) annotation system.
Identifications are significant when searches were performed in the human part of the SwissProt database.
Several actin isoforms can be identified from the peptides.
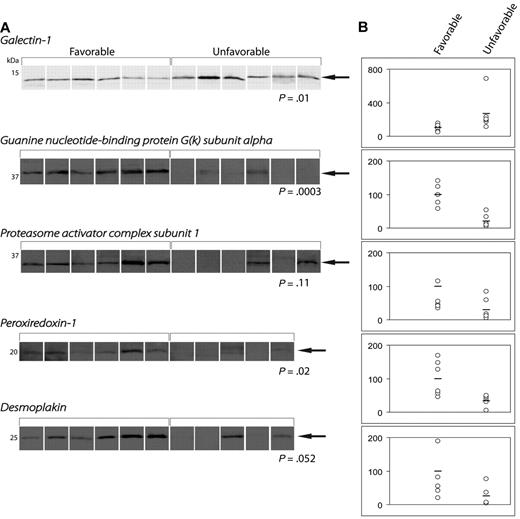
Validation of proteomic analysis by WB
Figure 3 shows WB analysis of the 5 proteins differentially expressed in the 2 patient cohorts: Gal-1, GNAI3, PRDX1, PSME1, and desmoplakin. Integrative analysis confirmed the differential expression of these 5 proteins in patient samples. However, Gal-1 was the only protein that was significantly up-regulated in treatment refractory patients (P = .01). The remaining 4 proteins showed either a significant (GNAI3, P = .0003; PRDX1, P = .02) or a trend-like nonsignificant (PSME1, P = .11; desmoplakin, P = .052) up-regulation in patients with favorable outcome.
Quantification of protein expression by Western blotting. (A) WBs of differentially expressed proteins. (B) Dot-plots demonstrating the band intensities according to treatment outcome.
Quantification of protein expression by Western blotting. (A) WBs of differentially expressed proteins. (B) Dot-plots demonstrating the band intensities according to treatment outcome.
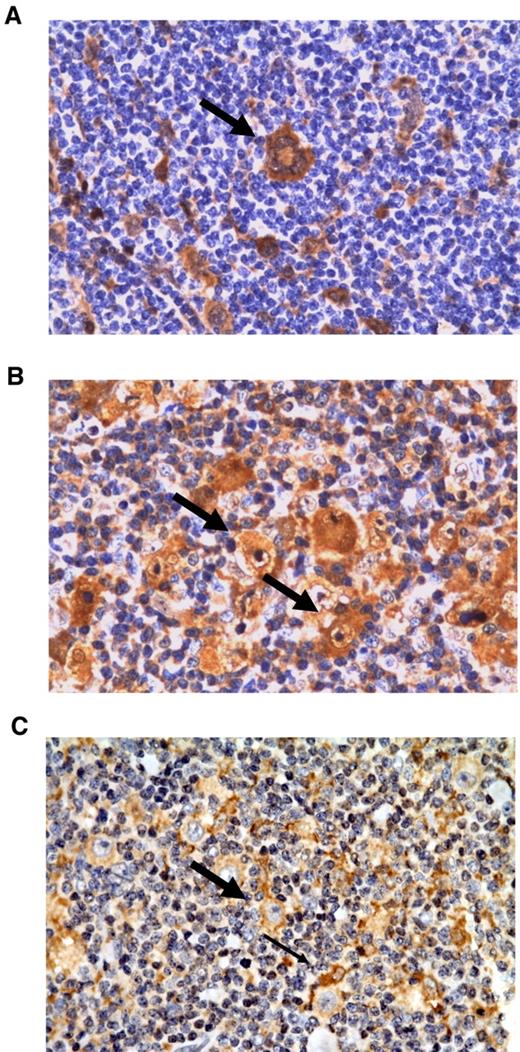
Immunohistochemical localization of Gal-1, PSME-1, and PRDX1 in the cHL microenvironment
Figure 4 illustrates representative immunohistochemical stainings for Gal-1, PSME-1, and PRDX1. Cytoplasmic and nuclear Gal-1 labeling were found in HRS cells, lymphoma-associated macrophages and endothelial cells in line with previous reports12,13 (Figure 4A). Furthermore, we also noted a positive nuclear/cytoplasmic reaction in some plasma cells. Most tumor-infiltrating lymphoid cells were Gal-1 negative. PSME1 immunostaining demonstrated a combined cytoplasmic/nuclear positivity in both tumor and nonneoplastic by-stander cells (Figure 4B). PRDX1 was also found to be weakly expressed by HRS cells. The signal localization was primarily cytoplasmic. With regard to the tumor microenvironment, PRDX1 was predominantly expressed in lymphoma-associated macrophages (Figure 4C).
Representative immunohistochemical stainings of the differentially expressed proteins Gal-1, PSME-1, and PRDX-1. (A) Gal-1 staining of a multinuclear HRS cell (thick arrow). (B) PSME-1 staining of HRS cells (thick arrow) and tumor-infiltrating cells. (C) PRDX1 staining showing a positive HRS cell (thick arrow) among PRDX-1–positive macrophages (thin arrow). All magnifications are 400×.
Representative immunohistochemical stainings of the differentially expressed proteins Gal-1, PSME-1, and PRDX-1. (A) Gal-1 staining of a multinuclear HRS cell (thick arrow). (B) PSME-1 staining of HRS cells (thick arrow) and tumor-infiltrating cells. (C) PRDX1 staining showing a positive HRS cell (thick arrow) among PRDX-1–positive macrophages (thin arrow). All magnifications are 400×.
Clinicopathologic features of TMA cohort patients
The main clinical and histopathologic features of the study cohort are summarized in Table 3. The patient population had a median age of 35 years and a male/female ratio of 1.8. The median follow-up was 7 years (range: 0.2-18.6 years). The majority of the patients (60%) had stage III-IV disease, whereas 40% presented with a lower clinical stage (I-II) but with concomitant B symptoms as a probable sign of more advanced disease. The large majority of the patients in the study cohort (82%) had B-symptoms at presentation and the median IPS score was 2. Approximately one-third of the patients had EBV-positive tumors. The large majority (84%) displayed histologic features consistent with the nodular sclerosis subtype.
Correlation of demographic and clinicopathological parameters with Gal-1 expression in HRS cells and in the tumor microenvironment
| Characteristic . | Total no. . | % . | HRS Gal-1 . | Non-HRS Gal-1, cell profile Gal-1 count/mm2 . | |||
|---|---|---|---|---|---|---|---|
| Gal-1 neg . | Gal-1 pos . | P† . | Median . | P‡ . | |||
| 143 | |||||||
| Age, y (n = 143) | |||||||
| < 45 | 91 | 64 | 46 | 45 | .11 | 1361 | .27 |
| ≥ 45 | 52 | 36 | 19 | 33 | 1496 | ||
| Sex (n = 143) | |||||||
| Male | 92 | 64 | 43 | 49 | .68 | 1387 | .30 |
| Female | 51 | 36 | 22 | 29 | 1361 | ||
| Ann Arbor stage (n = 143) | |||||||
| I-II | 57 | 40 | 23 | 34 | .32 | 1511 | .62 |
| III-IV | 86 | 60 | 42 | 44 | 1358 | ||
| B symptoms (n = 143) | |||||||
| Yes | 117 | 82 | 46 | 71 | .002 | 1482 | .03 |
| No | 26 | 18 | 19 | 7 | 1138 | ||
| Bulky disease, > 10 cm (n = 135)* | |||||||
| Yes | 49 | 36 | 23 | 26 | .76 | 1228 | .20 |
| No | 86 | 64 | 38 | 48 | 1377 | ||
| Histologic type (n = 143) | |||||||
| NSI | 78 | 55 | 37 | 41 | .03 | 1523 | .49 |
| NSII | 42 | 29 | 13 | 29 | 1302 | ||
| MC | 22 | 15 | 15 | 7 | 1387 | ||
| cHL, NOS | 1 | 1 | 0 | 1 | 1523 | ||
| IPS (n = 125)* | |||||||
| ≤ 2 | 68 | 54 | 36 | 32 | .11 | 1369 | .51 |
| > 2 | 57 | 46 | 22 | 35 | 1361 | ||
| EBV status (n = 143) | |||||||
| Positive | 48 | 34 | 28 | 20 | .03 | 1345 | .54 |
| Negative | 95 | 66 | 37 | 58 | 1384 | ||
| Characteristic . | Total no. . | % . | HRS Gal-1 . | Non-HRS Gal-1, cell profile Gal-1 count/mm2 . | |||
|---|---|---|---|---|---|---|---|
| Gal-1 neg . | Gal-1 pos . | P† . | Median . | P‡ . | |||
| 143 | |||||||
| Age, y (n = 143) | |||||||
| < 45 | 91 | 64 | 46 | 45 | .11 | 1361 | .27 |
| ≥ 45 | 52 | 36 | 19 | 33 | 1496 | ||
| Sex (n = 143) | |||||||
| Male | 92 | 64 | 43 | 49 | .68 | 1387 | .30 |
| Female | 51 | 36 | 22 | 29 | 1361 | ||
| Ann Arbor stage (n = 143) | |||||||
| I-II | 57 | 40 | 23 | 34 | .32 | 1511 | .62 |
| III-IV | 86 | 60 | 42 | 44 | 1358 | ||
| B symptoms (n = 143) | |||||||
| Yes | 117 | 82 | 46 | 71 | .002 | 1482 | .03 |
| No | 26 | 18 | 19 | 7 | 1138 | ||
| Bulky disease, > 10 cm (n = 135)* | |||||||
| Yes | 49 | 36 | 23 | 26 | .76 | 1228 | .20 |
| No | 86 | 64 | 38 | 48 | 1377 | ||
| Histologic type (n = 143) | |||||||
| NSI | 78 | 55 | 37 | 41 | .03 | 1523 | .49 |
| NSII | 42 | 29 | 13 | 29 | 1302 | ||
| MC | 22 | 15 | 15 | 7 | 1387 | ||
| cHL, NOS | 1 | 1 | 0 | 1 | 1523 | ||
| IPS (n = 125)* | |||||||
| ≤ 2 | 68 | 54 | 36 | 32 | .11 | 1369 | .51 |
| > 2 | 57 | 46 | 22 | 35 | 1361 | ||
| EBV status (n = 143) | |||||||
| Positive | 48 | 34 | 28 | 20 | .03 | 1345 | .54 |
| Negative | 95 | 66 | 37 | 58 | 1384 | ||
NSI indicates nodular sclerosis type I; NSII, nodular sclerosis type II; MC, mixed cellularity; NOS, not otherwise specified; and IPS, international prognostic score.
Parameter not available in all 143 patients.
χ2 test.
Wilcoxon rank-sum test or Kruskal-Wallis equality-of-populations rank test where appropriate.
Correlation of Gal-1 expression in neoplastic and nonneoplastic cells with demographic and clinicopathologic features
A total of 78 patients (55%) showed Gal-1 positivity in HRS cells. Interestingly, we found that cases with a high expression of Gal-1 in HRS cells frequently presented with B symptoms at diagnosis (P = .002). Gal-1 expression in HRS cells was correlated with histologic subtype. Furthermore, with respect to the expression of Gal-1 in nonneoplastic cells, only 1 significant correlation was found: high expression of Gal-1 was correlated with the presence of B symptoms (P = .03; Table 3).
Correlation of Gal-1 expression in neoplastic and nonneoplastic cells with patient survival in Hodgkin lymphoma
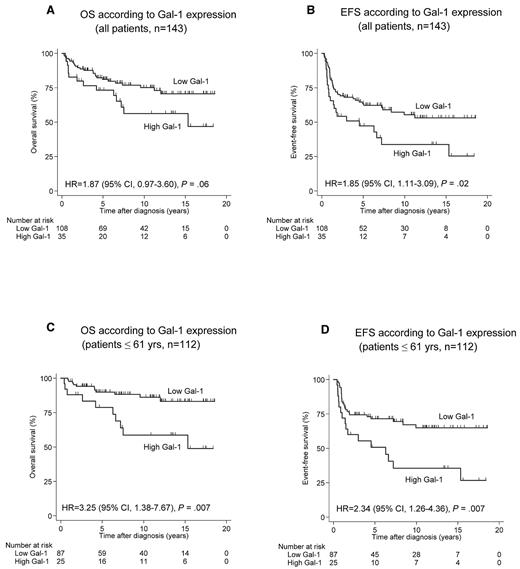
Patient survival (OS as well as EFS) was correlated with age, with higher survival in the group of patients < 45 years. Several other clinicopathologic parameters showed no correlation with survival. With respect to Gal-1 expression, there was no correlation between Gal-1 expression in HRS cells and OS or EFS (results not shown). Interestingly, a high Gal-1 expression in the tumor microenvironment was significantly correlated with poor EFS (P = .02; Table 4). Among the younger, transplant-eligible patient population (< 61 years), the adverse impact of a high number of Gal-1–positive cell profiles on outcome was more pronounced. EFS and OS for low versus high Gal-1 expression were 90% compared with 78% (P = .007) for the former and 69% compared with 49% (P = .007) for the latter end point. Survival curves are shown in Figure 5. In this younger patient population, high Gal-1 values retained their significant impact on EFS (P = .005) at the multivariate level. The predictive value of Gal-1 expression in younger patients (≤ 61 years) persisted also in terms of both EFS and OS despite the concomitant presence of macrophage-related parameters such as high CD68 (P values: .009 and .02, respectively) or high CD163 expression (P values: .02 and .003, respectively) in the Cox regression model.
Correlation of clinicopathological parameters and Gal-1 expression in nonneoplastic cells with survival
| Characteristic . | OS . | EFS . | ||||||
|---|---|---|---|---|---|---|---|---|
| 5-year OS, % . | HR . | 95% CI . | P . | 5-Year EFS, % . | HR . | 95% CI . | P . | |
| Age, y | ||||||||
| < 45 | 92 | 1 | ref | < .001 | 64 | 1 | ref | .005 |
| ≥ 45 | 57 | 5.36 | 2.86-10.06 | 48 | 1.91 | 1.21-3.01 | ||
| Sex | ||||||||
| Male | 78 | 1 | ref | .16 | 55 | 1 | ref | .09 |
| Female | 80 | 0.64 | 0.35-1.20 | 63 | 0.66 | 0.41-1.07 | ||
| Ann Arbor stage | ||||||||
| I-II | 80 | 1 | ref | .87 | 63 | 1 | ref | .33 |
| III-IV | 77 | 0.95 | 0.53-1.72 | 55 | 1.26 | 0.79-2.01 | ||
| B symptoms | ||||||||
| No | 79 | 1 | ref | .45 | 52 | 1 | ref | .84 |
| Yes | 79 | 1.37 | 0.61-3.07 | 60 | 0.94 | 0.54-1.66 | ||
| Bulky disease > 10 cm | ||||||||
| No | 82 | 1 | ref | .15 | 60 | 1 | ref | .27 |
| Yes | 71 | 1.57 | 0.85-2.92 | 54 | 1.31 | 0.81-2.11 | ||
| Histologic type | ||||||||
| NSI | 84 | 1 | ref | .21 | 64 | 1 | ref | .13 |
| NSII | 70 | 1.43 | 0.72-2.82 | 56 | 1.19 | 0.69-2.04 | ||
| MC | 77 | 1.11 | 0.45-2.72 | 49 | 1.47 | 0.77-2.79 | ||
| cHL, NOS | 33 | 4.50 | 1.05-19.35 | 0 | 3.77 | 1.16-12.3 | ||
| EBV status | ||||||||
| Negative | 78 | 1 | ref | .77 | 57 | 1 | ref | .86 |
| Positive | 79 | 1.09 | 0.59-2.03 | 60 | 0.96 | 0.59-1.55 | ||
| Gal-1 (all) | ||||||||
| Low | 81 | 1 | ref | .06 | 62 | 1 | ref | .02 |
| High | 72 | 1.87 | 0.97-3.60 | 45 | 1.85 | 1.11-3.09 | ||
| Gal-1 < 61 y | ||||||||
| Low | 90 | 1 | ref | 69 | 1 | ref | ||
| High | 78 | 3.25 | 1.38-7.67 | .007 | 49 | 2.34 | 1.26-4.36 | .007 |
| Characteristic . | OS . | EFS . | ||||||
|---|---|---|---|---|---|---|---|---|
| 5-year OS, % . | HR . | 95% CI . | P . | 5-Year EFS, % . | HR . | 95% CI . | P . | |
| Age, y | ||||||||
| < 45 | 92 | 1 | ref | < .001 | 64 | 1 | ref | .005 |
| ≥ 45 | 57 | 5.36 | 2.86-10.06 | 48 | 1.91 | 1.21-3.01 | ||
| Sex | ||||||||
| Male | 78 | 1 | ref | .16 | 55 | 1 | ref | .09 |
| Female | 80 | 0.64 | 0.35-1.20 | 63 | 0.66 | 0.41-1.07 | ||
| Ann Arbor stage | ||||||||
| I-II | 80 | 1 | ref | .87 | 63 | 1 | ref | .33 |
| III-IV | 77 | 0.95 | 0.53-1.72 | 55 | 1.26 | 0.79-2.01 | ||
| B symptoms | ||||||||
| No | 79 | 1 | ref | .45 | 52 | 1 | ref | .84 |
| Yes | 79 | 1.37 | 0.61-3.07 | 60 | 0.94 | 0.54-1.66 | ||
| Bulky disease > 10 cm | ||||||||
| No | 82 | 1 | ref | .15 | 60 | 1 | ref | .27 |
| Yes | 71 | 1.57 | 0.85-2.92 | 54 | 1.31 | 0.81-2.11 | ||
| Histologic type | ||||||||
| NSI | 84 | 1 | ref | .21 | 64 | 1 | ref | .13 |
| NSII | 70 | 1.43 | 0.72-2.82 | 56 | 1.19 | 0.69-2.04 | ||
| MC | 77 | 1.11 | 0.45-2.72 | 49 | 1.47 | 0.77-2.79 | ||
| cHL, NOS | 33 | 4.50 | 1.05-19.35 | 0 | 3.77 | 1.16-12.3 | ||
| EBV status | ||||||||
| Negative | 78 | 1 | ref | .77 | 57 | 1 | ref | .86 |
| Positive | 79 | 1.09 | 0.59-2.03 | 60 | 0.96 | 0.59-1.55 | ||
| Gal-1 (all) | ||||||||
| Low | 81 | 1 | ref | .06 | 62 | 1 | ref | .02 |
| High | 72 | 1.87 | 0.97-3.60 | 45 | 1.85 | 1.11-3.09 | ||
| Gal-1 < 61 y | ||||||||
| Low | 90 | 1 | ref | 69 | 1 | ref | ||
| High | 78 | 3.25 | 1.38-7.67 | .007 | 49 | 2.34 | 1.26-4.36 | .007 |
OS indicates overall survival; EFS, event-free survival; HR, hazard ratio; CI, confidence interval; ref, reference; NSI, nodular sclerosis type I; NSII, nodular sclerosis type II; MC, mixed cellularity; NOS, not otherwise specified; NLPHL, nodular lymphocyte predominant Hodgkin lymphoma; and IPS, international prognostic score.
OS and EFS according to Gal-1 expression. (A-B) Analysis including all patients (n = 143). (C-D) Analysis restricted to younger patients (< 61 years, n = 112). HR indicates hazard ratio.
OS and EFS according to Gal-1 expression. (A-B) Analysis including all patients (n = 143). (C-D) Analysis restricted to younger patients (< 61 years, n = 112). HR indicates hazard ratio.
Correlation between Gal-1 expression in the nonneoplastic cells and tumor-infiltrating inflammatory cells of Hodgkin lymphoma patients
We further examined the correlation between Gal-1 expression and the tumor inflammatory infiltrate characteristic of cHL. With regard to the tumor microenvironment, Gal-1 was positively correlated with the level of macrophage marker expression, in particular the M2 macrophage marker CD163 (P = .0007) and to a lesser extent the pan-macrophage marker CD68 (P = .052). This result is relevant in terms of the ability of Gal-1 to foster the differentiation of alternatively activated macrophages and tolerogenic dendritic cells.27-29 Furthermore, a significant correlation between Gal-1 and granzyme B+ cytotoxic T lymphocytes was found (P = .02). Conversely, no correlation with FoxP3+ T-regulatory cells was detected (P = .26) in spite of the abundant expression of Gal-1 in FoxP3+ T-regulatory cells.30
Discussion
The Human Genome Project has revealed that the number of proteins in the human proteome exceeds by far the number of protein-coding genes in the human genome (∼ 400 000 proteins versus ∼ 30 000 genes).31 Because proteins are the functional end product, it seems more appropriate to search for potential tumor biomarkers at the protein level rather than at the gene level. Previous studies in lymph nodes and lymphoma samples have shown that the 2D gel-based proteomics approach was feasible and reproducible.32 Furthermore, it was demonstrated that this technique yielded similar results, irrespective of whether fresh biopsies or frozen samples that had been stored up to 1 year were used. Subsequently, this methodology was applied to compare mantle cell lymphoma samples and inflammatory lymph node tissue, resulting in the identification of several differentially expressed proteins.33 In our study, we were able to demonstrate the feasibility of using archival whole-tumor frozen tissue from cHL patients for proteomic analysis. Although some of these samples had been stored for > 10 years, they still yielded technically satisfactory results. Proteomic-based expression profiles of the 2 patient groups selected on the basis of their response to treatment successfully identified proteins of prognostic value that were differentially expressed between these 2 groups.
The glycan-binding protein Gal-1 has been previously implicated as a modulator of cHL. Expression of this protein by Reed Sternberg cells has been demonstrated to confer immune privilege to cHL cells, creating an appropriate niche for tumor progression. On the basis of gene-expression profiling, Gal-1 was found to be overexpressed in HRS cell lines and its secretion favored a Th2-type immune response characterized by high IL-4 and IL-13 secretion and promoted the expansion of CD4+CD25+FoxP3+ regulatory T cells.12 Moreover, exposure to recombinant Gal-1 inhibited proliferation and IFN-γ secretion by EBV-specific T cells,11 and T cells specific for EBV-LMP1 were much more susceptible to Gal-1–mediated immunosuppression than conventional T cells,34 suggesting a critical role for Gal-1 in compromising cHL-specific immunity. Supporting these findings, Gal-1-deficient (Lgals1−/−) mice were found to be highly susceptible to Th1- and Th17-dependent inflammation compared with their wild-type counterparts.35 Subsequent studies have shown that Gal-1 expression is not only seen in HRS cells, but is also abundant in the tumor microenvironment.13 The latter observation was confirmed by our study, although a correlation between Gal-1 expression and FoxP3 up-regulation could not be demonstrated at the level of primary biopsies.
An important finding of the present study was the identification by proteomic analysis of Gal-1 among the proteins that are differentially expressed in treatment-refractory disease. This observation was further substantiated by immunohistochemical analysis, which unambiguously showed a correlation between Gal-1 expression within the tumor microenvironment and poor clinical outcome. This impact was much more evident among younger patients (≤ 61 years), and persisted for EFS after a multivariate analysis. Considering the fact that Gal-1 is expressed in macrophages and that macrophages have recently been shown by our group to predict a poor prognosis,5,36,37 it seemed relevant to test whether the adverse prognostic impact of Gal-1 also would persist in the presence, within the Cox regression model, of parameters associated with the number of macrophages (ie, CD68 or CD163 positivity). The predictive value of Gal-1 expression in younger patients (≤ 61 years) was retained in terms of both EFS and OS despite the concomitant presence of high CD68 or high CD163 expression, suggesting that the prognostic impact of high Gal-1 expression is not a mere reflection of the number of macrophages in the tumor tissue.
In addition to Gal-1, we identified in the pilot study PRDX1 and GNAI3 among proteins that were substantially up-regulated in patients with a favorable outcome. PRDX1 is a member of the peroxiredoxin family of antioxidant enzymes, which reduces hydrogen peroxide and alkyl hydroperoxides. In addition to its role as an antioxidant, it has been shown to enhance the cytotoxicity of natural killer cells.38 Interestingly, PRDX1-null mice develop hemolytic anemia (due to the accumulation of reactive oxygen species) and a variety of malignancies including lymphomas. For this reason, it has been suggested that this protein plays an antioxidant protective role and may function as a tumor suppressor.39
GNAI3 belongs to the guanine nucleotide-binding proteins (G-proteins) that are important regulators of Akt-mediated cell migration,40 a critical process that contributes to shaping the tumor microenvironment. Many cytokines and chemoattractants signal through the Akt/PKB pathway at the leading edge of migrating cells.41 All differentially expressed proteins identified on the basis of the proteomic analysis are currently being investigated in a validation setting. Therefore, while the results of the validation studies are pending, it is premature to formulate prognostic statements with regard to these additional differentially expressed proteins.
In summary, our study confirms the feasibility of using archival frozen tissues from cHL patients for proteomic analysis. The 2 clinical groups analyzed showed relevant differences in their protein expression profiles. Whereas the immunoregulatory glycan-binding protein Gal-1 was up-regulated in patient subsets with poorer outcome, PRDX1, a member of a family of antioxidant enzymes, and GNAI3, a cell migration–associated protein, were identified among proteins that were substantially up-regulated in patients with a favorable outcome. In cHL (as in certain non-Hodgkin lymphoma subtypes), there is now substantial evidence to show that the tumor microenvironment plays an important role in the biology and prognosis of the disease.42,43 Our study contributes further evidence to support this hypothesis identifying Gal-1 as a potential novel biomarker that may be useful in the prognostic and predictive analysis of cHL. It also underlines the advantages of proteomics in providing important clues to identifying new therapeutic, diagnostic, and prognostic targets in hematologic malignancies.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Mette Hjøllund Christiansen, Kristina Lystlund Lauridsen, Mona Britt Hansen, Inge Kjærgaard Nielsen, and Tom Nordfeld for technical assistance; collaborating pathologic departments for retrieval of tissue samples; and Jens Overgaard and the Department of Experimental Oncology for scientific inspiration and logistic support. The authors are also grateful to Professor Margaret Shipp, Dana-Farber Cancer Institute, Boston, MA, for scientific advice.
This work was supported by Aarhus University, the Karen Elise Jensen Foundation, the Max and Inger Wørzner Foundation, the Foundation in Commemoration of Eva and Henry Frænkel, the Aase and Ejner Danielsen Foundation, the Poul and Ellen Hertz Foundation, the Else and Mogens Wedell-Wedellsborg Foundation, the A. P. Møller and Chastine Mc-Kinney Møller Foundation for General Purposes, the Danish Cancer Research Foundation, Gangsted Foundation, the John & Birthe Meyer Foundation, Aarhus University Research Foundation, and the Medical Research Council. The Centre for Stochastic Geometry and Advanced Bioimaging is supported by the Villum Foundation.
Authorship
Contribution: P.K., M.L., K.B., S.H.-D., G.A.R., B.H., J.R.N., and F.d.A. contributed to the design of the study; K.B., S.H.-D., and M.B.M. retrieved the tissue samples; P.K., K.B., S.H.-D., and M.B.M. reviewed all cases; G.A.R. contributed vital reagents; P.K., M.L., and B.H. performed the laboratory work; P.K., M.L., B.H., J.R.N., and F.d.A. participated in the data analysis and interpretation; P.K., M.L., B.H., and F.d.A. wrote the manuscript; and all authors read, gave comments, and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Peter Kamper, MD, Department of Hematology, Aarhus University Hospital, Tage Hansens Gade 2, DK-8000 Aarhus C, Denmark; e-mail: petekamp@rm.dk.
References
Author notes
P.K. and M.L. contributed equally to this work.
B.H. and F.d.A. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal