Abstract
Prospective identification of patients whose chronic myeloid leukemia (CML) will progress to blast crisis is currently not possible. PP2A is a phosphatase and tumor suppressor that regulates cell proliferation, differentiation, and survival. Cancerous inhibitor of PP2A (CIP2A) is a recently described inhibitor of PP2A in breast and gastric cancer. The aim of this study was to investigate whether CIP2A played a role in CML and whether PP2A or its inhibitor proteins CIP2A or SET could predict clinical outcome. At the time of diagnosis of CML, patients who will later progress to blast crisis have significantly higher levels of CIP2A protein (P < .0001) than patients who do not progress, suggesting that PP2A is functionally inactive. We show that the potential mechanism for disease progression is via altered phosphorylation of the oncogene c-Myc. Knockdown of CIP2A results in increased PP2A activity, decreased c-Myc levels, and a decrease in BCR-ABL1 tyrosine kinase activity. We demonstrate that CIP2A levels at diagnosis can consistently predict patients who will progress to blast crisis. The data show that CIP2A is biologically and clinically important in CML and may be a novel therapeutic target.
Introduction
Chronic myeloid leukemia (CML) is a malignant disease of the primitive hematologic cell, characterized by inappropriate expansion of myeloid cells. Although this disease is readily controlled by imatinib, approximately one-third of patients will eventually fail treatment1,2 ; and a significant proportion of these will progress toward blast crisis (BC), which is usually rapidly fatal. Poor response to imatinib and progression to BC have been linked to high BCR-ABL1 tyrosine kinase activity,3 but why one patient can remain in well-controlled chronic phase for decades whereas another may rapidly progress to BC is poorly understood.
The BCR-ABL1 tyrosine kinase in CML is responsible for growth and survival of the malignant cells through activation of signaling pathways, such as the mitogen-activated protein kinase cascade and the PI3K pathway.4,5 A major cellular serine/threonine phosphatase working to down-regulate activation of these pathways is the tumor suppressor protein phosphatase 2A (PP2A).6 In CML cells, PP2A is a key target of BCR-ABL1 signaling; this protein becomes inactivated in these cells because BCR-ABL1 stimulates prevention of its auto-dephosphorylation at tyrosine307.7,8 Maintenance of pY307-PP2A levels in CML cells feeds back to BCR-ABL1 and facilitates increased and sustained kinase activity. Inhibition of BCR-ABL1 by imatinib results in reactivation of PP2A, inducing both suppression of growth and enhanced apoptosis of the leukemic cells.8 However, it is unknown whether Y307 in PP2A is a direct substrate of BCR-ABL1, and the mechanism regulating phosphorylation of PP2A at this site is not clearly defined.
One proposal for the mechanism through which BCR-ABL1 regulates PP2A activity in CML cells involves expression of the PP2A inhibitor protein SET. In CML cell lines, increased expression of BCR-ABL1 increases expression of SET through a process that is mediated by JAK2.9 Furthermore, Neviani et al8 also observed that SET levels rose at evolution of BC in chronic phase and BC primary CML samples, and that incubation with imatinib decreased SET levels in primary CML cells from a single patient undergoing BC.
Two additional proteins have been shown to inhibit PP2A activity. Overexpression of SET binding protein 1 (SETBP1) protects SET from protease cleavage and permits formation of a SETBP1-SET-PP2A complex, which inhibits PP2A phosphatase activity. However, although one report has indicated that high levels of SETBP1 expression predict adverse outcome in elderly patients with acute myeloid leukemia (AML),10 we have recently demonstrated that SETBP1 expression does not correlate with clinical outcome in CML.11 Thus, SETBP1 expression is probably not the inhibitory block on PP2A function in CML. The second protein of interest, called cancerous inhibitor of PP2A (CIP2A), functions in a cancer setting to prevent PP2A-mediated dephosphorylation of c-Myc at serine 62. pS62-Myc is stabilized against degradation, and CIP2A therefore promotes deregulated cell growth.12 High expression levels of CIP2A correlate with aggressiveness of breast and gastric cancers,13-15 and CIP2A depletion decreases growth of cells from these cancers.13,15 In mouse neural progenitor cells, overexpression of CIP2A increases progenitor cell self-renewal and proliferation.16 CIP2A has recently been shown to be overexpressed in AML patients and correlated with relapse after treatment.17 A single case study has reported a CIP2A-MLL translocation in infant AML.18 Nothing is known about CIP2A expression and function in CML.
Here we have investigated the expression of PP2A and its inhibitory proteins SET and CIP2A in CML patients at diagnosis and at 12 months after diagnosis or at disease progression. We show that the levels of these proteins are distinctly different according to the later outcome of the patient. In particular, we identify CIP2A as a potential predictive biomarker of disease progression because CIP2A protein levels are significantly higher in CML patients who later progress to BC than in patients who do not. Furthermore, we also show that high CIP2A levels in primary CML cells correlate with high levels of pS62-Myc, suggesting a link between high CIP2A expression, increased growth potential, and genetic instability. Finally, we support our clinical observations with mechanistic data; we show that small interfering RNA (siRNA)–mediated knockdown of CIP2A expression in K562 cells results in increased PP2A activity, decreased c-Myc levels, and a decrease in BCR-ABL1 tyrosine kinase activity. To our knowledge, this is the first report of CIP2A dictating outcome in any hematologic malignancy.
Methods
Patient cohort
All 31 CML patients included in this study were diagnosed in chronic phase at our center. The study was approved by the Liverpool Local Research Ethics Committee, and informed consent in accordance with the declaration of Helsinki was obtained from each patient. All were 18 or more years of age at diagnosis and were positive by metaphase cytogenetic analysis for the t(9;22) (Philadelphia) translocation. Quantitative RT-PCR was used to quantify BCR-ABL1 gene fusion transcripts as previously described.19 Patient characteristics are described in Table 1. All cases started imatinib treatment within 4 weeks of first presentation.
Patient characteristics
| . | No. of patients . | Male/female . | Average age, y . | Sokal score . | |||
|---|---|---|---|---|---|---|---|
| Low . | Intermediate . | High . | Unknown . | ||||
| CCR | 14 | 7/7 | 45 | 6 | 1 | 4 | 3 |
| No-CCR | 11 | 7/4 | 48 | 2 | 2 | 5 | 2 |
| BC | 6 | 5/1 | 37 | 1 | 2 | 2 | 1 |
| Total | 31 | 31 | 44 | 9 | 5 | 11 | 6 |
| . | No. of patients . | Male/female . | Average age, y . | Sokal score . | |||
|---|---|---|---|---|---|---|---|
| Low . | Intermediate . | High . | Unknown . | ||||
| CCR | 14 | 7/7 | 45 | 6 | 1 | 4 | 3 |
| No-CCR | 11 | 7/4 | 48 | 2 | 2 | 5 | 2 |
| BC | 6 | 5/1 | 37 | 1 | 2 | 2 | 1 |
| Total | 31 | 31 | 44 | 9 | 5 | 11 | 6 |
CCR indicates complete cytogenetic response; No-CCR, patients who had achieved a complete hematologic response but not a CCR by 12 months and who had not progressed; and BC, patients presented in CP but subsequently progressed to BC.
Clinical response
Patients were stratified into 3 clinical outcomes:
CCR.
Complete cytogenetic response (CCR) = no Philadelphia-positive metaphases among at least 20 marrow metaphases after 12 months of imatinib treatment. In some cases, serial cytogenetic data were not available, and achievement of CCR is based on a BCR-ABL1/ABL1 transcript ratio of < 1%, which we have previously shown20 to be tightly correlated with cytogenetically defined CCR (n = 14).
No-CCR.
Patients who had achieved a complete hematologic response but not a CCR by 12 months and who had not progressed (n = 11).
BC.
Patients presented in chronic phase but who subsequently progressed into BC (n = 6).
Sample collection and preparation
Mononuclear cells (MNCs) were separated from peripheral blood at diagnosis and after 12 months of imatinib treatment or at disease progression, by density-dependent centrifugation (Lymphoprep Axis-Shield), washed in RPMI 1640 (BioSera), and resuspended in 10% DMSO/10% FCS serum (BioSera)/RPMI at 4°C and cryopreserved in liquid nitrogen. When required, the cells were thawed and resuspended in culture media as previously described.19 K562, LAMA84, KCL22, and KY01 cell lines were used as BCR-ABL1-positive cell lines. The gastric cancer cell line AGS was used as a CIP2A-positive BCR-ABL1-negative control.
Samples were enriched for CD34+ cells using CliniMACS (Miltenyi Biotec) according to the manufacturer's instructions. All CD34+ cells were collected at diagnosis and then grouped according to the patients' eventual clinical outcome: CCR (n = 3), No-CCR (n = 4), or BC (n = 3).
Measurement of PP2A, phosphorylated PP2A, SET, CIP2A, and PIM1
Flow cytometric assays were used for the detection of PP2A, phosphorylated PP2A, SET, CIP2A, and PIM1 as previously described,3 using anti-PP2A antibody (Millipore), PP2A Y307 (Epitomics), SET (Santa Cruz Biotechnology), CIP2A (Santa Cruz Biotechnology), PIM1 (Abcam), and anti–mouse and anti–rabbit AlexFluor-488 (Invitrogen). Supplemental Figure 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) shows the optimization of the flow cytometry assay. Western blotting was used to confirm all findings in both K562 and LAMA84 CML cell lines.
Measurement of CrKL phosphorylation
Levels of pCrKL and CrKL (CT10 regulator of kinase-like) were measured by FACS as previously described.3
TaqMan gene expression assay of PP2A (catalytic subunit), SET, and SETBP1
Predesigned TaqMan quantitative RT-PCR assays were used in a 384-well assay plate. Each assay consists of a forward and reverse primer at 900nM each and a 6-FAM dye-labeled TaqMan MGB probe at 250nM. The assay's ID numbers are: PP2A (catalytic subunit), Hs00427259m1, SET Hs00853870g1, SETBP1 Hs00210209m1, and GAPDH Hs99999905m1. The relative expression level was calculated by the comparative Ct method that uses the 2−ΔΔCt formula to achieve results for relative quantification.21
CIP2A mRNA expression
The expression of CIP2A was measured using quantitative RT-PCR on a LightCycler 1.5 using LightCycler FastStart DNA MasterPlus SYBR Green I (Roche Diagnostics). CIP2A primers and PCR conditions were as previously described.13
c-Myc protein and phosphorylation status
CIP2A siRNA treatment
A total of 1 × 106 K562 cells were resuspended in 100-μL solution V Nucleofector Kit V (Amaxa Biosystems); 100nM siRNA was added to solution V (CIP2A siRNA and control siRNA; Santa Cruz Biotechnology). In each case, samples were nucleofected using the Amaxa instrument preset program T16. After nucleofection, the cells were cultured for 72 hours in a 24-well plate before analysis. In addition, these experiments were repeated using a different CIP2A siRNA (Integrated DNA Technologies) and a different CML cell line LAMA84.
Statistical analysis
Statistical analysis and comparisons were performed by Mann-Whitney and Student t tests using SPSS Version 16.0 (SPSS).
Results
PP2A protein expression and phosphorylation are increased in MNCs and CD34+ cells from patients destined to progress to BC
In initial experiments, PP2A protein levels were compared between chronic phase and established BC. Significantly elevated levels were seen in BC cells (supplemental Figure 2).
To determine whether PP2A protein levels were indicative of disease progression, we analyzed PP2A protein in MNCs of 31 patients who were in chronic phase at diagnosis and again in the same patients after 12 months of treatment or at transformation. The clinical response of these patients was stratified into 3 groups depending on outcome: CCR, No-CCR, and BC. No BCR-ABL1 kinase domain mutations were detected in any patients. At diagnosis, the mean PP2A protein level in patients destined to achieve CCR was not significantly different from that in normal MNCs, whereas that in patients destined not to achieve CCR (No-CCR) was significantly lower than that in normal cells (P = .02; Figure 1A). In both groups of patients, PP2A levels increase after 12 months of treatment. In contrast to the CCR and No-CCR patients, PP2A levels in patients destined to progress into BC were much higher (P < .001 compared with the No-CCR group), and these levels did not change after treatment (Figure 1A). When the same experiment was performed on CD34+ cells from the same patients, similar results were obtained (Figure 1B); PP2A protein was elevated in the diagnostic CD34+ cells from patients who subsequently progress into BC compared with the CCR and No-CCR groups. These results suggest that increased PP2A protein expression at diagnosis indicates a high probability of disease progression.
PP2A expression and phosphorylation are increased in MNC and CD34+ cells from patients destined to progress to BC. The horizontal line represents the mean normal level observed in 10 healthy volunteers. (A) Levels of PP2A protein as assessed by flow cytometry. Mean level in 10 healthy subjects = 21.4 (range, 17.1-25.7). The level of PP2A is high in patients destined to progress to BC, compared with nonresponding patients (P = .001). *Statistically significant difference between No-CCR and normal (P = .002). CCR, n = 14; No-CCR, n = 11; and BC, n = 6. (B) Level of PP2A protein in CD34+ cells. The level of PP2A is high in CD34+ cells in patients destined to progress to BC. CCR, n = 3; No-CCR, n = 4; and BC, n = 3. (C) Level of phosphorylated (inactive) PP2A protein, as assessed by flow cytometry (mean level in 10 healthy subjects = 1.6; range, 0.8-2.3). Phosphorylated levels of PP2A were greater at diagnosis in CCR (P = .01), No-CCR (P = .002) patients who progressed into BC (P = .012), and at BC (P = .004) compared with normal MNCs. CCR, n = 14; No-CCR, n = 11; and BC, n = 6. (D) Level of phosphorylated (inactive) PP2A protein in CD43+ cells. The degree of phosphorylation is greater in the CD34+ from patients destined to progress into BC. CCR, n = 3; No-CCR, n = 4; and BC, n = 3.
PP2A expression and phosphorylation are increased in MNC and CD34+ cells from patients destined to progress to BC. The horizontal line represents the mean normal level observed in 10 healthy volunteers. (A) Levels of PP2A protein as assessed by flow cytometry. Mean level in 10 healthy subjects = 21.4 (range, 17.1-25.7). The level of PP2A is high in patients destined to progress to BC, compared with nonresponding patients (P = .001). *Statistically significant difference between No-CCR and normal (P = .002). CCR, n = 14; No-CCR, n = 11; and BC, n = 6. (B) Level of PP2A protein in CD34+ cells. The level of PP2A is high in CD34+ cells in patients destined to progress to BC. CCR, n = 3; No-CCR, n = 4; and BC, n = 3. (C) Level of phosphorylated (inactive) PP2A protein, as assessed by flow cytometry (mean level in 10 healthy subjects = 1.6; range, 0.8-2.3). Phosphorylated levels of PP2A were greater at diagnosis in CCR (P = .01), No-CCR (P = .002) patients who progressed into BC (P = .012), and at BC (P = .004) compared with normal MNCs. CCR, n = 14; No-CCR, n = 11; and BC, n = 6. (D) Level of phosphorylated (inactive) PP2A protein in CD43+ cells. The degree of phosphorylation is greater in the CD34+ from patients destined to progress into BC. CCR, n = 3; No-CCR, n = 4; and BC, n = 3.
We next investigated whether PP2A protein levels were a function of PP2A gene expression. Quantitative RT-PCR was used to measure expression of the gene coding for the PP2A catalytic subunit. The pattern of PP2A gene expression was similar to that of PP2A protein expression. Interestingly, PP2A gene expression in all 3 groups, regardless of whether the sample was taken at diagnosis or after treatment, was lower than that observed in normal cells. Because protein levels of PP2A in the MNCs from the BC group were higher than in normal MNCs, this suggests a role for posttranslational modification in the stabilization of PP2A protein expression in the BC group (supplemental Figure 3).
PP2A is inactive when phosphorylated at tyrosine 307, and this can be used to indicate its activity level.8,10,24 Phosphorylated levels of PP2A were significantly greater at diagnosis in the CCR (P = .01), No-CCR (P = .002), and patients who progressed into BC (P = .012) and at BC (P = .004), compared with normal MNCs (Figure 1C). We next analyzed PP2A phosphorylation status in CD34+ cells from patients in these response groups and found elevated levels of pY307-PP2A in cells from the CCR and No-CCR response groups but dramatically increased levels of this phosphoprotein in cells from patients destined to progress (Figure 1D). These data support the conclusions of a previous report indicating a role for PP2A inactivation in the pathology of CML.8 Taken together, these data demonstrate that PP2A proteins and phosphorylation levels are increased in the MNCs and CD34+ cells of patients destined to progress to BC.
SET and SETBP1 expression is not indicative of clinical outcome
SET and SETBP1 have, respectively been implicated in regulating PP2A activity in the leukemic cells of CML and AML.8,10 We therefore measured the expression of SET and SETBP1 at diagnosis and again after 12 months of treatment or at transformation. In line with our previously reported data on SETBP1 expression,11 we found that expression of SETBP1 did not differ between the CCR, No-CCR, and BC clinical groupings of CML (Figure 2A). Moreover, and unlike in the leukemic cells of AML,10 SETBP1 gene expression levels were lower in CML samples compared with those from normal subjects. Interestingly, SETBP1 expression increased after 12 months of treatment in the CCR and No-CCR groups.
SET and SETBP1 at diagnosis and follow-up. SET protein is low at diagnosis in patients who subsequently progress to BC. (A) mRNA expression of SETBP1. No significant differences in SETBP1 expression were observed between the 3 response groups. CCR, n = 14; No-CCR, n = 11; and BC, n = 6. (B) Levels of SET protein. SET protein levels are higher than in normal MNCs (mean level in 10 healthy subjects = 0.2; range, 0.1-0.3) in the CCR and No-CCR groups (P < .001 and P = .01, respectively). CCR, n = 14; No-CCR, n = 11; and BC, n = 6. (C) SET mRNA expression. CCR, n = 14; No-CCR, n = 11; and BC, n = 6. (D) Level of SET protein in CD34+ cells. The level of SET protein assessed at diagnosis does not correlate with the patients' eventual clinical outcome, although this outcome is based on only 3 patients who later progressed while on imatinib treatment. CCR, n = 3; No-CCR, n = 4; and BC, n = 3.
SET and SETBP1 at diagnosis and follow-up. SET protein is low at diagnosis in patients who subsequently progress to BC. (A) mRNA expression of SETBP1. No significant differences in SETBP1 expression were observed between the 3 response groups. CCR, n = 14; No-CCR, n = 11; and BC, n = 6. (B) Levels of SET protein. SET protein levels are higher than in normal MNCs (mean level in 10 healthy subjects = 0.2; range, 0.1-0.3) in the CCR and No-CCR groups (P < .001 and P = .01, respectively). CCR, n = 14; No-CCR, n = 11; and BC, n = 6. (C) SET mRNA expression. CCR, n = 14; No-CCR, n = 11; and BC, n = 6. (D) Level of SET protein in CD34+ cells. The level of SET protein assessed at diagnosis does not correlate with the patients' eventual clinical outcome, although this outcome is based on only 3 patients who later progressed while on imatinib treatment. CCR, n = 3; No-CCR, n = 4; and BC, n = 3.
SET protein levels (Figure 2B) were significantly higher at diagnosis in the CCR and No-CCR groups compared with normal (P < .001 and P = .01, respectively). In patients who subsequently progressed to BC, there was a trend for lower SET levels. This trend was also seen for SET mRNA levels (Figure 2C) because samples from patients who later progressed had lower levels of SET mRNA than did samples from either CCR or No-CCR patients. Interestingly, in BC samples at transformation, SET protein levels increased to become spread over a higher range than at diagnosis and may suggest that SET plays a role during disease progression. This observation contrasts with the fall in SET expression observed in samples from patients within the CCR and No-CCR groups after 12 months of treatment. When we examined SET protein expression in diagnostic CD34+ cells, there was no apparent difference between the 3 clinical groups, although all patients were receiving imatinib treatment at 12 months or at disease progression follow-up (Figure 2D). Taken together, these results are broadly in agreement with previously reported data8 but also suggest that SET may not be the only factor inhibiting PP2A in the malignant cells from patients destined to progress to BC (compared with those from patients who do not). This implies that another mechanism may be inhibiting PP2A in this group of patients.
CIP2A protein level in MNCs and CD34+ at diagnosis is predictive of BC
An additional protein shown to regulate PP2A activity in some cancers that may have a role to play in CML is CIP2A.12-14 Figure 3A shows that CIP2A gene expression is not significantly different in any of the 3 response groups from that in normal MNCs. No statistically significant difference was observed between any of the 3 response groups or at either time point, nor were any of these different from normal MNCs.
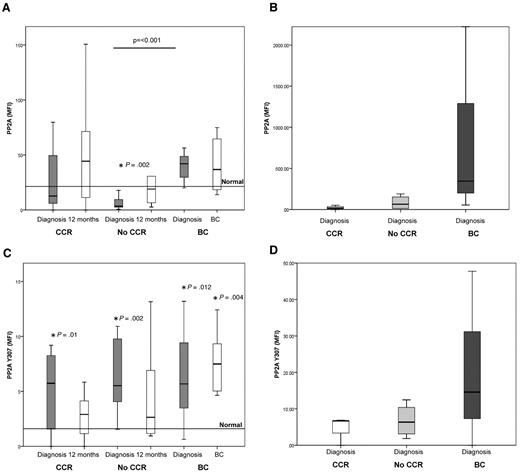
CIP2A protein level in MNCs and CD34+ at diagnosis is predictive of BC. (A) mRNA expression of CIP2A. No statistically significant difference was observed between any of the 3 response groups. CCR, n = 9; No-CCR, n = 9; and BC, n = 6. (B) Levels of CIP2A protein (mean level in 10 healthy subjects = 0.4; range, 0.17-0.63). CIP2A protein levels at diagnosis were significantly higher at diagnosis in patients who later progressed to BC than in CCR or No-CCR patients (P < .0001 and P = .01, respectively). As a patient progresses into BC, the CIP2A protein level increases further (P = .008). CCR, n = 11; No-CCR, n = 9; and BC, n = 6. (C) Level of CIP2A protein in diagnostic CD34+ cells stratified by the patients' clinical outcome. CIP2A is elevated in CD34 from patients destined to progress into BC. CCR, n = 3; No-CCR, n = 4; and BC, n = 3. (D) Kaplan-Meier plot of disease progression, stratified by CIP2A level at diagnosis. Patients with a high diagnostic CIP2A protein level (mean fluorescence intensity [MFI] > 5) have 100% probability of progressing to BC by 21 months (P < .0001). (E) CIP2A levels (mean ± SEM) in CML cell lines K562, KCL22, KY01, and a CIP2A-positive gastric cancer cell line AGS, before and after treatment with 5μM imatinib for 24 hours (n = 6).
CIP2A protein level in MNCs and CD34+ at diagnosis is predictive of BC. (A) mRNA expression of CIP2A. No statistically significant difference was observed between any of the 3 response groups. CCR, n = 9; No-CCR, n = 9; and BC, n = 6. (B) Levels of CIP2A protein (mean level in 10 healthy subjects = 0.4; range, 0.17-0.63). CIP2A protein levels at diagnosis were significantly higher at diagnosis in patients who later progressed to BC than in CCR or No-CCR patients (P < .0001 and P = .01, respectively). As a patient progresses into BC, the CIP2A protein level increases further (P = .008). CCR, n = 11; No-CCR, n = 9; and BC, n = 6. (C) Level of CIP2A protein in diagnostic CD34+ cells stratified by the patients' clinical outcome. CIP2A is elevated in CD34 from patients destined to progress into BC. CCR, n = 3; No-CCR, n = 4; and BC, n = 3. (D) Kaplan-Meier plot of disease progression, stratified by CIP2A level at diagnosis. Patients with a high diagnostic CIP2A protein level (mean fluorescence intensity [MFI] > 5) have 100% probability of progressing to BC by 21 months (P < .0001). (E) CIP2A levels (mean ± SEM) in CML cell lines K562, KCL22, KY01, and a CIP2A-positive gastric cancer cell line AGS, before and after treatment with 5μM imatinib for 24 hours (n = 6).
In sharp contrast, Figure 3B demonstrates that CIP2A protein levels in MNC taken at diagnosis were significantly higher in patients who later progressed to BC than in patients from the CCR and No-CCR groups (P < .0001 and P = .01, respectively). This observation was also confirmed in CD34+ cells; samples from patients destined to progress into BC had much higher levels of CIP2A than either CCR or No-CCR patients (Figure 3C). No correlation was seen between CIP2A expression and Sokal score (data not shown). These results suggest that CIP2A may be important in inhibiting PP2A function in CML patients who progress to BC. The importance of increased CIP2A protein expression in disease progression is further exemplified after 12 months of treatment; whereas CIP2A protein levels remain essentially unchanged in samples from the CCR and No-CCR groups, the level increases further at disease progression (P = .008, Figure 3B).
To test whether high expression of CIP2A protein is predictive of disease progression, progression-free survival was compared between patients with high (defined as a mean fluorescence intensity > 5) and low diagnostic CIP2A protein levels. Figure 3D shows that the median time to disease progression in patients with high CIP2A protein levels is 13 months, and their probability of progression is 100% at 21 months. Five of these 6 patients have died of their progression. In contrast, during the same time period, none of the patients with low diagnostic CIP2A levels progressed, and all remain progression-free and alive at their latest follow-up (average follow-up, 47 months). These data indicate that CIP2A protein expression is a biomarker of disease progression in CML.
We have previously demonstrated that BCR-ABL1 activity levels are also predictive of treatment response in CML.3 To see whether BCR-ABL1 was a factor controlling CIP2A protein levels, we treated the BCR-ABL1-positive cells lines K562, KCL22, and KY01 together with the BCR-ABL-negative gastric cancer cell line AGS (used as a CIP2A positive control)14 with the tyrosine kinase inhibitor imatinib for 24 hours. We found that imatinib treatment of the BCR-ABL1-positive cell lines resulted in a significant down-regulation of CIP2A protein levels (P = .002 for K562, P = .001 for KCL22, and P = .024 for KY01) but had no effect on CIP2A expression in AGS cells. Thus, BCR-ABL1 activity may be an important factor in the regulation of CIP2A protein expression (Figure 3E).
c-Myc protein is elevated at diagnosis in patients destined to progress to BC
One of the prescribed functions of CIP2A in cancer cells is to prevent PP2A-mediated dephosphorylation of c-Myc.12 To test whether c-Myc phosphorylation is a factor in CML pathophysiology, we used an ELISA to measure the levels of pS62-Myc in diagnostic samples. MNCs from patients destined to progress to BC had significantly higher levels of pS62-Myc than did those from patients within either the CCR or No-CCR groups (Figure 4A; P = .04). Because S62 phosphorylation of c-Myc stabilizes this protein against degradation,12 we also measured levels of total c-Myc protein in the same sample. We observed similar results; c-Myc protein levels were significantly higher in the cells of patients who subsequently progress to BC (Figure 4B; P = .002). These data demonstrate that high levels of c-Myc and phosphorylation at serine 62 indicate a high risk of disease progression. Because PP2A is the major phosphatase that dephosphorylates c-Myc, these results further suggest that PP2A function may be inhibited, possibly by high expression of CIP2A.
Assessing PP2A function. c-Myc is elevated at diagnosis in patients destined to progress into BC. Levels of c-Myc phosphorylation on serine 62 (A) and total c-Myc protein levels (B). Stratification by clinical outcome and abbreviations as for the previous figures. CCR, n = 10; No-CCR, n = 5; and BC, n = 5. (C) PIM1 is higher in the CD34+ cells from patients who will later progress into BC, confirming that PP2A function is inhibited. CCR, n = 3; No-CCR, n = 3; and BC, n = 3.
Assessing PP2A function. c-Myc is elevated at diagnosis in patients destined to progress into BC. Levels of c-Myc phosphorylation on serine 62 (A) and total c-Myc protein levels (B). Stratification by clinical outcome and abbreviations as for the previous figures. CCR, n = 10; No-CCR, n = 5; and BC, n = 5. (C) PIM1 is higher in the CD34+ cells from patients who will later progress into BC, confirming that PP2A function is inhibited. CCR, n = 3; No-CCR, n = 3; and BC, n = 3.
PIM1 is a protein kinase that can phosphorylate and stabilize c-Myc.23 PIM1 is itself phosphorylated, and in this state it is active and its expression is stable.25 However, this protein is a target for PP2A; and once it is dephosphorylated by PP2A, it is rapidly degraded in the proteasome.26 To further confirm that PP2A is functionally inactive in CML, we investigated PIM1 as another target of PP2A. Flow cytometry was used to measure PIM1 expression in CD34+ cells from diagnosis. PIM1 was elevated in patients destined to progress into BC compared with the CCR and No-CCR patients (Figure 4C). In addition, analysis undertaken in K562 cells revealed that activation of PP2A, either by addition of a PP2A activator or inhibition of CIP2A by imatinib, also decreased PIM1 levels (data not shown). Considering the role of PP2A in regulating PIM1 protein levels, these data provide further evidence that PP2A activity is suppressed in the CD34+ cells of patients at high risk of developing BC.
Finally, previous work by others has indicated that suppression of PP2A activity feeds back on BCR-ABL1 to facilitate increased and sustained kinase activity.8 One of the hallmarks of BCR-ABL1 activity is phosphorylation of CrkL,3,27 and we have previously shown that an increased pCrkL/CrkL ratio is indicative of a poor response to imatinib.3 We assessed the pCrkL/CrkL ratio on all (n = 10) available samples from the present cohort of patients. BCR-ABL1 activity was highest in patients destined to progress into BC (supplemental Figure 4).
CIP2A plays a key role in regulating PP2A in CML cells
To confirm a role for PP2A in regulating c-Myc and PIM1 expression, the CML cell line K562 was treated with imatinib. Figure 3E demonstrates that this reduces CIP2A protein expression. pS62-Myc and PIM1 levels were also reduced in the cells treated with imatinib (data not shown). These results are compatible with the notion that CIP2A expression regulated by BCR-ABL1 activity has a role in suppression of PP2A activity and in downstream activation of PIM1 and c-Myc.
To further investigate the effect of CIP2A on CML cells, K562 cells were treated with siRNA targeted at CIP2A (n = 5; Figure 5). After siRNA treatment, the mean knockdown of the CIP2A protein was 60% (P = .02; Figure 5A). Inhibition of CIP2A resulted in a 63% decrease in BCR-ABL1 tyrosine kinase activity as assessed by the pCrkL/CrkL ratio as previously described3 (P = .03; Figure 5B). PP2A protein levels remained unchanged (Figure 5C), but the degree of PP2A phosphorylation (at Y307, indicating inactivity) decreased by 53%, indicative of an increase in PP2A function (Figure 5D). In addition, SET and JAK2 decreased by 48% and 37%, respectively, with CIP2A siRNA treatment, indicating that CIP2A acts upstream of JAK2 and SET (Figure 5E-F). Phosphorylation of c-Myc at serine 62 and total c-Myc protein also decreased by approximately 50% (Figure 5G-H). Key results from these experiments were also confirmed by Western blotting, using an alternative siRNA sequence, and on the additional CML cell line LAMA84 (shown in supplemental Figure 5). These data confirm that PP2A is functionally inactivated by CIP2A and that decreasing the level of CIP2A protein removes the block on PP2A function and decreases BCR-ABL1 activity.
CIP2A plays a key role in regulating PP2A in CML cells. K562 cells were treated with CIP2A siRNA for 72 hours (n = 5). The effects are shown on: CIP2A protein levels as a control (A), BCR-ABL1 activity as assessed by the pCrkL/CrkL ratio (B), PP2A and phosphorylated PP2A (C-D), SET (E), and JAK2 (F). c-Myc phosphorylation at serine 62 and total c-Myc protein (G-H).
CIP2A plays a key role in regulating PP2A in CML cells. K562 cells were treated with CIP2A siRNA for 72 hours (n = 5). The effects are shown on: CIP2A protein levels as a control (A), BCR-ABL1 activity as assessed by the pCrkL/CrkL ratio (B), PP2A and phosphorylated PP2A (C-D), SET (E), and JAK2 (F). c-Myc phosphorylation at serine 62 and total c-Myc protein (G-H).
Discussion
Despite the dramatic effect of imatinib, CML remains a fatal disease for the proportion of patients who progress from chronic phase to BC. Recent data suggest that the second-generation TKI nilotinib decreases the rate of progression to BC in the first 12 months,28 but it remains to be seen whether this benefit is maintained at later time points. Our previously published biomarker data on the pCrKL/CrKL ratio and BCR-ABL1 transcript type can only predict patients likely to achieve a CCR and cannot predict patients destined to progress to BC.3,19 Various techniques have been used on diagnostic chronic phase samples to predict who will develop BC,29 and a variety of novel genomic lesions have been identified at BC.30 It is however not possible at diagnosis to reliably predict which patients will develop disease progression. This is in part because of a poor understanding of the factors facilitating development of the novel genomic lesions that lead to disease progression.
The present data suggest that CIP2A is an important determinant of future disease progression in CML. We show that high CIP2A protein levels are present in diagnostic MNCs and CD34+ cells of patients destined to progress into BC, and after imatinib treatment CIP2A levels increase further. The probability of disease progression is 100% at 21 months in patients with high CIP2A protein. Interestingly, no correlation was observed between CIP2A expression and Sokal score, suggesting that the presence of high levels of this protein is an independent biomarker of BC. In particular, of the 6 patients in this study who progressed to BC, only 2 had high Sokal scores (Table 1), whereas in the CCR group, 4 of the 14 patients had high Sokal but all achieved a CCR.
CIP2A functions as a regulator of PP2A activity, and high expression of this protein is reported to suppress the phosphatase activity of PP2A.12 We used specific siRNA and imatinib to reduce CIP2A levels in K562 cells and show that this results in a decrease in expression of 2 important PP2A targets, c-Myc and PIM1. These proteins are important because PIM1 is a kinase that acts to phosphorylate c-Myc and stabilize its expression.23,26 When PIM1 is dephosphorylated by PP2A, it becomes degraded within the proteasome.26 Similarly, c-Myc is also degraded in the proteosome once it is dephosphorylated by PP2A.12 The reductions in PIM1 and c-Myc (and concomitant reduction of pS62-Myc) expression therefore suggest that PP2A activity increases. This notion is supported by our observation that pY307-PP2A levels decreased without affecting PP2A protein levels in the treated cells. Phosphorylation of PP2A on Y307 deactivates this phosphatase, and removal of this phosphate by PP2A-mediated autodephosphorylation results in its reactivation.6 Taken together, the results of these experiments therefore suggest a mechanistic connection between high CIP2A expression, PP2A phosphorylation, and suppression of activity (illustrated by high levels of pY307-PP2A) and high levels of pS62-Myc and of c-Myc and PIM1 protein.
In the clinical samples we analyzed, we found that high CIP2A protein levels corresponded with high levels of Y307 and S62 phosphorylation of PP2A and c-Myc, respectively. Moreover, the patient samples containing high levels of CIP2A also had high levels of c-Myc and PIM1. Taken together with the cell line findings, these data connect CIP2A to c-Myc and PIM1 in primary CML cells and suggest that CIP2A, c-Myc, and PIM1 could be prognostic biomarkers.
High levels of pS62-Myc and c-Myc protein may have important consequences. c-Myc has a critical role in cell proliferation, and increased levels of this protein within cells promote entry into cell cycle. Thus, diagnostic chronic phase cells with high c-Myc probably have a greater proliferative potential. This notion is supported by studies of clinical gastric cancer biopsies in which aggressive (proliferative) disease is associated with high expression of CIP2A and c-Myc.10 Deregulated cell division is known to result in increased DNA mismatch repair errors and in genomic instability.31 This is also observed in CML because c-Myc may contribute to disease progression by promoting aneuploidy.31-34
Whether CIP2A-mediated inhibition of PP2A acts directly to block dephosphorylation of c-Myc or acts at a stage upstream of c-Myc remains to be determined. Junttila et al provide evidence suggesting that CIP2A binds directly to c-Myc and is recruited as a result of S62 phosphorylation.12 However, we found that the reduction of CIP2A resulted in a decrease in BCR-ABL1 activity, and a reduction in kinase activity would result in a decrease in the signals that are necessary to induce stable expression of c-Myc.
In the present study, patients from the CCR group had high levels of SET protein in samples taken at diagnosis. At the 12-month sampling, SET protein levels in the MNC from these patients were restored to almost normal levels. This is consistent with the in vitro findings of Neviani et al,8 which demonstrated that PP2A is inhibited by SET and that PP2A function can be restored by a reduction in SET levels caused by imatinib-induced inhibition of BCR-ABL1 and JAK2. Broadly similar data were seen in the No-CCR group, who also did not undergo disease progression. However, patients in the BC group had lower SET levels than did patients who do not progress. This suggests that PP2A activity may be regulated differently in cells of CCR and BC patients; in the former group of patients, PP2A activity is regulated predominantly by SET, whereas in the latter group, PP2A activity is regulated by SET and CIP2A. Figure 6 summarizes the present findings and postulates possible mechanisms of how PP2A is inhibited in CML. In patients who respond to imatinib therapy, PP2A activity in the malignant cells is suppressed by SET, and SET expression is regulated by BCR-ABL1 via JAK2.8 When imatinib is commenced, BCR-ABL1 activity decreases and this leads to removal of PP2A inhibition through a decrease in SET protein levels. However, in patients who progress to BC, CIP2A is present at a high level and contributes to PP2A suppression. Imatinib therapy does not affect CIP2A levels in the malignant cells of these patients and PP2A remains suppressed. c-Myc expression therefore remains at a higher level, increasing the probability of subsequent genetic damage through increased cell proliferation and thus disease progression to BC.
Model mechanisms by which CIP2A regulates PP2A and BCR-ABL1 signaling proteins. Possible mechanism of PP2A inhibition in CML. In most patients, PP2A is suppressed by SET, which is driven by BCR-ABL1 via JAK2. In patients destined to progress into BC, PP2A is inhibited by CIP2A both directly as well as through BCR-ABL.1
Model mechanisms by which CIP2A regulates PP2A and BCR-ABL1 signaling proteins. Possible mechanism of PP2A inhibition in CML. In most patients, PP2A is suppressed by SET, which is driven by BCR-ABL1 via JAK2. In patients destined to progress into BC, PP2A is inhibited by CIP2A both directly as well as through BCR-ABL.1
In conclusion, the present data demonstrate that CIP2A is an important determinant of future disease progression, and its use as a biomarker of BC needs to be tested both in animal models and prospectively in clinical samples. The data also suggest that CIP2A may be a useful therapeutic target in CML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Alison Holcroft and Joanna Middleton of the University of Liverpool Biobank for their help and support during this project as well as Dr Mark Glenn for advising on the siRNA work.
Authorship
Contribution: C.M.L., R.J.H., and R.E.C. designed the study; C.M.L., R.J.H., J.R.S., and R.E.C. wrote the manuscript; C.M.L. and A.G. performed the laboratory work for this study; and M.C. provided clinical samples.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard E. Clark, Department of Haematology, University of Liverpool, Liverpool, L69 3GA, United Kingdom; e-mail: clarkre@liv.ac.uk.


![Figure 3. CIP2A protein level in MNCs and CD34+ at diagnosis is predictive of BC. (A) mRNA expression of CIP2A. No statistically significant difference was observed between any of the 3 response groups. CCR, n = 9; No-CCR, n = 9; and BC, n = 6. (B) Levels of CIP2A protein (mean level in 10 healthy subjects = 0.4; range, 0.17-0.63). CIP2A protein levels at diagnosis were significantly higher at diagnosis in patients who later progressed to BC than in CCR or No-CCR patients (P < .0001 and P = .01, respectively). As a patient progresses into BC, the CIP2A protein level increases further (P = .008). CCR, n = 11; No-CCR, n = 9; and BC, n = 6. (C) Level of CIP2A protein in diagnostic CD34+ cells stratified by the patients' clinical outcome. CIP2A is elevated in CD34 from patients destined to progress into BC. CCR, n = 3; No-CCR, n = 4; and BC, n = 3. (D) Kaplan-Meier plot of disease progression, stratified by CIP2A level at diagnosis. Patients with a high diagnostic CIP2A protein level (mean fluorescence intensity [MFI] > 5) have 100% probability of progressing to BC by 21 months (P < .0001). (E) CIP2A levels (mean ± SEM) in CML cell lines K562, KCL22, KY01, and a CIP2A-positive gastric cancer cell line AGS, before and after treatment with 5μM imatinib for 24 hours (n = 6).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/24/10.1182_blood-2010-08-304477/4/m_zh89991172410003.jpeg?Expires=1765889872&Signature=qa5K9aXLND6xP2ROfEOgXZjHxBVtzR8P0Dr3ZoORSbwS2PqtlL4x5-TNwdV9Mg5tbiy2BezxSD72b3gqS6KkDkvLcDYyyMOXsFyX~aX793AutIJgxkIUI9ARihUutVMSjWzIohFjKglap2yHUo51F8~9tYlMo1FszP3-JuE8oISfilj44Kn6T~rTvYR2rary0CEANDy~yqby2fMMpB66EKhuNNwqGkbrROIOHGPEU~3QNrCY2SaKsITJ5o1tcICvxwsRQAZP0-B8oJt6xibyxT~nyGD0W37SF2BG~mkgl45salqC4s96eWj1B1G7x7By4ACrLHKJOAQcciPoGzBPvQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


