The Lunenburg Lymphoma Biomarker Consortium (LLBC) evaluated the prognostic value of IHC biomarkers in a large series of patients with diffuse large B-cell lymphoma (DLBCL). Clinical data and tumor samples were retrieved from 12 studies from Europe and North America, with patients treated before or after the rituximab era. Using tissue microarrays from 1514 patients, IHC for BCL2, BCL6, CD5, CD10, MUM1, Ki67, and HLA-DR was performed and scored according to previously validated protocols. Optimal cut points predicting overall survival of patients treated in the rituximab era could only be determined for CD5 (P = .003) and Ki67 (P = .02), whereas such cut points for BCL2, BCL6, HLA-DR, and MUM1 could only be defined in patients not receiving rituximab. A prognostic model for patients treated in the rituximab era identified 4 risk groups using BCL2, Ki67, and International Prognostic Index (IPI) with improved discrimination of low-risk patients. Newly recognized correlations between specific biomarkers and IPI highlight the importance of carefully controlling for clinical and biologic factors in prognostic models. These data demonstrate that the IPI remains the best available index in patients with DLBCL treated with rituximab and chemotherapy.
Introduction
The prediction of outcome in patients with lymphoma is an important challenge facing clinicians. After the description of various clinical prognostic factors, decisive progress was made > 15 years ago with the establishment of the International Prognostic Index (IPI) for diffuse large B-cell lymphoma (DLBCL), the result of a large, international cooperative effort.1 However, it was clearly assumed that the variables used to build this index were surrogates of DLBCL biology. This heterogeneity was already appreciated through various morphologic, immunologic, and cytogenetic features, resulting in numerous studies attempting to assess the prognostic value of single biomarkers.2 More recently, gene-expression profiling data revealed a greater level of molecular complexity in DLBCL, with the identification of specific gene-expression signatures such as germinal-center B-cell (GCB) or activated B-cell (ABC) signatures, which are associated with distinct genetic alterations and significantly different survival rates.3,–5 Given the difficulties in applying these molecular approaches in daily practice, an attempt was made to transfer this molecular classification into an IHC score.6 However, the prognostic value of this algorithm has provided very inconsistent results.7,,,,,–13 These discrepant data may be related to patient selection criteria and the limited sample sizes of some series, but also to variability in technical aspects such as antibodies, different scoring methods, criteria, and cut points. A validation study assessing the reproducibility of IHC marker scoring conducted by our consortium established that the staining and scoring methodologies should be normalized to obtain a reasonable level of confidence in interpreting these markers.14,15 Furthermore, it has been shown that the predictive value of markers may be dependent on treatment, especially with the use of anti-CD20 antibodies.16,17 Given these challenges, the Lunenburg Lymphoma Biomarker Consortium (LLBC) decided to launch a study based on a large series of patients with DLBCL uniformly treated with rituximab plus cyclophosphamide, hydroxydaunorubicin, oncovin, and prednisone/prednisolone (R-CHOP) within clinical studies. In this effort, controlled marker assessment as developed in the previous “validation study”14 was applied to determining the prognostic value of individual IHC markers or to explore a possible combined clinicobiologic index. Two additional series of CHOP-treated patients were also studied to provide further insight into potential biomarker cut points and interactions between biomarkers alone and with the IPI. The first CHOP patients were treated at the same time, because they represent the control arm of clinical trials comparing CHOP and R-CHOP, and therefore serve as the control for the R-CHOP series, which is biased toward an older population and higher-risk IPI. The second series was a larger cohort of CHOP-treated patients covering the complete spectrum of risk groups.
Methods
Patients
Prospective clinical studies organized by European and American collaborative groups or population-based reference centers were selected for this study based on (1) inclusion of patients with DLBCL treated with standard anthracycline based therapy, with at least 6 cycles of CHOP or CHOP-like chemotherapy with or without rituximab; (2) availability of complete clinical data at study registration and an updated follow-up of at least 3-years; and (3) the possibility of retrieving paraffin blocks from the diagnostic biopsy samples. Six of these studies—GELA 98-518 and 05-1 (NCT00135499), ECOG4494,19 BCCA,20 MINT,21 and HOVON-46 (NCT00028717)—performed between February 1998 and August 2005, included patients treated with R-CHOP in 2-armed studies with a control group treated with CHOP only or patients from registry data selected within 18 months of when rituximab was introduced (BCCA only). These cohorts are designated as rituximab-CHOP (r-CHOP) and control-CHOP (c-CHOP), respectively. The other studies (EORTC-20901, HOVON-25/26,22 GLHSG-B1/B2,23,24 and UK-Bart's & Manchester) included patients treated before the rituximab era, between August 1989 and February 2004; these patients are designated as earlier CHOP (e-CHOP) patients.
This study was approved by the ethical committees of all collaborating trial organizations and centers to comply with the Declaration of Helsinki. Data obtained from each corresponding group were electronically transferred to the LLBC statisticians and checked for data completeness and consistency with previous original publications of the clinical studies.
Sample collection, processing, IHC, and marker assessment
Tissue microarrays (TMAs) were constructed by each pathology reference center for all available samples using 0.6-mm core samples in triplicate, as described previously.14 Based on the results of the validation study, a policy of “one marker/one laboratory” was followed, implying that sections of all available TMAs for the complete series were stained for individual markers in one selected laboratory that demonstrated the most reliable results in the validation study. The same protocols and scoring criteria as in the validation study were used for CD20, CD5, CD10, BCL2, BCL6, HLA-DR, Ki67, and MUM1 IHC. Based on previously determined reproducibility levels, stained slides were circulated to be scored by 1 (CD20, CD10, and HLA-DR) or 3 (BCL2, BCL6, CD5, Ki67, and MUM1) of the participating pathologists.15 According to the LLBC validation study,14,15 2 score categories (negative vs positive) were used for CD20, CD10, and HLA-DR; 3 categories for BCL6 (no staining, weak/variable weak staining, and strong/variable strong staining) and CD5 (no staining, 1%-75% staining, and > 75% staining); 4 categories for Ki67 (< 25% staining, 26%-50% staining, 50%-75% staining, and > 75% staining); and 5 categories for BCL2 and MUM1 (0%-5% staining, 5%-25% staining, 26%-50% staining, 51%-75% staining, and > 75% staining). Scores for markers evaluated by 3 pathologists were determined using the following rules. When all 3 pathologists or 2 of 3 agreed, the score agreed upon was used. When no agreement was reached between the 3 pathologists, the median value was taken. If 2 pathologists agreed and 1 could not score a sample, these data on the patients from that TMA group were used to imput a value for this patient. With the imputed value, the final score for this patient was determined using the same rule. When 2 or 3 pathologists were unable to score a sample, this sample was considered as not scored. The adjusted algorithm to distinguish germinal center (GCB) versus nongerminal center (non-GCB) samples evaluated in this paper defines BCL6-positive as any staining and uses 25% as the cut point for MUM1. Although the original cut points used for this index were chosen arbitrarily and differ from the present one,6 based on our former validation study, agreement was significantly higher when the BCL6 (and CD10) descriptive categories of no staining, weak/variable weak staining, and strong/variable strong staining were used instead of percentages of patients scored. Therefore, this BCL6 scoring was implemented in this study.
Statistical methods
Univariate and multivariate analyses were used to assess the prognostic significance of biomarkers and the IPI for overall survival (OS), defined as time from treatment initiation to death or date of last follow-up. Correlations between biomarker and IPI were evaluated using the χ2 test adjusting for multiple comparisons using the Bonferroni method.
Because of the high percentage of cases with incomplete scores for all 8 markers (48%), classification and regression trees for survival outcomes (using the “rpart” function in R with 10-fold cross-validation) were implemented to determine prognostic models for the r-CHOP cohort using the IPI groups as well as the biomarkers. In this analysis each biomarker was included as originally scored. Models based on an optimal cut point identified from a univariate analysis were also evaluated. In the univariate analysis, the optimal cut point for each IHC biomarker was selected as the one with the maximum log-rank statistic to predict patient OS. Regression tree results are similar, with biomarkers included using the categories and the cut points.
The impact of the biomarkers and IPI on OS was also evaluated among the e-CHOP series, which included a broader cross-section of age and IPI groups. Given the large number of patients in the e-CHOP series, a test validation (2/3 to 1/3) design stratified by study group was implemented. Prognostic models were evaluated and applied to the c-CHOP and r-CHOP series of data.
The Cox proportional hazards (PH) regression model was used to estimate the relative risk for each of the prognostic groups. A global measure of fit, the Bayesian information criterion (BIC) was assessed, as well as a measure of discrimination, the area under the receiver operating characteristic curve for survival outcomes (the c-index).25,26 Low values of BIC indicate better fit and high values of c-index indicate better discrimination. A multiple imputation procedure (n = 5 datasets) was also used to evaluate the effect of missing scores. Similar results were obtained applying recursive partitioning and Cox PH regression to the multiple imputation datasets (results not shown). Analyses were performed using SAS Version 9 software and the R package Version 2.8.27
Results
Patient population and biomarker assessment
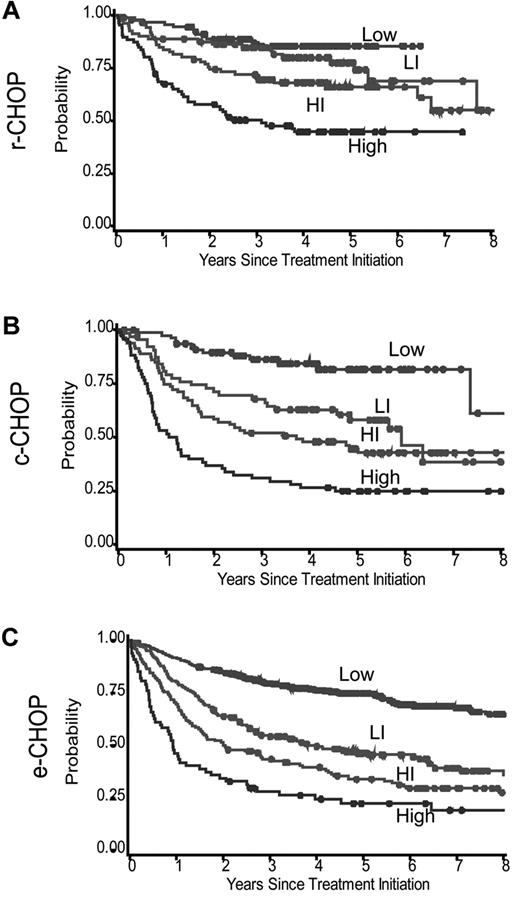
Clinical data were collected for 2451 patients with DLBCL from 12 studies (r-CHOP, 674 patients; c-CHOP, 620 patients; and e-CHOP, 1157 patients). Material for TMA was available for 1514 of these patients with 347 of 674 (52%) from r-CHOP, 289 of 620 (47%) from c-CHOP, and 878 of 1157 (76%) from e-CHOP. Individual IPI factors and IPI index were assessed for each cohort (not shown) and among those with available TMAs (Table 1). In these subsets, complete data for IPI were available for all c-CHOP and r-CHOP patients and for 845 of 878 (96%) e-CHOP patients. As a result of the inclusion criteria of the recent randomized R-CHOP versus CHOP clinical trials, a significantly higher percentage of patients were older than 60 years of age in the r-CHOP (n = 231, 67%) and c-CHOP (n = 211, 73%) cohorts compared with the e-CHOP cohort (n = 411, 47%; P < .001). Compared with those without TMA submission, patients with available TMA material were significantly younger (P < .001, 56% vs 69% > 60 years of age), had less-advanced disease (59% vs 72% stage III/IV), had fewer extranodal sites of involvement (20% vs 41% with >1 extranodal site), and were less likely to have elevated lactate dehydrogenase levels (47% vs 59% above normal value), resulting in a higher percentage of low-risk patients (41% vs 20%, P < .001). As expected, OS was significantly longer (P < .001) in the r-CHOP patients compared with the c-CHOP patients. In addition, highly significant differences in OS were detected by IPI risk groups (P < .0001 for all 3 cohorts; Table 1 and Figure 1).
Distribution of IPI and survival estimates in the 3 groups among the 1514 of 2451 patients for which blocks were available for construction of a TMA
| Analysis group . | All patients 4-year OS (95% CI) . | Age > 60 y No. of patients (%) . | IPI category low . | IPI category low/intermediate . | IPI category high/intermediate . | IPI category high . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of patients (%) . | 4-year OS (95% CI) . | No. of patients (%) . | 4-year OS (95% CI) . | No. of patients (%) . | 4-year OS (95% CI) . | No. of patients (%) . | 4-year OS (95% CI) . | |||
| r-CHOP (n = 347) | 69% (64,74%) | 231 (67%) | 94 (27%) | 85% (75, 91%) | 80 (23%) | 80% (69, 87%) | 87 (25%) | 68% (57, 77%) | 86 (25%) | 45% (34, 55%) |
| c-CHOP (n = 289) | 55% (49, 61%) | 211 (73%) | 79 (27%) | 84% (73, 91%) | 63 (22%) | 63% (50, 74%) | 79 (27%) | 48% (37, 58%) | 68 (24%) | 26% (17, 37%) |
| e-CHOP (n = 878)* | 59% (55, 62%) | 411 (47%) | 428 (50%) | 76% (72, 80%) | 208 (25%) | 49% (42, 56%) | 142 (17%) | 40% (31, 48%) | 67 (8%) | 24% (15, 35%) |
| Analysis group . | All patients 4-year OS (95% CI) . | Age > 60 y No. of patients (%) . | IPI category low . | IPI category low/intermediate . | IPI category high/intermediate . | IPI category high . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of patients (%) . | 4-year OS (95% CI) . | No. of patients (%) . | 4-year OS (95% CI) . | No. of patients (%) . | 4-year OS (95% CI) . | No. of patients (%) . | 4-year OS (95% CI) . | |||
| r-CHOP (n = 347) | 69% (64,74%) | 231 (67%) | 94 (27%) | 85% (75, 91%) | 80 (23%) | 80% (69, 87%) | 87 (25%) | 68% (57, 77%) | 86 (25%) | 45% (34, 55%) |
| c-CHOP (n = 289) | 55% (49, 61%) | 211 (73%) | 79 (27%) | 84% (73, 91%) | 63 (22%) | 63% (50, 74%) | 79 (27%) | 48% (37, 58%) | 68 (24%) | 26% (17, 37%) |
| e-CHOP (n = 878)* | 59% (55, 62%) | 411 (47%) | 428 (50%) | 76% (72, 80%) | 208 (25%) | 49% (42, 56%) | 142 (17%) | 40% (31, 48%) | 67 (8%) | 24% (15, 35%) |
Median follow-up was 4.4, 5.2, and 5.8 years among the r-CHOP, c-CHOP, and e-CHOP patients, respectively.
Thirty-three e-CHOP patients (4%) did not have IPI data.
OS of DLBCL patients. Data are from patients with TMA available according to the IPI for the 3 patient cohorts: r-CHOP 347 patients (panel 1a), c-CHOP 289 patients (panel 1b) and e-CHOP 878 patients (panel 1c). The log-rank P value was < .0001 for each cohort.
OS of DLBCL patients. Data are from patients with TMA available according to the IPI for the 3 patient cohorts: r-CHOP 347 patients (panel 1a), c-CHOP 289 patients (panel 1b) and e-CHOP 878 patients (panel 1c). The log-rank P value was < .0001 for each cohort.
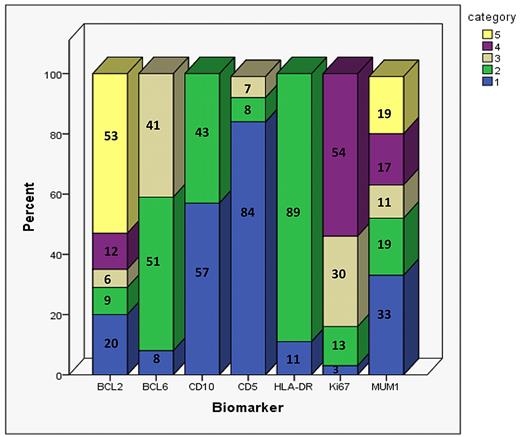
Of the 1514 patients, 793 (52%) had all 8 immunohistochemical markers scored, 320 (21%) patients had 7 of the 8 markers scored, and 137 (9%) had 6 of 8 markers scored. Markers could not be scored in individual cases for various reasons; for example, not representative or missing cores in the immunostained TMA section, inadequate staining results, or absence of an internal control staining precluding scoring as absence of staining in tumor cells. The proportion of patients whose TMA could be scored by each marker was as follows: 1306 (86%) for BCL2, 985 (65%) for BCL6, 1344 (89%) for CD10, 1366 (90%) for CD5, 1377 (91%) for HLA-DR, 1138 (75%) for Ki67, and 1249 (82%) for MUM1 (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). A detailed distribution of biomarkers scores in the r-CHOP cohort is provided in Figure 2.
Distribution of BCL2, BCL6, CD10, CD5, HLA-DR, Ki67, and MUM1 scoring results when using the previously validated methods in the 347 r-CHOP patients. Numbers on the bar graph correspond to the percentage of patients in each category. The categories are numbered from 1-5. For BCL2 and MUM1: 1 = 0%-5%, 2 = 5%-25%, 3 = 26%-50%, 4 = 51%-75%, and 5 = 76%-100%. For BCL6: 1 = no staining, 2 = weak/variable weak staining, and 3 = strong/variable strong staining. For CD10 and HLA-DR: 1 = negative and 2 = positive. For CD5: 1 = no staining, 2 = 1%-75% staining, and 3 = 76%-100% staining. For Ki67: 1 = 0%-25% staining, 2 = 26%-50% staining, 3 = 51%-75% staining, and 4 = 76%-100% staining. The number of r-CHOP patients scored for BCL2, BCL6, CD10, CD5, HLA-DR, Ki67, and MUM1 are 326, 212, 327, 333, 329, 239, and 293, respectively.
Distribution of BCL2, BCL6, CD10, CD5, HLA-DR, Ki67, and MUM1 scoring results when using the previously validated methods in the 347 r-CHOP patients. Numbers on the bar graph correspond to the percentage of patients in each category. The categories are numbered from 1-5. For BCL2 and MUM1: 1 = 0%-5%, 2 = 5%-25%, 3 = 26%-50%, 4 = 51%-75%, and 5 = 76%-100%. For BCL6: 1 = no staining, 2 = weak/variable weak staining, and 3 = strong/variable strong staining. For CD10 and HLA-DR: 1 = negative and 2 = positive. For CD5: 1 = no staining, 2 = 1%-75% staining, and 3 = 76%-100% staining. For Ki67: 1 = 0%-25% staining, 2 = 26%-50% staining, 3 = 51%-75% staining, and 4 = 76%-100% staining. The number of r-CHOP patients scored for BCL2, BCL6, CD10, CD5, HLA-DR, Ki67, and MUM1 are 326, 212, 327, 333, 329, 239, and 293, respectively.
Assessing the optimal cut point that predicted patient outcome for each biomarker
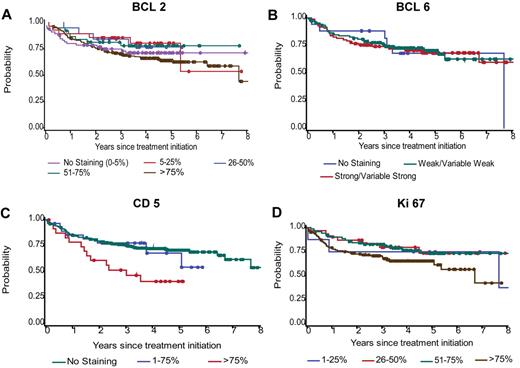
In a univariate analysis, we selected optimal cut points for OS based on the maximum log-rank statistic (Table 2). Among the r-CHOP patients, only CD5 (≤ 75% vs > 75%) and Ki67 (≤ 75% vs > 75%) could discriminate patient outcome (P < .05). A marginal difference (P = .09) was detected for BCL2 (≤ 75% vs > 75%) but not for BCL6 (Figure 3). Among the e-CHOP patients, which represent a broader range of age groups, the optimal cut points could be determined for BCL2 (≤ 75% vs > 75%), BCL6 (no staining vs any staining), CD5 (no staining vs any staining), HLA-DR (positive vs negative), and MUM1 (≤ 75% vs > 75%; Table 2). Several markers identified in the e-CHOP cohort retained their significance in the c-CHOP cohort (BCL2, BCL6, and marginally for MUM1). The optimal Ki67 cut point of ≤ 75% vs > 75% determined in the r-CHOP cohort was not found to be predictive in univariate analysis for the e-CHOP cohort (or for the c-CHOP cohort). No difference in OS was found for CD10 (positive vs negative; Table 2). Similar cut points were found using data obtained with imputation for missing scores (not shown).
Optimal cut points of IHC markers used to predict OS*
| Marker* . | Optimal cut point selected† . | r-CHOP . | e-CHOP . | c-CHOP . | |||
|---|---|---|---|---|---|---|---|
| HR (95% CI)‡ . | Log-rank P . | HR (95% CI)‡ . | Log-rank P . | HR (95% CI)‡ . | Log-rank P . | ||
| BCL2 | ≤ 75% vs > 75% | 1.4 (0.9, 2.2) | .09 | 1.7 (1.4, 2.1) | < .0001 | 1.6 (1.1, 2.3) | .02 |
| BCL6 | No staining vs staining | 1.1 (0.5, 2.7) | .76 | 1.7 (1.3, 2.3) | .0003 | 2.9 (1.7, 5.0) | < .0001 |
| CD5 | ≤ 75% vs > 75% | 2.4 (1.3, 4.3) | .003 | 1.6 (1.2, 2.2) | .004 | 1.0 (0.6, 1.9) | .9 |
| CD10 | Negative vs positive | 1.1 (0.8, 1.7) | .56 | 1.2 (0.9, 1.5) | .17 | 1.4 (1.0, 2.0) | .06 |
| HLA-DR | Negative vs positive | 1.3 (0.7, 2.4) | .36 | 1.6 (1.2, 2.1) | .001 | 1.3 (0.8, 1.9) | .33 |
| Ki67 | ≤ 75% vs > 75% | 1.8 (1.1, 2.9) | .02 | 0.8 (0.7, 1.0) | .11 | 1.0 (0.6, 1.5) | .86 |
| MUM1 | ≤ 75% vs > 75% | 1.4 (0.8, 2.3) | .22 | 1.3 (1.0, 1.7) | .03 | 1.5 (1.0, 2.4) | .06 |
| Marker* . | Optimal cut point selected† . | r-CHOP . | e-CHOP . | c-CHOP . | |||
|---|---|---|---|---|---|---|---|
| HR (95% CI)‡ . | Log-rank P . | HR (95% CI)‡ . | Log-rank P . | HR (95% CI)‡ . | Log-rank P . | ||
| BCL2 | ≤ 75% vs > 75% | 1.4 (0.9, 2.2) | .09 | 1.7 (1.4, 2.1) | < .0001 | 1.6 (1.1, 2.3) | .02 |
| BCL6 | No staining vs staining | 1.1 (0.5, 2.7) | .76 | 1.7 (1.3, 2.3) | .0003 | 2.9 (1.7, 5.0) | < .0001 |
| CD5 | ≤ 75% vs > 75% | 2.4 (1.3, 4.3) | .003 | 1.6 (1.2, 2.2) | .004 | 1.0 (0.6, 1.9) | .9 |
| CD10 | Negative vs positive | 1.1 (0.8, 1.7) | .56 | 1.2 (0.9, 1.5) | .17 | 1.4 (1.0, 2.0) | .06 |
| HLA-DR | Negative vs positive | 1.3 (0.7, 2.4) | .36 | 1.6 (1.2, 2.1) | .001 | 1.3 (0.8, 1.9) | .33 |
| Ki67 | ≤ 75% vs > 75% | 1.8 (1.1, 2.9) | .02 | 0.8 (0.7, 1.0) | .11 | 1.0 (0.6, 1.5) | .86 |
| MUM1 | ≤ 75% vs > 75% | 1.4 (0.8, 2.3) | .22 | 1.3 (1.0, 1.7) | .03 | 1.5 (1.0, 2.4) | .06 |
The number of the 1514 patients scored for BCL2, BCL6, CD5, CD10, HLA-dr, Ki67, MUM1, are 1306, 985, 1366, 1344, 1377, 1138, 1249, respectively
When optimal cut points were not determined using the r-CHOP series, the e-CHOP cut points are presented. Results obtained from applying the cut points to all cohorts are shown.
Reference groups: ≤ 75% staining for BCL2, CD5, MUM1, and Ki67; any staining for BCL6; positive for CD10 and HLA-DR.
OS by BCL2, BCL6, CD5, and Ki67 categories for the 347 r-CHOP patients. Categories are shown as originally scored. The number of r-CHOP patients scored for BCL2, BCL6, CD5, and Ki67 are 326, 212, 333, and 239, respectively.
OS by BCL2, BCL6, CD5, and Ki67 categories for the 347 r-CHOP patients. Categories are shown as originally scored. The number of r-CHOP patients scored for BCL2, BCL6, CD5, and Ki67 are 326, 212, 333, and 239, respectively.
The impact of combined markers that might define GCB versus non-GCB patients on OS was evaluated using an adjusted GCB/non-GCB algorithm (where BCL6 was dichotomized as no-staining/staining) and an optimized LLBC algorithm that uses optimal cut points determined from the univariate analysis (GCB/non-GCB LLBC with MUM1 dichotomized as ≤ 75% vs > 75% and BCL6 dichotomized as no-staining/staining).The adjusted GCB/non-GCB algorithm was not of prognostic value for c-CHOP or r-CHOP patients (hazard ratio [HR] = 1.3; 95% confidence interval [CI] 0.8-1.9 and HR = 1.1; CI 0.6-1.8, respectively, P > .25), but was of prognostic for the e-CHOP cohort (HR = 1.4; CI 1.1-1.7, P = .01; Figure 4). The GCB/non-GCB LLBC also lacked prognostic significance in the r-CHOP cohort (HR = 1.4; CI 0.8-2.6, P = .23 for r-CHOP), whereas it appeared to have a higher relative risk in the 2 CHOP patient cohorts (HR = 1.7; CI 1.3-2.2, P < .0001 for e-CHOP; HR = 1.8; CI 1.1-2.8, P = .01 for c-CHOP),
OS according to algorithms obtained by assessments of CD10, BCL6, and MUM1 scoring results. Algorithms were used to define an adjusted GCB/non-GCB index (with the MUM1 cut point defined as 25%) or the LLBC GCB/non-GCB index (with the MUM1 cut point defined as 75%) in the r-CHOP (A and D), c-CHOP (B and E), and e-CHOP (C and E), respectively. Among the 1046 patients scored using the adjusted GCB algorithm and with complete IPI data, 558 were CD10+ (GCB), 151 were CD10−/BCL6+/MUM1− (GCB), 242 were CD10−/BCL6+/MUM1+ (non-GCB), and 95 were CD10−/BCL6− (non-GCB). Among the 1046 patients scored using the LLBC GCB algorithm and with complete IPI data, 558 were CD10+ (GCB), 270 were CD10−/BCL6+/MUM1− (GCB), 123 were CD10−/BCL6+/MUM1+ (non-GCB), and 95 were CD10−/BCL6− (non-GCB).
OS according to algorithms obtained by assessments of CD10, BCL6, and MUM1 scoring results. Algorithms were used to define an adjusted GCB/non-GCB index (with the MUM1 cut point defined as 25%) or the LLBC GCB/non-GCB index (with the MUM1 cut point defined as 75%) in the r-CHOP (A and D), c-CHOP (B and E), and e-CHOP (C and E), respectively. Among the 1046 patients scored using the adjusted GCB algorithm and with complete IPI data, 558 were CD10+ (GCB), 151 were CD10−/BCL6+/MUM1− (GCB), 242 were CD10−/BCL6+/MUM1+ (non-GCB), and 95 were CD10−/BCL6− (non-GCB). Among the 1046 patients scored using the LLBC GCB algorithm and with complete IPI data, 558 were CD10+ (GCB), 270 were CD10−/BCL6+/MUM1− (GCB), 123 were CD10−/BCL6+/MUM1+ (non-GCB), and 95 were CD10−/BCL6− (non-GCB).
Pairwise correlation for prognostic biomarkers and the IPI categories
The large number of cases with available IHC data allowed assessment of the associations between the expression of each dichotomized biomarker and IPI category distribution using all 3 patient cohorts. As shown in Table 3, the percentage of patients expressing certain biomarkers differed between IPI categories. Patients with tumor cells that overexpressed BCL2 (> 75%) presented significantly more frequently with a higher IPI (P < .0001), whereas those that were either CD10 or HLA-DR positive had a significantly lower IPI at diagnosis (P = .008 and P = .04, respectively).
Biomarker expression and IPI category distribution in all patients with DLBCL
| Biomarker expression* . | No. of patients with biomarker scored and IPI† . | No. of patients with biomarker expression (%) . | Biomarker expression percentage by IPI categories . | Adjusted χ2P‡ . | |||
|---|---|---|---|---|---|---|---|
| Low . | Low/intermediate . | High/intermediate . | High . | ||||
| BCL2 > 75% | 1279 | 630 (49%) | 39 | 50 | 58 | 63 | < .0001 |
| BCL6 staining | 965 | 842 (87%) | 91 | 85 | 83 | 86 | NS |
| CD10 staining | 1316 | 558 (42%) | 49 | 43 | 35 | 37 | .008 |
| CD5 > 75% | 1339 | 115 (9%) | 7 | 10 | 9 | 10 | NS |
| HLA-DR staining | 1348 | 1180 (88%) | 91 | 89 | 84 | 82 | .04 |
| Ki67 > 75% | 1119 | 627 (56%) | 61 | 48 | 52 | 60 | .04 |
| MUM1 > 75% | 1225 | 264 (22%) | 20 | 20 | 25 | 26 | NS |
| GCB/non-GCB§ (adjusted) | 1046 | 709 (68%) | 72 | 67 | 61 | 66 | NS |
| GCB/non-GCB§ (LLBC) | 1046 | 828 (79%) | 83 | 79 | 72 | 79 | NS |
| Biomarker expression* . | No. of patients with biomarker scored and IPI† . | No. of patients with biomarker expression (%) . | Biomarker expression percentage by IPI categories . | Adjusted χ2P‡ . | |||
|---|---|---|---|---|---|---|---|
| Low . | Low/intermediate . | High/intermediate . | High . | ||||
| BCL2 > 75% | 1279 | 630 (49%) | 39 | 50 | 58 | 63 | < .0001 |
| BCL6 staining | 965 | 842 (87%) | 91 | 85 | 83 | 86 | NS |
| CD10 staining | 1316 | 558 (42%) | 49 | 43 | 35 | 37 | .008 |
| CD5 > 75% | 1339 | 115 (9%) | 7 | 10 | 9 | 10 | NS |
| HLA-DR staining | 1348 | 1180 (88%) | 91 | 89 | 84 | 82 | .04 |
| Ki67 > 75% | 1119 | 627 (56%) | 61 | 48 | 52 | 60 | .04 |
| MUM1 > 75% | 1225 | 264 (22%) | 20 | 20 | 25 | 26 | NS |
| GCB/non-GCB§ (adjusted) | 1046 | 709 (68%) | 72 | 67 | 61 | 66 | NS |
| GCB/non-GCB§ (LLBC) | 1046 | 828 (79%) | 83 | 79 | 72 | 79 | NS |
The number of the 1514 patients scored for BCL2, BCL6, CD5, CD10, HLA-DR, Ki67, MUM1, GCB (adjusted), and GCB (LLBC) are 1306, 985, 1366, 1344, 1377, 1138, 1249, 1067, and 1067, respectively.
The number of additional patients without IPI for BCL2, BCL6, CD5, CD10, HLA-DR, Ki67, MUM1, GCB (adjusted), and GCB (LLBC) are 27, 20, 18, 27, 29, 19, 24, 21, and 21, respectively.
χ2P values are adjusted for 9 multiple comparisons
Among the 1046 patients scored using the adjusted GCB algorithm and with complete IPI data, 558 were CD10+ (GCB), 151 were CD10−/BCL6+/MUM1-(GCB), 242 were CD10−/BCL6+/MUM1+ (non-GCB), and 95 were CD10−/BCL6− (non-GCB). Among the 1046 patients scored using the LLBC GCB algorithm and with complete IPI data, 558 were CD10+ (GCB), 270 were CD10−/BCL6+/MUM1− (GCB), 123 were CD10−/BCL6+/MUM1+ (non-GCB), and 95 were CD10−/BCL6− (non-GCB).
NS indicates nonsignificant.
Interestingly, pairs of markers also showed strong correlations. As expected, CD10 and BCL6 were correlated and CD10 and MUM1 inversely correlated. High BCL2 expression was also more frequently observed in patients who highly expressed MUM1. Expression of CD5 was also correlated with the high expression of BCL2 as well as lack of CD10. Finally, cases with high expression of MUM1 were found to be HLA-DR negative and had high expression of Ki67. (P < .002 for all; Table 4). These findings highlight the coordinated expression of biomarkers associated with the same impact on patient outcome and possibly a common biologic background.
Pairwise frequency of biomarkers
| . | Score . | BCL6 score . | CD10 score . | CD5 score . | HLA-DR score . | Ki67 score . | MUM1 score . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No staining . | Staining . | No staining . | Staining . | ≤75% . | >75% . | No staining . | Staining . | ≤75% . | >75% . | ≤75% . | >75% . | ||
| BCL2 | 0%-75% | 56 (12%) | 433 (88%) | 378 (59%) | 267 (41%) | 615 (95%)* | 33 (5%)* | 75 (12%) | 569 (88%) | 233 (42%) | 328 (58%) | 514 (85%)* | 89 (15%)* |
| > 75% | 58 (13%) | 381 (87%) | 351 (57%) | 265 (43%) | 554 (87%)* | 81 (13%)* | 86 (13%) | 553 (87%) | 246 (48%) | 267 (52%) | 410 (71%)* | 169 (29%)* | |
| BCL6 | No staining | 96 (82%)* | 21 (18%)* | 104 (87%) | 15 (13%) | 14 (11%) | 109 (89%) | 49 (44%) | 62 (56%) | 78 (68%) | 37 (32%) | ||
| Staining | 411 (49%)* | 422 (51%)* | 789 (93%) | 56 (7%) | 93 (11%) | 750 (89%) | 331 (42%) | 458 (58%) | 642 (79%) | 174 (21%) | |||
| CD10 | No staining | 669 (89%)* | 84 (11%)* | 109 (14%) | 643 (86%) | 298 (47%) | 332 (53%) | 503 (72%)* | 200 (28%)* | ||||
| Staining | 529 (95%)* | 29 (5%)* | 60 (11%) | 502 (89%) | 183 (39%) | 286 (61%) | 440 (89%)* | 56 (11%)* | |||||
| CD5 | 0%-75% | 154 (13%) | 1057 (87%) | 448 (44%) | 572 (56%) | 880 (79%) | 235 (21%) | ||||||
| > 75% | 15 (12%) | 105 (88%) | 40 (43%) | 54 (57%) | 77 (71%) | 31 (29%) | |||||||
| HLA-DR | No staining | 51 (37%) | 88 (63%) | 100 (66%)* | 52 (34%)* | ||||||||
| Staining | 442 (45%) | 534 (55%) | 858 (80%)* | 209 (20%)* | |||||||||
| Ki67 | 0%-75% | 408 (87%)* | 60 (13%)* | ||||||||||
| > 75% | 424 (69%)* | 187 (31%)* | |||||||||||
| . | Score . | BCL6 score . | CD10 score . | CD5 score . | HLA-DR score . | Ki67 score . | MUM1 score . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No staining . | Staining . | No staining . | Staining . | ≤75% . | >75% . | No staining . | Staining . | ≤75% . | >75% . | ≤75% . | >75% . | ||
| BCL2 | 0%-75% | 56 (12%) | 433 (88%) | 378 (59%) | 267 (41%) | 615 (95%)* | 33 (5%)* | 75 (12%) | 569 (88%) | 233 (42%) | 328 (58%) | 514 (85%)* | 89 (15%)* |
| > 75% | 58 (13%) | 381 (87%) | 351 (57%) | 265 (43%) | 554 (87%)* | 81 (13%)* | 86 (13%) | 553 (87%) | 246 (48%) | 267 (52%) | 410 (71%)* | 169 (29%)* | |
| BCL6 | No staining | 96 (82%)* | 21 (18%)* | 104 (87%) | 15 (13%) | 14 (11%) | 109 (89%) | 49 (44%) | 62 (56%) | 78 (68%) | 37 (32%) | ||
| Staining | 411 (49%)* | 422 (51%)* | 789 (93%) | 56 (7%) | 93 (11%) | 750 (89%) | 331 (42%) | 458 (58%) | 642 (79%) | 174 (21%) | |||
| CD10 | No staining | 669 (89%)* | 84 (11%)* | 109 (14%) | 643 (86%) | 298 (47%) | 332 (53%) | 503 (72%)* | 200 (28%)* | ||||
| Staining | 529 (95%)* | 29 (5%)* | 60 (11%) | 502 (89%) | 183 (39%) | 286 (61%) | 440 (89%)* | 56 (11%)* | |||||
| CD5 | 0%-75% | 154 (13%) | 1057 (87%) | 448 (44%) | 572 (56%) | 880 (79%) | 235 (21%) | ||||||
| > 75% | 15 (12%) | 105 (88%) | 40 (43%) | 54 (57%) | 77 (71%) | 31 (29%) | |||||||
| HLA-DR | No staining | 51 (37%) | 88 (63%) | 100 (66%)* | 52 (34%)* | ||||||||
| Staining | 442 (45%) | 534 (55%) | 858 (80%)* | 209 (20%)* | |||||||||
| Ki67 | 0%-75% | 408 (87%)* | 60 (13%)* | ||||||||||
| > 75% | 424 (69%)* | 187 (31%)* | |||||||||||
Biomarkers that are significantly (P < .002) correlated. Adjusting for multiple comparisons using the Bonferroni method, χ2P values < .05/21 are considered significant.
Developing a prognostic model using biomarkers
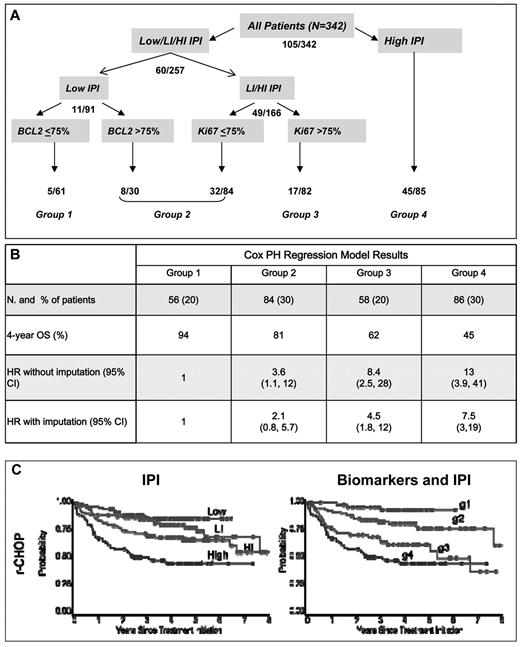
We developed a model using the biomarkers as originally scored on all r-CHOP patients with at least one marker scored (n = 342 of 347). The low-risk patients were separated by BCL2 and the low-intermediate/ high-intermediate patients were separated by Ki67 (Figure 5A). This model distinguished 4 groups (groups 1-4) of approximately equal size with improved model evaluation measures relative to the IPI (BIC = 1145 vs 1153; c-index = 0.69 vs 0.67; Table 5 and Figure 5B). The most favorable group comprised patients with a low IPI and BCL2 ≤ 75% and had an expected 4-year OS of 94%. Group 2 (81% 4-year OS) included low-IPI patients with BCL2 > 75% and low-intermediate/ high-intermediate IPI patients with Ki67 positivity in ≤ 75% of cells. Group 3 (62% 4-year OS) comprised low-intermediate/ high-intermediate patients with Ki67 > 75%, and group 4 comprised patients with a high IPI (4-year OS 45%; Figure 5C). When this model was applied to c-CHOP patients, the prognosis for group 2 and 3 appeared similar (Table 5).
Prognostic model using biomarkers and IPI. (A) Hierarchical tree model. Numbers indicate the number of deaths observed at each level in the population at risk. (B) Number of patients, 4-year OS, and HR for the risk of death (without and with imputation for samples with missing scores) in the r-CHOP cohort. (C) OS for r-CHOP patients according to the IPI and the biomarker and IPI model, respectively. Log-rank P values for both models were < .0001.
Prognostic model using biomarkers and IPI. (A) Hierarchical tree model. Numbers indicate the number of deaths observed at each level in the population at risk. (B) Number of patients, 4-year OS, and HR for the risk of death (without and with imputation for samples with missing scores) in the r-CHOP cohort. (C) OS for r-CHOP patients according to the IPI and the biomarker and IPI model, respectively. Log-rank P values for both models were < .0001.
Patient distribution and estimated relative risk of death for the prognostic model using biomarkers compared with the IPI in the r-CHOP cohort
| . | . | IPI groups . | Biomarkers and IPI* . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Low . | Low-intermediate . | High-intermediate . | High . | Group 1 . | Group 2 . | Group 3 . | Group 4 . | ||
| No. of patients (%)† | 94 (27) | 80 (23) | 87 (25) | 86 (25) | 56 (20) | 84 (30) | 58 (20) | 86 (30) | |
| r-CHOP | 4-year OS | 85% | 80% | 68% | 45% | 94% | 81% | 62% | 45% |
| HR (95% CI) | 1 | 1.6 (0.8, 3.4) | 2.5 (1.3, 4.9) | 5.1 (2.7, 9.6) | 1 | 3.6 (1.1, 12) | 8.4 (2.5, 28) | 13 (3.9, 41) | |
| No. of patients (%)† | 79 (27) | 63 (22) | 79 (27) | 68 (24) | 38 (16) | 92 (38) | 46 (19) | 68 (28) | |
| c-CHOP | 4-year OS | 84% | 63% | 48% | 26% | 95% | 62% | 54% | 26% |
| HR (95% CI) | 1 | 2.9 (1.5, 5.5) | 3.9 (2.1, 7.2) | 7.3 (4.0, 14) | 1 | 9.0 (2.2, 37) | 9.5 (2.2, 40) | 22 (5.5, 92) | |
| . | . | IPI groups . | Biomarkers and IPI* . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Low . | Low-intermediate . | High-intermediate . | High . | Group 1 . | Group 2 . | Group 3 . | Group 4 . | ||
| No. of patients (%)† | 94 (27) | 80 (23) | 87 (25) | 86 (25) | 56 (20) | 84 (30) | 58 (20) | 86 (30) | |
| r-CHOP | 4-year OS | 85% | 80% | 68% | 45% | 94% | 81% | 62% | 45% |
| HR (95% CI) | 1 | 1.6 (0.8, 3.4) | 2.5 (1.3, 4.9) | 5.1 (2.7, 9.6) | 1 | 3.6 (1.1, 12) | 8.4 (2.5, 28) | 13 (3.9, 41) | |
| No. of patients (%)† | 79 (27) | 63 (22) | 79 (27) | 68 (24) | 38 (16) | 92 (38) | 46 (19) | 68 (28) | |
| c-CHOP | 4-year OS | 84% | 63% | 48% | 26% | 95% | 62% | 54% | 26% |
| HR (95% CI) | 1 | 2.9 (1.5, 5.5) | 3.9 (2.1, 7.2) | 7.3 (4.0, 14) | 1 | 9.0 (2.2, 37) | 9.5 (2.2, 40) | 22 (5.5, 92) | |
Biomarkers and IPI: Group 1 = low IPI and BCL2 ≤ 75%; Group 2 = low IPI and BCL2 > 75% or low-intermediate IPI and Ki67 ≤ 75% or high-intermediate IPI and Ki67 ≤ 75%; Group 3 = low-intermediate IPI/high-intermediate IPI and Ki67 > 75%; Group 4 = high IPI.
Two hundred eighty-four of 347 r-CHOP and 244 of 289 c-CHOP patients had data available to compute biomarker and IPI index.
The impact of the biomarkers and IPI on OS was also evaluated among the e-CHOP series, which included a broader cross-section of ages and IPI groups. This analysis also resulted in the discrimination of low-IPI patients according to BCL2 and HLA-DR expression, with 4 separate risk groups identified (not shown). When evaluated in the r-CHOP cohort, this model, which was optimized for the e-CHOP series, did not result in substantial improvement of risk group definitions. Similar results were obtained by applying recursive portioning and Cox PH regression to the multiple imputation datasets.
Discussion
In the present study, the LLBC explored the validity of IHC markers as prognostic tools in patients with DLBCL. The availability of 3 large patient cohorts based on clinical studies with optimally documented and homogeneous treatments conducted in various countries resulted in a unique dataset not directly comparable to single institutions or single cooperative groups. This allowed for the evaluation of the impact of covariates and interrelations of biologic and clinical parameters. Although biases remain and low-risk R-CHOP–treated patients are relatively underrepresented, this series constitutes a very reasonable cross-section of all risk and age groups. The approach for staining and scoring IHC markers was standardized and based on previous reproducibility assessments.14,15 Although several markers could not be scored in all patients, this evaluation was performed on material collected from multiple prospective clinical trials and involved a panel of hematopathologists, and therefore is representative of clinical investigations. Individual cases stained and scored on whole sections at the time of diagnosis by individual pathologists may more closely reflect clinical practice, but are not practical for the evaluation of large series. Using a step-by-step statistical approach supported by the large number of available cases, several key findings were identified.
First, the optimal cut points for single markers that provided the best discrimination for patient outcome were identified on a rational basis and appeared to be different from those arbitrarily used in previous studies. Second, new correlations between individual biomarkers and IPI, as well as between biomarkers themselves, were identified. These were probably underestimated in previous studies and could likely account for some discordant results. The positive correlations between CD10 and BCL6 expression and the negative correlation between CD10 and MUM1 expression was expected, because they represent features consistent with the GCB subtype definition. Further, BCL2 overexpression has been reported in ABC-type DLBCL cases that overexpress MUM1.28 As described previously, a correlation among CD5, BCL2, and an inverse with CD10 was noted, which may represent a biologically separate, small subclass of CD5+ DLBCL.29,–31 These interrelations between biomarkers themselves and IPI parameters highlight the importance of carefully controlling for clinical and biologic characteristics when multiple markers are tested in prognostic models.
Among 347 patients treated with R-CHOP, this study demonstrates that only CD5 retained a prognostic significance. This is in contrast to the e-CHOP and c-CHOP cohorts in which BCL2, BCL6, and MUM1 and IHC algorithms related to gene expression in CHOP-treated patients were prognostic for OS. This difference clearly confirms that response to rituximab may differ in distinct biologic groups. It also emphasizes that the impact of prognostic factors may be strongly influenced by the nature of the therapy administered, and that immunotherapy may differ in that way from chemotherapy. The present r-CHOP (and c-CHOP) cohorts are, however, biased toward patients older than 60 years of age with adverse IPI scores, and such findings may need confirmation in larger r-CHOP cohorts. It should be noted that the GCB/non-GCB algorithms used in this study were not specifically designed as optimized surrogates for the biologic gold standard of GCB/ABC subtypes based on gene-expression analysis, but rather represent potential prognostic tools validated using IHC. However, the present data indicate that the GCB/non-GCB IHC-based algorithm based on CD10, BCL6, and MUM1 expression does not provide prognostic information in the rituximab era, in agreement with another large patient series reported recently.32 Overall, these results emphasize that prognostic indicators developed before the use of rituximab need to be thoroughly reassessed before any implementation in the current era of therapy may be considered.
Most importantly, this study demonstrates that variations in biologic features are largely overshadowed by the IPI for prognostic impact, and that biomarkers only allow subtle refinements of the IPI. This is demonstrated by the modest HR for death (ranging from 1.3 up to 1.8) conferred for individual biomarkers in the r-CHOP cohort (as well as in larger e-CHOP cohorts). Attempts to build an index using biomarkers alone, the GCB LLBC classification, or the combination of biomarkers with IPI individual factors (instead of IPI categories) did not result in an improved model fit or discrimination (data not shown). Several biomarkers appear to be able to further discriminate subgroups with distinct outcome within IPI categories. BCL2 was found to discriminate the outcome of low- or intermediate-risk patients treated with (and without) rituximab. This may be clinically relevant, because BCL2 overexpression is well known to confer chemotherapy resistance,33 and therapeutic targeting of this protein is currently under development.34 The adverse outcome associated with CD5 expression, also reported by others,35 may reflect the distinct origin of DLBCL expressing this antigen. Finally, in contrast to findings in patients treated without rituximab, Ki67 overexpression appears to confer a poor prognosis in patients treated with R-CHOP, which may be of importance for patients with intermediate IPI scores. The inability of biomarkers to further stratify outcome of high-IPI patients may reflect the importance of patient characteristics over biologic characteristics of tumor cells in this patient subset. However, in addition to their impact on patient prognosis, biomarkers may also contribute to defining more homogeneous biologic subsets of DLBCL for which targeted therapies are investigated.
Finally, this study indicates that attempts to validate IHC biomarkers for the prognostic stratification of patients clearly require large cohorts and reproducible methodology that allows for the control of cofactor interactions. In this regard, stratification based on biomarkers for guiding treatment options should be viewed cautiously. Various algorithms recently evaluated for their concordance with gene-expression classification of the cell of origin36 may also have to be evaluated in larger cohorts. The data in the present study demonstrate that the IPI remains the best available index in patients with DLBCL treated with rituximab and chemotherapy. Some progress may be possible with more reliable IHC markers,37,,,–41 cytogenetic markers,11,42,–44 or molecular markers,45,,,–49 but their assessment as prognostic factors need to be carefully evaluated to implement their routine use in clinical practice.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The Lunenburg Lymphoma Biomarker Consortium is a collaboration of 9 international lymphoma collaborative groups, each represented by clinical investigators and hematopathologists, supported by a team of data managers and statisticians, and with the active participation of other experts. Those include in particular: R. Gascoyne, M. Chhanabhai, L. Sehn (British Columbia Cancer Center); R. Gascoyne, S. Horning (ECOG); D. De Jong, M. van Glabbeke, H. Kluin-Nelemans. J. Raemaekers (EORTC Lymphoma Group); P. Gaulard, T. Molina, J. Briere, M. Fournier, B. Coiffier, H. Tilly, G. Salles (GELA); C. Thorns, A. Rosenwald, W. Klapper, G. Ott, S.H.-W. Bernd, L. Trumper, N. Schmitz, M. Pfreundschuh (German High Grade Non-Hodgkin Lymphoma Group–DSHNHL); P. Sonneveld, J. Doorduijn, P. Huijgens, L.F. Verdonck, G. van Imhoff, M. Steijaert I. Meulendijks, M. Testroote, W. van Putten, W. van der Holt, W. Graveland; A. Mulder, D. de Jong, K. Lam, J. van den Tweel (HOVON); B. Sander, E. Kimby (Nordic Lymphoma Study Group); J. Radford (Manchester, United Kingdom); M. Calaminici, A. Lee, A. Norton, A. Clear, A. Lister (St Bartholomew's Hospital); E. Campo, Independent pathology advisor (Barcelona, Spain); E. Weller, W. Xie, Y. He, B. Giblin (Dana-Farber Cancer Institute); M.-J. Kersten (Amsterdam, The Netherlands).
This study was supported by unrestricted grants from Bayer-Schering-Pharma, Eli-Lilly, Genentech, Millennium, and Roche.
Authorship
Contribution: G.S., D.d.J, A.R., R.D.G., and E.W designed the study, contributed and analyzed data, and wrote the manuscript; M. Chhanabhai, P.G., W.K., B.S., E.C., M.P., S.H., A. Lister, A. Lee, L.H.S., J.R., and A.H., designed the study and contributed and analyzed data; and W.X., M. Calaminici, C.T., and T.M contributed and analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gilles Salles, Hospices Civils de Lyon & Université Claude Bernard Lyon-1, UMR CNRS 5239, Lyon, France; e-mail: gilles.salles@chu-lyon.fr.