Abstract
MLL encodes a histone methyltransferase that is critical in maintaining gene expression during embryonic development and hematopoiesis. 11q23 translocations result in the formation of chimeric MLL fusion proteins that act as potent drivers of acute leukemia. However, it remains unclear what portion of the leukemic genome is under the direct control of MLL fusions. By comparing patient-derived leukemic cell lines, we find that MLL fusion-bound genes are a small subset of that recognized by wild-type MLL. In an inducible MLL-ENL model, MLL fusion protein binding and changes in H3K79 methylation are limited to a specific portion of the genome, whereas wild-type MLL distributes to a much larger set of gene loci. Surprisingly, among 223 MLL-ENL–bound genes, only 12 demonstrate a significant increase in mRNA expression on induction of the fusion protein. In addition to Hoxa9 and Meis1, this includes Eya1 and Six1, which comprise a heterodimeric transcription factor important in several developmental pathways. We show that Eya1 has the capacity to immortalize hematopoietic progenitor cells in vitro and collaborates with Six1 in hematopoietic transformation assays. Altogether, our data suggest that MLL fusions contribute to the development of acute leukemia through direct activation of a small set of target genes.
Introduction
Chromosomal translocations often lead to creation of chimeric fusion genes that define cancer subtypes and act as initiating events in oncogenesis. Fusion genes can acquire novel biologic features via their fusion partners, yet retain important functions of the wild-type protein. When chromosomal rearrangements involve genes encoding DNA-binding proteins, chimeric fusion proteins can serve as aberrant transcription factors leading to genome-wide dysregulation in gene expression. Studies have shown that fusion transcription factors may gain novel specificity in target gene selection. The EWS/FLI1 oncogenic transcription factor modulates a specific set of genes that are not bound by wild-type FLI1 in vivo.1 The AML1-ETO fusion protein preferentially binds to duplicated AML-1 DNA-binding sequences.2 In acute promyelocytic leukemia (APL), the PML/RARα fusion protein is selectively targeted to regions containing PU.1 consensus and RARE (retinoic acid response elements) half sites. The majority of those sites do not correspond to canonical RAREs.3 Chromosomal translocations involving the MLL (mixed-lineage leukemia) gene result in the formation of a chimeric transcription factor. It remains unknown whether the MLL fusion protein shares the same set of target genes compared with the wild-type MLL protein.
The histone methyltransferase MLL gene is frequently targeted by chromosomal translocations in acute myeloid, lymphoid, and biphenotypic leukemias, and rearrangement of MLL is associated with a poor prognosis. Wild-type MLL is critical for the maintenance of expression of its target genes. This activity is mediated by its carboxyl-terminal SET domain, which acts as a histone 3 lysine-4 (H3K4) methyltransferase.4,5 The critical consequence of 11q23 chromosomal translocations is the formation of a chimeric oncogenic transcription factor that retains the N-terminus of MLL but replaces carboxyl-terminal domains with sequences from its partner proteins. As a result of 11q23 gene rearrangements, MLL fuses in frame with > 60 different partner proteins.6 The most common partners are AF4, AF9, ENL, AF10, and ELL, which together account for > 85% of all MLL-rearranged leukemias. AF4, ENL, AF9, and AF10 form a complex that promotes H3K79 methylation through recruitment of histone methyltransferase DOT1L.7,8 Aberrant H3K79 methylation has been shown to be a key molecular mechanism in MLL fusion-induced dysregulation of gene expression.9,10
MLL is targeted to a specific set of gene loci, presumably via its own DNA binding domain, recognition of local histone modifications, and/or recruitment by sequence-specific transcription factors.11 However, the target genes of MLL fusion proteins compared with wild-type MLL remain poorly understood. In MLL-AF4–expressing mouse acute lymphoid leukemia cells, thousands of gene promoters exhibited increased levels of H3K79 methylation in comparison to normal control lymphocytes, suggesting MLL-AF4 has widespread targets in the leukemic genome.12 In contrast, by looking for co-occupancy of MLL and AF4 in one MLL-rearranged cell line, another study only identified approximately 169 gene regions that are bound by MLL-AF4.9 Independent expression profiling studies on primary patient samples and cellular leukemic models have revealed a smaller number of genes (∼ 100) which are differentially expressed in MLL-rearranged leukemias.12-14 Collectively, these studies do not provide a clear picture regarding what portion of the genome is directly controlled by MLL fusion proteins. To address these important issues in the present study, we undertook 2 parallel strategies, including mapping MLL binding in multiple human leukemia patient-derived cell lines, and a combined location and expression profiling analysis in an MLL-ENL–inducible system. Using this combined approach, we found that MLL fusions are targeted to a small set of genomic loci occupied by wild-type MLL. Strikingly, a large fraction of MLL fusion-bound genes do not exhibit a change in mRNA expression on inactivation of MLL-ENL, whereas a small set of target genes are up-regulated by MLL-ENL. Among target genes whose expression is strongly influenced by MLL-ENL, we have identified key developmental regulators that potentially contribute to the development of MLL-associated leukemia. This includes HOXA9 and its cofactor MEIS1, which have previously been shown to induce leukemic transformation. In addition, we identified the transcription factors EYA1, SIX1, and SIX4 as direct MLL-ENL targets. EYA proteins form heterodimers with members of the SIX protein family. We show that EYA1 is able to immortalize hematopoietic progenitor cells (HPCs) and collaborates with SIX1 in transformation assays. Taken together, we demonstrate that MLL fusion proteins, particularly MLL-ENL, directly target multiple transcription factor genes that are capable of transforming HPCs, suggesting that the transforming capacity of MLL fusion proteins is not limited to the specification of HOXA/MEIS1 gene targets.
Methods
The use of BM cells derived from mice for these studies was approved by the University of Chicago Animal Care and Use Committee.
Cell culture
MV4;11, THP-1, ML-2, HL60 and U937 cells were cultured in RPMI 1640 medium supplemented with 10% FBS. MLL-ENL–inducible cell line (csh2) was obtained from Dr Robert Slany (University Erlangen, Germany). Cells grew in RPMI 1640 medium supplemented with 10% FBS, with 5 ng/mL IL-3, IL-6, GM-CSF 50 ng/mL SCF, and 100nM 4-hydroxy-tamoxifen (4-OHT), as described.13
Hematopoietic immortalization assay
The Eya1 and Six1 cDNA fragments were cloned in the murine stem cell virus (MSCV) retroviral expression vector. Infection of lineage-depleted progenitor cells and culture of the transduced progenitors in methylcellulose culture were as previously described.15 The transduced murine hematopoietic progenitor cells were cultured in the presence of IL-3 (10 ng/mL), IL-6 (10 ng/mL), GM-CSF (10 ng/mL), SCF (100 ng/mL).
ChIP, PCR analysis, and ChIP-chip analysis
The ChIP was performed as described.16,17 Abs used were as follows: MLL (Bethyl A300-086A, A300-374A), the MLL antisera as described,18 H3K79me2 (Abcam ab3594), and rabbit IgG (Santa Cruz Biotechnology, sc2027). Ab-precipitated DNA fragments were purified. Eluted DNA fragments were used for qPCR or array analysis.
Analysis of ChIP-chip data
CisGenome was used to determine the binding/methylation locations of MLL, and/or H3K79 proteins in each sample.19 We selected a more suitable scale by log2 transforming the intensities and then removed possible systematic biases by quantile normalizing the data. Significant peaks representing the most likely locations of TF binding or H3K79 methylation were detected20 (see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Affymetrix microarray expression analysis
Genome-wide gene expression analysis was performed using Affymetrix Mouse Exon 1.0 ST Array. We used GeneBASE to estimate gene expression from the Affymetrix exon arrays.21 With the gene-level estimates, we applied the Limma package to determine differential gene expression (see supplemental Methods).22
All data can be accessed by using accession number GSE24794 in the Gene Expression Omnibus.
Results
Mapping MLL binding in MLL-rearranged and wild-type human leukemic cells
Primary leukemic samples with different MLL fusions have an indistinguishable expression pattern, suggesting that distinct MLL fusion proteins share common target genes.23 We mapped MLL binding in 3 MLL-rearranged (ML-2–expressing MLL-AF6, MV4;11–expressing MLL-AF4, and THP-1–expressing MLL-AF9) and 2 non-MLL-rearranged leukemia cell lines (U937 and HL60; Figure 1 left panel). The MLL antisera are expected to recognize both wild-type MLL and MLL fusion proteins, as they were raised against an amino-terminal fragment of MLL present in all fusion proteins.18 The observed binding pattern in MV4;11 and THP-1 can be attributed to either wild-type or to MLL fusion gene products in 11q23 leukemia cells. In contrast, the results obtained from ML-2 cells are specific for the MLL fusion protein because ML-2 cells lack wild-type MLL. The 2 non-MLL–rearranged myeloid leukemia cells, U937 and HL60, permit us to analyze the binding patterns contributed by the MLL wild-type protein.
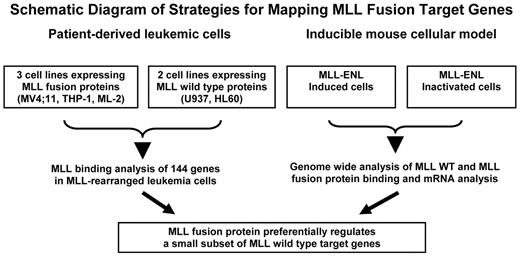
Schematic diagram of strategies for mapping MLL fusion target genes. All 5 human cell lines (top left) are derived from acute leukemia patients and represent early progenitor cells of myeloid origin. MV4;11 (expressing MLL-AF4) and THP-1 (expressing MLL-AF9) have one rearranged MLL allele and one wild-type (WT) MLL allele. ML-2 cells possess 2 alleles of MLL-AF6 but no WT MLL allele. U937 and HL60 cells contain 2 copies of WT MLL. The inducible mouse cells express MLL-ENL in the presence of the inducer. The MLL-ENL fusion protein is inactivated on withdrawal of the inducer. Target genes of MLL wild-type and MLL fusion proteins were obtained from MLL binding analysis in both human leukemic cells and the mouse inducible system.
Schematic diagram of strategies for mapping MLL fusion target genes. All 5 human cell lines (top left) are derived from acute leukemia patients and represent early progenitor cells of myeloid origin. MV4;11 (expressing MLL-AF4) and THP-1 (expressing MLL-AF9) have one rearranged MLL allele and one wild-type (WT) MLL allele. ML-2 cells possess 2 alleles of MLL-AF6 but no WT MLL allele. U937 and HL60 cells contain 2 copies of WT MLL. The inducible mouse cells express MLL-ENL in the presence of the inducer. The MLL-ENL fusion protein is inactivated on withdrawal of the inducer. Target genes of MLL wild-type and MLL fusion proteins were obtained from MLL binding analysis in both human leukemic cells and the mouse inducible system.
To map MLL binding in leukemic cells, we used ChIP. Previously, we generated MLL antisera which are specific for the detection of MLL proteins by Western blotting and immunoprecipitation.18 To test their suitability in ChIP assays, we compared the performance of these antisera with several commercially available ChIP quality Abs (supplemental Figure 1). We further compared the MLL-binding pattern identified in our ChIP-chip analysis with that from an earlier report by Guenther et al.24 At the 125-kb HOXA cluster region, a largely overlapping set of MLL-bound DNA sequences were identified in U937 cells by these 2 independent studies (supplemental Figure 2). Altogether, these data suggest that the MLL antisera are sensitive and specific in detecting MLL binding at selected genomic loci.
MLL fusion-bound genes are a subset of that recognized by the wild-type proteins in human leukemic cells
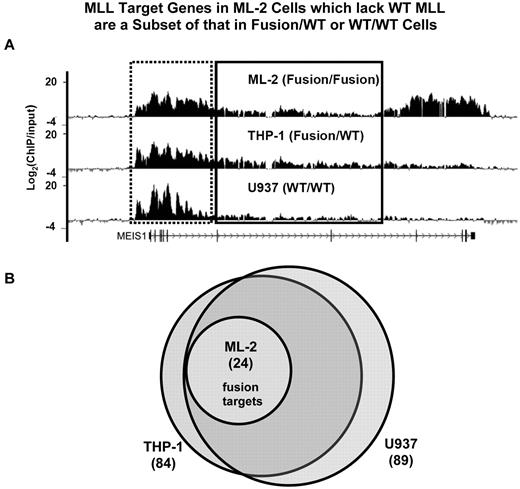
To examine whether MLL wild-type and fusion proteins regulate a similar or distinct set of genes, we used ChIP-chip (ChIP coupled with microarray) approach to interrogate ∼ 48 824 kb of genomic sequence in 5 different leukemic cell lines. We designed a custom array that contains the entire genomic loci of 144 genes which we and others have previously found to have altered expression in MLL-rearranged leukemias (supplemental Table 1). We searched for MLL binding in each gene locus between 2 kb upstream of TSS (transcription start site) and 2 kb downstream of TES (transcription end site). Genes with the presence of any MLL-binding peak (FDR < 0.25) in this defined area are referred to as “MLL targets” (see supplemental Methods). We found that MLL proteins bind to an extended domain at the promoter and 5′ gene region (Figure 2A). These analyses revealed a largely overlapping set of genes bound by MLL in WT/WT (U937: 89 genes, HL60: 72 genes) and Fusion/WT cells (MV4;11: 59 genes, THP-1: 84 genes; Figure 2B, supplemental Figure 3). Surprisingly, the MLL-bound genes in Fusion/Fusion (ML-2) cells (24 genes) are a small subset of that found in 3 of the other 4 cell lines, and overlap extensively with MLL targets identified in HL60 cells (Figure 2B, supplemental Figure 3). We refer to the set of 24 genes, defined by MLL-AF6 binding in ML-2 cells, as “24 MLL-AF6 target genes.” These data show that MLL-AF6 fusion proteins in ML-2 cells may be disproportionately distributed to a very limited set of gene loci.
MLL target genes in Fusion/Fusion cells are a subset of that in Fusion/WT or WT/WT cells. (A) Detection of MLL binding using ChIP-chip at the MEIS1 locus. Cross-linked chromatin from ML-2, U937, THP-1 cells was immunoprecipitated with the MLL antisera separately. The Ab precipitated DNA and the input DNA were labeled with Cy5 and Cy3, respectively, and hybridized to a customized NimbleGen array containing the entire gene loci of 144 genes (see “Analysis of ChIP-chip data”). The level of MLL protein enrichment is shown as Log2(ChIP/input). The promoter and 5′ gene region of MEIS1 is boxed by a dotted line, and the 3′ gene region is boxed by solid line. (B) The genomic loci of 144 human genes were examined for enrichment of MLL protein binding using the customized NimbleGen array. MLL target genes identified in ML-2 (Fusion/Fusion), THP-1(Fusion/WT), and U937 (WT/WT) cells were compared.
MLL target genes in Fusion/Fusion cells are a subset of that in Fusion/WT or WT/WT cells. (A) Detection of MLL binding using ChIP-chip at the MEIS1 locus. Cross-linked chromatin from ML-2, U937, THP-1 cells was immunoprecipitated with the MLL antisera separately. The Ab precipitated DNA and the input DNA were labeled with Cy5 and Cy3, respectively, and hybridized to a customized NimbleGen array containing the entire gene loci of 144 genes (see “Analysis of ChIP-chip data”). The level of MLL protein enrichment is shown as Log2(ChIP/input). The promoter and 5′ gene region of MEIS1 is boxed by a dotted line, and the 3′ gene region is boxed by solid line. (B) The genomic loci of 144 human genes were examined for enrichment of MLL protein binding using the customized NimbleGen array. MLL target genes identified in ML-2 (Fusion/Fusion), THP-1(Fusion/WT), and U937 (WT/WT) cells were compared.
MLL fusion-bound gene regions exhibit an aberrant distribution pattern of MLL in human leukemic cells
As our custom array was designed to interrogate the entire genomic loci of 144 potential MLL target genes, we examined the possibility that MLL fusion proteins may exhibit an aberrant distribution pattern. At the promoter and 5′ gene region of MEIS1, 1 of 24 MLL fusion target genes identified, MLL protein enrichment was highly similar across 4 different cell lines which are known to actively express MEIS1. However, at the genomic region near the 3′ end of the gene, we observed a higher level of MLL enrichment in MLL fusion cell lines (ML-2, MV4;11 and THP-1) than that in U937 non-MLL–rearranged leukemia (Figure 2A, and data not shown). We further confirmed the protein distribution pattern of MLL in both THP-1 and U937 cells using ChIP-qPCR (supplemental Figure 4).
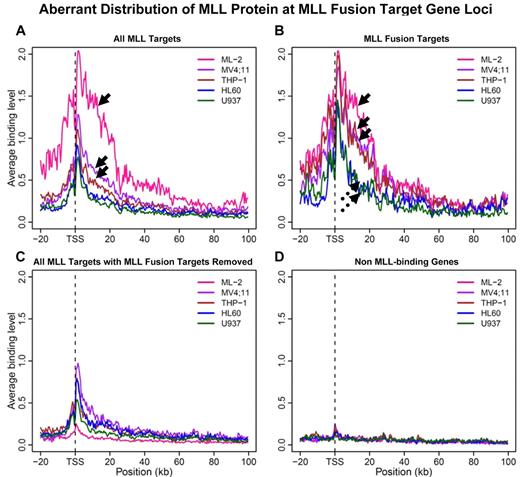
To determine whether the distribution of MLL at the MEIS1 locus is a common feature at MLL fusion target loci, we further made composite plots averaging binding signal from MLL target gene regions (Figure 3). When all MLL target genes in each of the 5 cell lines are considered, the MLL binding signals in ML-2 cells, solely derived from MLL-AF6, are significantly stronger than that in MV4;11 (expressing MLL-AF4) and THP-1 cells (expressing MLL-AF9), in which binding signals are attributable to both fusion and WT target genes (Figure 3A arrows, P = .007). Strikingly, when only fusion targets, as defined by MLL-AF6 binding in ML-2 cells, are taken into account, the averaged binding strength among all 3 fusion protein-expressing cells becomes indistinguishable (Figure 3B solid arrows, P = .5), while 2 non-MLL–rearranged cell lines share an almost identical distribution pattern (Figure 3B broken arrows, P = .8). For this set of fusion target genes, MLL-binding strength in MLL fusion cells, as a group, are significantly higher than that in the group of non-MLL–rearranged cells (P = .00006278). In contrast, when fusion target genes were removed from the total MLL target set, averaged binding strength is largely similar across all 4 cell lines (Figure 3C and P > .2), except that U937 cells show a lower level of protein enrichment. Minimal signal was detected at non-MLL target gene loci for all 5 cell lines (Figure 3D). Similar results were obtained when we performed the analysis with gene length normalization (supplemental Figure 5). Despite inclusion of different MLL fusion partner genes (AF6, AF4, or AF9), all 3 fusion cell lines exhibit a similar aberrant MLL protein distribution at identified MLL fusion target gene loci.
Aberrant distribution of the MLL protein at MLL fusion target gene loci. A 500-bp sliding window was used to scan the region spanning −20 kb to 100 kb from transcription start site (TSS) for each gene. Binding signals within each window were averaged for the respective set of genes. (A) MLL binding curve was plotted for each of the 5 cell lines when all MLL protein target genes (wild-type and fusion) were considered. Arrows point to MLL binding curves obtained from 3 MLL-rearranged cell lines with ML-2 cells on the top. (B) Average MLL binding curve for “24 MLL-AF6 target genes” (Figure 2B) were considered. Solid arrows point to MLL binding curves obtained from 3 MLL-rearranged cell lines; broken arrows 2 MLL nonrearranged cell lines. (C) All MLL protein targets excluding “24 MLL-AF6 target genes” (Figure 2B) were considered. (D) Average signals of non-MLL target genes for each cell line were plotted as negative controls.
Aberrant distribution of the MLL protein at MLL fusion target gene loci. A 500-bp sliding window was used to scan the region spanning −20 kb to 100 kb from transcription start site (TSS) for each gene. Binding signals within each window were averaged for the respective set of genes. (A) MLL binding curve was plotted for each of the 5 cell lines when all MLL protein target genes (wild-type and fusion) were considered. Arrows point to MLL binding curves obtained from 3 MLL-rearranged cell lines with ML-2 cells on the top. (B) Average MLL binding curve for “24 MLL-AF6 target genes” (Figure 2B) were considered. Solid arrows point to MLL binding curves obtained from 3 MLL-rearranged cell lines; broken arrows 2 MLL nonrearranged cell lines. (C) All MLL protein targets excluding “24 MLL-AF6 target genes” (Figure 2B) were considered. (D) Average signals of non-MLL target genes for each cell line were plotted as negative controls.
MLL fusions target a limited number of loci bound by wild-type MLL in an inducible MLL-ENL model
Analysis of our custom array of 144 MLL target gene loci suggests that MLL fusion proteins localize to a limited portion of genomic loci occupied by the MLL wild-type protein. To test this hypothesis in a more systematic way, we examined an inducible MLL-ENL cellular model (Figures 1 right panel, 4A).13
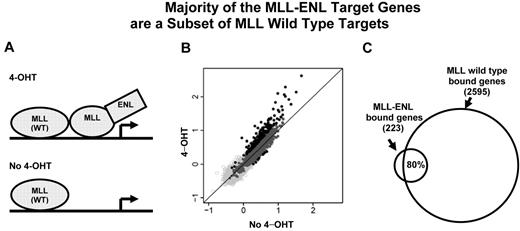
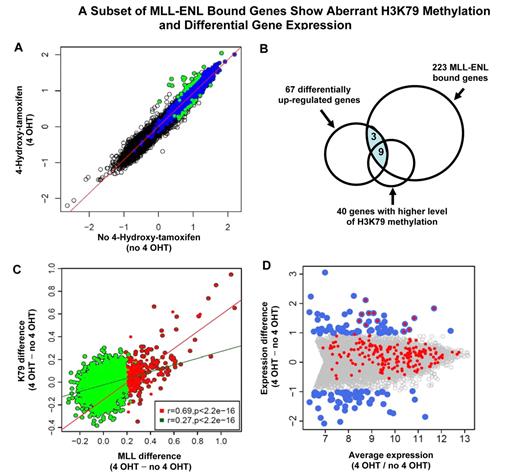
The majority of the MLL-ENL target genes are a subset of MLL wild-type targets. (A) A cartoon of the inducible system shows MLL wild-type and MLL-ENL binding at a target gene. MLL-ENL is fused to the ligand-binding domain of the estrogen receptor (not shown). On binding to a synthetic estrogen derivative 4-hydroxy-tamoxifen (4-OHT), MLL-ENL is released and activates its downstream targets by binding to specific genomic regions (top panel). In the absence of 4-hydroxy-tamoxifen (no 4-OHT), MLL-ENL is retained in a complex with heat shock protein (not shown). Only MLL wild-type protein is bound at the target gene locus (bottom panel). (B) MLL binding intensity under 4-OHT and no 4-OHT conditions were plotted for each of 16 644 genes tiled on the array. Dark gray and black dots represent 2513 wild-type MLL-bound genes. Black dots above and below the diagonal line represent MLL-bound genes with increased binding signal under 4-OHT (223 genes) or no 4-OHT (8 genes) conditions, respectively. (C) Venn diagram shows that ∼ 80% (176) of the 223 MLL differentially binding genes overlap with wild-type MLL target genes.
The majority of the MLL-ENL target genes are a subset of MLL wild-type targets. (A) A cartoon of the inducible system shows MLL wild-type and MLL-ENL binding at a target gene. MLL-ENL is fused to the ligand-binding domain of the estrogen receptor (not shown). On binding to a synthetic estrogen derivative 4-hydroxy-tamoxifen (4-OHT), MLL-ENL is released and activates its downstream targets by binding to specific genomic regions (top panel). In the absence of 4-hydroxy-tamoxifen (no 4-OHT), MLL-ENL is retained in a complex with heat shock protein (not shown). Only MLL wild-type protein is bound at the target gene locus (bottom panel). (B) MLL binding intensity under 4-OHT and no 4-OHT conditions were plotted for each of 16 644 genes tiled on the array. Dark gray and black dots represent 2513 wild-type MLL-bound genes. Black dots above and below the diagonal line represent MLL-bound genes with increased binding signal under 4-OHT (223 genes) or no 4-OHT (8 genes) conditions, respectively. (C) Venn diagram shows that ∼ 80% (176) of the 223 MLL differentially binding genes overlap with wild-type MLL target genes.
To determine the target genes bound by the wild-type MLL protein, we performed genome-wide location analysis in these cells with MLL-ENL inactivated. As our binding analysis of entire gene regions showed that the most prominent MLL enrichment is at the promoter and 5′ coding region (Figures 2A, 3), we designed a custom array containing (−1, +3.5) kb of the TSS, to capture binding information representing each of the 16 644 NCBI reference genes (see supplemental Methods). This analysis revealed 2513 MLL wild-type–bound genes, corresponding to 15% of 16 644 genes tiled on the array. Eighty-three percent (2077) of the MLL-bound genes had detectable mRNA transcripts (supplemental Figure 6A). The level of MLL binding positively correlated with the level of gene expression for the overlapping set of 2077 genes (Pearson correlation r = 0.14, P = .000000000073, supplemental Figure 6B). Interestingly, the 2077 MLL-bound genes only account for 26% of expressed genes. These data are consistent with an earlier report that wild-type MLL is only targeted to a subset of actively expressed genes.25
To determine the set of genes that are specifically bound by the MLL fusion protein, we searched for gene regions that exhibit a significant higher level of MLL signal in the presence of the inducer 4-hydroxy-tamoxifen (4-OHT; Figure 4A). While MLL protein enrichment remained unchanged for most genes in the genome, 223 genes (Figure 4B) showed a pattern of binding increase with activation of MLL-ENL (4-OHT). Nearly 80% (176) of those 223 MLL-ENL bound genes are a subset of MLL wild-type target genes (Figure 4C). We compared the 223 genes with the 24 fusion target genes (supplemental Figure 3) identified in the human ML-2 cell line, 10 genes were shared between this 2 sets. We refer to this set of 223 genes as “223 MLL-ENL target genes” (supplemental Table 2). In contrast, only 8 genes (Figure 4B) had a slightly higher level of protein binding at the MLL-ENL–inactivated condition (no 4-OHT). These data are consistent with an inducible system in which a tightly controlled MLL fusion protein can be released to act on downstream genes.
The 223 MLL-ENL target genes have aberrant H3K79 methylation and are highly expressed in primary human leukemic cells
A key mechanism in MLL-ENL–associated gene activation is stimulation of H3K79 methylation.7,8 We then examined how this histone mark was affected by MLL-ENL. In general, the level of MLL occupancy is positively correlated with the degree of H3K79 methylation (supplemental Figure 7). Surprisingly, H3K79 methylation at most of the 16 644 gene loci tiled on the array was largely unaffected by the induction of the MLL fusion protein (Figure 5A). We identified 40 genes (K79High_4HT) which have significantly higher H3K79 methylation level in the MLL-ENL–induced condition, and 20 genes (K79High_no4HT) with increased levels of this histone mark when MLL-ENL was inactivated (FDR < 0.25; Figure 5A). Sixty percent (24 of 40) of the K79High_4HT genes overlap with the 223 MLL-ENL–bound genes (Figure 5B), while none of the 20 K79High_no4HT genes are bound by MLL-ENL. These data suggest that the observed H3K79 changes at K79High_4HT gene loci are likely to be under direct control of the MLL-ENL protein. Furthermore, as MLL-ENL has been shown to promote H3K79 methylation, we reasoned that MLL fusion-bound genes would have a strong response in H3K79 methylation on MLL-ENL activation. Indeed, increased MLL binding is strongly associated with a higher level of H3K79me2 difference at the genomic regions of 223 MLL-bound genes (Pearson correlation r = 0.69, P < .00000000000000022). In contrast, MLL wild-type–bound genes show a much weaker response in H3K79 methylation (Pearson correlation r = 0.27, P < .00000000000000022; Figure 5C). These observations likely reflect the intrinsic property of MLL fusion proteins in promoting H3K79 methylation at its target gene loci.
A subset of MLL-ENL–bound genes exhibits aberrant H3K79 methylation and differential gene expression. (A) Identification of genes with differential methylation level of H3K79 in MLL-ENL activated and inactivated conditions. The level of H3K79 methylation under 4-OHT and no 4-OHT conditions is plotted for each of 16 644 genes tiled on the array. Blue dots represent 2568 H3K79 target genes identified in the MLL-ENL inactivated condition. Green dots indicate the subset of H3K79 target genes that show differential methylation level of this histone modification (FDR < 0.25). Green dots above the diagonal line represent the 40 genes with significantly higher level of H3K79 methylation in the MLL-ENL–induced condition (FDR < 0.25) that are referred to as H3K79High_4HT genes. The green dots below the diagonal line represent the 20 genes with significantly increased levels of H3K79 methylation when MLL-ENL was inactivated (FDR < 0.25), that are referred to as H3K79High_no4HT genes. (B) The set of 223 MLL-ENL–bound genes (Figure 4B) is compared with differentially up-regulated genes (panel D) and genes with higher level of H3K79 methylation (Figure 5A) in the MLL-ENL–induced condition. The shaded area indicates 12 MLL-ENL–bound genes with differential expression. (C) Correlation between MLL differential binding and differential H3K79 methylation. For each MLL target gene, the difference in MLL binding strength and the differences in the level of H3K79 methylation between 4-OHT and no 4-OHT conditions were calculated. Red dots represent 223 MLL-ENL target genes (Figure 4B), with the red line showing the regression line of MLL binding difference to methylation difference for 223 MLL-ENL target genes (Pearson correlation, r = 0.69). Green dots are 2513 MLL wild-type–bound genes eliminating 223 MLL-ENL target genes, with the green line showing the regression line (Pearson correlation, r = 0.27). (D) Identification of 12 MLL fusion-regulated genes. Average gene expression is plotted against the difference in gene expression between MLL-ENL induced and inactivated conditions. All 7858 expressed genes detected by Affymetrix Mouse Exon 1.0 ST Array are displayed. Blue dots represent 124 differentially expressed genes (67 up-regulated, 57 down-regulated, FDR < 0.05) on induction of MLL-ENL. The set of 223 MLL fusion-bound genes, shown as the red dots, are overlaid with 124 differentially expressed genes, and the overlapped blue and red dots indicate 12 MLL fusion target genes exhibiting significant increase in mRNA expression in the presence of MLL-ENL.
A subset of MLL-ENL–bound genes exhibits aberrant H3K79 methylation and differential gene expression. (A) Identification of genes with differential methylation level of H3K79 in MLL-ENL activated and inactivated conditions. The level of H3K79 methylation under 4-OHT and no 4-OHT conditions is plotted for each of 16 644 genes tiled on the array. Blue dots represent 2568 H3K79 target genes identified in the MLL-ENL inactivated condition. Green dots indicate the subset of H3K79 target genes that show differential methylation level of this histone modification (FDR < 0.25). Green dots above the diagonal line represent the 40 genes with significantly higher level of H3K79 methylation in the MLL-ENL–induced condition (FDR < 0.25) that are referred to as H3K79High_4HT genes. The green dots below the diagonal line represent the 20 genes with significantly increased levels of H3K79 methylation when MLL-ENL was inactivated (FDR < 0.25), that are referred to as H3K79High_no4HT genes. (B) The set of 223 MLL-ENL–bound genes (Figure 4B) is compared with differentially up-regulated genes (panel D) and genes with higher level of H3K79 methylation (Figure 5A) in the MLL-ENL–induced condition. The shaded area indicates 12 MLL-ENL–bound genes with differential expression. (C) Correlation between MLL differential binding and differential H3K79 methylation. For each MLL target gene, the difference in MLL binding strength and the differences in the level of H3K79 methylation between 4-OHT and no 4-OHT conditions were calculated. Red dots represent 223 MLL-ENL target genes (Figure 4B), with the red line showing the regression line of MLL binding difference to methylation difference for 223 MLL-ENL target genes (Pearson correlation, r = 0.69). Green dots are 2513 MLL wild-type–bound genes eliminating 223 MLL-ENL target genes, with the green line showing the regression line (Pearson correlation, r = 0.27). (D) Identification of 12 MLL fusion-regulated genes. Average gene expression is plotted against the difference in gene expression between MLL-ENL induced and inactivated conditions. All 7858 expressed genes detected by Affymetrix Mouse Exon 1.0 ST Array are displayed. Blue dots represent 124 differentially expressed genes (67 up-regulated, 57 down-regulated, FDR < 0.05) on induction of MLL-ENL. The set of 223 MLL fusion-bound genes, shown as the red dots, are overlaid with 124 differentially expressed genes, and the overlapped blue and red dots indicate 12 MLL fusion target genes exhibiting significant increase in mRNA expression in the presence of MLL-ENL.
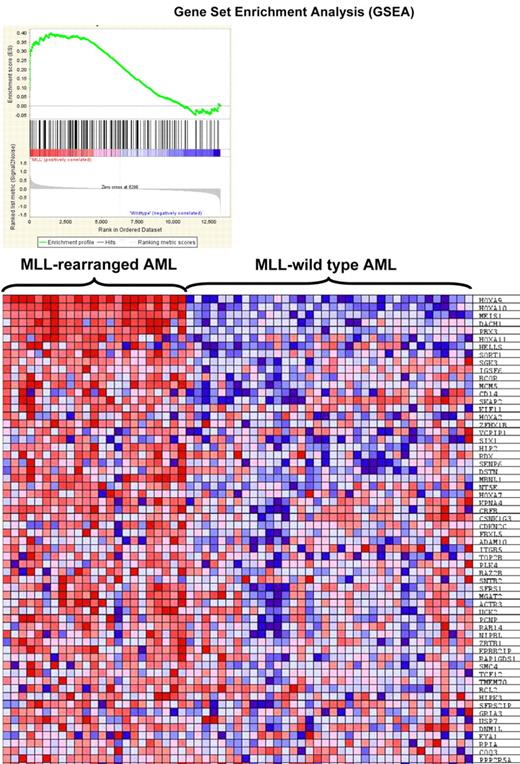
To determine whether identified MLL fusion binding genes are important in MLL-associated leukemia, we wondered whether the 223 fusion target genes are overexpressed in leukemia patients using a publicly available data consisting primary AML samples with or without MLL rearrangement.26 We found significant enrichment of the set of 223 genes in the expression signature unique for MLL-rearranged AML (ES = 0.4, P = .006, Figure 6).
Gene set enrichment analysis. Gene set enrichment analysis (GSEA) of gene expression in human MLL-rearranged AML (n = 23) compared with MLL–wild-type AML (n = 56; Ross et al26 ) using 223 MLL fusion protein-bound genes. (Top) GSEA enrichment plot, ES = 0.4, P = .006. (Bottom) The top 60 genes showing increased expression in human MLL-rearranged leukemias.
Gene set enrichment analysis. Gene set enrichment analysis (GSEA) of gene expression in human MLL-rearranged AML (n = 23) compared with MLL–wild-type AML (n = 56; Ross et al26 ) using 223 MLL fusion protein-bound genes. (Top) GSEA enrichment plot, ES = 0.4, P = .006. (Bottom) The top 60 genes showing increased expression in human MLL-rearranged leukemias.
Only a small percent of the 223 MLL-ENL–bound genes are susceptible to a change in mRNA expression
Having showed that MLL fusion-bound genes are expressed at a higher level as a whole in patient samples (Figure 6), we were interested in identifying the specific subset of genes whose expression is strongly influenced by the presence of MLL fusion proteins. Among 16 557 genes on the Affymetrix Mouse Exon 1.0 ST Array, 7858 genes are expressed as indicated by detectable mRNA transcripts (see supplemental Methods). We first compared the expression profiles between MLL-ENL–activated and –inactivated conditions. While the expression for most of the 16 557 genes on the array remained unchanged (Figure 5D gray dots), we identified 124 differentially expressed genes (67 up-regulated and 57 down-regulated genes, FDR < 0.05) on induction of MLL-ENL (Figure 5D blue dots). We intersected the set of 223 “MLL fusion-bound” genes (Figure 4B) with 124 differential expressed genes (Figure 5D). Only 12 of the 223 fusion target genes exhibit significant changes in mRNA expression in the presence of MLL-ENL (Figure 5D overlapped blue and red dots). These include Meis1, Hoxa9, Hoxa10, Hoxa11, Six1, Six4, Eya1, Cdkn2c, Hpgd, Gria3, Fut8, and 9630013D21Rik. They are referred to as “12 MLL fusion-regulated” genes. Nine of these 12 genes also show a significant increase in H3K79 methylation on activation of the MLL fusion (Meis1, Hoxa9, Hoxa10, Six1, Six4, Eya1, Cdkn2c, Fut8, and 9630013D21Rik; Figure 5B). These genes include key regulators in transcriptional regulation, cellular differentiation and cell-cycle control, as well as Hoxa9 and Meis1, which are known to be essential for the development of MLL leukemia.
As all 12 genes exhibit a significant increase in both MLL fusion protein binding and mRNA expression on MLL-ENL activation in this inducible cellular model, we hypothesized that they would be up-regulated in MLL-associated leukemia. These 12 MLL fusion-regulated genes are significantly enriched in the expression signature defined by human MLL-rearranged leukemia (ES = 0.81, P = .01, data not shown).26 When we extracted published mRNA expression data from a mouse MLL leukemia model, 7 of 12 MLL fusion-regulated genes are significantly overexpressed in MLL-ENL leukemia, 9 of 12 in MLL-AF1p leukemia, 8 of 12 in MLL-AF10 leukemia, and 6 of 12 in MLL-AF9 leukemia (supplemental Table 3).27 These data indicate that the 12 fusion-regulated genes identified in the MLL-ENL–inducible system are important for disease development in vivo.
EYA1 immortalizes hematopoietic progenitor cells and cotransduction with Six1 potentiates the transforming capacity of EYA1
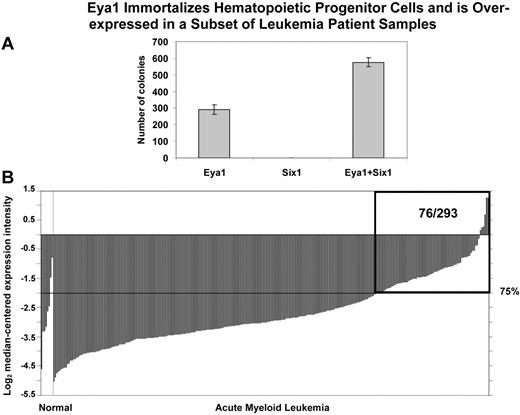
Two of the transcription factor genes identified in our analysis, EYA1 and SIX1, form a heterodimeric complex involved in the regulation of developmental pathways. To assess the potential of EYA1 and SIX1 to immortalize hematopoietic progenitor cells (HPCs), we transduced primary mouse HPCs with retroviruses encoding EYA1, SIX1, or MSCV vector. Murine HPCs were obtained from 5-FU–pretreated mice and “spinoculated” with retroviral supernatants containing either Eya1, Six1, Eya1, combined with Six1, and the MSCV vector alone as a control. The HPCs transduced with the MSCV vector alone did not become immortalized. After serial passage in methylcellulose, hematopoietic cells transduced with Eya1 produced tertiary colonies (Figure 7A), indicating that EYA1 has the potential to transform primary mouse BM cells. In contrast, transduction of HPCs with Six1 did not result in tertiary colony formation. However, cotransduction of HPCs with both Eya1 and Six1 resulted in an increase in tertiary colony formation compared with cells transduced with Eya1 alone (Figure 7A), indicating that SIX1 potentiates the transforming capacity of EYA1.
Eya1 immortalizes hematopoietic progenitor cells and is over-expressed in a subset of leukemia patient samples. (A) In vitro colony-forming assays. Primary mouse hematopoietic progenitor cells (HPCs) were transduced with retroviruses encoding Eya1, Six1, and both Eya1 and Six1. Numbers of colonies per dish at the third round of plating are shown (mean ± SD). After serial passage in methylcellulose, HPCs transduced with Eya1 produced tertiary colonies. Although transduction of Six1 by itself was not sufficient to immortalize HPCs, cotransduction with Eya1 resulted in increased tertiary colony formation compared with HPCs transduced with Eya1 alone. (B) Cancer outlier profile analysis (COPA) revealed EYA1 as a gene with outlier expression profile at the 75th percentile in the Valk et al acute myeloid leukemia dataset (n = 293).34 EYA1 expression is shown from all profiled samples in this dataset. The microarray data indicate that EYA1 is highly over-expressed in a subset of AML samples (76/293). The boxed region indicates 76 AML samples with overexpression of EYA1 among 293 patients examined. Visualization tools incorporated in Oncomine were used to generate graphical displays.28 “Normal” group includes 3 CD34+ selected and 5 unselected marrow samples; “Acute Myeloid Leukemia” group includes 285 samples.
Eya1 immortalizes hematopoietic progenitor cells and is over-expressed in a subset of leukemia patient samples. (A) In vitro colony-forming assays. Primary mouse hematopoietic progenitor cells (HPCs) were transduced with retroviruses encoding Eya1, Six1, and both Eya1 and Six1. Numbers of colonies per dish at the third round of plating are shown (mean ± SD). After serial passage in methylcellulose, HPCs transduced with Eya1 produced tertiary colonies. Although transduction of Six1 by itself was not sufficient to immortalize HPCs, cotransduction with Eya1 resulted in increased tertiary colony formation compared with HPCs transduced with Eya1 alone. (B) Cancer outlier profile analysis (COPA) revealed EYA1 as a gene with outlier expression profile at the 75th percentile in the Valk et al acute myeloid leukemia dataset (n = 293).34 EYA1 expression is shown from all profiled samples in this dataset. The microarray data indicate that EYA1 is highly over-expressed in a subset of AML samples (76/293). The boxed region indicates 76 AML samples with overexpression of EYA1 among 293 patients examined. Visualization tools incorporated in Oncomine were used to generate graphical displays.28 “Normal” group includes 3 CD34+ selected and 5 unselected marrow samples; “Acute Myeloid Leukemia” group includes 285 samples.
EYA1 and SIX1 are overexpressed in cases of human acute leukemia
To determine whether EYA1 and SIX1 are relevant to human leukemogenesis, we searched the Oncomine expression database and found that EYA1 is expressed at elevated levels in 76 of 293 leukemia samples as detected by Cancer Outlier Profile Analysis (COPA; Figure 7B, supplemental Figure 8).28 In particular, 6 of 17 human AML cases with 11q23 translocations exhibited overexpression of EYA1. We found that EYA1 overexpression is more frequently observed than that of SIX1 in hematologic malignancies, with COPA analysis demonstrating that SIX1 has a significant outlier profile in 7 leukemia datasets, whereas EYA1 exhibited a significant outlier profile in 14 leukemia datasets (supplemental Figure 8). Consistent with the finding that Eya1, but not Six1 alone, has the potential to transform murine HSCs, Eya1 is overexpressed in various mouse models of MLL-rearranged leukemia compared with normal BM, while Six1 up-regulation was not consistently observed (supplemental Table 3).
Discussion
MLL fusion target genes are a small set of that bound by MLL wild-type proteins
Chromosomal translocations involving the MLL gene represent a critical event in the initiation of a subset of acute leukemia. MLL fusion proteins, which are formed as a result of these translocations, contribute to leukemogenesis by defining an aberrant transcription program. It is not well understood, however, to which portion of the genome the MLL fusion proteins are specifically targeted. More importantly, it is not clear what changes in the massive dysregulation of gene expression and histone modification are under the direct control of MLL fusion proteins. Through integrative analyses of MLL protein binding, H3K79 methylation, and expression profiling, we demonstrate that MLL fusion proteins preferentially bind to a small portion of the leukemic genome in multiple patient-derived MLL leukemic cell lines and also in an inducible MLL-ENL cellular model. At these targeted loci, oncogenic MLL fusion proteins exhibit higher levels of protein enrichment with an extended distribution pattern across large genomic intervals. Surprisingly, only a small percent (5.5%) of MLL fusion-bound genes are dependent on the oncogenic protein for their increased levels of gene expression, consistent with a recent report which showed that only 3% of wild-type MLL target genes (as indicated by the presence of H3K4me3 peaks) display a reduction in gene expression on loss of Mll1.29
We examined leukemic cellular models expressing different MLL fusion proteins, including MLL-AF6 (ML-2 cells), MLL-AF9 (THP-1 cells), MLL-AF4 (MV4;11 cells), and MLL-ENL (inducible cell line). Together with AF10 and ELL, these frequent fusion partners account for > 85% of all MLL-positive leukemias.6 As controls, we used HL60 and U937 cells. HOXA cluster genes are up-regulated in U937 but not in HL60. ML-2 cells carry 2 alleles of MLL-AF6, and do not express the wild-type MLL protein. Thus, the ML-2 cell line serves as a unique model to explore the protein binding pattern solely contributed by MLL fusions. However, we cannot exclude the possibility that MLL protein binding in ML-2 cells might differ from that observed in most MLL-rearranged leukemia cells, which typically express both the fusion and wild-type proteins. MLL fusion protein binding behavior may also be affected by the fact that AF6 acts as a dimerization domain while other major fusion partners are nuclear elongation factors (AF4, AF9, ELL). However, expression signatures are very similar among primary leukemic samples with different MLL fusions.23 Specifically, MLL fusions to AF1p or AF6, which contain dimerization domains, show an indistinguishable expression pattern from those partners involving components of elongation factor complexes as fusion partners, suggesting that different fusion partners exert a similar influence on target gene selection of the MLL fusion protein.23-27 Consistent with this notion, MLL-AF6 targets are a small subset of those bound by MLL wild-type and fusion protein (MLL-AF4 or MLL-AF9) in human patient-derived leukemic cell lines (Figure 2B, supplemental Figure 3). Similarly, MLL-ENL–bound genes account for ∼ 7% of MLL wild-type targets in the mouse inducible system (Figure 4C). Results from these 2 independent biologic systems support our conclusion that MLL fusions are targeted to a small set of genes bound by wild-type MLL proteins.
Key target genes in MLL-rearranged leukemia
Among 223 fusion-bound genes, we further identified a core group of 12 genes which express at a significantly high level in the presence of MLL-ENL and appear to exhibit a strong correlation between a change in MLL-ENL binding and a change in gene expression. Interestingly, this short list of genes includes several transcription factors. This includes Hoxa9, Hoxa10, and Meis1, which are known critical targets in MLL-rearranged leukemia.13,30 Our analysis also identifies Eya1, Six1, and Six4 as novel direct target genes of MLL-ENL (Figure 7), which have not previously been shown to be involved in MLL-rearranged leukemia. Eya1, Six1, and Six4 belong to the retinal determination gene network based on their requirement for Drosophila eye development. EYA and SIX family members form a heterodimeric complex that plays a critical role in various developmental processes.31 EYA proteins contain transactivation and phosphatase domains whereas DNA binding is mediated by SIX family members.
We found that EYA1 and/or SIX1 are overexpressed in a subset of leukemia patients (Figure 7B, supplemental Figure 8) using COPA (Cancer Outlier Profile Analysis) analysis.32,33 Review of the Oncomine database showed that 6 of 17 human AML cases with 11q23 translocations exhibited increased expression of EYA1. EYA1 is also up-regulated in some cases of other subgroups of acute leukemia including CBFβ-MYH11 and PML-RARα.34 In a recent murine leukemia model of NUP98-PHD fusions, the 2 genes that exhibited the highest level of up-regulation in hematopoietic stem/progenitor cells were Hoxa9 and Eya1.35 Review of gene expression data from mouse MLL-fusion leukemia models, shows that Eya1 is expressed at a significantly increased level compared with normal BM.27 In human T-cell prolymphocytic leukemia, SIX1 is found to be significantly overexpressed in comparison to normal T cells.36 A role for these genes in carcinogenesis is also supported by previous reports which showed overexpression of both SIX1 and EYA1 in cancers of the lung, breast, prostate, cervix, and kidney (Rhodes et al28 and supplemental Figure 9). Similarly, overexpression of HOXA9 and MEIS1 is not limited to MLL-rearranged leukemias. Together, these data suggest that EYA1 and SIX1 may be aberrantly activated by distinct mechanisms in MLL-rearranged and nonrearranged leukemia, as well as in solid tumors.
MLL fusion proteins bind directly to target genes that form 2 sets of heterodimeric transcription factor complexes: HOXA9/MEIS1 and EYA1/SIX1. HOXA9 transforms hematopoietic cells in vitro, and transduction of murine hematopoietic cells with HOXA9 results in the development of acute myeloid leukemia in vivo. Although enforced expression of MEIS1 by itself is not sufficient to induce the development of leukemia, coexpression with HOXA9 accelerates the development of leukemia compared with HOXA9 alone.37 We now demonstrate the capacity of EYA1 to transform hematopoietic progenitor cells in vitro. Similar to MEIS1, transduction of SIX1 alone does not immortalize hematopoietic cells. However, in cells cotransduced with both genes, SIX1 collaborates with EYA1 to increase tertiary colony formation. However, we cannot exclude the possibility that SIX1 might immortalize HPCs under different assay conditions (eg, method of selection of HPCs or cytokines for in vitro culture), or might transform cells of different hematopoietic lineages. We also cannot exclude the possibility that a certain level of SIX1 expression is essential to the transforming properties of EYA1 or that other SIX family members might provide functional redundancy in the formation of a heterodimeric complex with EYA1. Taken together, these studies demonstrate that the leukemic properties of MLL fusion proteins are not restricted to the activation of HOXA cluster genes and MEIS1. These data are consistent with recent reports which showed that the MLL fusion protein, but not HOXA9/MEIS1, can fully transform committed progenitor cells.38,39 Activation of an additional set of genes, including those involved in the Wnt/β-catenin pathway, is required to induce leukemia.40 These findings, together with our results, have important therapeutic implications as strategies focused on targeting HOXA9/MEIS1 might not be sufficient to inhibit the aberrant transcriptional program mediated by MLL fusions.
Target gene selection by MLL fusions
Our data suggest that MLL fusion proteins directly activate a subset of genes that are regulated by MLL wild-type proteins. These findings are consistent with a recent report that AML1-ETO preferentially regulates selected AML1 target genes.41 It remains unclear; however, what is the underlying mechanism that leads to the observed differences in target gene selection between MLL wild-type and fusion proteins. Wild-type MLL is a 3969 amino acid protein that contains multiple domains involved in DNA binding and recognition of histone marks.42 The N-terminus of MLL includes AT hook, a minor groove DNA-binding motif,43 and the CxxC domain that can specifically bind to unmethylated cytosine guanine dinucleotides.44,45 The plant homeodomain (PHD) finger motifs, lost in the MLL fusion proteins, are involved in the recruitment of MLL to H3K4 methylated regions.11,46 Specific recruitment of MLL to the HOXA9 locus is mediated by the CxxC and PHD domain through direct interactions with the polymerase-associated factor (PAF) elongation complex and H3K4 methylation.47,48 Interestingly, MLL1 fusion proteins, which lack the PHD fingers, require prebinding of a wild-type MLL1 complex and CxxC domain for stable HOXA9 association.47 Furthermore, evidence has emerged for an important role of sequence-specific transcription factors in the recruitment of MLL to specific genomic loci.11 In this regard, it is worth noting that the carboxyl-terminal transactivating domain (TAD), only present in the wild-type MLL protein, is involved in the interaction with transcription factor c-Myb.49,50 These structural and functional studies shed light on the overlapping yet distinct mechanisms in targeting MLL wild-type and MLL fusion proteins in the genome. Loss of the PHD fingers and the carboxyl-terminal TAD and SET domains in MLL fusion proteins coupled with the acquisition of domains within the partner proteins that recruit transcription elongation complexes may explain, at least in part, the more restricted distribution of the MLL fusion protein observed in our study.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Robert K. Slany (University Erlangen) for providing the MLL-ENL–inducible cell line, Dr Heide Ford (University of Colorado) for sharing the Eya1 and Six1 cDNA, and Dr Qianben Wang (Ohio State University) for his help with the ChIP experiments.
This research was supported by National Institutes of Health grant CA105049 (M.J.T.), a Leukemia & Lymphoma Society SCOR grant (M.J.T.), the family of Robert A. Chapski, and by the Young Investigator Award from Cancer Research Foundation (Q.-f.W.). This work was also supported by 100 Talents Program from Chinese Academy of Sciences (to Q.-f.W.) and the National Natural Science Foundation of China (grant 81070442 [Q.-f.W.], grant 31071140 [S.M.], and grant 81000220 [F.H.]).
National Institutes of Health
Authorship
Contribution: Q.-f.W. designed research, performed most of the experiments, contributed to data analysis, and wrote the paper; G.W. analyzed data; S.M. performed PCR experiments, and contributed to data analysis and manuscript preparation; F.H. and J.W. analyzed the data and prepared figures and tables; J.D. and R.T.L. performed hematopoietic immortalization assay; R.M. and J.J.K. performed DNA cloning and plasmid preparation; S.P. contributed to the design of custom array for ChIP-chip analysis; H.J. helped study design and supervised data analysis; and M.J.T. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael J. Thirman, MD, Section of Hematology/Oncology, University of Chicago, 5841 South Maryland Ave, MC2115, Chicago, IL 60637; e-mail: mthirman@medicine.bsd.uchicago.edu.