We have recently reported inactivation of the tyrosine phosphatase PTPN2 (also known as TC-PTP) through deletion of the entire gene locus in ∼ 6% of T-cell acute lymphoblastic leukemia (T-ALL) cases. T-ALL is an aggressive disease of the thymocytes characterized by the stepwise accumulation of chromosomal abnormalities and gene mutations. In the present study, we confirmed the strong association of the PTPN2 deletion with TLX1 and NUP214-ABL1 expression. In addition, we found cooperation between PTPN2 deletion and activating JAK1 gene mutations. Activating mutations in JAK1 kinase occur in ∼ 10% of human T-ALL cases, and aberrant kinase activity has been shown to confer proliferation and survival advantages. Our results reveal that some JAK1 mutation–positive T-ALLs harbor deletions of the tyrosine phosphatase PTPN2, a known negative regulator of the JAK/STAT pathway. We provide evidence that down-regulation of Ptpn2 sensitizes lymphoid cells to JAK1-mediated transformation and reduces their sensitivity to JAK inhibition.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive leukemia occurring in children and adults that is characterized by the massive production of undifferentiated thymocytes of clonal origin. T-ALL arises from the neoplastic transformation of a lymphoid progenitor cell that has accumulated multiple genetic lesions that alter its differentiation and self-renewal properties and provide proliferation and survival advantages.1 The aberrant activation of transcription factors such as TAL1, TLX1, TLX3, MLL, and HOXA is thought to be the driving force of the transformation process and, based on the identity of the causal transcription factor, T-ALLs can be delineated in oncogenic subgroups with a characteristic immunophenotype and expression profile.2,3 In addition to the ectopic expression of these transcription factors, several other lesions have been identified, including mutations of NOTCH1 and FBXW7, inactivation of PTEN, deletion of CDKN2A (p16), mutations of PHF6, and activation of LCK, JAK1, and ABL1 tyrosine kinases.4,,,,,,,–12
Mutations in JAK1 and episomal fusion of NUP214 to ABL1 are known oncogenic events that support the proliferation and survival of T-ALL cells.9,10,13,14 Whereas the chimeric NUP214-ABL1 fusion gene occurs specifically in the cortical subtype characterized by aberrant expression of homeobox transcription factors TLX1 and TLX3, mutated JAK1 kinase appears in different T-ALL subgroups. Three independent studies have reported the occurrence of JAK1 mutations in T-ALL. In 2008, Flex et al studied a cohort of Italian T-ALL patients and found the frequency of JAK1 mutations in children and adults to be 2% and 18%, respectively. That study also found a clinical association with older age at diagnosis, poor treatment response, and overall predicted outcome.10 Around the same time, a second group sequenced exons 5, 14, and 17 of JAK1 in 11 adult Korean T-ALL patients and identified 3 missense mutations in the coding region of JAK1 (27%).14 However, the latest study performed by Asnafi et al could not confirm the frequencies reported previously. Sequence analysis of the JAK1 gene in 108 adult French T-ALL patients identified only 4 individuals with JAK1 mutations.9 Whether JAK1 mutations occur in adult T-ALL patients at different frequencies depending on ethnic origin remains to be elucidated in future studies.
Protein tyrosine phosphorylation is a highly dynamic process that is kept in balance by the antagonistic function of protein tyrosine kinases and protein tyrosine phosphatases. Therefore, enhanced signal traffic can arise from either up-regulated protein tyrosine kinase function or loss of negative regulation. We have recently identified acquired deletions of the T-cell protein tyrosine phosphatase gene PTPN2 in ∼ 6% of T-ALL cases.15 Loss of PTPN2 was restricted to an immature T-ALL subset characterized by deregulated expression of the homeobox transcription factor TLX1. Four cases also harbored the oncogenic fusion gene NUP214-ABL1, and we identified NUP214-ABL1 as novel, bona fide substrate of PTPN2. Absence of the PTPN2 protein evidently enhanced NUP214-ABL1 kinase activity, thereby sensitizing cells to leukemogenic transformation. These genetic and functional data indicate that the loss of PTPN2 enhances the oncogenic properties of NUP214-ABL1 and suggest that this loss could also potentiate other oncogenic kinases such as JAK1, a known substrate of PTPN2.16,–18
Methods
Cell lines
HEK293T and Ba/F3 cells were obtained from DSMZ. Cells were grown in DMEM (HEK293T) or RPMI 1640 (Ba/F3) medium supplemented with 10% FBS. The medium of cytokine-dependent Ba/F3 cells was supplemented with 1 ng/mL of recombinant IL-3 (Peprotech).
Patient samples
T-ALL samples were collected at various institutions and were obtained after informed consent following the Declaration of Helsinki and with approval from the ethical committees of all participating institutions. Diagnosis of T-ALL was based on morphology, cytochemistry, and immunophenotyping according to World Health Organization and European Group for the Immunologic Characterization of Leukemias criteria. Molecular data on 13 T-ALL cases were described previously.15 Array comparative genomic hybridization (aCGH) profiles of additional T-ALLs were generated on SNIP-6 Affymetrix arrays in a collaboration between the Necker Hospital and the Pasteur-Cerba platform in France.
Quantitative PCR
Quantitative real-time PCR was performed on a LightCycler 480 PCR instrument (Roche). Reactions were performed in a total volume of 15 μL: 2× SYBR Green I Master Mix (7.5 μL), 5pmol forward primer (1.25 μL), 5pmol reverse primer (1.25 μL), and 10ng genomic DNA template (5 μL). RNMT and CDH2 were used as control genes and results were normalized to a control individual with a normal PTPN2 copy number using the comparative ct (δδ ct) method. Primer sequences were as follows: PTPN2 for 5′-TGAGAGAATCTGGCTCCTTGAAC, reverse 5′-GCCCAATGCCTGCACTACA, CDH2 for 5′-TGAGAGAATCTGGCTCCTTGAAC, reverse 5′-GCCCAATGCCTGCACTACA, and RNMT for TGCAGTTGTCAGTTTGTCTGTCA, reverse CACGCATTTCTCAGCATCATG. To determine knockdown efficiency of Ptpn2 in Ba/F3 cells, the following primer pairs were used: mHprt1 for 5′-CATTATGCCGAGGATTTGG, reverse 5′-GCAAGTCTTTCAGTCCTGT; mPtpn2 for 5′-GTGATCCATTGCAGTGCG and 5′-TTCCATCAGAACAAGACAGGTAT. All primers were purchased from Integrated DNA Technologies.
Sequence analysis of PTPN2 and JAK1
Sequence analysis of both PTPN2 and JAK1 were performed at the genomic DNA level. The initial screen was done with whole-genome–amplified DNA as a template. Positive hits were confirmed in an independent PCR from primary patient material. PCR composition and primer sequences for the mutational analysis of PTPN2 were described previously.15 All exons of JAK1 were amplified with Taq DNA polymerase (Promega), and PCR products were directly sequenced with internal primers at the VIB Genetics Service Facility (Antwerp, Belgium). Primer sequences will be provided upon request.
Plasmids
Constructs for ectopic expression of JAK1 wild-type and mutants have been described previously.15,19 All mutants were cloned into pMSCV-puro and pMSCV-GFP (Clontech). JAK1 mutant p.K648N was generated through the exchange of a 700-bp fragment using the unique restriction sites RsrII and NsiI. For knockdown of Ptpn2 in mouse cells, 4 shRNAs were cloned into pMSCV-GFP constructs containing a mir30-flanking cassette. Two control constructs containing either an shRNA-targeting human ALK (shRNA-control1) or a nontargeting shRNA (shRNA-control2) were generated and used as controls. The ALK gene is not expressed in the hematopoietic system and therefore represents a good control shRNA. The nontargeting shRNA contains a sequence that does not target any known human or mouse mRNA, and was used as a second control in some experiments. The sequences used were: shRNA-control1: CGGAAGGAATATTCACTTCTAA and TTAGAAGTGAATATTCCTTCCA; shRNA-control2: CTCGCTTGGGCGAGAGTAA and TTACTCTCGCCCAAGCGAG; shRNA-Ptpn2-D: GTGTGAAGCTCTTATCTGA and TCAGATAAGAGCTTCACAC; shRNA-Ptpn2-C: GCTCTTATCTGAAGATGTA and TACATCTTCAGATAAGAGC; shRNA-Ptpn2-B: ACTGAATATGAGAAAGTAT and ATACTTTCTCATATTCAGTA; and shRNA-Ptpn2-A: CACAAAGAAGTTACATCTT and AAGATGTAACTTCTTTGTG. ShRNA sequence Ptpn2-B was obtained from Delphine Desprez and Michel L. Tremblay (Goodman Cancer Research Center, Department of Biochemistry, McGill University, Montreal, QC).
Retroviral transduction
Viral vectors were produced in HEK293T cells using an EcoPack packaging plasmid and TurboFect transfection reagent (Fermentas). Virus was harvested after 48 hours and Ba/F3 cells were transduced as described previously.20 Ba/F3 cells were retrovirally transduced with MSCV-puro vectors containing different JAK1 cDNAs or empty pMSCV-puro vector, and selected with puromycin before cotransduction with pMSCV-GFP-shRNA constructs. Transduction efficiency was assessed after 48 hours and ranged between 90% and 95%.
Transformation experiments
Ba/F3 cells were washed twice in PBS to remove all traces of cytokines and seeded out in triplicate. The Vi-CELL XR cell viability analyzer (Beckman Coulter) was used to determine cell numbers and viability at the indicated time points. All experiments were terminated at day 8 and cell lines showing no sign of cell growth were declared to be nontransforming.
siRNA experiments
Ba/F3 cells were electroporated as described previously using small interfering RNAs (siRNAs) from the TriFECTA Kit (MMC.RNAI.N008977.10; Integrated DNA Technologies).21 Knock-down efficiency of Ptpn2 was confirmed on the RNA and protein levels using quantitative real-time PCR and protein blot analysis, respectively.
Inhibitor experiments
JAK1-dependent Ba/F3 cells (which are cytokine independent) were seeded out in triplicate in 24-well plates at a density of 0.1 × 106 cells. Absolute cell numbers were determined 48 hours later using the cell-viability analyzer. For dose-response curves, JAK1(K648N)–expressing Ba/F3 cells were treated with increasing concentrations of JAK inhibitor I (Calbiochem) or DMSO as a control, and cell proliferation and viability were assessed after 24 hours. JAK inhibitor I was serially diluted to determine the IC50, the concentration of inhibitor that gave a 50% inhibition. For all other experiments, JAK inhibitor I was used at a concentration of 100nM.
Western blotting
Cells were lysed in complete cell lysis buffer (Cell Signaling Technology) supplemented with complete protease inhibitors (Roche). Protein concentrations were determined with the Bio-Rad protein assay. Western blot samples (30 μg) were prepared in NuPAGE LDS sample buffer (Invitrogen), and electrophoresis was performed with a 4%-12% gradient gel (Invitrogen). Separated proteins were transferred to a PVDF membrane, blocked in 5% nonfat milk, and detected with the respective antibodies. The following antibodies were used in this study: Ptpn2 (3E2) (from MédiMabs); JAK1 (HR-785), phospho-JAK1 (Tyr1022/1023), STAT5 (L-20), and ERK (C-16) (all from Santa Cruz Biotechnology); β-actin (AC-15; from Sigma-Aldrich); and STAT1 and STAT3 (both from Cell Signaling Technology). A phospho-STAT antibody sampler kit was used for the detection of STAT proteins (Cell Signaling Technology).
AlphaScreen SureFire kinase assay
Stat5 phosphorylation levels of Ba/F3 cells were determined using the AlphaScreen SureFire STAT5 (p-Tyr695;Tyr699) kinase assay kit (PerkinElmer). Cells (25 × 104) were lysed through the addition of 5× lysis buffer in a total volume of 100 μL. Lysates were simultaneously analyzed for phosphorylation levels of Stat5 and total Erk protein (loading control). All assays were performed according to the manufacturer's instructions.
Statistical analysis
Significant differences were determined using Prism software Version 5 (GraphPad). The mean of 2 groups was compared with the Student t test. Normality tests were used to test the assumption of a normal distribution.
Results
Deletion of PTPN2 is specifically found in TLX1-positive T-ALL
We have recently identified focal deletions of the tyrosine phosphatase gene PTPN2 in ∼ 6% of T-ALL.15 Remarkably, deletion of the PTPN2 gene was strongly associated with aberrant expression of the homeobox transcription factors TLX1 (12 of 13 cases) and TLX3 (1 of 13 cases; Table 1). In addition, 30% of the PTPN2-deleted cases also harbored the NUP214-ABL1 fusion gene.15 Among the 13 identified PTPN2-deleted T-ALL individuals, 8 featured biallelic loss and 5 monoallelic loss of PTPN2. Individuals with heterozygous loss of PTPN2 showed reduced PTPN2 expression levels compared with T-ALL, along with normal PTPN2 copy numbers,15 strongly suggesting that in those cases with incomplete deletion, the loss of one copy of PTPN2 was a driving oncogenic event.
Characterization of T-ALL individuals with PTPN2 inactivation
| Individual . | Sex . | Age, y . | PTPN2 deletion* . | PTPN2 mutation . | Deregulated transcription factor† . | NUP214-ABL1‡ . | JAK1 status . |
|---|---|---|---|---|---|---|---|
| 1§ | F | 30 | PTPN2−/− | NA | TLX1 | − | wt |
| 2§ | M | 24 | PTPN2−/− | NA | TLX1 | + | wt |
| 3§ | F | 27 | PTPN2−/− | NA | TLX1 | − | wt |
| 4§ | M | 21 | PTPN2−/− | NA | TLX1 | − | wt |
| 5§ | M | 45 | PTPN2−/− | NA | TLX1 | + | wt |
| 6§ | M | 8 | PTPN2−/− | NA | TLX3 | − | c.1944A > C p.K648N |
| 7§ | M | 43 | PTPN2−/− | NA | TLX1 | − | wt |
| 8§ | M | 20 | PTPN2−/− | NA | TLX1 | − | wt |
| 9§ | F | 4 | PTPN2+/− | wt | TLX1 | + | wt |
| 10§ | M | 11 | PTPN2+/− | wt | TLX1 | − | wt |
| 11§ | F | 35 | PTPN2+/− | wt | TLX1 | − | wt |
| 12§ | F | 9 | PTPN2+/− | wt | TLX1 | + | wt |
| 13§ | F | 49 | PTPN2+/− | c.862C > T, p.R288Stop | TLX1 | − | c.1953T > C p.Y652H |
| 14 | M | 34 | PTPN2−/− | NA | TLX1 | − | wt |
| 15 | F | 42 | PTPN2+/− | wt | TLX1 | − | wt |
| 16 | F | 38 | PTPN2+/− | wt | TLX1 | + | wt |
| 17 | M | 53 | PTPN2−/− | NA | TLX1 | − | wt |
| 18 | M | 28 | PTPN2−/− | NA | TLX1 | + | wt |
| Individual . | Sex . | Age, y . | PTPN2 deletion* . | PTPN2 mutation . | Deregulated transcription factor† . | NUP214-ABL1‡ . | JAK1 status . |
|---|---|---|---|---|---|---|---|
| 1§ | F | 30 | PTPN2−/− | NA | TLX1 | − | wt |
| 2§ | M | 24 | PTPN2−/− | NA | TLX1 | + | wt |
| 3§ | F | 27 | PTPN2−/− | NA | TLX1 | − | wt |
| 4§ | M | 21 | PTPN2−/− | NA | TLX1 | − | wt |
| 5§ | M | 45 | PTPN2−/− | NA | TLX1 | + | wt |
| 6§ | M | 8 | PTPN2−/− | NA | TLX3 | − | c.1944A > C p.K648N |
| 7§ | M | 43 | PTPN2−/− | NA | TLX1 | − | wt |
| 8§ | M | 20 | PTPN2−/− | NA | TLX1 | − | wt |
| 9§ | F | 4 | PTPN2+/− | wt | TLX1 | + | wt |
| 10§ | M | 11 | PTPN2+/− | wt | TLX1 | − | wt |
| 11§ | F | 35 | PTPN2+/− | wt | TLX1 | − | wt |
| 12§ | F | 9 | PTPN2+/− | wt | TLX1 | + | wt |
| 13§ | F | 49 | PTPN2+/− | c.862C > T, p.R288Stop | TLX1 | − | c.1953T > C p.Y652H |
| 14 | M | 34 | PTPN2−/− | NA | TLX1 | − | wt |
| 15 | F | 42 | PTPN2+/− | wt | TLX1 | − | wt |
| 16 | F | 38 | PTPN2+/− | wt | TLX1 | + | wt |
| 17 | M | 53 | PTPN2−/− | NA | TLX1 | − | wt |
| 18 | M | 28 | PTPN2−/− | NA | TLX1 | + | wt |
Determined by FISH, quantitative PCR, or aCGH; −/− indicates a biallelic deletion; and +/−, a monoallelic deletion.
Positivity for TLX1 or TLX3 expression.
Positivity for NUP214-ABL1 expression or rearrangement; + indicates positive for NUP214-ABL1; and −, negative for NUP214-ABL1.
Patient results have been published previously and are consecutively numbered.15
NA indicates not applicable; and wt, wild-type.
To confirm our previous findings, we screened a new set of T-ALL cases (n = 74) for deletions of the PTPN2 gene. We identified 5 cases with altered copy number status of PTPN2 (Figure 1A and Table 1), and subsequent aCGH analysis confirmed the presence of focal deletions of the entire PTPN2 gene in all cases. Two individuals featured monoallelic deletion and the 3 others showed biallelic loss of PTPN2 (Figure 1A). Sequence analysis of the coding region of PTPN2 in individuals with monoallelic deletion did not provide evidence of any nonsense, splice site, or frameshift mutation in the retained allele. In agreement with our previous observations, newly identified cases with deletion of PTPN2 belonged to the TLX1-positive T-ALL subgroup, and 2 PTPN2-deleted leukemias also harbored the NUP214-ABL1 fusion gene (Table 1). These data confirm our previous findings on the frequency of PTPN2 deletions in T-ALL, the restriction of PTPN2 inactivation to the TLX1 subgroup, and the repeated occurrence of loss of the PTPN2 gene in NUP214-ABL1–positive leukemias.
Activating JAK1 mutations and inactivation of PTPN2 occur together in T-ALL patients. (A) Quantitative PCR analysis confirmed an abnormal copy number of PTPN2 in 5 T-ALL cases. Patients 14, 17, and 18 displayed biallelic loss and patients 15 and 16 monoallelic loss of PTPN2. RNMT was used as control gene and values were normalized to a control individual (○). Patient 1 is shown as positive control and was described previously (red circles).15 *Analyzed diagnosis sample of patient 16 contained only 60% leukemic cells. (B) Sequence analysis of T-ALL patients with PTPN2 inactivation (n = 18) identified a heterozygous point mutation in the pseudokinase domain of JAK1 (exon 14) in patient 13. The c.1953T > C change results in the amino acid substitution p.Y652H. Patient 6 harbored a novel point mutation in one allele of JAK1 (c.1944A > C), which resulted in the exchange of amino acid p.K648N.
Activating JAK1 mutations and inactivation of PTPN2 occur together in T-ALL patients. (A) Quantitative PCR analysis confirmed an abnormal copy number of PTPN2 in 5 T-ALL cases. Patients 14, 17, and 18 displayed biallelic loss and patients 15 and 16 monoallelic loss of PTPN2. RNMT was used as control gene and values were normalized to a control individual (○). Patient 1 is shown as positive control and was described previously (red circles).15 *Analyzed diagnosis sample of patient 16 contained only 60% leukemic cells. (B) Sequence analysis of T-ALL patients with PTPN2 inactivation (n = 18) identified a heterozygous point mutation in the pseudokinase domain of JAK1 (exon 14) in patient 13. The c.1953T > C change results in the amino acid substitution p.Y652H. Patient 6 harbored a novel point mutation in one allele of JAK1 (c.1944A > C), which resulted in the exchange of amino acid p.K648N.
Loss of PTPN2 can be found in JAK1 mutation–positive T-ALL
As shown previously15 and by the results of the current study, among T-ALL individuals with deletion of PTPN2 (n = 18), one-third (6 of 18) also expressed the NUP214-ABL1–activated tyrosine kinase. Further, we have shown previously that loss of PTPN2 increases NUP214-ABL1 signaling. We speculated that cases with loss of PTPN2 but lacking NUP214-ABL1 expression could possibly express another activated tyrosine kinase that would gain further oncogenic activity due to the PTPN2 deletion. PTPN2 is an important modulator of JAK1 kinase activity, and its loss enhances JAK1-mediated cytokine receptor signaling.15,16 Therefore, we investigated whether some of the cases with deletion of PTPN2 harbored activating mutations in JAK1, a gene known to be mutated in 3%-20% of T-ALL cases.
Sequence analysis of the entire coding region of JAK1 in PTPN2-deleted cases (n = 18) identified mutations in the JAK1 kinase gene in 2 NUP214-ABL1–negative cases. One case of leukemia (individual 13) harbored the previously described c.1953T > C change in JAK1, corresponding to a p.Y652H amino acid substitution (Figure 1B and Table 1).9 The other case (individual 6) harbored a novel missense mutation (p.K648N) affecting the pseudokinase domain of JAK1 (Figure 1B and Table 1). Our results found JAK1 mutations in 2 of 18 T-ALL cases with deletion of PTPN2, suggesting a possible cooperation between oncogenic JAK1 kinase function and PTPN2 loss-of-function in these T-ALL cases.
JAK1-overexpressing but not JAK2-overexpressing Ba/F3 cells can be transformed to IL-3–independent growth after down-regulation of Ptpn2
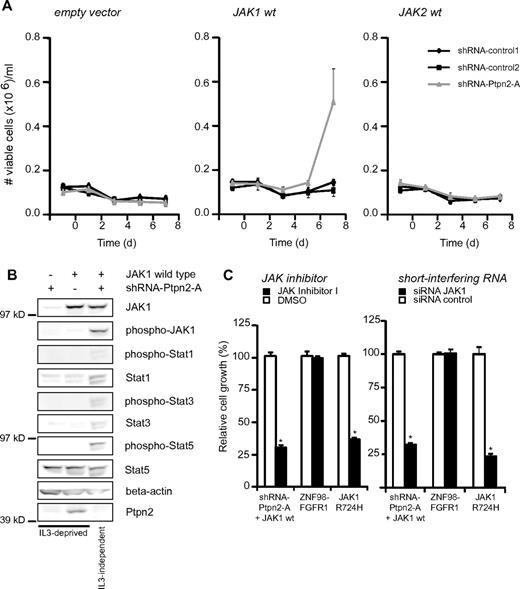
We next looked for a functional relationship between loss of Ptpn2 and JAK1 activation in the IL-3–dependent pro B-cell line Ba/F3. This cytokine-dependent cell line is frequently used to evaluate the oncogenic potential of mutated kinases,22 and was therefore chosen as a model system with which to analyze a possible link between Ptpn2 expression and JAK1 activation. It was reported previously that JAK1 mutants can easily transform Ba/F3 cells to growth factor–independent proliferation, but that wild-type JAK1 does not possess such activity.9 In addition, shRNA-mediated knockdown of Ptpn2 alone did not transform the cells to cytokine independence (Figure 2A). However, overexpression of wild-type JAK1 in Ba/F3 cells combined with knockdown of Ptpn2 transformed Ba/F3 cells to IL-3–independent growth, illustrating a clear relationship between JAK1 signaling and Ptpn2 protein levels (Figure 2A). Western blot analysis confirmed the constitutive activation of JAK1 and downstream Stat proteins in transformed cells. In the case of Stat1 and Stat3, continuous phosphorylation in the absence of cytokines was associated with increased levels of total Stat1 and Stat3 protein in transformed cells compared with cytokine-dependent Ba/F3 cells (Figure 2B). Transformed Ba/F3 cells were sensitive to a JAK inhibitor and to a JAK1-targeting siRNA, illustrating that JAK1 activation was the driving force for their proliferation and survival (Figure 2C). To determine whether the observed cooperative effect was specific for JAK1 kinase, we performed an identical experiment with Ba/F3 cells ectopically expressing JAK2 wild-type, a sister kinase of JAK1, and no direct substrate of PTPN2.23 We observed no transformation of JAK2 wild-type either alone or in combination with Ptpn2 shRNA–mediated knockdown (Figure 2A).
Analysis of transforming capabilities of Ptpn2 knockdown. (A) Transformation assay of Ba/F3 cells with or without knockdown of Ptpn2 and stably transduced with empty control vector, JAK1 wild-type, or JAK2 wild-type. Only JAK1-positive cells with knockdown of Ptpn2 transformed to cytokine independence. Ba/F3 cells carrying the indicated constructs were deprived of IL-3 and cell numbers were recorded in triplicate every other day. The y-axis displays the number of viable cells. Data shown are representative of 3 independent experiments. *P < .005. (B) Western blot analysis of Ba/F3 cells with knockdown of Ptpn2, overexpression of JAK1 (wild-type), or both. Growth factor–dependent Ba/F3 cells were cytokine depleted for 6 hours before cell lysis. Shown is 1 representative result of 3 independent experiments. (C) Ba/F3 cells transformed by JAK1 wild-type with knockdown of Ptpn2 or JAK1(R724H) mutant (positive control) or ZNF98-FGFR1 (negative control) were treated with a JAK inhibitor or with JAK1 siRNA and cell numbers were recorded after 48 hours. Values are displayed as relative cell growth (%). Graph represents mean values ± SEM. Significant differences in cell growth compared with those of control siRNA are indicated (n = 3). *P < .05; **P < .005.
Analysis of transforming capabilities of Ptpn2 knockdown. (A) Transformation assay of Ba/F3 cells with or without knockdown of Ptpn2 and stably transduced with empty control vector, JAK1 wild-type, or JAK2 wild-type. Only JAK1-positive cells with knockdown of Ptpn2 transformed to cytokine independence. Ba/F3 cells carrying the indicated constructs were deprived of IL-3 and cell numbers were recorded in triplicate every other day. The y-axis displays the number of viable cells. Data shown are representative of 3 independent experiments. *P < .005. (B) Western blot analysis of Ba/F3 cells with knockdown of Ptpn2, overexpression of JAK1 (wild-type), or both. Growth factor–dependent Ba/F3 cells were cytokine depleted for 6 hours before cell lysis. Shown is 1 representative result of 3 independent experiments. (C) Ba/F3 cells transformed by JAK1 wild-type with knockdown of Ptpn2 or JAK1(R724H) mutant (positive control) or ZNF98-FGFR1 (negative control) were treated with a JAK inhibitor or with JAK1 siRNA and cell numbers were recorded after 48 hours. Values are displayed as relative cell growth (%). Graph represents mean values ± SEM. Significant differences in cell growth compared with those of control siRNA are indicated (n = 3). *P < .05; **P < .005.
Knock-down of Ptpn2 potentiates the transformation capacity of JAK1 mutants and alters their sensitivity to JAK inhibition
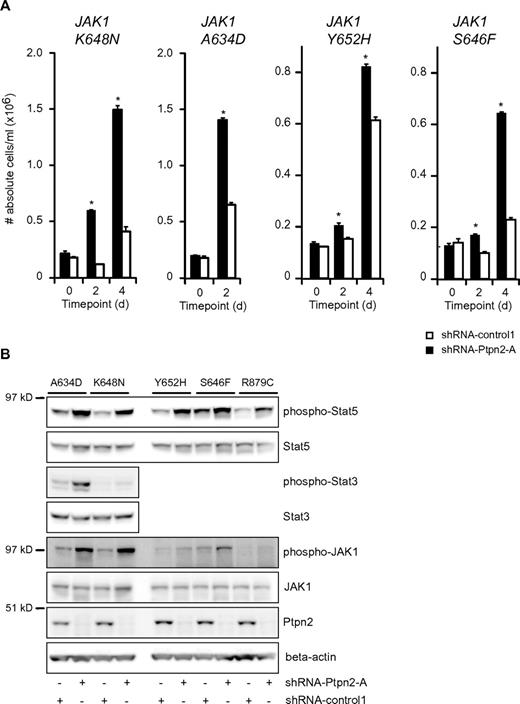
Because we found co-occurrence of PTPN2 inactivation and mutation of JAK1 in T-ALL, we next determined the effect of Ptpn2 knockdown on the capacity of various JAK1 mutants to transform Ba/F3 cells. Knock-down of Ptpn2 clearly facilitated the transformation to cytokine-independent growth of Ba/F3 cells expressing various JAK1 mutants (Figure 3A). Transformation of the cells was accompanied by increased phosphorylation of JAK1 and Stat5 (Figure 3B). Constitutive activation of Stat3 was identified in Ba/F3 cells expressing the strong JAK1(A634D) and JAK1(K648N) mutants, and in both instances increased Stat3 phosphorylation was detected after knockdown of Ptpn2 (Figure 3B).
Reduction of Ptpn2 expression levels accelerates the transformation process of activating JAK1 mutants. (A) Transformation assay of Ba/F3 cells expressing various mutant forms of JAK1. Expression of 4 different JAK1 mutants conferred cytokine-independent growth on Ba/F3 cells. Simultaneous reduction of Ptpn2 expression (shRNA-Ptpn2-A; black bars) enhanced the transformation process compared with control cells (shRNA-control1; white bars). The y-axis displays the number of viable cells. Cell numbers ± SEM were recorded in triplicate. Data shown are representative of 3 independent experiments. *P < .05. (B) Western blot analysis of cytokine-independent Ba/F3 cells. shRNA-mediated knockdown of Ptpn2 resulted in increased phosphorylation of JAK1 and Stat5 compared with control cells (shRNA-control1). Five different JAK1 mutants are shown. Efficient knockdown was confirmed using a Ptpn2 antibody. β-actin was used as an independent loading control.
Reduction of Ptpn2 expression levels accelerates the transformation process of activating JAK1 mutants. (A) Transformation assay of Ba/F3 cells expressing various mutant forms of JAK1. Expression of 4 different JAK1 mutants conferred cytokine-independent growth on Ba/F3 cells. Simultaneous reduction of Ptpn2 expression (shRNA-Ptpn2-A; black bars) enhanced the transformation process compared with control cells (shRNA-control1; white bars). The y-axis displays the number of viable cells. Cell numbers ± SEM were recorded in triplicate. Data shown are representative of 3 independent experiments. *P < .05. (B) Western blot analysis of cytokine-independent Ba/F3 cells. shRNA-mediated knockdown of Ptpn2 resulted in increased phosphorylation of JAK1 and Stat5 compared with control cells (shRNA-control1). Five different JAK1 mutants are shown. Efficient knockdown was confirmed using a Ptpn2 antibody. β-actin was used as an independent loading control.
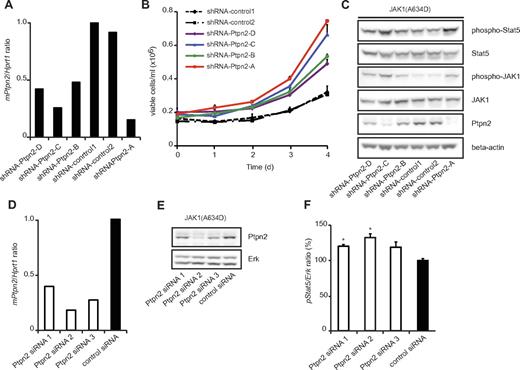
To confirm the specificity of our observation that the oncogenic capacity of JAK1 mutants is modulated by Ptpn2 expression, we used 4 different Ptpn2-targeting shRNAs with various knockdown efficiencies (Figure 4A), and studied the proliferation of Ba/F3 cells expressing the JAK1(A634D) mutant after cytokine removal. We observed a clear correlation between knockdown efficiency and the effect on proliferation of the Ba/F3 cells. Cells expressing the JAK1(A634D) mutant with the lowest Ptpn2 expression proliferated faster than cells with less knockdown of Ptpn2, and cells with normal Ptpn2 levels (shRNA-control1 or shRNA-control2) grew at the slowest speeds (Figure 4B). By monitoring the phosphorylation levels of JAK1 and Stat5, we also found a clear inverse correlation between activation of the JAK/Stat signaling pathway and Ptpn2 protein levels in transformed cells (Figure 4C). We obtained similar results with siRNA-mediated knockdown of Ptpn2. Ba/F3 cells transformed by JAK1(A634D) were electroporated with 3 different Ptpn2-targeting siRNAs or a nontargeting siRNA, and the Stat5 phosphorylation status was quantified using the AlphaScreen SureFire assay system. This assay also showed an inverse correlation between activation levels of Stat5 and Ptpn2 expression (Figure 4D-F).
Ptpn2 modulates the oncogenic transformation of JAK1(A634D)-expressing Ba/F3 cells in a dose-dependent manner. (A) Quantitative real-time PCR analysis of Ptpn2 transcript levels in JAK1(A634D) mutation–positive Ba/F3 cells carrying 6 different shRNA constructs. Hprt1 was used for normalization of expression values and the y-axis displays Ptpn2/Hprt1 ratios. (B) JAK1 mutation–positive Ba/F3 cells cotransduced with the indicated shRNA constructs were deprived of IL-3 and cell growth was monitored every 24 hours for 4 days. Independent of the knockdown efficiency of Ptpn2, cell lines with reduced Ptpn2 expression (shRNA-Ptpn2) transformed appreciably faster to cytokine independence compared with the 2 control cell lines (shRNA-control). Differences in latency among the 4 cell lines carrying Ptpn2-targeting shRNA constructs were correlated with knockdown efficiency. (C) Protein blot analysis of transformed cell lines showed enhanced JAK/Stat activation in cells with a reduced amount of Ptpn2 protein. β-actin was used as an independent loading control. (D) Quantitative real-time PCR analysis of Ptpn2 transcript levels in JAK1(A634D) mutation–positive Ba/F3 cells after 48 hours of electroporation with various siRNAs. Hprt1 was used for normalization of expression values. The x-axis displays Ptpn2/Hprt1 ratios. (E) Knock-down efficiency of applied siRNAs was confirmed by protein blot analysis. Erk is shown as loading control. (F) STAT5 phosphorylation was determined in JAK1(A634D) Ba/F3 cells electroporated with different Ptpn2-targeting siRNAs or a control siRNA. Absorption values were normalized for total Erk. Bar graph shows the relative changes compared with control cells (mean values ± SEM). *P < .05.
Ptpn2 modulates the oncogenic transformation of JAK1(A634D)-expressing Ba/F3 cells in a dose-dependent manner. (A) Quantitative real-time PCR analysis of Ptpn2 transcript levels in JAK1(A634D) mutation–positive Ba/F3 cells carrying 6 different shRNA constructs. Hprt1 was used for normalization of expression values and the y-axis displays Ptpn2/Hprt1 ratios. (B) JAK1 mutation–positive Ba/F3 cells cotransduced with the indicated shRNA constructs were deprived of IL-3 and cell growth was monitored every 24 hours for 4 days. Independent of the knockdown efficiency of Ptpn2, cell lines with reduced Ptpn2 expression (shRNA-Ptpn2) transformed appreciably faster to cytokine independence compared with the 2 control cell lines (shRNA-control). Differences in latency among the 4 cell lines carrying Ptpn2-targeting shRNA constructs were correlated with knockdown efficiency. (C) Protein blot analysis of transformed cell lines showed enhanced JAK/Stat activation in cells with a reduced amount of Ptpn2 protein. β-actin was used as an independent loading control. (D) Quantitative real-time PCR analysis of Ptpn2 transcript levels in JAK1(A634D) mutation–positive Ba/F3 cells after 48 hours of electroporation with various siRNAs. Hprt1 was used for normalization of expression values. The x-axis displays Ptpn2/Hprt1 ratios. (E) Knock-down efficiency of applied siRNAs was confirmed by protein blot analysis. Erk is shown as loading control. (F) STAT5 phosphorylation was determined in JAK1(A634D) Ba/F3 cells electroporated with different Ptpn2-targeting siRNAs or a control siRNA. Absorption values were normalized for total Erk. Bar graph shows the relative changes compared with control cells (mean values ± SEM). *P < .05.
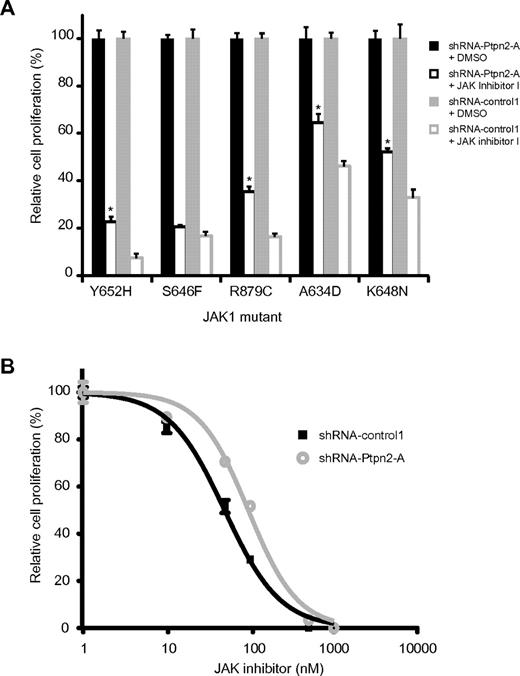
Because knockdown of PTPN2 in NUP214-ABL1–transformed lymphoid cells increases NUP214-ABL1 signaling and reduces the cells' sensitivity to the ABL1 inhibitor imatinib,15 we also investigated whether the same is true for JAK1-transformed Ba/F3 cells. Indeed, JAK1-dependent Ba/F3 cells expressing lower levels of Ptpn2 were less sensitive to JAK inhibitor I (Figure 5A). Furthermore, determination of the IC50 value in the JAK1(K648N) mutant revealed a reduced JAK inhibitor response after knock-down of Ptpn2 (49.3 vs 94.9 nm; Figure 5B).
Ptpn2 protein levels alter sensitivity to JAK1 inhibition of transformed Ba/F3 cells. (A) IL-3–independent Ba/F3 cells were treated with either JAK inhibitor I (100 nM, white) or DMSO (black). Silencing of Ptpn2 consistently reduced the sensitivity of JAK1-dependent Ba/F3 cells to inhibition of the JAK/Stat pathway. Different JAK1 mutants are shown on the x-axis. The number of the respective untreated cells was set as 100% and the y-axis displays relative cell growth (%). Results shown are the average of 3 determinations. Bars indicate SEM. *P < .05. Black bars indicate shRNA-Ptpn2-A; gray bars, shRNA-control1. The blot shown is representative of 3 independent experiments. (B) Differences in Ptpn2 expression levels affected the sensitivity of JAK1-dependent Ba/F3 cells to JAK inhibition. Reduction of the Ptpn2 protein resulted in a shift of the dose-response curve from 49.3 to 94.8nM. A dose-response curve was obtained by exposure of the cells to increasing concentrations of JAK inhibitor I. Cell numbers were determined 24 hours after inhibitor addition. The y-axis displays cell growth relative to DMSO-treated control cells (%). Each data point is an average of 3 calculations. Error bars represent SEM.
Ptpn2 protein levels alter sensitivity to JAK1 inhibition of transformed Ba/F3 cells. (A) IL-3–independent Ba/F3 cells were treated with either JAK inhibitor I (100 nM, white) or DMSO (black). Silencing of Ptpn2 consistently reduced the sensitivity of JAK1-dependent Ba/F3 cells to inhibition of the JAK/Stat pathway. Different JAK1 mutants are shown on the x-axis. The number of the respective untreated cells was set as 100% and the y-axis displays relative cell growth (%). Results shown are the average of 3 determinations. Bars indicate SEM. *P < .05. Black bars indicate shRNA-Ptpn2-A; gray bars, shRNA-control1. The blot shown is representative of 3 independent experiments. (B) Differences in Ptpn2 expression levels affected the sensitivity of JAK1-dependent Ba/F3 cells to JAK inhibition. Reduction of the Ptpn2 protein resulted in a shift of the dose-response curve from 49.3 to 94.8nM. A dose-response curve was obtained by exposure of the cells to increasing concentrations of JAK inhibitor I. Cell numbers were determined 24 hours after inhibitor addition. The y-axis displays cell growth relative to DMSO-treated control cells (%). Each data point is an average of 3 calculations. Error bars represent SEM.
Discussion
T-ALL is an aggressive T-cell malignancy that is most common in children and adolescents. Current treatment for T-ALL consists mainly of multi-agent combination chemotherapy. Improvements in treatment regimens has led to significant increases in survival, but the long-term survival rates for adult T-ALL patients are still below 40% for those < 60 years of age and even lower in older patients.24 These observations indicate that, especially for older T-ALL patients, new and less toxic drugs are needed to improve overall survival. JAK1 may be an interesting drug target in this subgroup of T-ALL patients. JAK1 is mutated in up to 20% of older T-ALL patients,10 and new JAK inhibitors are under clinical development.25,26 In addition to JAK1 mutations, we have recently identified deletion of the tyrosine phosphatase PTPN2 in 6% of T-ALL cases. PTPN2 is a negative regulator of the JAK/STAT pathway, and its loss sensitizes T cells to cytokine stimulation.15 The results of the present study confirm the relationship between loss of PTPN2 and JAK1 by demonstrating that down-regulation of Ptpn2 makes mouse lymphoid cells more susceptible to transformation.
It is currently unclear whether the loss of PTPN2 is an independent oncogenic event or if it occurs along with other mutations in signaling proteins. We have shown herein that the loss of PTPN2 occurs in a subset of JAK1 mutation–positive T-ALL cases and that down-regulation of Ptpn2 increases the activation status of both wild-type and mutant JAK1, resulting in increased activation of the JAK/STAT pathway and increased cytokine-independent cell proliferation. Therefore, inactivation of PTPN2 in T-ALL may cause increased JAK/STAT signaling and, especially in JAK1 mutation–positive T-ALL, the loss of PTPN2 may further assist in activation of proliferation and survival signals within leukemic cells. This increased activation of JAK1 is associated with reduced responsiveness to JAK inhibition, suggesting that T-ALL individuals with JAK1 mutation in combination with loss of PTPN2 may respond less well to JAK inhibitors. The direct implication of JAK1 mutations in the pathogenesis of leukemia, together with our current results, warrant further studies into the use of JAK inhibitors for the treatment of leukemia.
We provide genetic and functional evidence for the concomitant appearance of mutations in JAK1 kinase with inactivation of its negative regulator PTPN2, with each potentiating the oncogenic capacity of the other. These data, together with our previous findings in NUP214-ABL1-positive PTPN2-negative T-ALL individuals, suggest that PTPN2 inactivation may appear mainly in association with deregulated tyrosine kinase signaling, the oncogenic activity of which is further increased by the absence of the negative regulator PTPN2. The presence and identity of other oncogenic kinases in T-ALLs featuring loss of PTPN2 but negative for both the NUP214-ABL1 fusion and activating JAK1 mutations remain to be determined. We speculate that other tyrosine phosphatases may be mutated or lost in T-ALL cases with JAK1 mutation, because PTPN2 is only lost in a small subset of JAK1 mutation–positive T-ALLs. This is also to be expected, because deletions of PTPN2 are specifically found in TLX1-positive cases, whereas JAK1 mutations are not restricted to any particular subtype of T-ALL.
Constitutively activated tyrosine kinases activate a variety of signaling pathways that provide proliferation and survival advantages to cancer cells. Both our current results and those from a previous study15 suggest that cancer cells may have the pressure to lose additional negative regulators of oncogenic kinases to further strengthen these proliferation and survival signals, and that cells with such increased proliferation capacity may be selected for during the development and progression of tumors. We have provided evidence for such evolution during the development of T-ALL, but this is likely to also occur in most, if not in all, tumor types. Full genome sequencing of tumors is likely to provide further insights into this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Delphine Desprez and Michel L. Tremblay (Goodman Cancer Research Center, Department of Biochemistry, McGill University, Montreal, QC) for providing shRNA sequence Ptpn2-B.
This work was supported by the Fonds voor Wetenschappelijk Onderzoek (National Fund for Scientific Research)–Vlaanderen (G.0287.07 to J.C.), by the Foundation against Cancer (SCIE2006-34 to J.C.), by a European Research Council Starting Grant (to J.C.), by grants from the Katholieke Universiteit Leuven (concerted action grant to J.C. and P.V.), by the Interuniversity Attraction Poles granted by the Federal Office for Scientific, Technical and Cultural Affairs, Belgium (to J.C., P.V., and S.C.), by a grant from the French program Carte d'Identite des Tumeurs (Ligue Contre le Cancer) and from Canceropole d'Ile de France (to J.S.), by the Associazione Italiana per la Ricerca sul Cancro IG 2009-8803 (to M.T.), by Actions de Recherche Concertées, Communauté Francaise de Belgique, and Direction de la Recherche Scientifique (contract no. ARC 09/14-021 to T.H.). Work in the Macintyre laboratory was supported by grants from the “association Laurette Fugain” and the regional committee of the “Ligue contre le Cancer,” and was performed in collaboration with H. Mossafa, Pasteur Cerba FR. P.V. is a senior clinical investigator of Fonds voor Wetenschappelijk Onderzoek-Vlaanderen.
Authorship
Contribution: M.K. designed the study, performed experiments, analyzed data, and wrote the manuscript; V.A. and N.M. performed experiments and analyzed data; J.S., T. Hornakova, L.K., S.C., F.S., J.P.M., P.V., M.T., R.F., E.M., and T. Haferlach provided reagents and wrote the manuscript; and J.C. designed the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jan Cools, Center for Human Genetics, Gasthuisberg O&N1, Herestraat 49-box 602, 3000 Leuven, Belgium; e-mail: jan.cools@cme.vib-kuleuven.be.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal