Abstract
Regulated by histone acetyltransferases and deacetylases (HDACs), histone acetylation is a key epigenetic mechanism controlling chromatin structure, DNA accessibility, and gene expression. HDAC inhibitors induce growth arrest, differentiation, and apoptosis of tumor cells and are used as anticancer agents. Here we describe the effects of HDAC inhibitors on microbial sensing by macrophages and dendritic cells in vitro and host defenses against infection in vivo. HDAC inhibitors down-regulated the expression of numerous host defense genes, including pattern recognition receptors, kinases, transcription regulators, cytokines, chemokines, growth factors, and costimulatory molecules as assessed by genome-wide microarray analyses or innate immune responses of macrophages and dendritic cells stimulated with Toll-like receptor agonists. HDAC inhibitors induced the expression of Mi-2β and enhanced the DNA-binding activity of the Mi-2/NuRD complex that acts as a transcriptional repressor of macrophage cytokine production. In vivo, HDAC inhibitors increased the susceptibility to bacterial and fungal infections but conferred protection against toxic and septic shock. Thus, these data identify an essential role for HDAC inhibitors in the regulation of the expression of innate immune genes and host defenses against microbial pathogens.
Introduction
The innate immune system plays an essential role in antimicrobial defenses. Detection of microbial pathogens is carried out by sentinel cells of the innate immune system that are located in tissues (macrophages and dendritic cells [DCs]) in close contact with the host's natural environment or that are rapidly recruited to the site of infection (neutrophils). Recognition of invasive pathogens by immune cells relies on their capacity to detect microbial- or pathogen-associated molecular patterns, such as endotoxin, peptidoglycan, lipopeptides, glucans or mannans, flagellin, and nucleic acids. This process involves the coordinated actions of soluble and cellular molecules composing components of the complement system, acute phase proteins, and membrane-associated or intracellular pattern-recognition molecules, including Toll-like receptors (TLRs), nucleotide-binding oligomerization domain-like receptors, retinoic acid-inducible gene I like receptors, C-type lectin receptors, and scavenger receptors. Ligand-activated receptors trigger the mitogen-activated protein kinase, nuclear factor-κB (NF-κB), and interferon-related factor (IRF) signal transduction pathways that induce the transcription and production of immune genes, including cytokines that are critical for the activation of innate and adaptive immunity.1,2
Chromatin structure plays a central role in regulating gene expression. Acetylation of histones is an essential epigenetic mechanism controlling chromatin structure, DNA accessibility for transcription factors, and gene expression. The net state of acetylation of the ϵ amino groups of lysine residues of histones is regulated by the opposing actions of histone acetyltransferases and histone deacetylases (HDACs). Acetylation of histones relaxes the chromatin structure promoting gene transcription, whereas deacetylation of histones compacts the chromatin structure favoring gene silencing. HDACs have been classified into 4 subclasses based on their homology with yeast HDACs, their subcellular localization, and their enzymatic activity.3 Beside histones, nonhistones proteins (such as α-tubulin, heat shock protein 90, steroid receptors, and regulators of nuclear import) are also modified by reversible acetylation.4,5 Therefore, histone acetyltransferases and HDACs affect diverse biologic functions, principally cell differentiation, growth, and survival.6-8
HDACs are at the center of great interest for 2 major reasons. First, dysregulated HDAC expression or activity has been linked to the pathogenesis of cancer and inflammatory and autoimmune diseases. Second, small-molecule inhibitors of class I, II, and IV HDACs have been shown to exhibit anticancer activity with good safety profiles notably in patients with hematologic malignancies. Thus, these drugs are among the most promising anticancer agents under development.6,8-10 Here we analyzed the impact of HDAC inhibitors on gene expression profiles in macrophages and DCs in vitro and on the host response to bacteria and fungi in vivo. We found that HDAC inhibitors exerted profound inhibitory effects on the host innate immune antimicrobial defense response, down-regulating the expression of innate immune receptors, interfering with transcriptome remodeling after stimulation with TLR agonists, and inhibiting the expression of key antimicrobial cytokines and accessory molecules in whole blood, macrophages, DCs, and splenocytes. Consistent with these immunosuppressive effects, HDAC inhibitors enhanced the susceptibility of mice to bacterial and fungal infections. Conversely, HDAC inhibitors protected mice from septic shock.
Methods
Ethics statement
All animal procedures were approved by the Office Vétérinaire du Canton de Vaud (authorizations 876.5, 876.6, and 877.5) and performed according to the institution guidelines for animal experiments.
Mice, cells, and reagents
Eight- to 12-week-old female BALB/c mice (Charles River Laboratories) were housed under specific pathogen–free conditions. Bone marrow derived macrophages (BMDMs), thioglycollate-elicited peritoneal macrophages, and RAW 264.7 macrophages (ATCC TIB-71) were cultured as previously described.11 Bone marrow–derived DCs (BMDCs) were obtained by culturing bone marrow cells in Iscove modified Dulbecco medium containing 10% fetal calf serum (FCS; Sigma-Aldrich), 50μM 2-mercaptoethanol, and granulocyte-macrophage colony-stimulating factor. Splenocytes were cultured in RPMI medium containing 2mM l-glutamine and 10% FCS and 50μM 2-mercaptoethanol. Human myeloid DCs (moDCs) were produced as described previously.12 Human whole blood assay was performed as described previously.13
Cells were exposed to Salmonella minnesota ultra pure lipopolysaccharide (LPS; List Biologicals Laboratories), Pam3CSK4 (EMC Microcollections), CpG oligonucleotide (CpG ODN; Invivogen), toxic shock syndrome toxin-1 (Toxin Technology), staphylococcal enterotoxin B, concanavalin A (Sigma-Aldrich), or heat-inactivated Escherichia coli O18 (E coli), Staphylococcus aureus, and Candida albicans. Trichostatin A (TSA), suberoylanilide hydroxamic acid (SAHA), and valproic acid (VPA) were from Sigma-Aldrich. The concentrations of TSA and VPA used in vitro did not affect the viability of BMDMs (> 85% cell recovery after 18 hours). Orfiril (Desitin Pharmaceuticals), a commercial injectable solution of sodium valproate, was used for in vivo experiments.
Microarray analysis and quantitative real-time PCR
For each experimental condition, 2 independent samples were processed in parallel. Low RNA input fluorescent linear amplification kit (Agilent Technologies) was used for cDNA synthesis and cRNA amplification. Experimental samples were labeled with cyanine 5-CTP, whereas a control Universal mouse RNA mixture (Stratagene) was labeled using cyanine 3-CTP (PerkinElmer). Labeled cRNA was hybridized onto high-density oligonucleotide microarrays containing approximately 20 000 60-mer (Mouse Development Oligo Microarray Kit, reference G4120A, and Mouse Oligo Microarray Kit, V2, reference G4121B, Agilent Technologies). Slides were scanned using a Microarray Scanner G2565AA system (Agilent Technologies) at a resolution of 5 μm. For data analysis, local background-subtracted signals were calculated using Feature Extraction software (Agilent Technologies, Version A6.1.1). To ensure spot quality, features and their respective background, which were not uniform in pixel fluorescence intensity distribution in both channels, were flagged (nonuniformity outlier flagging algorithm). Data were imported in GeneSpring, Version 7.0 (Agilent Technologies) and then normalized using both per spot (signal channel divided by the corresponding control channel and generation of log10 ratio) and per chip (to the 50th percentile). The microarray dataset has been deposited in the Gene Expression Omnibus database (GEO; National Center for Biotechnology Information; accession numbers GPL7291, GSE22409).
Real-time polymerase chain reaction (RT-PCR) was performed with a 7500 Fast Real-Time PCR System using the Power SYBR Green PCR Master Mix (Applied Biosystems) and primer pairs (supplemental Table 4, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).14 Samples were tested in triplicates. A standard made of successive dilutions of a reference cDNA was processed in parallel. Gene-specific expression levels were assessed relative to the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Hprt and reported in arbitrary units. In selected experiments, standards consisted of serial dilutions of a plasmid containing the target gene.
Cytokine measurements
Tumor necrosis factor (TNF) and interleukin-6 (IL-6) concentrations were measured by bioassay,15 IL-12p40, and interferon-γ (IFN-γ) by enzyme-linked immunosorbent assay (ELISA; BD Biosciences), and a broad screening of cytokines and chemokines production was performed using the Luminex technology (Luminex Corporation).14
Proliferation
Splenocytes (1.5 × 105) were cultured for 48 hours in 96-well culture plates. Proliferation was monitored by measuring 3H-thymidine incorporation over 18 hours.
Flow cytometry
BMDCs were incubated with 2.4G2 monoclonal antibody (mAb) and mAbs (BD Biosciences) specific for mouse major histocompatibility class II (14-4-4S-fluorescein isothiocyanate), CD11c (HL3-phycoerythrin [PE]), and CD40 (3/23-biotin revealed with CyChrome-conjugated streptavidin; BD Biosciences). moDCs were incubated with the lineage Cocktail 1 (fluorescein isothiocyanate–conjugated mAbs specific for CD3, CD14, CD16, CD19, CD20, and CD56) and mAbs specific for human CD11c (B-ly6-PE-Cy5), HLA-DR (L243-APC) and CD40 (5C3-PE), CD80 (L307.4-PE), CD86 (FUN-1-PE), or CC-chemokine receptor 7 (CCR7; 3D12-PE).12 Data were analyzed using FlowJo Version 8.5.3 software (TreeStar).
Electrophoretic mobility shift assay
A total of 2 μg of nuclear extracts was incubated for 15 minutes at room temperature with a radiolabeled consensus NF-κB probe (Santa Cruz Biotechnology) and analyzed by electrophoretic mobility shift assay.16
Western blot analyses
Cell lysates were electrophoresed through polyacrylamide gels and transferred onto nitrocellulose membranes.14 Membranes were incubated with antibodies specific for phosphorylated (phospho)-extracellular signal-regulated kinase (ERK)1/2, total-ERK1/2, phospho-p38, total-p38, phospho-IRF3, (Cell Signaling Technology), c-jun, Mi2b (Santa Cruz Biotechnology), SNF2β/BRG1 (Millipore), IRF7 (Invitrogen), phospho-signal-transducer and activator of transcription protein (STAT1; BD Biosciences), and α-tubulin (Sigma-Aldrich), and then revealed with secondary horse-radish peroxidase-conjugated goat antirabbit IgG and the ECL Western blotting analyses system (GE Healthcare). Acid-soluble proteins were extracted and analyzed by Western blotting using antiacetylated histone H3 and H4 antibodies (Cell Signaling Technology).13
Chromatin immunoprecipitation
Chromatin immunoprecipitation analysis was performed using the ChIP assay kit (Millipore) using antiacetylated histone H4 (06-866, Upstate Biotechnology) or Mi2b (sc-11378X, Santa Cruz Biotechnology) rabbit polyclonal antisera and normal rabbit IgG (sc-2027, Santa Cruz Biotechnology) as described previously.13 Immunoprecipitated DNA was amplified by PCR using primers described in supplemental Table 4.
siRNA silencing in RAW 264.7 mouse macrophages
RAW 264.7 macrophages (6 × 104 cells per well) were seeded in 24-well plates and transfected the next day with 187.5 ng of Mi-2b or Brg1 siRNA duplexes or negative control (sequences available in supplemental Table 4) and HiPerFect transfection reagent (QIAGEN) according to manufacturer's instruction. After 3 days, cells were stimulated for 4 hours with 0 to 10 ng/mL LPS. Gene expression was analyzed by real-time PCR. Mi-2b and Brg1 mRNA levels were decreased by 60% in cells transfected with the specific siRNAs.
In vivo models
Klebsiella pneumonia sepsis.
A total of 10 CFU of a clinical isolate of K pneumoniae was injected intranasally into mice treated 15 minutes earlier with a single dose of valproate (Orfiril, 200 mg/kg intraperitoneally).17
Systemic candidiasis.
Mice were challenged through the tail vein with 1.2 × 105 CFU of a clinical isolate of C albicans. Valproate treatment (200 mg/kg intraperitoneally) was administrated 15 minutes before C albicans challenge and repeated daily during 24 days.
Pam3CSK4-induced shock.
Mice were injected with D-galactosamine (2 g/kg intraperitoneally) followed immediately after by Pam3CSK4 (1.6 mg/kg intraperitoneally).11 Valproate injections (200 mg/kg intraperitoneally every 12 hours) were started 2 days before D-galactosamine injection and discontinued 48 hours after Pam3CSK4 challenge.
CLP.
Animals were pretreated intraperitoneally 1 hour before cecal ligation and puncture (CLP) with valproate (200 mg/kg) and injected subcutaneously every 12 hours with gentamicin (10 mg/kg), clindamycin (30 mg/kg), and buprenorphine (0.1 mg/kg).
Doses of valproate were selected based on previous publications and adjusted to the specific conditions of the sepsis models.
Statistical analysis
Comparisons among treatment groups were performed using the Fisher exact test for categorical data and the Mann-Whitney tests for continuous variables. The Kaplan-Meier method was used for survival, and differences were analyzed by the log-rank sum test. The analyses were performed using Prism software Version 5.03 (GraphPad). All reported P values are 2-sided, and values less than .05 were considered to indicate statistical significance. For microarray analyses, statistical significance of differentially expressed genes (2-fold changes) was evaluated by analysis of variance using the Benjamini and Hochberg false discovery rate correction (5%).
Results
Inhibition of HDACs down-regulates the expression of innate immune genes in macrophages
Transcriptome analyses.
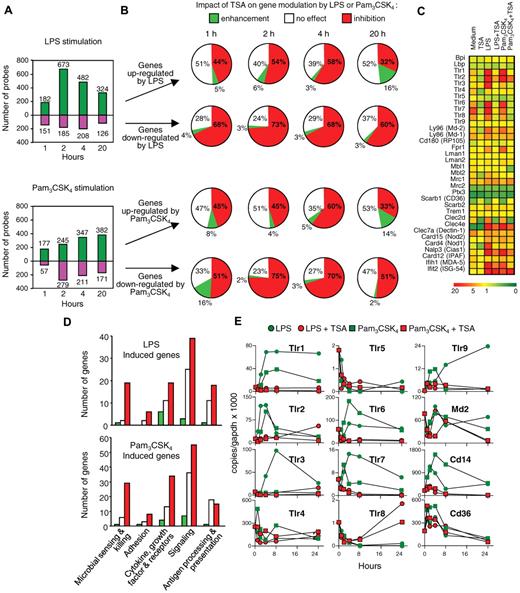
The transcriptome of BMDMs was examined using the Agilent high-density Mouse Development Oligoarrays, which contained 20 280 unique 60-mer corresponding predominantly to expressed sequence tags and with the Mouse Oligo Microarray Kit (V2) containing 20 156 indexed gene probes. At baseline, TSA (t = 4 hours of incubation), a prototypical broad-spectrum inhibitor of class I, II, and IV HDACs, modified the expression of 1594 macrophage genes (7.7% of the transcriptome), of which 772 (3.6%) were found to be down-regulated and 822 (4.1%) up-regulated. HDAC inhibitors significantly reduced the expression of numerous pattern recognition molecules and immune receptors involved in the sensing of a broad range of microbial products, including bacterial lipopeptides, lipoteichoic acid, peptidoglycan, endotoxin, flagellin, viral nucleic acids, and fungal β-glucan (supplemental Table 1). We then performed time-course analyses of gene expression profiles of BMDMs preincubated for 1 hour with TSA before stimulation for 1, 2, 4, and 20 hours with TLR1/TLR2 (Pam3CSK4 lipopeptide) or with TLR4 (LPS) agonists (Figure 1A-B). TSA inhibited the up-regulation of 32% to 58% and 33% to 60% of the genes induced by LPS or Pam3CSK4, respectively. Conversely, TSA counter-regulated the down-regulation of 60% to 73% and 51% to 75% of the genes repressed by LPS or Pam3CSK4. In contrast, TSA potentiated the effects of LPS and Pam3CSK4 on only a small proportion of genes (2%-16%). The genes modulated by TSA included signal transduction, immunoregulation, cytoskeleton and cell structure, metabolism and cell cycle, growth, and apoptosis (complete listing of the genes: Figure 1C-D, supplemental Tables 2-3).
Trichostatin A inhibits the expression of innate immune genes in macrophages. BMDMs were preincubated for 1 hour with or without TSA (100nM) before exposure (C-D, 4 hours) to LPS (100 ng/mL) or Pam3CSK4 (100 ng/mL). Transcriptome was analyzed with Agilent Mouse Development Oligoarrays (A-B) or Mouse Oligo Microarray Kit V2 (C-D). (A) Number of genes either up-regulated or down-regulated by LPS and Pam3CSK4 without preincubation with TSA (fold change > 2 vs medium). (B) Effect of 1-hour preincubation with TSA on genes (expressed in percentage) either up-regulated or down-regulated by LPS or Pam3CSK4 (fold change > 2 vs medium). White represents no change; red, inhibition; and green, increase by TSA compared with stimulation with LPS or Pam3CSK4 alone. (C) Heat map of selected pattern recognition molecules (medium, TSA, LPS and Pam3CSK4: 4 hours of incubation, LPS + TSA and Pam3CSK4 + TSA; 1-hour preincubation with TSA followed by 4-hour incubation with LPS and Pam3CSK4). (D) Effect of 1-hour preincubation with TSA (fold changes were calculated vs LPS or Pam3CSK4 alone) on a selection of genes up-regulated by LPS or Pam3CSK4 and grouped into various categories based on their biologic functions (microbial sensing and killing; adhesion; cytokine, growth factor and receptors; signaling; antigen processing and presentation). White represents no change; red, inhibition; and green, increase by TSA compared with stimulation with LPS or Pam3CSK4 alone. (E) Tlr1-9, Md-2, Cd14, and Cd36 mRNA copy number was determined by RT-PCR and expressed relative to that of GAPDH. Data are representative of 2 independent experiments.
Trichostatin A inhibits the expression of innate immune genes in macrophages. BMDMs were preincubated for 1 hour with or without TSA (100nM) before exposure (C-D, 4 hours) to LPS (100 ng/mL) or Pam3CSK4 (100 ng/mL). Transcriptome was analyzed with Agilent Mouse Development Oligoarrays (A-B) or Mouse Oligo Microarray Kit V2 (C-D). (A) Number of genes either up-regulated or down-regulated by LPS and Pam3CSK4 without preincubation with TSA (fold change > 2 vs medium). (B) Effect of 1-hour preincubation with TSA on genes (expressed in percentage) either up-regulated or down-regulated by LPS or Pam3CSK4 (fold change > 2 vs medium). White represents no change; red, inhibition; and green, increase by TSA compared with stimulation with LPS or Pam3CSK4 alone. (C) Heat map of selected pattern recognition molecules (medium, TSA, LPS and Pam3CSK4: 4 hours of incubation, LPS + TSA and Pam3CSK4 + TSA; 1-hour preincubation with TSA followed by 4-hour incubation with LPS and Pam3CSK4). (D) Effect of 1-hour preincubation with TSA (fold changes were calculated vs LPS or Pam3CSK4 alone) on a selection of genes up-regulated by LPS or Pam3CSK4 and grouped into various categories based on their biologic functions (microbial sensing and killing; adhesion; cytokine, growth factor and receptors; signaling; antigen processing and presentation). White represents no change; red, inhibition; and green, increase by TSA compared with stimulation with LPS or Pam3CSK4 alone. (E) Tlr1-9, Md-2, Cd14, and Cd36 mRNA copy number was determined by RT-PCR and expressed relative to that of GAPDH. Data are representative of 2 independent experiments.
We then focused our analyses on innate immune gene families known to play a critical role in the host antimicrobial defense response. TSA exerted prominent inhibitory effects on LPS- or Pam3CSK4-induced genes (Figure 1C-D). Indeed, TSA inhibited LPS- and/or Pam3CSK4-induced up-regulation of genes encoding for molecules involved in the sensing of microbial compounds, such as Tlrs, Cd14, Md-2, scavenger receptors (Scarb2), cytosolic microbial sensors (Aim1, Mda-5, Nlrp3, Nod1, Nod2, Eif2ak2/Pkr, Pycard/Asc, Mevf/Pyrin, and RIG-I), c-type lectins (Clecs and Msr1), formyl peptide receptors (Fprs), IgE and IgG Fc receptors, complement and complement receptors (C1qa, C1r, and Cfb/H2-Bf), and adhesion molecules (Icam1, integrins, and Vcam1; Table 1). The effect of TSA on the expression of Tlrs, Cd14, Cd36, and Md-2 was confirmed by real-time PCR (Figure 1E). TSA also inhibited the expression of adaptor molecules (MyD88 and Ticam2), kinases (Iraks, Jaks, Lck, Map3ks, Ripk2, Syk, Tank, Tbk1, and Traf1), phosphatases, and transcription modulators (Atfs, Cebps, Irfs, Junb, Nfkbs, Spic, Stats, and Socs1; Table 1). In addition, TSA down-regulated a wide range of LPS- and/or Pam3CSK4-stimulated mediators involved in chemotaxis, inflammation, tissue repair, and antigen processing and presentation. This list of genes included cytokines (Il1a, Il1rn, Il6, Il12a, Il12b, Il15, Il18, Il23a, Il27, Ltb, Tnf, Tnfaip3, Tnfsf4, and Tnfsf9), chemokines (Ccl4, Ccl7-9, Ccl12, Ccl17, Ccl22, Ccl24, Cklfsf3, Cklfsf6, Cklfsf7, Cxcl2, Cxcl5, Cxcl12, Cxcl16, and Cx3cl1), growth factors (Csf2, Edn1, and Tmpo), and their receptors (Il1rl1, Il2rg, Il4ra, Il10rb, Il13ra1, Crlf3, Cxcl12, Cxcl16, Pdfrl, Tnfrsf1a, Tnfrsf14, Ifnar1, Ifnar2, Ifngr2, Csf3r, and Ednrb), cathelicidin antimicrobial peptide (Calmp), matrix metalloproteinases, ubiquitins, proteasome subunits and molecules involved in autophagy (Atg16l), antigen transport (Tap1 and Tap2), and peptide presentation (H-2D, H-2E, and H-2Q; Table 1). Therefore, TSA strongly affected transcriptome remodeling of BMDMs stimulated with microbial products exerting predominantly inhibitory effects, indicating that acetylation of histones or nonhistone proteins is required for optimal transcription of a large number of macrophage genes involved in microbial sensing and host defenses.
Selection of genes whose up-regulation by LPS or Pam3CSK4 is inhibited by TSA in BMDMs
| Adhesion molecule . | Innate receptor . | Signal transduction . | Phosphatase . | Transcription regulator . | Cytokine, chemokine, and growth factor . | Cytokine, chemokine, and growth factor receptor . | Antigen processing and presentation . | Ubiquiti-nation . | Others . |
|---|---|---|---|---|---|---|---|---|---|
| Alcam | Aim1 | Csnk1a1 | Dusp1 (Mkp-1) | Atf3 | Ccl4 | Ccrl2 | Atg16l | Ubc | Calmp |
| Cd47 | Birc (cIAP2) | Irak2 | Dusp4 | Atf4 | Ccl7 | Crlf3 | Ctsz | Ube1l | C1qa |
| Icam1 | Card4 (Nod1) | Irak3 | Dusp16 | Atf5 | Ccl8 | Csf3r | H2-DMb1 | Ube2c | C1r |
| Itga4 | Carc15 (Nod2) | Jak1 | Ptpn6 | Batf | Ccl9 | Ednrb | H2-Ea | Ube2d3 | H2-Bf |
| Itga5 | Cd14 | Jak2 | Ptpn9 | Bcl3 | Ccl12 | Ifnar1 | H2-Eb1 | Ube2e2 | F10 |
| Itgal | Cias1 (Nlrp3) | Lck | Ptpn12 | Bcl7c | Ccl17 | Ifnar2 | H2-M3 | Ube2j1 | Mmp9 |
| Itgax | Clec2d | Map3k7ip2 | Bcl10 | Ccl22 | Ifngr2 | H2-Q10 | Ube2j2 | Mmp14 | |
| Itgb3 | Clec4a2 | Map3k8 (Tpl2) | Cebpb | Ccl24 | Il1rl1 | H2-Q7 | Ube2l6 | Mmp25 | |
| Vcam1 | Clec4d | Mapkapk2 | Cebpd | Ccrl2 | Il2rg | Psma3 | Ube2m | Nos2 | |
| Clec4e | Myd88 | Cited2 | Csf2 | Il4ra | Psma4 | Ube2r2 | Pde4b | ||
| Eif2ak2 (Pkr) | Pias1 | Hmgb2 | Cklfsf3 | Il10rb | Psma5 | Ube2v1 | Pld2 | ||
| Fcer1g | Prkr | Ikbke | Cksfs6 | Il13ra1 | Psma6 | Ubl3 | Ptger4 | ||
| Fcgr2b | Ptk2 | Irf1 | Cklfsf7 | Pdfrl | Psmb7 | Ubtd1 | Ptges | ||
| Frp1 | Ripk2 | Irf2 | Cxcl2 | Ptafr | Psmb8 | Ufm1 | Ptgir | ||
| Fpr-rs2 | Socs1 | Irf5 | Cxcl5 | Tnfrsf1a | Psmb9 | Usp12 | Ptgs2 | ||
| Ifih1 (Mda5) | Stk19 | Irf7 | Cxcl12 | Tnfrsf14 | Psmb10 | Usp18 | Sod2 | ||
| Ly96 (Md-2) | Syk | Junb | Cxcl16 | Psme1 | Usp24 | Txn1 | |||
| Mefv (Pyrin) | Tank | Klf6 | Cx3cl1 | Psme2b | Usp42 | Tnfrsf5 (Cd40) | |||
| Msr1 | Tbk1 | Nfkb1 | Edn1 | Tap1 | |||||
| Pycard | Ticam2 | Nfkb2 | Il1a | Tap2 | |||||
| Scarb2 | Traf1 | Nfkbib | Il1rn | Tapbp | |||||
| Ddx58 (RIG-l) | Trim30 | Nfkbie | Il6 | Tapbpl | |||||
| Tlr1 | Nfkbiz | Il12a | |||||||
| Tlr2 | Pias1 | Il12b | |||||||
| Tlr3 | Rel | Il13ra1 | |||||||
| Tlr6 | Rela | Il15 | |||||||
| Tlr7 | Relb | Il18 | |||||||
| Tlr9 | Spic | Il23a | |||||||
| Stat3 | Il27 | ||||||||
| Stat5a | Inhba | ||||||||
| Ltb | |||||||||
| Pbef1 | |||||||||
| Tnf | |||||||||
| Tmpo | |||||||||
| Tnfaip3 (A20) | |||||||||
| Tnfsf4 (OX-40L) | |||||||||
| Tnfsf9 (4-1BBL) |
| Adhesion molecule . | Innate receptor . | Signal transduction . | Phosphatase . | Transcription regulator . | Cytokine, chemokine, and growth factor . | Cytokine, chemokine, and growth factor receptor . | Antigen processing and presentation . | Ubiquiti-nation . | Others . |
|---|---|---|---|---|---|---|---|---|---|
| Alcam | Aim1 | Csnk1a1 | Dusp1 (Mkp-1) | Atf3 | Ccl4 | Ccrl2 | Atg16l | Ubc | Calmp |
| Cd47 | Birc (cIAP2) | Irak2 | Dusp4 | Atf4 | Ccl7 | Crlf3 | Ctsz | Ube1l | C1qa |
| Icam1 | Card4 (Nod1) | Irak3 | Dusp16 | Atf5 | Ccl8 | Csf3r | H2-DMb1 | Ube2c | C1r |
| Itga4 | Carc15 (Nod2) | Jak1 | Ptpn6 | Batf | Ccl9 | Ednrb | H2-Ea | Ube2d3 | H2-Bf |
| Itga5 | Cd14 | Jak2 | Ptpn9 | Bcl3 | Ccl12 | Ifnar1 | H2-Eb1 | Ube2e2 | F10 |
| Itgal | Cias1 (Nlrp3) | Lck | Ptpn12 | Bcl7c | Ccl17 | Ifnar2 | H2-M3 | Ube2j1 | Mmp9 |
| Itgax | Clec2d | Map3k7ip2 | Bcl10 | Ccl22 | Ifngr2 | H2-Q10 | Ube2j2 | Mmp14 | |
| Itgb3 | Clec4a2 | Map3k8 (Tpl2) | Cebpb | Ccl24 | Il1rl1 | H2-Q7 | Ube2l6 | Mmp25 | |
| Vcam1 | Clec4d | Mapkapk2 | Cebpd | Ccrl2 | Il2rg | Psma3 | Ube2m | Nos2 | |
| Clec4e | Myd88 | Cited2 | Csf2 | Il4ra | Psma4 | Ube2r2 | Pde4b | ||
| Eif2ak2 (Pkr) | Pias1 | Hmgb2 | Cklfsf3 | Il10rb | Psma5 | Ube2v1 | Pld2 | ||
| Fcer1g | Prkr | Ikbke | Cksfs6 | Il13ra1 | Psma6 | Ubl3 | Ptger4 | ||
| Fcgr2b | Ptk2 | Irf1 | Cklfsf7 | Pdfrl | Psmb7 | Ubtd1 | Ptges | ||
| Frp1 | Ripk2 | Irf2 | Cxcl2 | Ptafr | Psmb8 | Ufm1 | Ptgir | ||
| Fpr-rs2 | Socs1 | Irf5 | Cxcl5 | Tnfrsf1a | Psmb9 | Usp12 | Ptgs2 | ||
| Ifih1 (Mda5) | Stk19 | Irf7 | Cxcl12 | Tnfrsf14 | Psmb10 | Usp18 | Sod2 | ||
| Ly96 (Md-2) | Syk | Junb | Cxcl16 | Psme1 | Usp24 | Txn1 | |||
| Mefv (Pyrin) | Tank | Klf6 | Cx3cl1 | Psme2b | Usp42 | Tnfrsf5 (Cd40) | |||
| Msr1 | Tbk1 | Nfkb1 | Edn1 | Tap1 | |||||
| Pycard | Ticam2 | Nfkb2 | Il1a | Tap2 | |||||
| Scarb2 | Traf1 | Nfkbib | Il1rn | Tapbp | |||||
| Ddx58 (RIG-l) | Trim30 | Nfkbie | Il6 | Tapbpl | |||||
| Tlr1 | Nfkbiz | Il12a | |||||||
| Tlr2 | Pias1 | Il12b | |||||||
| Tlr3 | Rel | Il13ra1 | |||||||
| Tlr6 | Rela | Il15 | |||||||
| Tlr7 | Relb | Il18 | |||||||
| Tlr9 | Spic | Il23a | |||||||
| Stat3 | Il27 | ||||||||
| Stat5a | Inhba | ||||||||
| Ltb | |||||||||
| Pbef1 | |||||||||
| Tnf | |||||||||
| Tmpo | |||||||||
| Tnfaip3 (A20) | |||||||||
| Tnfsf4 (OX-40L) | |||||||||
| Tnfsf9 (4-1BBL) |
Cytokine production.
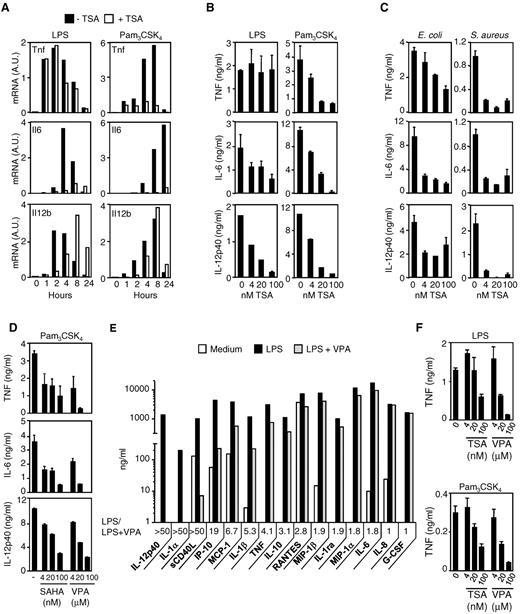
To validate the observations generated from microarray profiling, we quantified the production of cytokines (ie, TNF, IL-6, and IL-12p40) in BMDMs exposed to LPS, Pam3CSK4, E coli, or S aureus. Real-time PCR analyses (Figure 2A), bioassay, and ELISA measurements (Figure 2B-C) confirmed that TSA strongly inhibited TNF, IL-6, and IL-12p40 production in a time- and dose-dependent manner. Yet, TSA did not inhibit LPS-induced Tnf mRNA and protein (Figure 2A-B). Similar results were obtained in thioglycolate-elicited peritoneal macrophages (data not shown).
HDAC inhibitors inhibit cytokine release by macrophages exposed to microbial products and bacteria. BMDMs were preincubated for 1 hour with or without TSA (100nM unless specified) before exposure to LPS (100 ng/mL), Pam3CSK4 (100 ng/mL), and heat-killed E coli or S aureus (107 CFU/mL). (A-C) TNF, IL-6, and IL-12p40 mRNA (A) and protein (B-C) production by BMDMs. TNF, IL-6, and IL-12p40 mRNA levels were analyzed by RT-PCR, and results are expressed as the ratio of cytokines to GAPDH mRNA levels. Data are representative of 3 independent experiments. Cytokine were quantified in cell culture supernatants collected after 8 hours (TNF) and 18 hours (IL-6 and IL-12p40). Data are mean ± SD of triplicate samples from one experiment representative of 3 independent experiments. A.U. indicates arbitrary units. (D) BMDMs were preincubated for 1 hour with or without SAHA (4, 20, and 100nM) or VPA (4, 20, and 100μM) before exposure to Pam3CSK4 (100 ng/mL). TNF, IL-6, and IL-12p40 were quantified in cell culture supernatants collected after 8 hours (TNF) and 18 hours (IL-6 and IL-12p40). Data are mean ± SD of triplicate samples from 1 experiment representative of 3 independent experiments. (E-F) Human whole blood was incubated for 18 hours with VPA (E, 100μM) or TSA together with either LPS (10 ng/mL) or Pam3CSK4 (100 ng/mL). Cytokine and chemokine production was assessed by the Luminex technology (“Cytokine measurements”), and LPS/LPS + VPA ratios were calculated (E). TNF was quantified by bioassay. Data are mean ± SD of triplicate samples from one donor and are representative of 2 independent experiments (F).
HDAC inhibitors inhibit cytokine release by macrophages exposed to microbial products and bacteria. BMDMs were preincubated for 1 hour with or without TSA (100nM unless specified) before exposure to LPS (100 ng/mL), Pam3CSK4 (100 ng/mL), and heat-killed E coli or S aureus (107 CFU/mL). (A-C) TNF, IL-6, and IL-12p40 mRNA (A) and protein (B-C) production by BMDMs. TNF, IL-6, and IL-12p40 mRNA levels were analyzed by RT-PCR, and results are expressed as the ratio of cytokines to GAPDH mRNA levels. Data are representative of 3 independent experiments. Cytokine were quantified in cell culture supernatants collected after 8 hours (TNF) and 18 hours (IL-6 and IL-12p40). Data are mean ± SD of triplicate samples from one experiment representative of 3 independent experiments. A.U. indicates arbitrary units. (D) BMDMs were preincubated for 1 hour with or without SAHA (4, 20, and 100nM) or VPA (4, 20, and 100μM) before exposure to Pam3CSK4 (100 ng/mL). TNF, IL-6, and IL-12p40 were quantified in cell culture supernatants collected after 8 hours (TNF) and 18 hours (IL-6 and IL-12p40). Data are mean ± SD of triplicate samples from 1 experiment representative of 3 independent experiments. (E-F) Human whole blood was incubated for 18 hours with VPA (E, 100μM) or TSA together with either LPS (10 ng/mL) or Pam3CSK4 (100 ng/mL). Cytokine and chemokine production was assessed by the Luminex technology (“Cytokine measurements”), and LPS/LPS + VPA ratios were calculated (E). TNF was quantified by bioassay. Data are mean ± SD of triplicate samples from one donor and are representative of 2 independent experiments (F).
To confirm the findings obtained with TSA, we tested the effects of 2 other HDAC inhibitors: SAHA, a hydroxamate, and VPA, a short chain fatty acid. Like TSA, SAHA and VPA used at clinically relevant concentrations (4-100nM and 4-100μM) dose-dependently inhibited TNF, IL-6, and IL-12p40 production in BMDMs stimulated with Pam3CSK4 (Figure 2D). VPA also markedly reduced (up to 50-fold) the production of 13 of 15 mediators induced by LPS or Pam3CSK4 in whole blood (Figure 2 E-F). HDAC inhibitors down-regulated the production of IL-1ra and IL-10, suggesting that the reduced expression of proinflammatory cytokines was not the result of an increased expression of anti-inflammatory cytokines.
Expression of IFNs and IFN-dependent genes.
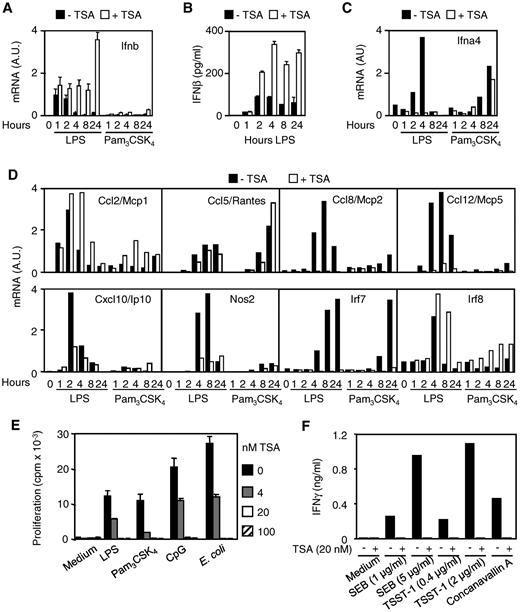
Together with cytokines and chemokines, type I interferons (IFN-α and IFN-β) are central mediators of innate and adaptive immune responses against viral and bacterial infections.18,19 In response to LPS stimulation, macrophages produce copious amounts of IFN-β that stimulates the transcription of Ifna, Ccl2, Ccl8, Ccl12, Cxcl10, and Nos2 (encoding for iNOS), Irf7, and Irf8.20,21 LPS, but not Pam3CSK4, induced rapid (1 hour) Ifnb mRNA expression and sustained IFN-β secretion by BMDMs, whereas LPS or Pam3CSK4 induced early (2 hours and 4 hours, LPS) or late (8 hours and 24 hours, Pam3CSK4) Ifna4 mRNA up-regulation (Figure 3A-C). Interestingly, TSA up-regulated and markedly prolonged (up to 24 hours) IFN-β mRNA and protein expression after LPS exposure (Figure 3A-B), confirming microarray data (supplemental Table 2). In contrast, it completely inhibited the up-regulation of Ifna4 mRNA (Figure 3C). TSA also inhibited the expression of LPS-induced Ccl8, Ccl12, Cxcl10, Nos2, and Irf7 mRNA (Figure 3D) and of numerous other LPS-induced IFN-β-dependent genes (supplemental Figure 1). Yet, TSA did not reduce LPS-induced Ccl2 and Irf8 mRNA and late (24 hours) IFN-β-independent Ifna4 mRNA (Pam3CSK4) and Ccl5 mRNA (LPS and Pam3CSK4). Thus, the massive accumulation of IFN-β induced by TSA in BMDMs did not overcome its inhibition of IFN-β-dependent gene expression. Of interest, TSA dose-dependently suppressed the proliferation of splenocytes induced by LPS, Pam3CSK4, CpG ODN, or E coli (Figure 3E). It also inhibited the release of IFN-γ by splenocytes exposed to staphylococcal enterotoxin B, toxic shock syndrome toxin-1, or concanavalin A (Figure 3F).
HDAC inhibition enhances the production of IFN-β. (A-D) RNA and cell culture supernatants were collected from BMDMs preincubated for 1 hour with (+) or without (−) TSA (100nM) before exposure to LPS (100 ng/mL) or Pam3CSK4 (100 ng/mL). (A-B) IFN-β mRNA and protein expression was quantified by RT-PCR and ELISA. Results are expressed as the ratio of Ifnb mRNA level to that of GAPDH. Data are mean ± SD of triplicate samples from 1 experiment representative of 2 experiments. (C-D) Ifna4 (C), Ccl2, Ccl5, Ccl8, Ccl12, Cxcl10, Nos2, Irf7, and Irf8 (D) mRNA contents were quantified by RT-PCR. Results are expressed as the ratio of mRNA level of the gene of interest to that of GAPDH. Data are representative of 2 independent experiments. A.U. indicates arbitrary units. (E-F) Splenocytes were incubated with TSA and LPS (5 μg/mL), Pam3CSK4 (5 μg/mL), CpG ODN (CpG, 0.5μM), E coli (5 × 107 CFU/mL), staphylococcal enterotoxin B (SEB, 1-5 μg/mL), toxic shock syndrome toxin-1 (TSST-1, 0.4-2.0 μg/mL), and concanavalin A (5 μg/mL). (E) Proliferation was measured by 3H-thymidine incorporation. Data are mean ± SD of triplicate samples and are representative of 2 independent experiments. (F) IFN-γ production was quantified by ELISA in cell culture supernatants collected after 48 hours. Data are representative of 2 independent experiments.
HDAC inhibition enhances the production of IFN-β. (A-D) RNA and cell culture supernatants were collected from BMDMs preincubated for 1 hour with (+) or without (−) TSA (100nM) before exposure to LPS (100 ng/mL) or Pam3CSK4 (100 ng/mL). (A-B) IFN-β mRNA and protein expression was quantified by RT-PCR and ELISA. Results are expressed as the ratio of Ifnb mRNA level to that of GAPDH. Data are mean ± SD of triplicate samples from 1 experiment representative of 2 experiments. (C-D) Ifna4 (C), Ccl2, Ccl5, Ccl8, Ccl12, Cxcl10, Nos2, Irf7, and Irf8 (D) mRNA contents were quantified by RT-PCR. Results are expressed as the ratio of mRNA level of the gene of interest to that of GAPDH. Data are representative of 2 independent experiments. A.U. indicates arbitrary units. (E-F) Splenocytes were incubated with TSA and LPS (5 μg/mL), Pam3CSK4 (5 μg/mL), CpG ODN (CpG, 0.5μM), E coli (5 × 107 CFU/mL), staphylococcal enterotoxin B (SEB, 1-5 μg/mL), toxic shock syndrome toxin-1 (TSST-1, 0.4-2.0 μg/mL), and concanavalin A (5 μg/mL). (E) Proliferation was measured by 3H-thymidine incorporation. Data are mean ± SD of triplicate samples and are representative of 2 independent experiments. (F) IFN-γ production was quantified by ELISA in cell culture supernatants collected after 48 hours. Data are representative of 2 independent experiments.
HDAC inhibitors impair the response of DCs
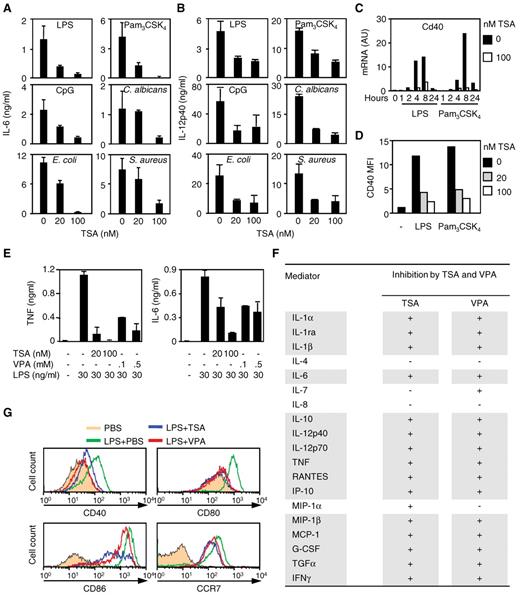
We then examined the effects of HDAC inhibitors on innate immune responses of mouse BMDCs and human moDCs. TSA inhibited the production of cytokines (IL-6 and IL-12p40) and the up-regulation of CD40 by BMDCs stimulated with LPS, Pam3CSK4, CpG ODN, E coli, S aureus, or C albicans (Figure 4A-D). Similarly, TSA and VPA inhibited proinflammatory and anti-inflammatory cytokines (TNF, IL-1α, IL-1β, IL-1ra, IL-6, IL-10, IL-12p40, and IL-12p70), chemokines (IP-10/CXCL10, MIP-1β/CCL4, MCP-1/CCL2, and RANTES/CCL5), IFN-γ, G-CSF, and TGF-α production by moDCs in response to LPS stimulation (Figure 4E-F). TSA and VPA also inhibited the up-regulation induced by LPS of the costimulatory molecules CD40, CD80, and CD86 and of CCR7 (Figure 4G). These results suggested that HDAC inhibitors affect biologic activities of DCs, playing a central role in orchestrating the innate and adaptive responses to infection.
HDAC inhibitors inhibit the response of DCs to microbial stimulation. (A-C) BMDCs were preincubated for 1 hour with TSA before stimulation for 18 hours or the indicated time with LPS (100 ng/mL), Pam3CSK4 (100 ng/mL), CpG oligonucleotide (CpG, 0.7μM), and heat-killed C albicans, E coli, or S aureus (107 CFU/mL). (A-B) IL-6 and IL-12p40 production. Data are mean ± SD of triplicate samples and are representative of 4 independent experiments. (C) Cd40 mRNA expression quantified by RT-PCR. Results are expressed as the ratio of Cd40 mRNA level to that of GAPDH. Data are mean ± SD of 1 experiment representative of 3 independent experiments. AU indicates arbitrary units. (D) CD40 mean fluorescence intensity (MFI) determined by flow cytometry. Data are representative of 3 independent experiments. (E-G) Human moDCs were preincubated for 1 hour with TSA (E-F, 100nM) or VPA (E-F, 100μM) before exposure to LPS (30 ng/mL) for 18 hours. (E) TNF and IL-6 production. Data are mean ± SD of triplicate samples and are representative of 2 independent experiments. (F) Effect of TSA and VPA on cytokine and chemokine production by 2 independent preparations of moDCs. Mediators were analyzed using the Luminex technology (“Cytokine measurements”). + indicates inhibition (fold change > 2); and −, no effect. (G) CD40, CD80, CD86, and CCR7 expression analyzed by flow cytometry. Data are representative of 2 independent experiments.
HDAC inhibitors inhibit the response of DCs to microbial stimulation. (A-C) BMDCs were preincubated for 1 hour with TSA before stimulation for 18 hours or the indicated time with LPS (100 ng/mL), Pam3CSK4 (100 ng/mL), CpG oligonucleotide (CpG, 0.7μM), and heat-killed C albicans, E coli, or S aureus (107 CFU/mL). (A-B) IL-6 and IL-12p40 production. Data are mean ± SD of triplicate samples and are representative of 4 independent experiments. (C) Cd40 mRNA expression quantified by RT-PCR. Results are expressed as the ratio of Cd40 mRNA level to that of GAPDH. Data are mean ± SD of 1 experiment representative of 3 independent experiments. AU indicates arbitrary units. (D) CD40 mean fluorescence intensity (MFI) determined by flow cytometry. Data are representative of 3 independent experiments. (E-G) Human moDCs were preincubated for 1 hour with TSA (E-F, 100nM) or VPA (E-F, 100μM) before exposure to LPS (30 ng/mL) for 18 hours. (E) TNF and IL-6 production. Data are mean ± SD of triplicate samples and are representative of 2 independent experiments. (F) Effect of TSA and VPA on cytokine and chemokine production by 2 independent preparations of moDCs. Mediators were analyzed using the Luminex technology (“Cytokine measurements”). + indicates inhibition (fold change > 2); and −, no effect. (G) CD40, CD80, CD86, and CCR7 expression analyzed by flow cytometry. Data are representative of 2 independent experiments.
HDAC inhibitors increase the expression and the DNA binding of the transcriptional repressor Mi-2β
We then examined whether HDAC inhibitors affected signal transduction pathways in the macrophage (Figure 5). Neither ERK1/2 or p38 phosphorylation nor NF-κB, c-jun, IRF3, or IRF7 nuclear translocation was inhibited by TSA, VPA, or SAHA in BMDMs, peritoneal macrophages, or RAW 264.7 macrophages (Figure 5A-C; and data not shown). Consistent with the fact that it enhanced LPS-induced IFN-β expression (Figure 3B), TSA markedly prolonged STAT1α/β phosphorylation (Figure 5D). Thus, HDAC inhibitors did not inhibit mitogen-activated protein kinases, NF-κB, IRFs, and STAT1 signal transduction pathways induced by microbial products in macrophages.
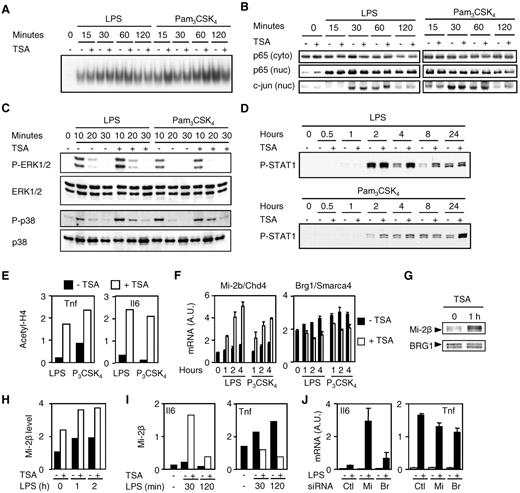
HDAC inhibitors increase Mi-2β expression and recruitment to TSA-sensitive promoter. (A-I) BMDMs were preincubated for 1 hour with (+) or without (−) TSA (100nM unless specified) and exposed to LPS (100 ng/mL) or Pam3CSK4 (100 ng/mL) for the indicated time or 1 hour (E). NF-κB DNA binding activity and NF-κB p65, c-jun, phosphorylated (P-), and total ERK1/2 and p38 and P-STAT1 expression were analyzed by electrophoretic mobility shift assay (A) and Western blot (B-D) using nuclear (nuc) and cytosolic (cyto) extracts. The retarded complex detected by electrophoretic mobility shift assay was dose-dependently inhibited by cold wild-type but not mutant NF-κB oligonucleotide, and supershifted using anti-p65 antibody (data not shown). Acetylation of histone H4 (E) and Mi-2β recruitment (I) to Tnf and Il6 promoters were analyzed by chromatin immunoprecipitation. Mi-2β and BRG1 mRNA and protein levels were quantified by real-time PCR (F) and Western blot (G) with densitometric analyses (H). Data are representative of 2 to 5 independent experiments. (J) RAW 264.7 macrophages transfected with control (Ctl), Mi-2β (Mi), or BRG1 (Br) siRNAs. After 3 days, cells were incubated for 4 hours with (+) or without (−) 10 ng/mL of LPS. Il6 and Tnf mRNA levels were analyzed by RT-PCR and results expressed as the ratio of cytokine to GAPDH mRNA levels. Data are representative of triplicate determinations from 1 experiment.
HDAC inhibitors increase Mi-2β expression and recruitment to TSA-sensitive promoter. (A-I) BMDMs were preincubated for 1 hour with (+) or without (−) TSA (100nM unless specified) and exposed to LPS (100 ng/mL) or Pam3CSK4 (100 ng/mL) for the indicated time or 1 hour (E). NF-κB DNA binding activity and NF-κB p65, c-jun, phosphorylated (P-), and total ERK1/2 and p38 and P-STAT1 expression were analyzed by electrophoretic mobility shift assay (A) and Western blot (B-D) using nuclear (nuc) and cytosolic (cyto) extracts. The retarded complex detected by electrophoretic mobility shift assay was dose-dependently inhibited by cold wild-type but not mutant NF-κB oligonucleotide, and supershifted using anti-p65 antibody (data not shown). Acetylation of histone H4 (E) and Mi-2β recruitment (I) to Tnf and Il6 promoters were analyzed by chromatin immunoprecipitation. Mi-2β and BRG1 mRNA and protein levels were quantified by real-time PCR (F) and Western blot (G) with densitometric analyses (H). Data are representative of 2 to 5 independent experiments. (J) RAW 264.7 macrophages transfected with control (Ctl), Mi-2β (Mi), or BRG1 (Br) siRNAs. After 3 days, cells were incubated for 4 hours with (+) or without (−) 10 ng/mL of LPS. Il6 and Tnf mRNA levels were analyzed by RT-PCR and results expressed as the ratio of cytokine to GAPDH mRNA levels. Data are representative of triplicate determinations from 1 experiment.
Given that histone acetylation is associated with active gene transcription, we then analyzed by chromatin immunoprecipitation the extent of histone H4 acetylation of the Tnf and Il6 promoters in BMDMs stimulated with LPS or Pam3CSK4 (Figure 5E). TSA increased histone H4 acetylation of both promoters, suggesting that there was no correlation between the status of histone acetylation and the observed down-regulatory effect of TSA on gene transcription (Figure 2A).
The Mi-2/NuRD and SWI/SNF (also called BAF in mammals) ATP-dependent remodeling complexes play a central role in regulating gene expression.22 Mi-2β (CHD4) acts as a transcriptional repressor, whereas BRG1 (SMARCA4, the catalytic subunit of the BAF complex) acts as a transcriptional activator of secondary LPS-induced genes in J77.4 macrophages.23 Given that HDAC inhibitors impaired the expression of secondary (ie, Il6) but not primary (ie, Tnf) LPS-induced genes in BMDMs, we hypothesized that TSA mediated its effects by affecting the expression of the Mi-2β and BRG1 dyad. Interestingly, TSA markedly up-regulated the expression of Mi-2β mRNA in BMDMs exposed to LPS and Pam3CSK4 (Figure 5F). In constrast, TSA had a modest inhibitory effect on the expression of BRG1. In line with these findings, TSA increased basal and LPS-induced Mi-2β protein expression (Figure 5G-H). Chromatin immunoprecipitation studies revealed that TSA strongly increased Mi-2β recruitment to the Il6 promoter in BMDMs exposed to LPS, whereas it reduced the binding of Mi-2β to the Tnf promoter (Figure 5I). siRNA-mediated silencing of Mi-2β in RAW 264.7 mouse macrophages greatly enhanced LPS-induced Il6 mRNA expression without a significant effect on Tnf mRNA levels (Figure 5J). Conversely, BRG1 silencing only had a very modest effect on both Il6 and Tnf mRNA expression. Altogether, these data suggest that TSA may inhibit cytokine production via an increased expression of the transcriptional repressor Mi-2β in macrophages.
HDAC inhibitors impair innate immune responses in vivo
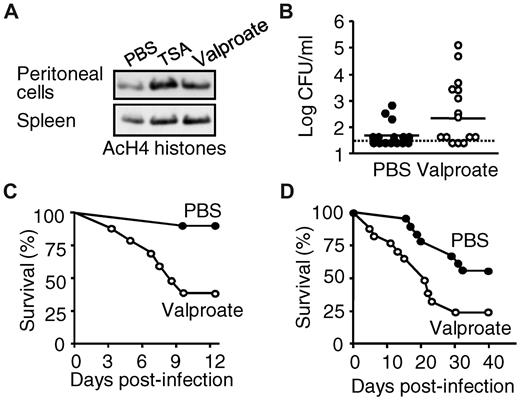
We next investigated the effects of HDAC inhibitors in experimental models of bacterial and fungal sepsis or toxic shock titrated to cause either mild or severe infections or shock. We first verified that valproate enhanced the acetylation of histone H4 in vivo in peritoneal exudate cells and splenocytes (Figure 6A). In an otherwise nonsevere, acute K pneumoniae pneumonia model, valproate increased the proportion (53% vs 80%) and magnitude of bloodstream infections (Figure 6B; P = .04) and mortality (from 6% to 60%, P = .0004; Figure 6C). Consistent with valproate-induced impaired cytokine production by C albicans–infected BMDMs and BMDCs (Figure 4; and data not shown), valproate treatment was associated with accelerated (mean time to death: 21.5 days vs > 40 days) and increased mortality (44% vs 75%, P = .02) in a model of chronic Candida infection (Figure 6D). Thus, inhibition of HDACs impairs host defenses in vivo, increasing the susceptibility to and mortality of bacterial and fungal sepsis.
HDAC inhibition increases mortality to nonsevere infection with K pneumonia and C albicans. BALB/c mice were injected intraperitoneally with valproate (Orfiril, 200 mg/kg) or phosphate-buffered saline (PBS). (A) Peritoneal exudate cells and splenocytes were collected after 1 hour. Histone H4 acetylation (AcH4) was analyzed by Western blotting. (B-C) Mice were infected intranasally with 10 CFU of K pneumoniae 15 minutes after valproate. (B) Circulating bacterial counts 3 days after infection and (C) survival (n = 15 mice per treatment groups; P = .04 and .0004 for bacterial counts and survival, respectively). The dashed line represents the lower limit of detection. (D) Survival of BALB/c mice injected with 1.2 × 105 CFU of C albicans and treated with valproate or PBS daily (n = 18 mice per treatment group; P = .02).
HDAC inhibition increases mortality to nonsevere infection with K pneumonia and C albicans. BALB/c mice were injected intraperitoneally with valproate (Orfiril, 200 mg/kg) or phosphate-buffered saline (PBS). (A) Peritoneal exudate cells and splenocytes were collected after 1 hour. Histone H4 acetylation (AcH4) was analyzed by Western blotting. (B-C) Mice were infected intranasally with 10 CFU of K pneumoniae 15 minutes after valproate. (B) Circulating bacterial counts 3 days after infection and (C) survival (n = 15 mice per treatment groups; P = .04 and .0004 for bacterial counts and survival, respectively). The dashed line represents the lower limit of detection. (D) Survival of BALB/c mice injected with 1.2 × 105 CFU of C albicans and treated with valproate or PBS daily (n = 18 mice per treatment group; P = .02).
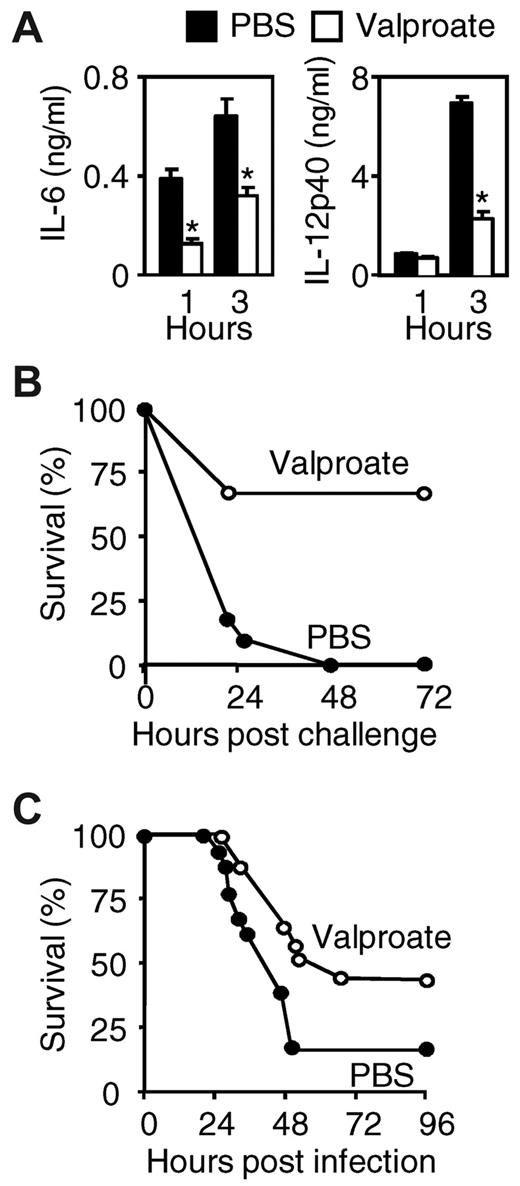
Severe sepsis and septic shock are characterized by an early overwhelming inflammatory response to microbial invasion, and inhibition of proinflammatory mediators confers protection in sepsis models.24,25 We therefore tested whether valproate might exert protective effects in models of fulminant toxic shock induced by Pam3CSK4 or CLP. Administration of valproate caused a 2- to 3-fold reduction of IL-6 and IL-12p40 circulating levels (Figure 7A) and a notable increase in survival (0%-64%, P < .001) in the Pam3CSK4 toxic shock model (Figure 7B). Similarly, valproate treatment increased survival from 17% to 42% (P = .04) in the CLP model (Figure 7C).
HDAC inhibition protects from lethal toxic shock and severe sepsis. (A-B) BALB/c mice were injected intraperitoneally with valproate (Orfiril, 200 mg/kg started 2 days before and discontinued 2 days after induction of shock) or PBS. Animals were sensitized with D-galactosamine and injected intraperitoneally with Pam3CSK4. (A) Plasma levels of IL-6 and IL-12p40 were determined 1 hour and 3 hours after challenge with Pam3CSK4. Data are mean ± SD of 8 mice per treatment group. *P < .001 for valproate versus PBS. (B) Survival of BALB/c mice subjected to Pam3CSK4-induced shock (n = 11 mice per treatment group; P = .001). (C) Survival of BALB/c mice subjected to CLP and treated with either valproate or PBS (every 12 hours, starting 30 minutes after surgery; n = 16-18 mice per treatment group). P = .04.
HDAC inhibition protects from lethal toxic shock and severe sepsis. (A-B) BALB/c mice were injected intraperitoneally with valproate (Orfiril, 200 mg/kg started 2 days before and discontinued 2 days after induction of shock) or PBS. Animals were sensitized with D-galactosamine and injected intraperitoneally with Pam3CSK4. (A) Plasma levels of IL-6 and IL-12p40 were determined 1 hour and 3 hours after challenge with Pam3CSK4. Data are mean ± SD of 8 mice per treatment group. *P < .001 for valproate versus PBS. (B) Survival of BALB/c mice subjected to Pam3CSK4-induced shock (n = 11 mice per treatment group; P = .001). (C) Survival of BALB/c mice subjected to CLP and treated with either valproate or PBS (every 12 hours, starting 30 minutes after surgery; n = 16-18 mice per treatment group). P = .04.
Discussion
These studies identify an essential role for acetylation of histones and nonhistone proteins in the regulation of inflammatory and innate immune gene expression, and in host defensive responses against microbes. Genome-wide microarray analyses revealed a critical role for HDACs in the expression of host defense genes, including pattern-recognition receptors, adaptor molecules, kinases, transcription regulators, complement factors, cytokines, chemokines, and growth factors. HDAC inhibitors exert both immunosuppressive (a predominant effect) and immunostimulatory activities. Located at the forefront of the host defenses against microbial invasion, macrophages and DCs are an important source of cytokines. Inhibition of HDACs caused a marked reduction of cytokine and chemokine production induced by both cell types after exposure to TLR agonists. Of note, HDAC inhibitors also suppressed the release of IFN-α and IFN-γ by macrophages and splenocytes. Likewise, TSA inhibited IFN-α–stimulated gene activation and late IFN-β induction by Sendai virus and doubled-stranded RNA, and prevented IFN-α–mediated inhibition of the cytopathic effects of vesicular stomatitis virus.26,27 Unexpectedly, we observed that HDAC inhibitors sustained LPS-induced IFN-β production in BMDMs. Nevertheless, the expression of numerous IFN-β/STAT1-dependent genes was strongly inhibited by TSA and VPA, indicating that the increased production of IFN-β did not overcome the potent inhibitory effects of HDAC inhibitors.
Phagocytes and professional antigen-presenting cells help bridge innate and adaptive immunity. Macrophages and DCs are key producers of IL-12 and IL-23 promoting the generation of protective Th1 and Th17 responses against intracellular and extracellular pathogens.28,29 HDAC inhibitors were found to be potent inhibitors of IL-12 and IL-23 production by macrophages and DCs as well as CCR7, which is critical for the migration of DCs to secondary lymphoid organs (present data).30-32 TSA inhibited the expression of several genes involved in antigen processing and presentation. Moreover, HDAC inhibitors have been reported to inhibit DC-stimulated allogeneic T-cell proliferation and Th1-cell activation32-34 and to block the differentiation of IL-17–producing T cells.35 Thus, inhibition of HDACs impacts on several key macrophages and DCs functions likely to affect the production of critical cytokines and the mounting of protective Th1 and Th17 immune responses. Of note, TSA did not inhibit TNF production by BMDMs and peritoneal macrophages stimulated with LPS. In contrast, TSA exerted potent inhibitory effects on whole blood, DCs, and BMDMs stimulated with microbial products other than LPS (Pam3CSK4, E coli, and S aureus). Similar results were obtained using SAHA and VPA. These observations suggest that HDAC inhibitors differentially affect gene expression according to the cell type or the stimulus studied. This could account for the discrepant effects of HDAC inhibitors on TNF production reported in the literature.30-33,36 Indeed, delaying TSA treatment until 1 hour after the addition of Pam3CSK4 to BMDMs, or inhibiting protein synthesis at the time of preincubation with TSA, abrogated TSA-mediated inhibition of Pam3CSK4-induced TNF production. Thus, inhibition of stimulus-induced Tnf mRNA expression by TSA requires de novo protein synthesis, which cannot overcome a rapid induction of gene expression in BMDMs stimulated with LPS.
HDAC inhibitors have been reported to interfere with the activation of the mitogen-activated protein kinases, IRFs, STAT1, AP-1, or NF-κB signal transduction pathways. However, these findings have been inconsistent and controversial.26,30,31,36,37 In BMDMs, we did not observe any impact of 3 HDAC inhibitors (TSA, VPA, and SAHA) on ERK1/2 or p38 phosphorylation or on NF-κB, c-jun, IRF3, or IRF7 nuclear translocation induced by LPS or Pam3CSK4. Yet, we found that TSA markedly inhibited the recruitment of NF-κB p65 and of RNA polymerase to the Il6 promoter (data not shown). This finding is consistent with the notion that acetylation of the NF-κB subunits themselves or of molecules involved in the NF-κB signal transduction pathway controls the extent, potency, and duration of NF-κB–mediated transcriptional activity.38
Transcriptional repression mediated by HDAC inhibitors may also rely on acetylation-dependent recruitment of transcriptional corepressors or changes in chromatin architecture.38 In line with this hypothesis, we have previously shown that TSA inhibits the expression of the proinflammatory cytokine macrophage migration inhibitory factor via a local deacetylation of chromatin impairing the recruitment of the basal transcriptional machinery to the macrophage migration inhibitory factor promoter.13,39 In the present study, we provide data suggesting that HDAC inhibitors induce the expression of Mi-2β and the activity of the Mi-2/NuRD complex, which acts as a transcriptional repressor of secondary LPS-induced cytokines, such as IL-6. Although little is known about the in vivo function of Mi-2β, mice deficient in metastasis-associated protein 2, a component of the Mi-2/NuRD complex, develop a lupus-like syndrome characterized by the hypersecretion of cytokines.40
A main finding of our study is the fact that HDAC inhibitors impair the host natural defenses against microbial pathogens. Administration of valproate increased the susceptibility of mice to bacterial and fungal infections converting a nonsevere bacterial pneumonia into a highly lethal infection and markedly increasing the mortality of invasive candidiasis. Moreover, we have also observed that HDAC inhibitors reduced the expression of phagocytic receptors and phagocytosis and killing of bacteria by macrophages (M.M., J.L., T.C., T.R., manuscript in preparation). These results clearly demonstrate that the inhibitory effects of HDAC inhibitors on innate immune cells in vitro translate into robust immunosuppressive effects in vivo that negatively impact on the susceptibility to and outcome of infections. Several arguments led us to believe that these observations may have clinical implications. A vast amount of data indicate that interfering with critical mediators of innate or adaptive immunity (eg, TNF or IL-1) increases the risk of infections.41 Indeed, treatment of patients with TNF antagonists has been associated with an overall increased risk of bacterial (tuberculosis, nontuberculous mycobacteriosis, listeriosis, and salmonellosis) and fungal (candidiasis) infections.42,43 HDAC inhibitors are currently used for the treatment of hematologic malignancies and solid tumors often in combination with other cancer or immune suppressive therapies increasing the risk of infection. In phase 1 and 2 clinical trials, patients treated with valproate, SAHA, MS-275, and ITF2357 have developed severe infections, even without neutropenia, that could be the result of the immune-suppressive activities of these HDAC inhibitors.44-49 These data suggest that monitoring of infections may be warranted in clinical trials of HDAC inhibitors.
Taken advantage of their broad anti-inflammatory and immunomodulatory properties targeting several key mediators implicated in the pathogenesis of septic shock (such as TLRs, MyD88, signal transducing molecules, and proinflammatory cytokines), we reasoned that HDAC inhibitors may prove to be beneficial as adjunctive therapy for septic shock. Consistent with this assumption, valproate exhibited remarkable protective effects in a toxic shock model induced by Pam3CSK4 and in the CLP peritoneal sepsis model. In a recent fascinating article, Xu et al detected histones in the circulation of septic patients and baboons and showed that these proteins played a pathogenic role in sepsis.50 Purified histones H3 and H4 were found to be toxic for endothelial cells, to cause microvascular thrombosis, and to be lethal when injected intravenously into mice. Notably, antihistone H4 antibodies rescued mice from lethal shock induced by LPS, TNF, or CLP. Given that histones appear to act as noxious danger-associated endogenous molecular patterns, one may wonder whether the acetylation status of extracellular histones correlates with toxicity. If so, deacetylation of histones released by necrotic and apoptic cells could exert detoxifying and cytoprotective effects.
Taken together, the present data identify protein acetylation as a key mechanism regulating the expression of important innate immune genes critically implicated in the sensing of and host responses to microbial pathogens. Inhibition of HDACs impairs essential biologic functions of innate immune cells, reducing their capacity to induce a proinflammatory response, to engulf and kill pathogens, and to mount an adaptive response increasing the susceptibility to infection. On the other hand, the broad anti-inflammatory and immune-suppressive properties of HDAC inhibitors were found to be beneficial as adjunctive therapy for septic shock. Thus, these results suggest that the use of HDAC inhibitors as anticancer agents may increase the risk of infection and sepsis, whereas they may offer new therapeutic options for the management of patients with septic shock.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by research funding from the Swiss National Science Foundation (310000-114073/1 and 310030-132744/1, T.R.; 310030-118266, T.C.; 3100A0-112370/1, J.S.; and 3100A0-116075, P.F.), by the Swiss Society for Infectiology (Merck Sharp & Dohme–Chibret AG Award; T.R.), and by the Leenaards Foundation (T.R., J.S.).
Authorship
Contribution: T.R. conceived and supervised the studies, performed experiments, and wrote the paper; J.L., G.G., X.C.D., M.M., A.-L.C., M.K.R., and I.M. carried out in vitro experiments; D.L.R. performed in vivo experiments; T.K., P.F., and J.S. performed microarray analyses; and T.C. discussed the study results and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thierry Roger, Infectious Diseases Service, Department of Medicine, Centre Hospitalier Universitaire Vaudois and University of Lausanne, BH 19-111, rue du Bugnon 46, CH-1011 Lausanne, Switzerland; e-mail: Thierry.Roger@chuv.ch.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal