Abstract
The chemokine CXCL12, via its receptor CXCR4, promotes increased survival of chronic lymphocytic leukemia (CLL) B cells that express high levels of ζ-chain–associated protein (ZAP-70), a receptor tyrosine kinase associated with aggressive disease. In this study, we investigated the underlying molecular mechanisms governing this effect. Although significant differences in the expression or turnover of CXCR4 were not observed between ZAP-70+ and ZAP-70− cell samples, CXCL12 induced greater intracellular Ca2+ flux and stronger and more prolonged phosphorylation of extracellular signal-regulated kinase (ERK) and mitogen-activated protein kinase/ERK kinase (MEK) in the ZAP-70+ CLL cells. The CXCL12-induced phosphorylation of ERK and MEK in ZAP-70+ CLL cells was blocked by sorafenib, a small molecule inhibitor of RAF. Furthermore, ZAP-70+ CLL cells were more sensitive than ZAP-70− CLL cells to the cytotoxic effects of sorafenib in vitro at concentrations that can readily be achieved in vivo. The data suggest that ZAP-70+ CLL cells may be more responsive to survival factors, like CXCL12, that are elaborated by the leukemia microenvironment, and this sensitivity could be exploited for the development of new treatments for patients with this disease. Moreover, sorafenib may have clinical activity for patients with CLL, particularly those with ZAP-70+ CLL.
Introduction
Chronic lymphocytic leukemia (CLL) is a disease characterized by the accumulation of mature monoclonal B cells in the blood, secondary lymphoid tissue, and marrow.1,2 Regardless of their apparent longevity in vivo, CLL B cells undergo apoptosis in vitro unless rescued by monocyte-derived nurse-like cells (NLCs) or marrow stromal cells.3-6 In line with this hypothesis, the marrow is invariably infiltrated with CLL cells in patients, and the extent of infiltration correlates with clinical stage and prognosis.5,7 These accessory cells also protect CLL cells from drug-induced apoptosis in vitro.8 Thus, it has been postulated that CLL cells receive survival signals from these accessory cells, which constitute part of the CLL B-cell microenvironment in secondary lymphoid tissue and marrow.6 Such niches could protect leukemia cells from spontaneous or drug-induced apoptosis in vivo, motivating the current study to better understand the survival pathways triggered by the microenvironment.
Accessory cells such as NLCs protect CLL cells from apoptosis in vitro in part through the secretion of the stromal cell-derived factor-1α (renamed as CXCL12).9,10 CXCL12 is a highly conserved chemokine that signals through the chemokine receptor CXCR4, which is expressed at high levels by CLL cells.3,10,11 Although most noted for its role in directing cell migration, CXCL12 also provides survival stimuli to CLL cells and partially protects them from spontaneous or drug-induced apoptosis or both in vitro.3,9 Further, the enhanced viability of these cells in the presence of CXCL12 can be blocked by antibodies to CXCL123 or peptide inhibitors of CXCR4.8
In prior studies, it was found that treatment of CLL cells with CXCL12 induced activation of extracellular signal-regulated kinase (ERK).8,12 In this study, we further examined the survival and signaling responses of CLL cells to CXCL12 to characterize the mechanism for the survival benefit. In addition, we compared the CXCL12-induced responses of CLL cells from 2 subgroups of patients, with high or low expression levels of ζ-chain–associated protein of 70 kDa (ZAP-70), a tyrosine kinase whose high-level expression is correlated with increased risk of early disease progression and relatively short survival 12,13 .
Methods
Preparation of CXCL12
CXCL12 was prepared as previously described.14 Briefly, CXCL12 was expressed as a His-tag fusion protein and purified from inclusion bodies in BL21 Escherichia coli. Bacterial pellets were resuspended in 10mM Tris (tris(hydroxymethyl)aminomethane) pH 8.0, 1mM MgCl2 with 200 μg of DNAse, and complete protease inhibitor cocktail (free of EDTA [ethylenediaminetetraacetic acid]; Roche Applied Science) and then sonicated and washed with deoxycholate. Pellets were solubilized in 6M Guanadine-HCl, 100mM sodium phosphate, 10mM Tris-Cl, pH 8.0, with the use of a dounce homogenizer. The protein was then filtered and purified over a nickel-nitriloacetic acid column and refolded with Hampton Fold-It Buffer no. 8 (Hampton Research). After refolding, CXCL12 was dialyzed and concentrated with the use of Amicon Ultra centrifugal concentrators (molecular weight cutoff = 5000). The His-tag was removed by cleaving with enterokinase (NEB) at a 1:100 000 molar ratio overnight at room temperature. CXCL12 was then purified by high-performance liquid chromatography, and the purity was assessed by mass spectrometry. Transwell migration assays (Corning) with Jurkat cells were performed to verify the functionality of the purified protein.
Isolation of CLL B cells, cell culture, and reagents
Blood samples were collected from patients at the University of California San Diego (UCSD) Moores Cancer Center who satisfied diagnostic and immunophenotypic criteria for common B-cell CLL after providing written informed consent in compliance with the Declaration of Helsinki1 and the institutional review board of UCSD. Peripheral blood mononuclear cells (PBMCs) were isolated from patients with CLL by density centrifugation with Ficoll-Hypaque (GE Healthcare) and were resuspended in 90% fetal calf serum and 10% dimethylsulfoxide (DMSO) for viable storage in liquid nitrogen. If not otherwise indicated, the CLL cells were isolated from thawed PBMCs by negative selection with the use of anti-CD2 and anti-CD14 magnetic beads (Miltenyi Biotech).
The B-RAF and C-RAF inhibitor KG5 and the control kinase inhibitor KG1 were kindly provided by Dr D. Cheresh (UCSD). The RAF inhibitor GW5074 was purchased from Sigma. Sorafenib and sunitinib were purchased from LC Laboratories. All inhibitors were solubilized in DMSO, which was used in all experiments as a negative control.
Generation of NLCs
PBMCs were isolated from the blood of healthy volunteers (San Diego Blood Bank) over a Ficoll (GE Healthcare) density gradient. CD14+ monocytes were isolated from PBMCs by positive selection with the use of anti-CD14 beads (Miltenyi Biotech) following the manufacturer's instructions. To generate NLCs, 1.25 × 105/well CD14+ cells were cocultured with 3 × 106/well purified CLL B cells in 1 mL of media in a 24-well plate (BD) in culture media (RPMI 1640 supplemented with 10mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid]; Invitrogen), penicillin (100 U/mL) and streptomycin (100 μg/mL; Invitrogen), 50uM Beta-mercaptoethanol, and 10% pooled human serum (Omega Scientific) for 12 days. At this point CLL B cells were gently washed off, and the adherent NLCs were used for coculture experiments with the use of freshly purified CLL cells.
Analysis of ZAP-70 expression by flow cytometry
The expression levels of ZAP-70 in CLL cells were determined by flow cytometry as previously described. The cutoff was set at 20%: > 20% of CLL cells expressing ZAP-70 were defined as ZAP-70+ and < 20% were defined as ZAP-70−.13
CXCR4 expression turnover measurement
CLL cells were incubated with 30nM CXCL12 for 30 minutes and either collected immediately and stained for CXCR4 expression (0 minutes) to assess receptor down-modulation or washed and recultured for another 30, 60, or 240 minutes and then collected and stained for CXCR4 expression, to assess receptor reexpression after ligand removal. CLL cells (1 × 105) were incubated for 20 minutes at 4°C in 100μL of phosphate-buffered saline (PBS)/5% fetal calf serum/0.1% sodium azide (staining buffer) with phycoerythrin–conjugated immunoglobulin G specific for human CXCR4, or the appropriate phycoerythrin-conjugated isotype control (R&D Systems). Cells were then washed 4 times with staining buffer, fixed in 3.7% formaldehyde in PBS (pH 7.2-7.4), and examined by flow cytometry with the use of a FACSCalibur (Beckon Dickinson). Data were analyzed with the FlowJo 7.2.2 software (TreeStar Inc).
Calcium flux measurements
Calcium flux assays were performed with the Molecular Devices Calcium 4 assay kit and measurements were acquired with a Molecular Devices FlexStation3 (Molecular Devices Corporation). Purified CLL B cells were washed twice in 0.5% bovine serum albumin/PBS and then suspended at 2.5 × 106 cells/mL in assay buffer (1× Hanks Balanced salt solution, 20mM HEPES, pH 7.4, 0.1% bovine serum albumin). One hundred microliters of cell suspension plus 10 μL of calcium 4 dye were distributed into wells of a 96-well Biocoat plate (BD Biosciences) and incubated at 37°C/5%CO2 for 1 hour before analysis. CXCL12 was diluted in ddH2O and distributed into wells of a 300-μL V-bottom 96-well plate (Corning Inc) and 50 μL of CXCL12 (8nM) or ddH2O was distributed into the assay plate with cells at the start of measurements with the use of the liquid handling capabilities of the FlexStation3. Calcium flux measurements were averaged between triplicate data points and normalized against the ddH2O control calcium flux response. CLL cells from 12 separate patients were analyzed for calcium flux, and, although there is variability in the overall maximum fluorescence signal observed between runs, the trend was the same as the one shown for 3 ZAP-70+ and 3 ZAP-70− CLL cells, which were analyzed together in one run. All statistics were determined with GraphPad Prism software Version 5.00 for Windows (GraphPad Software).
Immunoblot
CLL cells stimulated with 30nM CXCL12 were lysed for 20 minutes on ice in RIPA lysis buffer (10mM Tris pH 7.4, 150mM NaCl, 1% TritonX-100, 0.1% sodium deoxycholate, 0.1% sodium dodecylsulfate (SDS), 5mM EDTA supplemented with 1mM phenylmethylsulfonyl fluoride, Halt phosphatase inhibitor (Thermo Fisher Scientific), 1mM sodium vanadate, 1mM sodium fluoride, and complete protease inhibitor cocktail (Roche). Protein concentration was determined with the DC (Detergent Compatible) protein assay (Bio-Rad). The lysates were snap-frozen and stored at −80°C. Equal amounts of protein lysates (∼ 30 μg) were separated by gel electrophoresis with the use of a NuPAGE Novex 4%-12% Bis-Tris Midi Gel (Invitrogen) and transferred to polyvinylidene fluoride membranes (Bio-Rad). Membranes were washed with 1× TBST (Tris-Buffered Saline Tween-20) and blocked for 1 hour at room temperature in 5% milk/TBST. Membranes were probed overnight for phospho p44/p42 (ERK1/2; Thr202/Tyr 204), phospho (p)–mitogen-activated protein kinase/ERK kinase 1 and 2 (MEK1/2; Ser 217/221), MEK1/2, p44/p42 (ERK1/2), and β-actin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH), using antibody from Cell Signaling Technology. The next day, membranes were washed with 1× TBST and incubated with goat anti–rabbit or anti–mouse horseradish peroxidase–conjugated secondary antibodies (Santa Cruz Biotechnology) diluted to 1:5000 and 1:2000, respectively, in 5%milk/TBST for 1 hour at room temperature. Antibodies were detected with the use of either an enhanced chemiluminescence detection kit (GE Healthcare) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific).
p-ERK and p-MEK enzyme-linked immunoabsorbent assay
For each patient, the induction of p-MEK or p-ERK after CXCL12 stimulation (30nM) was measured with p-MEK1 (Ser217/221) and p-p44/42 mitogen-activated protein kinase (Thr202/Tyr204) sandwich enzyme-linked immunoabsorbent assay kits (Cell Signaling). The kinetics are displayed as the amount of p-MEK measured (optical density [OD] at 450 nm) for each time point after CXCL12 stimulation (0, 3, 10, 30, and 60 minutes.). For each patient, the overall p-MEK and p-ERK expression kinetics were compared by calculating the area under the curve with the use of GraphPad Prism software.
Measurement of cell viability
Purified CLL cells were cultured at 1 × 106 cells/mL in 24-well plates (BD) under various conditions. Determination of CLL cell viability was based on the analysis of mitochondrial transmembrane potential (ΔΨm) with the use of 3,3′-dihexyloxacarbocyanine iodide (DiOC6; Invitrogen) and cell membrane permeability to propidium iodide (PI; Sigma). For viability assays, 100 μL of the cell culture was collected at the indicated days and transferred to polypropylene tubes containing 100 μL of 40μM DiOC6 and 10 μg/mL PI in culture media. The cells were then incubated at 37°C for 15 minutes and analyzed within 30 minutes by flow cytometry with the use of a FACSCalibur (Becton Dickinson). Fluorescence was recorded at 525 nm for DiOC6 and at 600 nm for PI. Data were analyzed with the FlowJo 7.2.2 software (TreeStar). The percentage of viable cells was determined by gating on PI-negative and DiOC6-bright cells.
Statistical analysis
Unless indicated otherwise, data are presented as either the mean ± standard deviation (SD) or the median ± SD. Statistical significance was determined with the use of paired or unpaired Student t test or 2-way analysis of variance. P values < .05 were considered significant.
Results
Influence of CXCL12 on calcium flux and receptor turnover in ZAP-70+ CLL cells versus ZAP-70− CLL cells
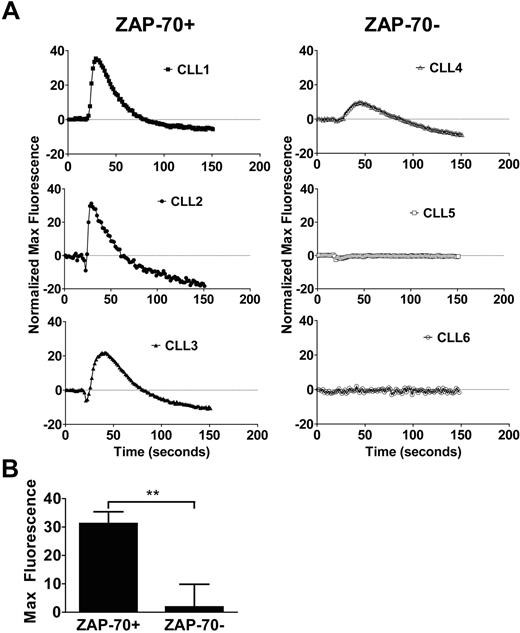
The goal of this study was to understand differences in signaling in CLL cells from patients with aggressive versus indolent diseases. Because expression of high levels of the receptor tyrosine kinase ZAP-70 is associated with aggressive disease,13 ZAP-70 expression is used to segregate the 2 groups of patients (see “Methods”). Consequently, in referring to the cells as being ZAP-70+ and ZAP-70−, we refer to the disease category not the exact expression levels of ZAP-70 in individual cells. We previously showed that CXCL12 could enhance the survival of CLL cells in vitro.3,9 Furthermore, subsequent studies showed that CLL cells which expressed high levels of ZAP-70 appeared more responsive to the survival stimulus provided by CXCL12 than ZAP-70− CLL cells.12 Because of this difference, we examined the capacity of CXCL12 to induce intracellular Ca2+ flux in ZAP-70+ versus ZAP-70− CLL cells in vitro, because this is a common response of chemokine receptors to their ligands. Whereas CXCL12 could induce a robust intracellular Ca2+ flux in ZAP-70+ CLL cells, it induced only modest-to-poor calcium flux in ZAP-70− CLL cells (Figure 1A). The maximum calcium flux signal induced by CXCL12 in ZAP-70+ CLL cells (31 ± 3, median maximal fluorescence ± SD; n = 3) was significantly greater than that observed in ZAP-70− CLL cells (2 ± 3, median maximal fluorescence ± SD; n = 3; P = .0069) (Figure 1B). Within 90 seconds after exposure to the chemokine, the calcium flux returned back to baseline levels.
CXCL12 confers stronger calcium flux in ZAP-70+ CLL cells. (A) Representative calcium flux profiles of 3 ZAP-70+ (filled symbols) and 3 ZAP-70− (open symbols) CLL cells in response to 8nM CXCL12 stimulation. Measurements represent an average of triplicate data points that have been normalized to buffer controls. (B) The maximal fluorescence signal from the calcium flux was averaged between the ZAP-70+ (n = 3) and ZAP-70− (n = 3) CLL cells and was found to be significantly different on the basis of unpaired Student t test (P < .01). Data shown are median ± SD.
CXCL12 confers stronger calcium flux in ZAP-70+ CLL cells. (A) Representative calcium flux profiles of 3 ZAP-70+ (filled symbols) and 3 ZAP-70− (open symbols) CLL cells in response to 8nM CXCL12 stimulation. Measurements represent an average of triplicate data points that have been normalized to buffer controls. (B) The maximal fluorescence signal from the calcium flux was averaged between the ZAP-70+ (n = 3) and ZAP-70− (n = 3) CLL cells and was found to be significantly different on the basis of unpaired Student t test (P < .01). Data shown are median ± SD.
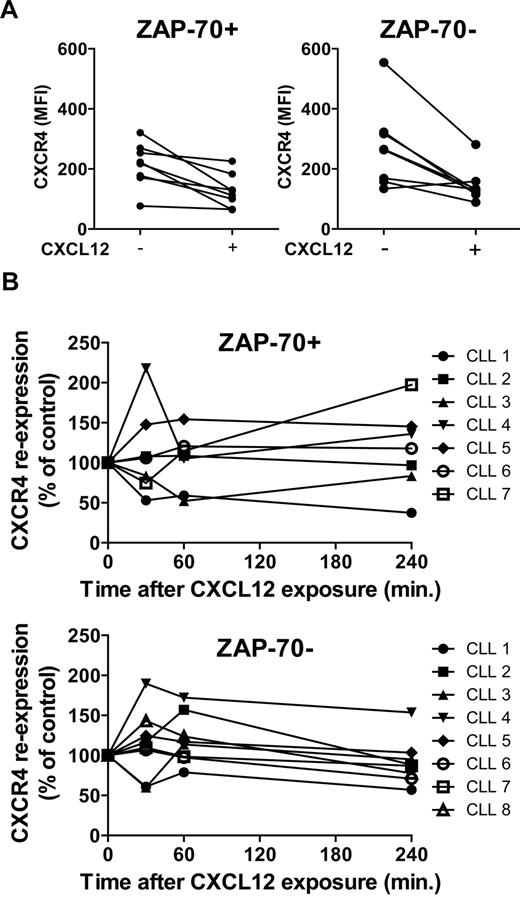
The difference in calcium flux could be caused by differences in receptor expression levels or turnover kinetics. Thus, we examined ZAP-70+ and ZAP-70− CLL cells for expression of CXCR4 and CXCR7, which are the only known receptors for CXCL12.15,16 In agreement with previous work, the cells from each patient expressed CXCR417 and the expression levels of CXCR4 on ZAP-70+ CLL cells were similar to the levels on ZAP-70− CLL cells, as described12 (data not shown). However, CLL cells did not express detectable surface levels of CXCR7 in contrast to normal B cells18 (data not shown). Therefore, we examined the relative capacity of CXCL12 to induce down-modulation of CXCR4 (Figure 2A) and whether CXCR4 reexpression on the cell surface, through de novo synthesis and recycling (Figure 2B), differed between CLL cells from the 2 groups. A similar rapid down-modulation of CXCR4 was observed on both CLL populations (Figure 2A), and the reexpression of CXCR4 on the surface of both cell populations was not significantly different after removal of CXCL12 (Figure 2B). Therefore, the differences in the response of ZAP-70+ and ZAP-70− CLL cells must be because of differences in downstream signaling.
CXCR4 expression and down-modulation in response to CXCL12. Purified CLL cells from ZAP-70+ CLL samples (n = 7) or ZAP-70− CLL samples (n = 8) were incubated with or without CXCL12 (80nM) for 30 minutes. The CLL cells were either collected immediately and stained to assess CXCR4 down-modulation after incubation with its ligand (A) or washed and recultured for another 30, 60, or 240 minutes and then collected and stained to study CXCR4 reexpression overtime in the absence of the ligand (B). Cells were analyzed by flow cytometry, and the data shown depict CXCR4 expression as mean fluorescence intensity (MFI). (A) CXCR4 expression level is compared in the presence or absence of CXCL12 for 30 minutes. (B) CXCR4 reexpression is expressed as a percentage of control, which is the level of CXCR4 remaining on the surface after 30 minutes of CXCL12 stimulation, and corresponds to time 0.
CXCR4 expression and down-modulation in response to CXCL12. Purified CLL cells from ZAP-70+ CLL samples (n = 7) or ZAP-70− CLL samples (n = 8) were incubated with or without CXCL12 (80nM) for 30 minutes. The CLL cells were either collected immediately and stained to assess CXCR4 down-modulation after incubation with its ligand (A) or washed and recultured for another 30, 60, or 240 minutes and then collected and stained to study CXCR4 reexpression overtime in the absence of the ligand (B). Cells were analyzed by flow cytometry, and the data shown depict CXCR4 expression as mean fluorescence intensity (MFI). (A) CXCR4 expression level is compared in the presence or absence of CXCL12 for 30 minutes. (B) CXCR4 reexpression is expressed as a percentage of control, which is the level of CXCR4 remaining on the surface after 30 minutes of CXCL12 stimulation, and corresponds to time 0.
Intracellular signaling in response to CXCL12
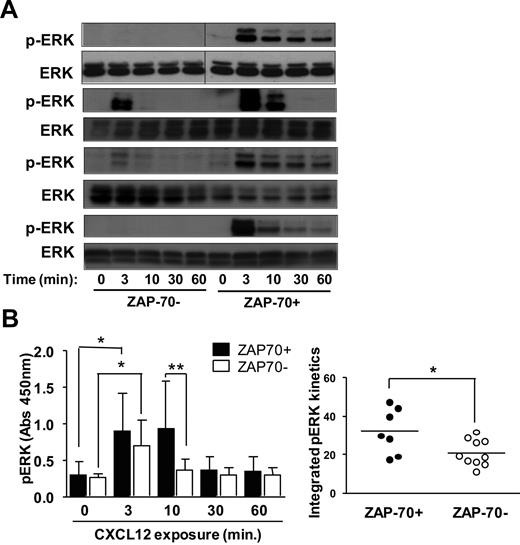
Prior studies have shown that CXCL12 could induce activation of ERK in CLL cells.3,9 Because activation of this pathway can enhance cell survival, we reasoned that ERK activation could account for the enhanced survival of ZAP-70+ CLL cells after stimulation with CXCL12. First, we examined the relative magnitude and duration of ERK phosphorylation in ZAP-70+ versus ZAP-70− CLL cells in response to CXCL12 via immunoblot analyses. CXCL12 induced increased and prolonged ERK phosphorylation in ZAP-70+ CLL cells compared with that observed with ZAP-70− CLL cells (Figure 3A). Such differences were also observed with enzyme-linked immunoabsorbent assay–based measurements (Figure 3B). Although increased ERK activation was noted at 3 minutes in both ZAP-70+ (P = .007) and ZAP-70− CLL samples (P = .003), at 10 minutes after exposure, significantly higher levels of p-ERK were measured in ZAP-70+ CLL cells (n = 7) compared with ZAP-70− CLL cells (n = 10) (P < .05) (Figure 3B left). In addition to comparing the ERK response at the individual time points we also examined the complete ERK response to CXCL12, taking into account the intensity and duration of ERK activation. For this comparison we measured the integrated response over time (see “Methods”) and observed that ZAP-70+ CLL cells showed an increased amplitude of ERK activation (36 ± 16; n = 8) than ZAP-70− CLL cells (20 ± 7; n = 10; P = .015) (Figure 3B right). The individual kinetics of CXCL12-induced p-ERK for each patient are shown in supplemental Figure 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
CXCL12 induces increased and prolonged ERK activation in ZAP-70+ CLL cells. CLL cells were exposed to 30nM CXCL12 for 0-60 minutes at which point the cells were harvested, lysed, and analyzed for p-ERK. (A) Immunoblots were probed with anti–p-ERK and total ERK (ERK) antibody. Representative data of 4 ZAP-70+ and 4 ZAP-70− CLL samples are shown. Vertical lines have been inserted to indicate repositioned gel lanes to align ZAP-70− and ZAP-70+ samples. (B) To obtain quantitative results, cell lysates were analyzed by a p-ERK–specific enzyme-linked immunoabsorbent assay. (Left) Depicted is the absorbance measured at 450 nm. Data are shown as mean ± SD of ZAP-70− CLL samples (n = 9) and ZAP-70+ CLL samples (n = 7). * indicates a statistically significant difference; P < .05 paired Student t test. Two-way analysis of variance was used for the comparison ZAP-70+ vs ZAP-70− CLL samples at 10 minutes. (Right) The integral under the curve was measured as described in “Methods” and is depicted for all ZAP-70+ and ZAP-70− CLL cases shown on the left. Data shown are median ± SD; * indicates a statistically significant difference; P < .05 unpaired Student t test.
CXCL12 induces increased and prolonged ERK activation in ZAP-70+ CLL cells. CLL cells were exposed to 30nM CXCL12 for 0-60 minutes at which point the cells were harvested, lysed, and analyzed for p-ERK. (A) Immunoblots were probed with anti–p-ERK and total ERK (ERK) antibody. Representative data of 4 ZAP-70+ and 4 ZAP-70− CLL samples are shown. Vertical lines have been inserted to indicate repositioned gel lanes to align ZAP-70− and ZAP-70+ samples. (B) To obtain quantitative results, cell lysates were analyzed by a p-ERK–specific enzyme-linked immunoabsorbent assay. (Left) Depicted is the absorbance measured at 450 nm. Data are shown as mean ± SD of ZAP-70− CLL samples (n = 9) and ZAP-70+ CLL samples (n = 7). * indicates a statistically significant difference; P < .05 paired Student t test. Two-way analysis of variance was used for the comparison ZAP-70+ vs ZAP-70− CLL samples at 10 minutes. (Right) The integral under the curve was measured as described in “Methods” and is depicted for all ZAP-70+ and ZAP-70− CLL cases shown on the left. Data shown are median ± SD; * indicates a statistically significant difference; P < .05 unpaired Student t test.
We next examined differences between ZAP-70+ versus ZAP-70− CLL cells in the CXCL12-induced activation of kinases that can contribute to ERK activation, starting with p-MEK. At 3 minutes after stimulation with CXCL12, ZAP-70+ CLL cells (n = 8) had significantly higher levels of p-MEK (P = .01) than did ZAP-70− CLL cells (n = 7; P < .01) (Figure 4 left). The response pattern closely mimicked the induction of p-ERK, with the strongest signal at 3 minutes. Furthermore, as with p-ERK, the CXCL12-induced MEK phosphorylation was prolonged in ZAP-70+ CLL cells compared with ZAP-70− CLL cells (Figure 4 left). When investigating the amplitude of the p-MEK response as a function of time (see “Methods”) we found that ZAP-70+ CLL cells expressed significantly higher levels of p-MEK (79 ± 35; n = 8) than ZAP-70− CLL cells (45 ± 15; n = 7; P = .03) (Figure 4 right). The individual kinetics of CXCL12-induced p-MEK for each patient are represented in supplemental Figure 2.
CXCL12 induces pronounced MEK activation in ZAP-70+. CLL cells were exposed to 30nM CXCL12 for 0-60 minutes at which point the cells were harvested, and the lysates were analyzed for p-MEK protein expression by enzyme-linked immunoabsorbent assay. Depicted is the absorbance measured at 450 nm. Results are shown as mean ± SD of ZAP-70− CLL samples (n = 7) and ZAP-70+ CLL (n = 8); * indicates a statistically significant difference; P < .05 paired Student t test. Two-way analysis of variance was used for the comparison ZAP-70+ CLL vs ZAP-70− CLL at 3 minutes. (Right) The integral under the time curve was measured as described in “Methods” and is depicted for all ZAP-70+ and ZAP-70− CLL cases shown on the left. Data shown are median ± SD; * indicates a statistically significant difference; P < .05 unpaired Student t test.
CXCL12 induces pronounced MEK activation in ZAP-70+. CLL cells were exposed to 30nM CXCL12 for 0-60 minutes at which point the cells were harvested, and the lysates were analyzed for p-MEK protein expression by enzyme-linked immunoabsorbent assay. Depicted is the absorbance measured at 450 nm. Results are shown as mean ± SD of ZAP-70− CLL samples (n = 7) and ZAP-70+ CLL (n = 8); * indicates a statistically significant difference; P < .05 paired Student t test. Two-way analysis of variance was used for the comparison ZAP-70+ CLL vs ZAP-70− CLL at 3 minutes. (Right) The integral under the time curve was measured as described in “Methods” and is depicted for all ZAP-70+ and ZAP-70− CLL cases shown on the left. Data shown are median ± SD; * indicates a statistically significant difference; P < .05 unpaired Student t test.
CXCL12-mediated MEK activation in ZAP-70+ CLL cells is RAF dependent
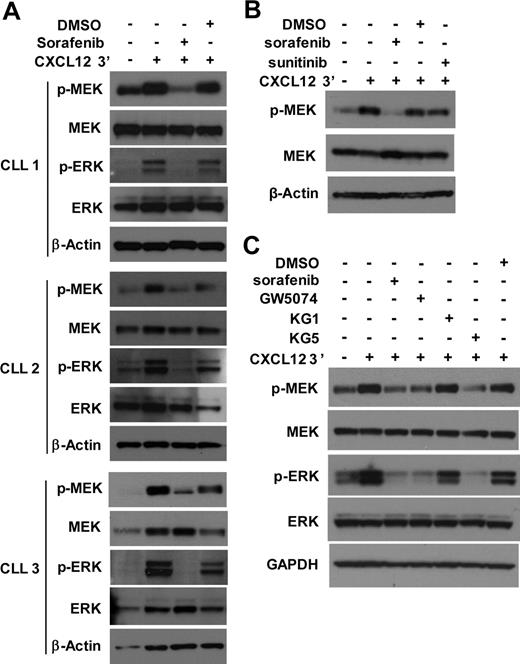
Because phosphorylation of MEK could depend on the activity of RAF, we evaluated whether sorafenib, a small molecule inhibitor of RAF,19 could block CXCL12-induced activation of CLL cells in vitro. Because ZAP-70− CLL cells did not show significant MEK activation, we focused on ZAP-70+ CLL cells. Sorafenib,19 was found to block CXCL12-induced activation of MEK and ERK in ZAP-70+ CLL cells (Figure 5A). Because sorafenib also targets non-RAF kinases,19 additional RAF inhibitors were tested. Consistent with the notion that the effect of sorafenib was because of its action on RAF, CXCL12-induced activation of MEK/ERK was not affected by sunitinib (Figure 5B). Sunitinib is an inhibitor of non-RAF kinases that are also inhibited by sorafenib,20 including vascular epidermal growth factor receptors, platelet-derived growth factor receptors, Flt3 and c-Kit, but it does not target RAF.21 As a positive control, the activity of sunitinib in CLL cells was demonstrated by its capacity to block CXCL12-induced activation of cAMP response element-binding in the same experiment (data not shown). Thus, these shared kinase targets are not involved in the activation of MEK after treatment with CXCL12.
CXCL12-mediated MEK and ERK activation in ZAP-70+ CLL cells is RAF dependent. (A) CLL cells were pretreated with 10μM sorafenib or DMSO as solvent control for 30 minutes before the addition of CXCL12 or media control. After exposure of CLL cells to CXCL12 for 3 minutes, the cells were harvested, lysed, and analyzed for the presence of the indicated proteins by immunoblot. Results are shown from 3 ZAP-70+ CLL samples. (B) ZAP-70+ CLL cells were pretreated with 10μM sorafenib, 10μM sunitinib, or DMSO as solvent control for 30 minutes before the addition of CXCL12 or media control. After exposure of CLL cells to CXCL12 for 3 minutes, cells were harvested and analyzed as above. Results are shown from 1 representative of 3 ZAP-70+ CLL samples, all showing similar responses. (C) ZAP-70+ CLL cells were pretreated with sorafenib, GW5074, KG1, KG5 (all inhibitors were used at 10μM), or DMSO as solvent control for 30 minutes before the addition of CXCL12 or media control. After exposure of CLL cells to CXCL12 for 3 minutes, cells were harvested and analyzed as above. Results are shown from 1 representative of 6 ZAP-70+ CLL samples; all but 1 sample showed similar responses. GAPDH indicates glyceraldehyde-3-phosphate dehydrogenase.
CXCL12-mediated MEK and ERK activation in ZAP-70+ CLL cells is RAF dependent. (A) CLL cells were pretreated with 10μM sorafenib or DMSO as solvent control for 30 minutes before the addition of CXCL12 or media control. After exposure of CLL cells to CXCL12 for 3 minutes, the cells were harvested, lysed, and analyzed for the presence of the indicated proteins by immunoblot. Results are shown from 3 ZAP-70+ CLL samples. (B) ZAP-70+ CLL cells were pretreated with 10μM sorafenib, 10μM sunitinib, or DMSO as solvent control for 30 minutes before the addition of CXCL12 or media control. After exposure of CLL cells to CXCL12 for 3 minutes, cells were harvested and analyzed as above. Results are shown from 1 representative of 3 ZAP-70+ CLL samples, all showing similar responses. (C) ZAP-70+ CLL cells were pretreated with sorafenib, GW5074, KG1, KG5 (all inhibitors were used at 10μM), or DMSO as solvent control for 30 minutes before the addition of CXCL12 or media control. After exposure of CLL cells to CXCL12 for 3 minutes, cells were harvested and analyzed as above. Results are shown from 1 representative of 6 ZAP-70+ CLL samples; all but 1 sample showed similar responses. GAPDH indicates glyceraldehyde-3-phosphate dehydrogenase.
We also examined whether other inhibitors of RAF could block CXCL12-induced MEK activation in CLL cells with the use of KG5, a kinase inhibitor of RAF signaling through B-RAF and C-RAF in addition to platelet-derived growth factor receptors α and β, Flt3 and Kit.22 As a control we used KG1, a kinase inhibitor that targets all of these kinases except B- and C-RAF.22 Whereas KG5 blocked MEK/ERK activation, KG1 did not (Figure 5C). Finally, we tested GW5074, an inhibitor of B-RAF and C-RAF, and found that it also inhibited CXCL12-induced MEK activation in ZAP-70+ CLL cells (Figure 5C). Collectively, these data imply that CXCL12-induced activation of MEK/ERK in CLL depends on the activity of RAF.
Sorafenib causes enhanced apoptosis in ZAP-70+ CLL cells
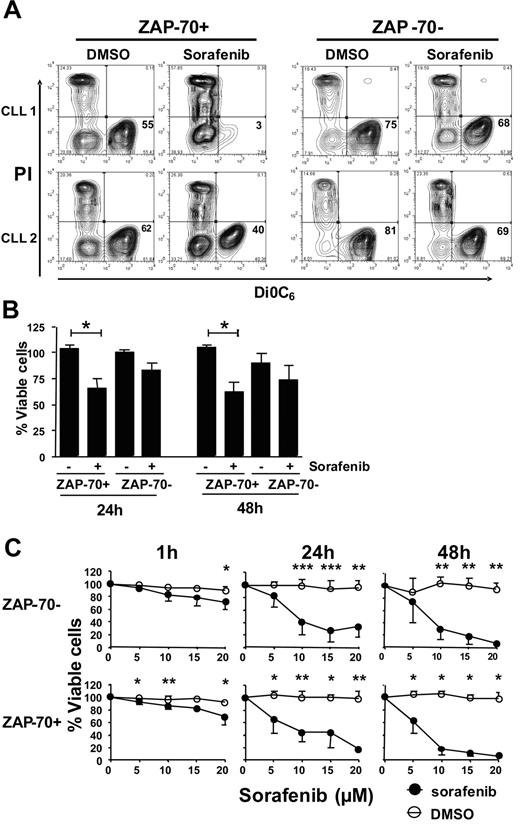
Because sorafenib could inhibit activation of MEK/ERK in ZAP-70+ CLL cells, we examined whether it was selectively cytotoxic for ZAP-70+ CLL cells in vitro. Indeed, 5μM sorafenib induced significantly higher levels of apoptosis in ZAP-70+ CLL cells than in ZAP-70− CLL cells (Figure 6A-B). After 24 hours the mean viability (± SD) of the ZAP-70+ CLL cells in the presence of sorafenib was 65% (± 22%; n = 5; P = .02 vs DMSO) compared with 82% (± 17%; n = 6; P = NS vs DMSO) for the ZAP-70− CLL cells. After 48 hours, these numbers dropped to 62% (± 18%; n = 5; P = .03 vs DMSO) and 73% (± 33%; n = 6; P = NS vs DMSO) for the ZAP-70+ and ZAP-70− CLL cells, respectively. Thus, at both time points the ZAP-70+ CLL cells showed enhanced sorafenib-induced apoptosis compared with the ZAP-70− CLL cells (Figure 6B). Although we noticed increased sensitivity to sorafenib at 5μM (Figure 6A-B), higher concentrations were cytotoxic to all CLL samples examined, following a dose- and time-dependent pattern (Figure 6C; supplemental Figure 4). However, ZAP-70+ CLL cells were more sensitive than ZAP-70− CLL cells to sorafenib at concentrations ≤ 5μM.
Sorafenib causes increased apoptosis in ZAP-70+ CLL cells. CLL cells were cultured in the presence of sorafenib or DMSO control, added once at the beginning of the culture. CLL cells were harvested at the indicated times and stained with DiOC6/PI and analyzed by flow cytometry. (A) Presented are contour maps from 2 representative ZAP-70+ and 2 ZAP-70− CLL samples treated with DMSO or 5μM sorafenib for 24 hours. The relative DiOC6 and PI fluorescence intensities are depicted on the x- and y-axis, respectively. Cells in the lower right quadrant, which are DiOC6 bright and PI negative, are viable, and those numbers were used for the generation of the plots shown below. (B) CLL cells were cultured in the presence of 5μM sorafenib or DMSO control and harvested after 24 and 48 hours for analysis of viability as above. Results are represented relative to untreated control at day 0, which was set as 100%. Data shown are mean ± SD from ZAP-70− (n = 5) and ZAP-70+ CLL cells (n = 5); * indicates a statistically significant difference; P < .05 paired Student t test. (C) CLL cells were cultured in increasing doses of sorafenib and analyzed for viability as above.
Sorafenib causes increased apoptosis in ZAP-70+ CLL cells. CLL cells were cultured in the presence of sorafenib or DMSO control, added once at the beginning of the culture. CLL cells were harvested at the indicated times and stained with DiOC6/PI and analyzed by flow cytometry. (A) Presented are contour maps from 2 representative ZAP-70+ and 2 ZAP-70− CLL samples treated with DMSO or 5μM sorafenib for 24 hours. The relative DiOC6 and PI fluorescence intensities are depicted on the x- and y-axis, respectively. Cells in the lower right quadrant, which are DiOC6 bright and PI negative, are viable, and those numbers were used for the generation of the plots shown below. (B) CLL cells were cultured in the presence of 5μM sorafenib or DMSO control and harvested after 24 and 48 hours for analysis of viability as above. Results are represented relative to untreated control at day 0, which was set as 100%. Data shown are mean ± SD from ZAP-70− (n = 5) and ZAP-70+ CLL cells (n = 5); * indicates a statistically significant difference; P < .05 paired Student t test. (C) CLL cells were cultured in increasing doses of sorafenib and analyzed for viability as above.
Sorafenib causes apoptosis of CLL cells in the presence of NLCs
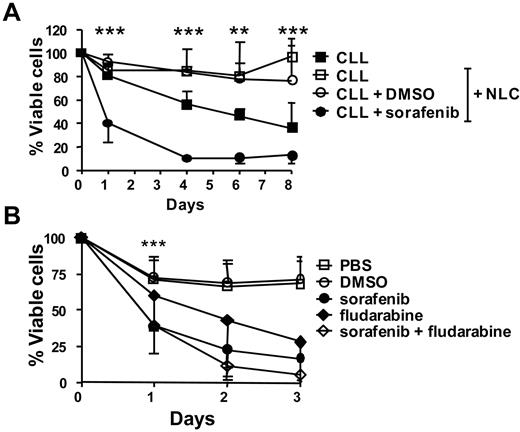
Because cells of the microenvironment can protect CLL cells from spontaneous and drug-induced apoptosis,8 we examined whether sorafenib could kill CLL cells even when cocultured with NLCs, which presumably exist in the CLL microenvironment and can protect them from apoptosis in vitro. As expected NLCs protected CLL cells from spontaneous apoptosis in vitro (Figure 7A). However, the addition of sorafenib to these NLC/CLL cocultures rapidly and significantly reduced the viability of the CLL cells. After 4 days, most CLL cells were dead, using 10μM sorafenib in the presence of NLCs (Figure 7A; supplemental Figure 5 shows individual patient responses).
Sorafenib causes apoptosis of CLL cells in the presence of NLCs. (A) CLL cells were cultured for 8 days alone or in the presence of NLCs with or without 10μM sorafenib or DMSO control added only once at the beginning of the culture. Viability was measured and analyzed as above. Data shown are mean ± SD from 4 different CLL samples. The mean viability ± SD of CLL cells cultured in presence of NLCs + sorafenib was 40% ± 16% (day 1), 10% ± 3% (day 4), 11% ± 6% (day 6), and 12% ± 6% (day 8). The * indicates a statistically significant difference; P < .05 paired Student t test. (B) CLL cells were cultured for 3 days in the presence of NLCs with or without 10μM sorafenib and 10μM fludarabine alone or in combination added only once at the beginning of the culture. Viability was measured and analyzed as above. Data shown are mean ± SD from 4 different CLL samples. The * indicates a statistically significant difference between sorafenib and fludarabine at day 1 (P = .0002, paired Student t test).
Sorafenib causes apoptosis of CLL cells in the presence of NLCs. (A) CLL cells were cultured for 8 days alone or in the presence of NLCs with or without 10μM sorafenib or DMSO control added only once at the beginning of the culture. Viability was measured and analyzed as above. Data shown are mean ± SD from 4 different CLL samples. The mean viability ± SD of CLL cells cultured in presence of NLCs + sorafenib was 40% ± 16% (day 1), 10% ± 3% (day 4), 11% ± 6% (day 6), and 12% ± 6% (day 8). The * indicates a statistically significant difference; P < .05 paired Student t test. (B) CLL cells were cultured for 3 days in the presence of NLCs with or without 10μM sorafenib and 10μM fludarabine alone or in combination added only once at the beginning of the culture. Viability was measured and analyzed as above. Data shown are mean ± SD from 4 different CLL samples. The * indicates a statistically significant difference between sorafenib and fludarabine at day 1 (P = .0002, paired Student t test).
Because the frontline therapy for CLL is fludarabine,23 we compared fludarabine and sorafenib for their potential to induce the apoptosis of CLL cells in the presence of NLCs and asked whether sorafenib could enhance fludarabine-induced CLL cell death. When tested at the same dose of 10μM, CLL cell viability was lower in the presence of sorafenib (38 ± 19%) compared with fludarabine (60 ± 23%; P = .0002) after 24 hours (Figure 7B). Although increased apoptosis by sorafenib compared with fludarabine was still observed after 2 and 3 days, the differences were not significant. A combination of sorafenib and fludarabine caused a small but insignificant additional increase in apoptosis (Figure 7B).
Discussion
It has been postulated that CLL cells receive survival signals from their microenvironment and that these signals limit the activity of antileukemia drugs in vivo. Such microenvironmental interactions could be a major impediment to the eradication of minimal residual disease in patients with CLL using current therapies. Therefore, identifying the mechanism(s) whereby CLL cells respond to microenvironmental survival signals could reveal novel targets for therapy. One of the survival factors elaborated in the microenvironment is CXCL12,3,9 a chemokine that enhances the survival of CLL cells in vitro, particularly CLL cells that express high levels of ZAP-70 which is associated with aggressive disease13 compared with ZAP-70− CLL cells.12 Our goal was to understand differences in signaling in CLL cells from patients segregated as having aggressive versus indolent diseases on the basis of their ZAP-70 status.
Cell surface expression of CXCR4 does not differ between ZAP-70+ and ZAP-70− CLL cells,12 and we found no significant differences in the extent of receptor down-regulation or surface reexpression of CXCR4 of both, implicating downstream signaling events in the differential effects observed on CXCL12-induced survival of these 2 subgroups of cells. The effects of CXCL12 are mediated exclusively by CXCR4, because the other receptor for CXCL12, CXCR7,15,16 is not expressed on the CLL cells18 (data not shown). A genetic analysis of CXCR4 sequences in CLL showed no common variations in the sequences that correlate with risk of CLL (data not shown).24 Therefore, signaling differences are not likely to be because of receptor mutations but rather downstream events. Prior studies showed that CXCL12 signaling causes ERK activation in CLL cells.9,12 However, because the overall signaling cascade linking CXCL12 to ERK activation and ultimately to the survival of CLL cells has not fully explored, it became the focus of our studies.
We found that CXCL12 induced high levels of intracellular calcium flux in ZAP-70+ CLL cells, whereas most ZAP-70− CLL cells showed little or no intracellular calcium flux. Calcium-induced ERK activation has been described in B cells25 and T cells.26 We have previously shown that aberrant expression of ZAP-70 in CLL cells leads to increases in intracellular calcium flux in response to B-cell receptor engagement.27 Furthermore, ionomycin, a calcium ionophore that opens calcium channels, induced MEK activation in ZAP-70+ CLL cells (data not shown). Taken together, these data suggest that increased CXCL12-mediated ERK signaling might be attributed to increased calcium flux, which in turn may lead to increased phospholipase C activity in ZAP-70+ CLL cells relative to ZAP-70− CLL cells, a hypothesis currently under investigation.
In concordance with findings by Richardson et al,12 we found that CXCL12 caused increased and prolonged activation of ERK in CLL cells, particularly those CLL cells that express ZAP-70. Extending these observations, we showed that CXCL12 induces increased RAF-dependent p-MEK levels in ZAP-70+ CLL cells compared with ZAP-70− CLL cells, a finding that could explain the increased levels of p-ERK downstream of MEK. Although originally developed as a RAF inhibitor, sorafenib was subsequently found to inhibit other tyrosine kinases, including vascular epidermal growth factor receptor 2 and 3, platelet derived growth factor receptor-β, Flt3, and c-Kit.19 That these kinases do not play a role in the MEK signaling induced by CXCL12 is indicated by the lack of activity on the MEK/ERK pathways of another drug, sunitinib, which inhibits these other kinases,21 but not RAF. Two other RAF inhibitors also blocked CXCL12-induced activation of MEK/ERK in CLL cells, providing additional support for this model.
Conceivably, CLL cells from patients with ZAP-70+ or ZAP-70− may have differential expression of other kinases, phosphatases, or adapter proteins that play a role in the signaling cascade triggered by exposure to CXCL12. For example, the differences in MEK activation between ZAP-70+ and ZAP-70− CLL cells could be attributed to higher levels or activity of phosphatases in the ZAP-70− CLL cells. One possible candidate is the SH2-containing inositol 5′-phosphate (SHIP) phosphatase, which has been implicated in suppression of RAF and ERK activation in mouse pre-B cells28 and in signaling lymphocyte activation molecule–mediated ERK activation in a B-cell line.29 Furthermore, increased SHIP-1 protein levels were reported in ZAP-70− CLL cells, and it was found to be constitutively tyrosine phosphorylated to a greater extent than in ZAP-70+ CLL cells.30 Thus, increased SHIP-1 levels in ZAP-70− CLL cells could be responsible for the short or absent CXCL12-induced ERK activation.
In any case, the relative sensitivity of ZAP-70+ CLL cells to CXCL12 compared with CLL cells lacking ZAP-70 appears rooted in the differential CXCL12-induced activation of the RAF/MEK/ERK signaling pathway, making this pathway an attractive target for development of new treatments for patients with CLL, particularly those with ZAP-70+ CLL who have a tendency for relatively rapid disease progression and shorter survival. To this end, we found that sorafenib blocked CXCL12-induced activation of the RAF/MEK/ERK pathway and sorafenib induced more pronounced apoptosis in ZAP-70+ CLL than in ZAP-70− CLL cells, suggesting an increased dependence on CLL cells from patients with ZAP-70+ on this pathway for survival. However, at higher doses sorafenib dramatically reduces CLL cell survival in both ZAP-70+ and ZAP-70− CLL cells.
Importantly, a single dose of 10μM sorafenib induced apoptosis of most CLL cells even when cocultured with NLCs, which can protect CLL cells from fludarabine-induced apoptosis in vitro.8 These results suggest that sorafenib could be an effective novel therapeutic for CLL. Although our data support the involvement of RAF in CXCL12-mediated activation of MEK/ERK, the sorafenib-induced apoptosis of CLL cells could be a consequence of inhibition of other sorafenib targets in addition to MEK/ERK.
In summary, our data show that the extent and duration of MEK and ERK activation and subsequent survival of CLL cells after treatment with CXCL12 is significantly greater in CLL cells from patients with ZAP-70+ than in CLL cells from patients with ZAP-70−. Furthermore, the RAF-dependent MEK activation suggests a new method for the treatment of CLL with the inhibitor sorafenib. Conceivably, ZAP-70+ CLL cells might be more sensitive/responsive than ZAP-70− CLL cells to other survival factors produced by the microenvironment in addition to CXCL12; thus, further identification of such factors and their effects on CLL may provide fertile ground for the development of additional strategies to improve the outcome of patients with CLL, particularly those with an aggressive form of the disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Laura Z. Rassenti (Chronic Lymphocytic Research Consortium) for assistance with CLL samples and Lloyd Howard Wang for his excellent technical assistance.
This work was supported by the Lymphoma Research Foundation (grant CLL-07-029; D.M., T.M.H., and T.J.K.), RO1-AI37113 (T.M.H.), and “Le Fond de la Recherche en Santé du Québec” (J.-F.F).
National Institutes of Health
Authorship
Contribution: D.M. designed the research, supervised the study, analyzed the data, designed figures, and wrote the paper; J.-F.F. and M.O. performed experiments, analyzed data, designed figures, and revised the manuscript; I.S.B. performed experiments, analyzed data, and designed figures; T.M.H. designed experiments, analyzed data, and revised the manuscript; T.J.K. provided patient samples, contributed to scientific discussion, and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Davorka Messmer, Moores UCSD Cancer Center, 3855 Health Science Dr no. 0820, La Jolla, CA 92093-0820; e-mail: dmessmer@ucsd.edu.
References
Author notes
D.M., J.-F.F., and M.O. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal