Abstract
We previously demonstrated that outcome of pediatric 11q23/MLL-rearranged AML depends on the translocation partner (TP). In this multicenter international study on 733 children with 11q23/MLL-rearranged AML, we further analyzed which additional cytogenetic aberrations (ACA) had prognostic significance. ACAs occurred in 344 (47%) of 733 and were associated with unfavorable outcome (5-year overall survival [OS] 47% vs 62%, P < .001). Trisomy 8, the most frequent specific ACA (n = 130/344, 38%), independently predicted favorable outcome within the ACAs group (OS 61% vs 39%, P = .003; Cox model for OS hazard ratio (HR) 0.54, P = .03), on the basis of reduced relapse rate (26% vs 49%, P < .001). Trisomy 19 (n = 37/344, 11%) independently predicted poor prognosis in ACAs cases, which was partly caused by refractory disease (remission rate 74% vs 89%, P = .04; OS 24% vs 50%, P < .001; HR 1.77, P = .01). Structural ACAs had independent adverse prognostic value for event-free survival (HR 1.36, P = .01). Complex karyotype, defined as ≥ 3 abnormalities, was present in 26% (n = 192/733) and showed worse outcome than those without complex karyotype (OS 45% vs 59%, P = .003) in univariate analysis only. In conclusion, like TP, specific ACAs have independent prognostic significance in pediatric 11q23/MLL-rearranged AML, and the mechanism underlying these prognostic differences should be studied.
Introduction
Pediatric acute myeloid leukemia (AML) is a clinically and genetically heterogeneous disease. In addition to the patient's initial response to treatment, its prognosis is largely determined by the presence of cytogenetic abnormalities and genetic lesions.1–6 Several recurrent cytogenetic abnormalities, such as 11q23/MLL-rearrangements, predict outcome in myeloid neoplasms and acute leukemia.7 So far, > 60 different translocation partners (TPs) have been identified, and new partners are still being reported to add to the diversity of MLL-rearranged leukemia.8,9 The authors of a recent international study10 highlighted the heterogeneity of 11q23/MLL-rearranged pediatric AML by demonstrating that outcome is dependent on TPs. This study also revealed that additional cytogenetic aberrations (ACAs) were an independent adverse prognostic factor,10 but so far, it is unknown which additional aberration(s) determine this unfavorable outcome signature.
The authors of a recent large study in an adult AML cohort11 showed that additional cytogenetic abnormalities in t(9;11)(p22;q23) AML did not affect outcome. However, the Berlin-Frankfurt-Münster group showed that children with t(9;11)(p22;q23) with additional aberrations had lower rates of overall survival (OS) than those with other subgroups of AML.6
To date, no large studies have been undertaken to study the prognostic relevance of specific ACAs in pediatric MLL-rearranged AML. In this multicenter international study, we retrospectively analyzed data from a large cohort (n = 733) to determine which ACAs contribute to the prognostic effect in pediatric MLL-rearranged AML.
Patients and methods
Patients
Patients' data collected in the retrospective international study by Balgobind et al10 were included in this study. In summary, data from 756 patients with 11q23/MLL-rearranged pediatric AML were collected from 11 collaborative study groups—the Berlin-Frankfurt-Münster Study Group (Germany and Austria); the Japanese Pediatric Leukemia/Lymphoma Study Group (Japan); the Leucémies Aiguës Myéloblastiques de l'Enfant Cooperative Group (France); the Czech Pediatric Hematology Working Group (Czech Republic); the St Jude Children's Research Hospital (United States); the Associazione Italiana Ematologia Oncologia Pediatrica (Italy); Research Center for Pediatric Oncology and Hematology (Belarus); the Children's Oncology Group (United States); the Nordic Society for Pediatric Hematology and Oncology (Denmark, Finland, Iceland, Norway, and Sweden); the Dutch Children's Oncology Group (The Netherlands); and 2 centers of the Medical Research Council (United Kingdom). Patients were treated by national/collaborative group AML trials.12–22 The treatment protocols were approved according to local law and guidelines and by the institutional review boards of each participating center, with informed consent obtained from the patients' parents or legal guardians in accordance with the Declaration of Helsinki.
Inclusion criteria for the current analyses were diagnosis between January 1, 1993, and January 1, 2005; younger than 18 years of age at diagnosis; and involvement of 11q23 or MLL as determined by G-, Q-, or R-banded karyotyping; FISH; or RT-PCR. Exclusion criteria were secondary AML after congenital BM failure disorders, aplastic anemia, previous chemotherapy or radiotherapy for other diseases, and previous myelodysplastic syndrome (MDS). Patients with Down syndrome were included if they met the other inclusion criteria. All clinical data obtained at initial diagnosis, data on treatment (therapy protocol, including HSCT), and all events during follow-up were checked for consistency and completeness.10
Cytogenetic analysis
All karyotypes were centrally reviewed by 2 cytogeneticists (J.H., S.C.R.) and assigned to 11q23/MLL-rearranged groups on the basis of TP.10 All karyotypes were designated according to the International System for Human Cytogenetic Nomenclature 2005.23
To analyze ACAs, data from all patients with incomplete karyotypes were excluded. For all cases included in the analysis, the number of aberrations was counted. Each aberration separated from the rest of the karyotype by a comma was counted as one abnormality (regardless of its complexity), every aberration was counted only once (if present in multiple clones), and constitutional aberrations were excluded. Triploidy and tetraploidy were counted as 1 aberration (1 event). In this cohort of 11q23/MLL-rearranged cases, ACAs cases were defined as having 2 or more aberrations, including the 11q23/MLL-rearrangement (n = 344). All cases with 3 or more aberrations were considered having a complex karyotype, consistent with previously used definitions.24,25 Numerical aberrations were defined as loss or gain of a full chromosome. Balanced translocations were defined as translocations in which no material seemed to be gained or lost as determined by conventional karyotyping. Structural aberrations were defined as aberrations resulting from breakpoints within a chromosome. In all unbalanced translocations we described which material was lost and gained and also whether 11q23 was involved. The presence of a balanced overall karyotype was defined as a karyotype with 2 complete copies of all autosomes and complete copies of sex chromosomes without any additional material (2n). Definitions used for cytogenetic classification are summarized in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Statistical analyses
Complete remission (CR) was defined as < 5% blasts in the BM, with regeneration of trilineage hematopoiesis plus absence of extramedullary disease.26 Early death was defined as any death within the first 6 weeks of treatment. Treatment of patients who did not obtain CR within the specified time in the protocol was considered a failure on day 0. OS was measured from the date of diagnosis to the date of last follow-up or death from any cause. Event-free survival (EFS) was calculated from the date of diagnosis to the first event or to the date of last follow-up. Events included nonremittance, relapse, secondary malignancy, or death from any cause. Cumulative incidence of relapse (CIR) was calculated from the date of CR to the first relapse. Refractory disease was included in the EFS and CIR analyses by arbitrarily setting the event date on day 0. For OS, EFS, and CIR analyses, patients who did not experience an event were censored at the time of last follow-up.
The Kaplan-Meier method was used to estimate the 5-year probabilities of OS and EFS, and survival estimates were compared by the log-rank test. The Gray test for competing risks was used for CIR analysis. Multivariate analyses were performed with the Cox proportional hazards model. Continuous variables known to be of prognostic value in AML were categorized according to cutoff points (eg, > 2 or 10 years of age, white blood cell [WBC] count < 20 × 109/L or > 100 × 109/L]). The χ2 or Fisher exact test was used to compare differences in proportions of variables among groups; the Mann-Whitney U test was used for continuous variables. All P values are descriptive and explorative and were considered significant if ≤ .05. All statistical data were analyzed by the use of SAS-PC, Version 9.1 (SAS Institute Inc).
Results
Distribution of ACAs
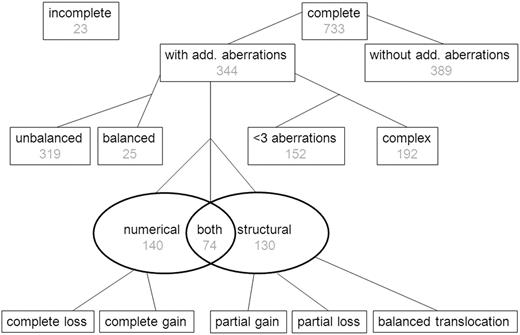
Of the 756 patients, 733 (97%) had complete karyotypes, and their data were included in the study (see flowchart in Figure 1). There were no significant differences in the patients included (n = 733) and not included (n = 23) in this study with respect to sex, age, WBC count, and TP group (data not shown). ACAs were found in 344 (47%) of 733 cases (Figure 1). The number of additional aberrations ranged from 0 to 15 (mean, 1.2 additional aberrations; supplemental Figure 1).
Flow chart showing the presence and type of ACAs in 756 pediatric patients with 11q23/MLL-rearranged AML. Complete karyotypes were not available for 23 patients, and they were therefore excluded from analyses. The presence or absence of ACAs was determined for 733 patients for whom complete karyotypes were available. In the cohort having ACAs balanced karyotype was coded for 25 patients; the remaining had an unbalanced karyotype. The types of aberrations were coded as numerical, structural, or both, and the number of aberrations was also coded. Losses and gains are further coded in other figures.
Flow chart showing the presence and type of ACAs in 756 pediatric patients with 11q23/MLL-rearranged AML. Complete karyotypes were not available for 23 patients, and they were therefore excluded from analyses. The presence or absence of ACAs was determined for 733 patients for whom complete karyotypes were available. In the cohort having ACAs balanced karyotype was coded for 25 patients; the remaining had an unbalanced karyotype. The types of aberrations were coded as numerical, structural, or both, and the number of aberrations was also coded. Losses and gains are further coded in other figures.
There were 3 or more aberrations (including the 11q23/MLL-rearrangement) in 192 of 733 (26%) cases, which were therefore defined as complex karyotypes. Of the 344 cases with ACAs, 140 (41%) had numerical ACAs only, 130 (38%) had structural ACAs only, and 74 (22%) had both numerical and structural ACAs (Figure 1). There were 25 (7%) cases of ACA that had only balanced structural abnormalities in their karyotypes (Figure 1).
Distribution of ACAs in clinically relevant groups
Tables 1 and 2 show the distribution of ACAs by TP group and clinically relevant parameters (sex, age, WBC count, and FAB [ie, French-American-British] subtype). TP groups 9p22 and 19p13 were characterized by a relatively high frequency of numerical ACAs, whereas groups 10p12, 10p11.2, and 4q21 showed greater prevalence of structural ACAs (P < .001; Table 1). Also, there were significant differences in the number of aberrations among TP groups: the 6q27 group had a relatively high number of ACAs (P = .002), whereas groups 9p22, 19p13, and 1q21 had a lower number of ACAs (Table 2).
Distribution of ACAs by translocation partner and clinically relevant parameters
| . | n . | ACAs, n (%) . | ACAs type . | ||
|---|---|---|---|---|---|
| Numerical, n (%) . | Structural, n (%) . | Both, n (%) . | |||
| TP group | |||||
| 9p22 | 316 | 148 (47) | 84 (57)* | 40 (27)* | 24 (16)* |
| 10p12 | 96 | 48 (50) | 13 (27)* | 26 (54)* | 9 (19)* |
| 6q27 | 35 | 17 (49) | 8 (47)* | 7 (41)* | 2 (12)* |
| 19p13 | 30 | 10 (33) | 6 (60)* | 1 (10)* | 3 (30)* |
| 19p13.1 | 34 | 13 (38) | 5 (38)* | 4 (31)* | 4 (31)* |
| 19p13.3 | 25 | 13 (52) | 5 (38)* | 4 (31)* | 4 (31)* |
| 1q21 | 24 | 6 (25) | 2 (33)* | 3 (50)* | 1 (17)* |
| 4q21 | 13 | 8 (62) | 2 (25)* | 4 (50)* | 2 (25)* |
| 10p11.2 | 12 | 7 (58) | 0* | 5 (71)* | 2 (29)* |
| 17q21 | 12 | 3 (25) | 1 (33)* | 1 (33)* | 1 (33)* |
| other | 136 | 71 (52) | 14 (20)* | 35 (49)* | 22 (31)* |
| 733 | |||||
| Sex | |||||
| Male | 358 | 171 (48) | 78 (46) | 63 (37) | 30 (18) |
| Female | 375 | 173 (46) | 62 (36) | 67 (39) | 44 (25) |
| 733 | |||||
| Age, y | |||||
| < 2 | 344 | 143 (42) | 45 (31)† | 68 (48)† | 30 (21)† |
| 2-9 | 219 | 115 (53) | 57 (50)† | 35 (30)† | 23 (20)† |
| ≥ 10 | 170 | 86 (51) | 38 (44)† | 27 (31)† | 21 (24)† |
| 733 | |||||
| WBC, ×109/L | |||||
| < 20 | 339 | 175 (52) | 84 (49)† | 51 (29)† | 40 (23)† |
| 20-99 | 203 | 87 (43) | 28 (32)† | 40 (46)† | 19 (22)† |
| ≥ 100 | 171 | 73 (43) | 25 (34)† | 35 (48)† | 13 (18)† |
| 713 | |||||
| FAB | |||||
| M0 | 23 | 12 (52) | 6 (50) | 3 (25) | 3 (25) |
| M1 | 39 | 20 (51) | 9 (45) | 7 (35) | 4 (20) |
| M2 | 32 | 12 (38) | 7 (58) | 4 (33) | 1 (8) |
| M4 | 134 | 49 (37) | 21 (43) | 21 (43) | 7 (14) |
| M5 | 446 | 217 (49) | 88 (41) | 83 (38) | 46 (21) |
| M7 | 19 | 15 (79) | 7 (47) | 2 (13) | 6 (4) |
| n.d. | 7 | 5 (71) | 0 | 2 (40) | 3 (60) |
| 700 | |||||
| Overall | 344 (47) | 140 (41) | 130 (38) | 74 (22) | |
| . | n . | ACAs, n (%) . | ACAs type . | ||
|---|---|---|---|---|---|
| Numerical, n (%) . | Structural, n (%) . | Both, n (%) . | |||
| TP group | |||||
| 9p22 | 316 | 148 (47) | 84 (57)* | 40 (27)* | 24 (16)* |
| 10p12 | 96 | 48 (50) | 13 (27)* | 26 (54)* | 9 (19)* |
| 6q27 | 35 | 17 (49) | 8 (47)* | 7 (41)* | 2 (12)* |
| 19p13 | 30 | 10 (33) | 6 (60)* | 1 (10)* | 3 (30)* |
| 19p13.1 | 34 | 13 (38) | 5 (38)* | 4 (31)* | 4 (31)* |
| 19p13.3 | 25 | 13 (52) | 5 (38)* | 4 (31)* | 4 (31)* |
| 1q21 | 24 | 6 (25) | 2 (33)* | 3 (50)* | 1 (17)* |
| 4q21 | 13 | 8 (62) | 2 (25)* | 4 (50)* | 2 (25)* |
| 10p11.2 | 12 | 7 (58) | 0* | 5 (71)* | 2 (29)* |
| 17q21 | 12 | 3 (25) | 1 (33)* | 1 (33)* | 1 (33)* |
| other | 136 | 71 (52) | 14 (20)* | 35 (49)* | 22 (31)* |
| 733 | |||||
| Sex | |||||
| Male | 358 | 171 (48) | 78 (46) | 63 (37) | 30 (18) |
| Female | 375 | 173 (46) | 62 (36) | 67 (39) | 44 (25) |
| 733 | |||||
| Age, y | |||||
| < 2 | 344 | 143 (42) | 45 (31)† | 68 (48)† | 30 (21)† |
| 2-9 | 219 | 115 (53) | 57 (50)† | 35 (30)† | 23 (20)† |
| ≥ 10 | 170 | 86 (51) | 38 (44)† | 27 (31)† | 21 (24)† |
| 733 | |||||
| WBC, ×109/L | |||||
| < 20 | 339 | 175 (52) | 84 (49)† | 51 (29)† | 40 (23)† |
| 20-99 | 203 | 87 (43) | 28 (32)† | 40 (46)† | 19 (22)† |
| ≥ 100 | 171 | 73 (43) | 25 (34)† | 35 (48)† | 13 (18)† |
| 713 | |||||
| FAB | |||||
| M0 | 23 | 12 (52) | 6 (50) | 3 (25) | 3 (25) |
| M1 | 39 | 20 (51) | 9 (45) | 7 (35) | 4 (20) |
| M2 | 32 | 12 (38) | 7 (58) | 4 (33) | 1 (8) |
| M4 | 134 | 49 (37) | 21 (43) | 21 (43) | 7 (14) |
| M5 | 446 | 217 (49) | 88 (41) | 83 (38) | 46 (21) |
| M7 | 19 | 15 (79) | 7 (47) | 2 (13) | 6 (4) |
| n.d. | 7 | 5 (71) | 0 | 2 (40) | 3 (60) |
| 700 | |||||
| Overall | 344 (47) | 140 (41) | 130 (38) | 74 (22) | |
ACAs (%) indicates number of cases with additional aberrations and percentage within this group; Numerical (%), number of cases with only numerical additional aberrations and percentage of specific group (row); Structural (%), number of cases with only structural additional aberrations and percentage of specific group (row); Both (%), number of cases with both numerical and structural additional aberrations and percentage of specific group (row); and TP group, site of translocation on partner chromosome
ACA indicates additional cytogenetic aberrations; dx, diagnosis; FAB, French American British morphology classification subtype; n.d., not determined; TP, translocation partner; and WBC, white blood cell count.
Values significantly different at the P < .01 level (χ2).
Values significantly different at the †P < .05 level (χ2).
Number of aberrations by 11q23 translocation partner and clinically relevant parameters
| . | Number of aberrations . | |||||||
|---|---|---|---|---|---|---|---|---|
| 0 . | 1 . | 2 . | 3 . | 4 . | 5 . | > 5 . | All . | |
| TP group | ||||||||
| 9p22 | 168 (44)* | 75 (49)* | 33 (41)* | 19 (43)* | 7 (28)* | 14 (33)* | 316 (43) | |
| 10p12 | 1 (14)* | 47 (12)* | 19 (13)* | 12 (15)* | 7 (16)* | 5 (20)* | 5 (12)* | 96 (13) |
| 6q27 | 1 (14)* | 17 (4)* | 7 (5)* | 1 (1)* | 9 (21)* | 35 (5) | ||
| 19p13 | 20 (5)* | 3 (2)* | 4 (5)* | 1 (2)* | 1 (4)* | 1 (2)* | 30 (4) | |
| 19p13.1 | 1 (14)* | 20 (5)* | 7 (5)* | 2 (3)* | 1 (2)* | 3 (12)* | 34 (5) | |
| 19p13.3 | 12 (3)* | 7 (5)* | 3 (7)* | 2 (8)* | 1 (2)* | 25 (3) | ||
| 1q21 | 18 (5)* | 3 (2)* | 1 (1)* | 1 (2)* | 1 (2)* | 24 (3) | ||
| 4q21 | 5 (1)* | 2 (1)* | 4 (5)* | 1 (2)* | 1 (4)* | 13 (2) | ||
| 10p11.2 | 5 (1)* | 2 (1)* | 3 (4)* | 2 (8)* | 12 (2) | |||
| 17q21 | 9 (2)* | 1 (1)* | 1 (1)* | 1 (2)* | 12 (2) | |||
| Other | 4 (57)* | 61 (16)* | 26 (17)* | 19 (24)* | 11 (25)* | 4 (16)* | 11 (26)* | 136 (19) |
| 733 | ||||||||
| Sex | ||||||||
| Male | 187 (49)† | 89 (59)† | 35 (44)† | 13 (30)† | 11 (44)† | 23 (53)† | 358 (49) | |
| Female | 7 (100)† | 195 (51)† | 63 (41)† | 45 (56)† | 31 (70)† | 14 (56)† | 20 (47)† | 375 (51) |
| 733 | ||||||||
| Age, y | ||||||||
| < 2 | 4 (57)* | 197 (52)* | 61 (40)* | 39 (49)* | 12 (27)* | 16 (64)* | 15 (35)* | 344 (47) |
| 2-9 | 104 (27)* | 49 (32)* | 29 (36)* | 22 (50)* | 5 (20)* | 10 (23)* | 219 (30) | |
| ≥ 10 | 3 (43)* | 81 (21)* | 42 (28)* | 12 (15)* | 10 (23)* | 4 (16)* | 18 (42)* | 170 (23) |
| 733 | ||||||||
| WBC, ×109/L | ||||||||
| < 20 | 5 (71) | 159 (42) | 76 (50) | 39 (49) | 23 (52) | 16 (64) | 21 (49) | 339 (46) |
| 20-99 | 1 (14) | 115 (30) | 38 (25) | 20 (25) | 14 (32) | 5 (20) | 10 (23) | 203 (28) |
| ≥ 100 | 1 (14) | 97 (25) | 34 (22) | 19 (24) | 7 (16) | 2 (8) | 11 (26) | 171 (23) |
| 713 (97) | ||||||||
| FAB | ||||||||
| M0 | 11 (3)* | 4 (3)* | 2 (3)* | 3 (7)* | 1 (4)* | 2 (5)* | 23 (3) | |
| M1 | 19 (5)* | 12 (8)* | 4 (5)* | 1 (2)* | 3 (7)* | 39 (5) | ||
| M2 | 1 (14)* | 19 (5)* | 7 (5)* | 2 (3)* | 3 (7)* | 32 (4) | ||
| M4 | 2 (29)* | 83 (22)* | 25 (16)* | 12 (15)* | 5 (11)* | 2 (6)* | 5 (12)* | 134 (18) |
| M5 | 4 (57)* | 225 (59)* | 97 (64)* | 54 (68)* | 24 (55)* | 17 (68)* | 25 (58)* | 446 (61) |
| M7 | 4 (1)* | 3 (2)* | 2 (3)* | 4 (9)* | 6 (14)* | 19 (3) | ||
| n.d. | 2 (1)* | 1 (1)* | 1 (1)* | 1 (2)* | 1 (4)* | 1 (2)* | 7 (1) | |
| 700 (95) | ||||||||
| Overall | 7 (1) | 382 (52) | 152 (21) | 80 (11) | 44 (6) | 25 (3) | 43 (6) | 733 |
| . | Number of aberrations . | |||||||
|---|---|---|---|---|---|---|---|---|
| 0 . | 1 . | 2 . | 3 . | 4 . | 5 . | > 5 . | All . | |
| TP group | ||||||||
| 9p22 | 168 (44)* | 75 (49)* | 33 (41)* | 19 (43)* | 7 (28)* | 14 (33)* | 316 (43) | |
| 10p12 | 1 (14)* | 47 (12)* | 19 (13)* | 12 (15)* | 7 (16)* | 5 (20)* | 5 (12)* | 96 (13) |
| 6q27 | 1 (14)* | 17 (4)* | 7 (5)* | 1 (1)* | 9 (21)* | 35 (5) | ||
| 19p13 | 20 (5)* | 3 (2)* | 4 (5)* | 1 (2)* | 1 (4)* | 1 (2)* | 30 (4) | |
| 19p13.1 | 1 (14)* | 20 (5)* | 7 (5)* | 2 (3)* | 1 (2)* | 3 (12)* | 34 (5) | |
| 19p13.3 | 12 (3)* | 7 (5)* | 3 (7)* | 2 (8)* | 1 (2)* | 25 (3) | ||
| 1q21 | 18 (5)* | 3 (2)* | 1 (1)* | 1 (2)* | 1 (2)* | 24 (3) | ||
| 4q21 | 5 (1)* | 2 (1)* | 4 (5)* | 1 (2)* | 1 (4)* | 13 (2) | ||
| 10p11.2 | 5 (1)* | 2 (1)* | 3 (4)* | 2 (8)* | 12 (2) | |||
| 17q21 | 9 (2)* | 1 (1)* | 1 (1)* | 1 (2)* | 12 (2) | |||
| Other | 4 (57)* | 61 (16)* | 26 (17)* | 19 (24)* | 11 (25)* | 4 (16)* | 11 (26)* | 136 (19) |
| 733 | ||||||||
| Sex | ||||||||
| Male | 187 (49)† | 89 (59)† | 35 (44)† | 13 (30)† | 11 (44)† | 23 (53)† | 358 (49) | |
| Female | 7 (100)† | 195 (51)† | 63 (41)† | 45 (56)† | 31 (70)† | 14 (56)† | 20 (47)† | 375 (51) |
| 733 | ||||||||
| Age, y | ||||||||
| < 2 | 4 (57)* | 197 (52)* | 61 (40)* | 39 (49)* | 12 (27)* | 16 (64)* | 15 (35)* | 344 (47) |
| 2-9 | 104 (27)* | 49 (32)* | 29 (36)* | 22 (50)* | 5 (20)* | 10 (23)* | 219 (30) | |
| ≥ 10 | 3 (43)* | 81 (21)* | 42 (28)* | 12 (15)* | 10 (23)* | 4 (16)* | 18 (42)* | 170 (23) |
| 733 | ||||||||
| WBC, ×109/L | ||||||||
| < 20 | 5 (71) | 159 (42) | 76 (50) | 39 (49) | 23 (52) | 16 (64) | 21 (49) | 339 (46) |
| 20-99 | 1 (14) | 115 (30) | 38 (25) | 20 (25) | 14 (32) | 5 (20) | 10 (23) | 203 (28) |
| ≥ 100 | 1 (14) | 97 (25) | 34 (22) | 19 (24) | 7 (16) | 2 (8) | 11 (26) | 171 (23) |
| 713 (97) | ||||||||
| FAB | ||||||||
| M0 | 11 (3)* | 4 (3)* | 2 (3)* | 3 (7)* | 1 (4)* | 2 (5)* | 23 (3) | |
| M1 | 19 (5)* | 12 (8)* | 4 (5)* | 1 (2)* | 3 (7)* | 39 (5) | ||
| M2 | 1 (14)* | 19 (5)* | 7 (5)* | 2 (3)* | 3 (7)* | 32 (4) | ||
| M4 | 2 (29)* | 83 (22)* | 25 (16)* | 12 (15)* | 5 (11)* | 2 (6)* | 5 (12)* | 134 (18) |
| M5 | 4 (57)* | 225 (59)* | 97 (64)* | 54 (68)* | 24 (55)* | 17 (68)* | 25 (58)* | 446 (61) |
| M7 | 4 (1)* | 3 (2)* | 2 (3)* | 4 (9)* | 6 (14)* | 19 (3) | ||
| n.d. | 2 (1)* | 1 (1)* | 1 (1)* | 1 (2)* | 1 (4)* | 1 (2)* | 7 (1) | |
| 700 (95) | ||||||||
| Overall | 7 (1) | 382 (52) | 152 (21) | 80 (11) | 44 (6) | 25 (3) | 43 (6) | 733 |
The number of aberrations indicates total number of aberrations in the karyotype, including 11q23/MLL-rearrangement, percentages per group shown in parentheses (per column).
dx indicates diagnosis; FAB, French American British morphology classification subtype; n.d., not determined; TP, translocation partner; and WBC, white blood cell count.
Significantly different at the P < .01 level (χ2).
Significantly different at the P < .05 level (χ2).
ACAs were less likely to occur in young children (< 2 years of age) than in children 2-9 years of age or 10 years or older (42% vs 53% vs 51%, P = .02; Table 1). However, structural ACAs were more frequent in children < 2 years of age than in children 2-9 years of age or 10-18 years of age (48% vs 30% vs 31%, P < .01; Table 1). There was a greater prevalence of highly complex karyotypes (> 5 aberrations) in children 10-18 years of age than those younger than 2 years or 2-9 years of age (11% vs 4% vs 5%, P = .02, Table 2).
Although the number of patients with FAB M7 was small, ACAs were more likely to occur in patients with AML FAB M7 compared with those with other FAB types (79% vs 46%, P = .008), whereas patients with AML FAB M2 and M4 had the lowest occurrence of ACAs (Table 1). Also, patients with AML FAB M7 seem to have a higher number of aberrations than those with other FAB morphologies (P = .003; Table 2).
Specific recurrent aberrations
Trisomy 8 was the most frequently occurring numerical abnormality (130/733, 18% of all cases and 38% of ACA cases, Figure 2A). In addition, trisomy 4, 6, 13, 19, and 21 were recurrent ACAs (at least 15 cases each). Two cases with Down syndrome were included in this study. However, because constitutional aberrations were not included in the additional aberrations, they were not included in the trisomy 21 group. Only 11 patients had losses of full chromosomes, collectively accounting for 25 monosomies (Figure 2A).
Frequency (number of cases) of numerical and structural ACAs. (A) Numerical ACAs. Gains are shown on the positive y-axis, and losses are shown on the negative y-axis. Chromosomes are on the x-axis. (B) Structural ACAs The short arms (p) of the chromosomes are shown on the positive y-axis and the long arms (q) on the negative y-axis. Lightest shades are used for losses, medium-shaded colors are used for gains, and the darkest-shaded colors for breakpoints of balanced translocations. Chromosomes are on the x-axis. Balanced 11q23 translocations are not included in the figure.
Frequency (number of cases) of numerical and structural ACAs. (A) Numerical ACAs. Gains are shown on the positive y-axis, and losses are shown on the negative y-axis. Chromosomes are on the x-axis. (B) Structural ACAs The short arms (p) of the chromosomes are shown on the positive y-axis and the long arms (q) on the negative y-axis. Lightest shades are used for losses, medium-shaded colors are used for gains, and the darkest-shaded colors for breakpoints of balanced translocations. Chromosomes are on the x-axis. Balanced 11q23 translocations are not included in the figure.
Figure 2B shows the collective analysis of structural ACAs per chromosome arm but does not include breakpoints involved in balanced 11q23/MLL-translocations. However, the figure includes unbalanced 11q23/MLL-translocations in which chromosomal material was lost or gained. Chromosomes 1 and 11 were most frequently involved in structural ACAs. Analysis of specific breakpoints showed that 11q23 was the only breakpoint found more than 10 times (data not shown).
Univariate analysis of the prognostic impact of ACAs on survival
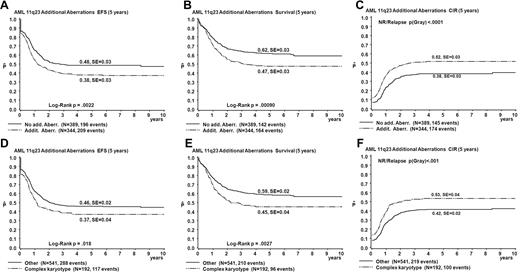
Table 3 summarizes results of the univariate analysis of survival parameters. The EFS and OS estimates of patients with ACAs were significantly lower than those without ACAs (EFS 38% vs 48%, P = .002; OS 47% vs 62%, P < .001; Figure 3). CIR estimates of patients with ACAs were significantly greater than for those without ACAs (52% vs 38%, P < .001; Figure 3). Patients with complex karyotypes had significantly worse outcomes than those without complex karyotypes (EFS 37% vs 46%, P = .02; OS 45% vs 59%, P = .003; CIR 53% vs 42%, P < .001; Figure 3).
Univariate survival analysis of the complete cohort (n = 733)
| . | Complete cohort . | ||||||
|---|---|---|---|---|---|---|---|
| n . | EFS . | P (log-rank) . | OS . | P (log-rank) . | CIR . | P (Gray) . | |
| Additional aberrations | .002† | < .001† | < .001† | ||||
| Absent | 389 | 0.48 | 0.62 | 0.38 | |||
| Present | 344 | 0.38 | 0.47 | 0.52 | |||
| No. of aberrations | < .001† | < .001† | .001† | ||||
| 2 | 152 | 0.39 | 0.50 | 0.50 | |||
| 3 | 80 | 0.45 | 0.53 | 0.48 | |||
| ≥ 3 | 192 | 0.37 | .018* | 0.45 | .003† | 0.53 | < .001† |
| 4 | 44 | 0.40 | 0.50 | 0.53 | |||
| 5 | 25 | 0.36 | 0.43 | 0.60 | |||
| > 5 | 43 | 0.18 | < .001† | 0.25 | 0.61 | ||
| Type | .001† | .003† | < .001† | ||||
| Numerical | 140 | 0.47 | 0.56 | 0.41 | |||
| Structural | 130 | 0.32 | 0.43 | 0.59 | |||
| Both | 74 | 0.31 | 0.40 | 0.59 | |||
| Trisomy | |||||||
| 4 | 20 | 0.43 | .72 | 0.52 | .87 | 0.52 | .93 |
| 6 | 36 | 0.35 | .43 | 0.35 | .029* | 0.54 | .65 |
| 8 | 130 | 0.53 | < .001† | 0.61 | .003† | 0.35 | < .001† |
| 13 | 18 | 0.49 | .52 | 0.64 | .41 | 0.40 | .37 |
| 19 | 37 | 0.17 | .003† | 0.24 | < .001† | 0.54 | .88 |
| 21 | 35 | 0.19 | .007† | 0.28 | .015* | 0.69 | .014* |
| . | Complete cohort . | ||||||
|---|---|---|---|---|---|---|---|
| n . | EFS . | P (log-rank) . | OS . | P (log-rank) . | CIR . | P (Gray) . | |
| Additional aberrations | .002† | < .001† | < .001† | ||||
| Absent | 389 | 0.48 | 0.62 | 0.38 | |||
| Present | 344 | 0.38 | 0.47 | 0.52 | |||
| No. of aberrations | < .001† | < .001† | .001† | ||||
| 2 | 152 | 0.39 | 0.50 | 0.50 | |||
| 3 | 80 | 0.45 | 0.53 | 0.48 | |||
| ≥ 3 | 192 | 0.37 | .018* | 0.45 | .003† | 0.53 | < .001† |
| 4 | 44 | 0.40 | 0.50 | 0.53 | |||
| 5 | 25 | 0.36 | 0.43 | 0.60 | |||
| > 5 | 43 | 0.18 | < .001† | 0.25 | 0.61 | ||
| Type | .001† | .003† | < .001† | ||||
| Numerical | 140 | 0.47 | 0.56 | 0.41 | |||
| Structural | 130 | 0.32 | 0.43 | 0.59 | |||
| Both | 74 | 0.31 | 0.40 | 0.59 | |||
| Trisomy | |||||||
| 4 | 20 | 0.43 | .72 | 0.52 | .87 | 0.52 | .93 |
| 6 | 36 | 0.35 | .43 | 0.35 | .029* | 0.54 | .65 |
| 8 | 130 | 0.53 | < .001† | 0.61 | .003† | 0.35 | < .001† |
| 13 | 18 | 0.49 | .52 | 0.64 | .41 | 0.40 | .37 |
| 19 | 37 | 0.17 | .003† | 0.24 | < .001† | 0.54 | .88 |
| 21 | 35 | 0.19 | .007† | 0.28 | .015* | 0.69 | .014* |
CIR, indicates 5-year cumulative incidence of relapse; EFS, 5-year event-free survival estimates; n, number of patients; OS, 5-year overall survival estimate; P (Gray), P value from the Gray test; and P (log-rank), P value from log-rank test.
Significant at P < .05 level.
Significant at P < .01 level.
Survival curves obtained from univariate analysis comparing patients with ACAs to patients without ACAs and comparing patients with complex karyotype with all patients with < 3 aberrations. (A-C) Patients with ACAs are compared to patients without ACAs. (D-F) Patients with complex karyotype are compared to patients with < 3 aberrations. EFS (A,D), OS (Survival; B,E), and CIR (C,F).
Survival curves obtained from univariate analysis comparing patients with ACAs to patients without ACAs and comparing patients with complex karyotype with all patients with < 3 aberrations. (A-C) Patients with ACAs are compared to patients without ACAs. (D-F) Patients with complex karyotype are compared to patients with < 3 aberrations. EFS (A,D), OS (Survival; B,E), and CIR (C,F).
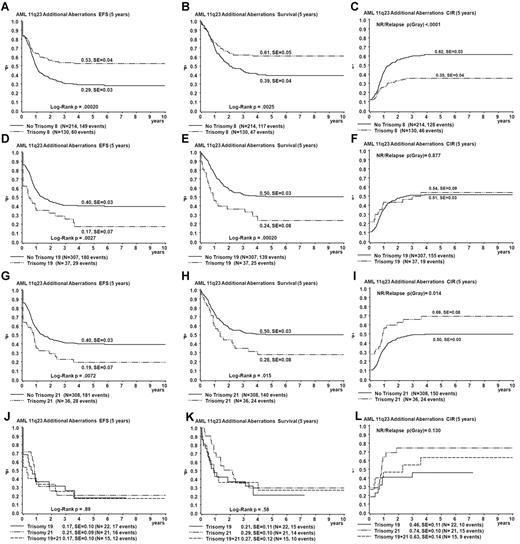
The presence of trisomy 8 (n = 130) was a favorable prognostic factor (EFS 53% vs 29% for patients without trisomy 8, P < .001; OS 61% vs 39% for patients without trisomy 8, P = .003; CIR 35% vs 62% for patients without trisomy 8, P < .001; Figure 4). Survival differences are mainly explained by reduced relapse rate in trisomy 8 patients (relapse rate 26% vs 49% for patients without trisomy 8, P < .001; Figure 4). The presence of trisomy 19 (n = 37) and trisomy 21 (n = 36) was an unfavorable prognostic factor (EFS 17% vs 40% for patients without trisomy 19, P = .003; OS 24% vs 50% for patients without trisomy 19, P < .001; CIR 54% vs 51% for patients without trisomy 19, P = .88; and EFS 19% vs 40% for patients without trisomy 21, P = .007; OS 28% vs 50% for patients without trisomy 21, P = .02; CIR 69% vs 50% for patients without trisomy 21, P = .01; Figure 4). Both trisomies 19 and 21 were present in 15 patients. Survival curves for patients with either trisomy 19 or 21 were not different from those for patients with both trisomies 19 and 21 (Figure 4). Combined trisomy 19 and trisomy 8 was present in 23 patients. These patients showed a survival curve intermediate to that of trisomy 8 and trisomy 19 cases (EFS 30%, data not shown). The survival disadvantage of patients with trisomy 19 seems to be determined by refractory disease (probability of CR 74% for patients with trisomy 19 vs 89% for patients with other ACAs, as calculated over the fraction of patients who survive beyond the first 6 weeks after diagnosis, P = .04) rather than relapse. In addition, patients with trisomy 19 had a significantly greater incidence of early death (16% vs 3.3% in other ACA cases, P = .004), which could not be explained by adverse clinical prognostic factors such as greater WBC or age. Structural aberrations were diverse and randomly distributed among TP groups and survival analysis of patients with specific breakpoints was not feasible because none of the breakpoints was involved > 10 times.
Comparison of survival curves obtained from univariate analysis for patients with trisomy 8, trisomy 19, and those with trisomy 21 and defined by strata of occurrence of trisomy 19 and trisomy 21. For curves A-I, patients with a specific trisomy are compared with patients with other ACAs. Patients with trisomy 8 are shown in parels A-C, patients with trisomy 19 in panels D-F, and patients with trisomy 21 in panels G-I. The strata of occurrence of trisomy 19 and trisomy 21 are shown in panels J-L. EFS (A,D,G,J), OS (Survival; B,E,H,K), and CIR (C,F,I,L).
Comparison of survival curves obtained from univariate analysis for patients with trisomy 8, trisomy 19, and those with trisomy 21 and defined by strata of occurrence of trisomy 19 and trisomy 21. For curves A-I, patients with a specific trisomy are compared with patients with other ACAs. Patients with trisomy 8 are shown in parels A-C, patients with trisomy 19 in panels D-F, and patients with trisomy 21 in panels G-I. The strata of occurrence of trisomy 19 and trisomy 21 are shown in panels J-L. EFS (A,D,G,J), OS (Survival; B,E,H,K), and CIR (C,F,I,L).
Multivariate analyses of the prognostic impact of ACAs on survival
Table 4 summarizes results of the multivariate survival analysis. Cox proportional hazards model for EFS, OS, and relapse incidence of the full cohort (n = 733) showed that trisomy 8 and trisomy 19 were independent prognostic factors at P < .05 for EFS (hazard ratio [HR] 0.57, P = .02; and HR 1.77, P = .01) and OS (HR 0.54, P = .03; and HR 2.11, P = .002; Table 4). Structural aberrations as a general finding predicted EFS (HR 1.39, P = .01; Table 4). The TPs identified by Balgobind et al10 (10p12, 6q27, 1q21, and 10p11.2) remained significant independent prognostic factors in these models. Trisomy 8, 19, and 21 were not significant factors in the model for the prediction of relapse incidence. Complexity of the karyotype, tested by different cutoff values (≥ 2 aberrations, ≥ 3 aberrations, and > 5 aberrations), was not a significant factor for outcome in all models and was therefore excluded from the final model. A separate analysis of t(9;11)(p22;q23) cases showed that they did not differ considerably from the complete cohort (supplemental Figure 2 and supplemental Table 2).
Multivariate survival analysis of the complete cohort by use of the Cox proportional hazards model
| . | Cox proportional hazards model . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| EFS . | OS . | Relapse incidence . | |||||||
| HR . | CI . | P . | HR . | CI . | P . | HR . | CI . | P . | |
| TP | |||||||||
| 9p22 | 1 | reference | 1 | reference | 1 | reference | |||
| other | 1.15 | (0.87-1.51) | .328 | 1.13 | (0.82-1.57) | .461 | 1.17 | (0.92-1.47) | .195 |
| 10p12 | 1.36 | (1.01-1.83) | .042* | 1.62 | (1.16-2.27) | .005† | 1.76 | (1.36-2.29) | .000† |
| 6q27 | 2.29 | (1.54-3.39) | .000† | 2.72 | (1.77-4.19) | .000† | 2.79 | (1.80-4.33) | .000† |
| 19p13 | 1.06 | (0.62-1.80) | .832 | 1.44 | (0.82-2.51) | .204 | 0.88 | (0.57-1.37) | .579 |
| 19p13.1 | 1.11 | (0.69-1.79) | .667 | 0.97 | (0.53-1.77) | .931 | 1.04 | (0.71-1.53) | .841 |
| 19p13.3 | 1.06 | (0.60-1.88) | .832 | 1.64 | (0.90-3.00) | .105 | 1.18 | (0.71-1.94) | .522 |
| 1q21 | 0.12 | (0.03-0.49) | .003† | 0.00 | 0.68 | (0.44-1.05) | .080 | ||
| 4q21 | 1.46 | (0.74-2.88) | .276 | 2.04 | (1.02-4.09) | .043* | 1.84 | (0.99-3.43) | .054 |
| 10p11.2 | 2.12 | (1.10-4.06) | .024* | 2.56 | (1.24-5.32) | .011* | 1.37 | (0.67-2.78) | .384 |
| 17q21 | 1.14 | (0.53-2.43) | .743 | 1.15 | (0.47-2.82) | .763 | 1.28 | (0.68-2.42) | .446 |
| Trisomy | |||||||||
| No trisomy | 1 | reference | 1 | reference | 1 | reference | |||
| 8 | 0.57 | (0.36-0.92) | .022* | 0.54 | (0.32-0.94) | .028* | 0.79 | (0.56-1.12) | .188 |
| 19 | 1.77 | (1.13-2.78) | .012* | 2.11 | (1.31-3.42) | .002† | 1.15 | (0.68-1.94) | .596 |
| 21 | 1.35 | (0.85-2.13) | .198 | 1.25 | (0.76-2.03) | .377 | 0.98 | (0.60-1.60) | .926 |
| Type | |||||||||
| No ACAs | 1 | reference | 1 | reference | 1 | reference | |||
| numerical | 1.16 | (0.83-1.63) | .376 | 1.17 | (0.84-1.62) | .353 | 1.09 | (0.81-1.47) | .588 |
| structural | 1.39 | (1.07-1.80) | .013* | 1.27 | (0.98-1.63) | .068 | 1.13 | (0.90-1.43) | .288 |
| . | Cox proportional hazards model . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| EFS . | OS . | Relapse incidence . | |||||||
| HR . | CI . | P . | HR . | CI . | P . | HR . | CI . | P . | |
| TP | |||||||||
| 9p22 | 1 | reference | 1 | reference | 1 | reference | |||
| other | 1.15 | (0.87-1.51) | .328 | 1.13 | (0.82-1.57) | .461 | 1.17 | (0.92-1.47) | .195 |
| 10p12 | 1.36 | (1.01-1.83) | .042* | 1.62 | (1.16-2.27) | .005† | 1.76 | (1.36-2.29) | .000† |
| 6q27 | 2.29 | (1.54-3.39) | .000† | 2.72 | (1.77-4.19) | .000† | 2.79 | (1.80-4.33) | .000† |
| 19p13 | 1.06 | (0.62-1.80) | .832 | 1.44 | (0.82-2.51) | .204 | 0.88 | (0.57-1.37) | .579 |
| 19p13.1 | 1.11 | (0.69-1.79) | .667 | 0.97 | (0.53-1.77) | .931 | 1.04 | (0.71-1.53) | .841 |
| 19p13.3 | 1.06 | (0.60-1.88) | .832 | 1.64 | (0.90-3.00) | .105 | 1.18 | (0.71-1.94) | .522 |
| 1q21 | 0.12 | (0.03-0.49) | .003† | 0.00 | 0.68 | (0.44-1.05) | .080 | ||
| 4q21 | 1.46 | (0.74-2.88) | .276 | 2.04 | (1.02-4.09) | .043* | 1.84 | (0.99-3.43) | .054 |
| 10p11.2 | 2.12 | (1.10-4.06) | .024* | 2.56 | (1.24-5.32) | .011* | 1.37 | (0.67-2.78) | .384 |
| 17q21 | 1.14 | (0.53-2.43) | .743 | 1.15 | (0.47-2.82) | .763 | 1.28 | (0.68-2.42) | .446 |
| Trisomy | |||||||||
| No trisomy | 1 | reference | 1 | reference | 1 | reference | |||
| 8 | 0.57 | (0.36-0.92) | .022* | 0.54 | (0.32-0.94) | .028* | 0.79 | (0.56-1.12) | .188 |
| 19 | 1.77 | (1.13-2.78) | .012* | 2.11 | (1.31-3.42) | .002† | 1.15 | (0.68-1.94) | .596 |
| 21 | 1.35 | (0.85-2.13) | .198 | 1.25 | (0.76-2.03) | .377 | 0.98 | (0.60-1.60) | .926 |
| Type | |||||||||
| No ACAs | 1 | reference | 1 | reference | 1 | reference | |||
| numerical | 1.16 | (0.83-1.63) | .376 | 1.17 | (0.84-1.62) | .353 | 1.09 | (0.81-1.47) | .588 |
| structural | 1.39 | (1.07-1.80) | .013* | 1.27 | (0.98-1.63) | .068 | 1.13 | (0.90-1.43) | .288 |
Results are of 3 independent analyses.
ACA indicates additional cytogenetic aberrations; CI, 95% confidence interval; EFS, event-free survival; HR, hazard ratio; and OS, overall survival.
Significant at P < .05 level.
Significant at P < .01 level.
Discussion
The heterogeneity of pediatric AML is mainly determined by specific karyotypes and molecular aberrations, which have become important prognosticators.1,3–6,8,11,27–33 In addition, within distinct groups such as 11q23/MLL-rearranged AML, we have reported that additional cytogenetic aberrations are of prognostic relevance.10 In the present exploratory study, we identified trisomy 8, trisomy 19, and trisomy 21 to be recurrent ACAs of prognostic significance in pediatric 11q23/MLL-rearranged AML. Multivariate analysis showed that only trisomy 8 and trisomy 19 as additional aberrations were of independent prognostic value. Notably, the adverse outcome for 11q23/MLL-rearranged AML patients harboring trisomy 19 was because of refractory disease and early death rather than an increased rate of relapse. Complex karyotype was a frequent finding (26%) and a negative prognostic factor in univariate analysis only.
Trisomy 19 in AML is an aberration that is rarely found as the sole aberration.34 In infants with AML it is associated with t(7;12)(q36;p13) and t(7;12)(q32;p13).35 In most of such cases it can seem to be the sole aberration because of the cryptic t(7;12).35 Trisomy 19 has been described as an additional aberration with adverse prognostic significance in adult AML.11 It has been postulated that a gene dosage effect of the DNA methyltransferase 1 located on 19p13.2 contributes to the hypermethylation found in patients with MDS and thereby to prognosis.36 Future studies may reveal whether this mechanism also contributes to aberrant methylation found in pediatric 11q23/MLL-rearranged AML.37
In our study, trisomy 8 was found to be an independent favorable prognostic factor. Kok et al38 identified a gene expression signature with high HOXA gene expression in adult AML patients with AML with trisomy 8 as the sole abnormality, which clustered together with patents with MLL-rearranged AML. This finding may suggest similarities in the biology of these diseases. In contrast, in pediatric MDS, trisomy 8 is recognized as a positive prognostic factor, possibly because of differences in apoptosis regulation between cells with trisomy 8 and cells with other abnormalities.39,40 To date, it is not clear how trisomy 8 influences the biology of MLL-rearranged AML.
Interestingly, in our study, although 26% of all cases of 11q23/MLL-rearranged had complex karyotypes, this ACA was not an independent prognostic factor. Although the use of definitions on complex karyotypes is not uniform, the occurrence of complex karyotypes in pediatric AML cohorts has been reported to range from 7% to 15%.2,6,14,41 A Cancer and Leukemia Group B study on adult de novo AML showed that patients with increased number of aberrations had significantly worse outcome than those with normal karyotypes.42 Recently, Göhring et al43 used a new definition of “structural complex karyotype,” defined as a karyotype with ≥ 3 chromosomal aberrations including at least one structural aberration. This specific karyotype independently predicted very poor survival in a cohort of 192 children with advanced MDS.43
Although all the cases of complex karyotype in our study fit their definition, we did not find the presence of such karyotype to be associated with the poor prognosis that was reported in pediatric advanced MDS.43 Only some studies have specifically shown a correlation between complexity of the karyotype and outcome in pediatric AML.2,6,14,33,44 EFS rates for patients with complex karyotype have ranged from 29% to 42% in these studies, which is comparable with the EFS obtained in our study. Alternatively, a strong negative association between monosomal karyotype, defined as a karyotype with at least 2 monosomies or 1 monosomy combined with at least 1 structural aberration, and outcome was described in adult AML.45 This monosomal karyotype was only present in 1.5% (n = 11) of our cases and therefore it was not possible to evaluate the predictive value in our pediatric 11q23/MLL-rearranged AML cohort.
Although we have added additional prognostic factors in our study, the multivariate models still point out that previously determined risk factors (among which the TPs) retain their independent prognostic significance irrespective of ACA status.
A limitation of our study is the variety of treatment regimens, although all protocols had a similar backbone, including intensive chemotherapy with cytarabine/anthracycline. Unfortunately, numbers were too small to do specific analyses for different protocols, or to draw any meaningful conclusion regarding provided treatment and outcome.
In separate analysis of t(9;11)(p22;q23) cases, we confirmed most of the findings from the complete cohort, regarding frequent recurrent aberrations and predictive factors. In addition, FAB M5 morphology was still recognized as independent favorable prognostic factor in this group of patients.
In conclusion, in this exploratory study we have identified specific ACAs as novel independent prognostic variables in pediatric 11q23/MLL-rearranged AML, which can be identified by conventional karyotyping. Future studies should be aimed to test the associations found in this study in different patient cohorts. Our findings may also guide further studies that unravel the biologic differences that determine outcome differences in 11q23/MLL-rearranged AML as well as future treatment stratification.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Brian V. Balgobind for data collection and Vani Shanker from the Department for Scientific Editing of the St Jude Children's Research Hospital for her input.
This work was funded by the Rotterdam Oncology Research Foundation KOCR (E.A.C.), the Parents' Foundation Giessen (J.H.), the Swedish Childhood Cancer Foundation (E.S.F.), and by a grant for Clinical Cancer Research from the Ministry of Health, Labor and Welfare, Japan (A.M., T.T., D.T.).
National Institutes of Health
Authorship
Contribution: E.A.C., S.C.R., J.H., R.P., C.M.Z., and M.M.v.d.H.-E. conveyed and planned the study, analyzed the data, and wrote the paper; M.Z. performed the statistical analyses and wrote the paper; and T.A.A., A.A., H.B.B., M.C., U.C., M.N.D., E.F., B.G., H.H., C.J.H., N.A.H., G.J.L.K., A.L., N.L., L.L.N., A.M., C.P., D.R., J.E.R., F.O.S., J.S., I.S., S.S., T.T., D.T., D.W., and Z.Z. participated in data collection and in critical review and final approval of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marry M. van den Heuvel-Eibrink, MD, PhD, Associate Professor in Pediatric Oncology/Hematology, Erasmus MC/Sophia Children's Hospital, Department of Pediatric Oncology/Hematology, Rm Sp2568, Dr. Molewaterplein 60, PO Box 2060, 3000 CB Rotterdam, The Netherlands; e-mail: m.vandenheuvel@erasmusmc.nl.
References
Author notes
S.C.R. and J.H. contributed equally to this article.
C.M.Z. and M.M.v.d.H.-E. contributed equally to this article.