Abstract
The DNA damage pathway plays a central role in chemoresistance in chronic lymphocytic leukemia (CLL), as indicated by the prognostic impact of TP53 and ATM loss/mutations. We investigated the function of the p53 axis in primary CLL samples by studying p53 and p21 responses to irradiation by FACS and RT-PCR. We observed a distinct response pattern for most cases with a 17p deletion (n = 16) or a sole TP53 mutation (n = 8), but not all cases with a p53 aberration were detected based on a number of different assays used. Samples with a small clone with a TP53 mutation remained undetected in all assays. Only 1 of 123 cases showed high expression of p53, which is suggestive of p53 aberration without proof of mutation of TP53. Samples with an 11q deletion showed a heterogeneous response, with only 13 of 30 showing an abnormal response based on cutoff. Nevertheless, the overall induction of p53 and p21 was impaired, suggesting a gene-dosage effect for ATM in the 11q-deleted samples. The detectability of p53 defects is influenced by clonal heterogeneity and sample purity. Functional assays of p53 defects will detect a small number of cases not detectable by FISH or TP53 mutational analysis. The clinical utility of functional p53 testing will need to be derived from clinical trials.
Introduction
Chronic lymphocytic leukemia (CLL) is a heterogeneous disease characterized by an accumulation of mainly nonproliferating B lymphocytes.1-5 Deletions of 17p13 (17p) and 11q23 (11q) in CLL are associated with early disease progression and shorter overall survival.6,7 The critical regions of 17p and 11q contain 2 prominent tumor-suppressor genes mutated at varying proportions: TP53 (tumor protein 53) and ATM (Ataxia telangiectasia mutated). Mutations of TP53 and ATM, even in the absence of a chromosomal deletion, have been identified and have adverse effects on patient survival.8-12 p53 and ATM proteins are central regulators of the DNA-damage–response pathway.13-15 Their activation leads to cell-cycle arrest and DNA repair, apoptosis, or senescence, depending on the cellular context.
Impaired p53 function through mutation and/or deletion is the best-characterized factor associated with chemoresistance in CLL.16,17 However, a considerable number (> 50%) of refractory CLL patients do not exhibit TP53 mutations or 17p deletion.18 Deregulation of other components of the DNA-damage response and repair pathways might be involved in CLL chemoresistance; for example, ATM, DNA-PK (DNA-dependent protein kinase), the tumor suppressors p16INK4a and p14ARF, and microRNA-34a.19-24
Based on the profound impact of p53 mutations and deletions, it is tempting to speculate that further “functional” defects in the p53 pathway could exist that cause refractory CLL. Therefore, a functional assessment of the DNA-damage–response pathway may help to elucidate the role of p53 and other putative regulators of drug response in CLL. Defects in the pathway could be identifiable in CLL patients that may escape fluorescence in situ hybridization (FISH; 17p and 11q) or mutational analysis.
In primary CLL cells, functional assays have been developed to detect p53 and ATM defects using reverse transcription–multiplex ligation-dependent probe amplification assay (RT-MLPA) of p53 targets25,26 and to detect p53 and p21 proteins (a central downstream target of p53) by Western blot or fluorescence-activated cell sorting (FACS) analysis after DNA damage or inhibition of MDM2-p53 interaction by Nutlin-3a.27-29 A recent study found a good relation between FACS abnormalities and 17p deletions in 8 patients, but also revealed a certain amount of heterogeneity and discordance among patients with the 11q deletion,30 suggesting that the precise correlation of 17p deletions, 11q deletions, and particularly TP53 mutations, with the p53/p21 assay is currently unclear. A recent report added another layer of complexity by suggesting an association between single-nucleotide polymorphisms in the p21 gene with impaired p21 up-regulation and intact p53 induction.31
While RT-MLPA and Western blot depend on samples with high tumor cell content or sorted tumor cells to specifically investigate gene expression or protein levels in the CLL cells, FACS analysis can also be applied to samples with low tumor load. In addition, FACS facilitates quantitative analysis. Nevertheless, intracellular staining may be challenging to standardize, as has been the case in the standardization process for ZAP-70 (zeta-chain–associated protein kinase 70) FACS analysis.32,33
We set up a FACS assay for investigating the levels of p53 and p21 and used the fluorescent intensities of the CD19-negative cells as internal controls, as described previously for ZAP-70 FACS in CLL samples.34 The focus of our study was to analyze the p53 pathway after DNA damage in a well-characterized, large CLL cohort. We combined the FACS analysis with TP53 mutational status, RT-MLPA gene expression data (p53 targets, apoptosis-related genes), expression of miR-34a, and clinical details. Our primary aim was to assess: (1) the number of dysfunctional cases that may escape FISH and TP53 mutational analysis and (2) the fidelity of the assay in detecting CLL with mutant TP53 to assess the potential clinical utility of the test. The results were validated in a separate cohort.
Methods
Patients
A cohort of 59 CLL patients was enrolled in this study after obtaining ethics approval from the University of Ulm institutional review board, with informed consent obtained in compliance with the Declaration of Helsinki. Patients were selected based on having a percentage of lymphocytes of more than 80%. Their age ranged from 35 to 83 years (mean, 64 years). The population was selected to represent meaningful numbers of patients from high-risk groups. An independent second cohort consisted of 64 samples from untreated patients before first-line treatment. While the genetic profile of this group was different from the first group, it more closely resembled the genetic profile of an unselected cohort with treatment indication. The median purity of the CD5/CD19-positive population was 77%.
Genetic analysis
FISH analysis and IGHV (immunoglobulin heavy-chain variable gene) sequencing were performed as described previously.6,35,36 A germline homology of 98% was used as a cutoff to distinguish between IGHV-mutated and -unmutated cases.
Mutations in the coding region of TP53 (exons 4-10) were identified using denaturing high-performance liquid chromatography. Assay conditions and sequencing details were as described previously.37
Cell culture and in vitro treatment
Lymphocytes were isolated from blood samples with Biocoll solution (Biochrom) and aliquots frozen viably at −196°C. For analysis, cells were thawed and cultured in RPMI 1640 medium supplemented with 10% fetal calf serum and glutamine at 37°C in the presence of 5% CO2. Ionizing radiation was performed by exposing the cells to a Cs137 source.
RNA isolation and real-time RT-polymerase chain reaction
Total RNA was isolated using the mirVana PARIS Kit (Ambion). Real-time RT-polymerase chain reaction (RT-PCR) for hsa-miR-34a was performed as described previously.23
For quantitative RT-PCR of p21, 100 ng of mRNA was reverse-transcribed using the QuantiTect reverse transcription kit following the manufacturer's instructions. Real-time PCR was performed using SYBR Green Rox Mix (ABgene) and the following primers: p21 forward 5′-CGC TAA TGG CGG GCT G-3′; p21 reverse 5′-CGG TGA CAA AGT CGA AGT TCC-3′; LaminB1 forward 5′-CTG GAA ATG TTT GCA TCG AAG A-3′; LaminB1 reverse 5′-GCC TCC CAT TGG TTG ATC C-3′.
RT-MLPA
For RT-MLPA, CLL cells of 18 patients were harvested 16 hours after irradiation and had a viability of 50%-93% (determined by Annexin). The median lymphocyte count in these patients was 94%.
The assay was performed as described previously.25 Briefly, 100 ng of total RNA was reverse-transcribed using a gene-specific probe mix. The resulting cDNA was annealed overnight at 60°C to the MLPA probes. Annealed oligonucleotides were covalently linked by Ligase-65 (MRC-Holland) at 54°C. Ligation products were amplified by PCR (33 cycles, 30 seconds at 95°C, 30 seconds at 60°C, and 1 minute at 72°C) using one unlabeled and one 6-carboxy-fluorescein–labeled primer (10 pM). PCR products were run on an ABI 3100 capillary sequencer in the presence of 1pM ROX 500 size standard (Applied Biosystems). Results were analyzed using the programs Genescan and Genotyper (Applied Biosystems). Category tables containing the area for each assigned peak (scored in arbitrary units) were compiled in Genotyper and exported for further analysis with Microsoft Excel spreadsheet software. Data were normalized by setting the sum of all signals at 100% and expressing individual peaks relative to the 100% value.
Western blot
Cell pellets were resuspended in ice-cold lysis buffer (10mM Tris base, pH 8, 1% Triton X-100, 130mM NaCl, 5mM EDTA [ethylenediaminetetraacetic acid], 100μM orthovanadate + protease inhibitors; Invitrogen). The cell debris was separated by centrifugation. The protein concentration was analyzed using Bradford reagent (Bio-Rad). For each lane of the Western blot, 15-20 μg of total protein was denatured and run on a gel using the NuPage Gel system (Invitrogen). Protein transfer to polyvinylidene fluoride membranes (Immobilon-P; Millipore) was performed via semidry blotting or wet blotting for large proteins. The membranes were blocked with 5% nonfat milk in phosphate-buffered saline containing 0.05% Tween 20. The primary antibodies anti–p53 (BD Pharmingen), anti–p21 (BD Pharmingen), and anti–β-actin (Santa Cruz Biotechnology) were incubated overnight at 4°C. The secondary antibodies contained peroxidase that was used for detection with chemiluminescence (Amersham Pharmacia Biotech). For quantification, the Western blot films were scanned and densitometrically analyzed using ImageJ Version 1.41 software.
p53/p21 flow cytometry
Based on the results of Pettitt et al and Carter et al, we set up a flow cytometric assay.27,28 Patient cells were harvested 24 hours after irradiation, stained with anti–CD19-PC7 antibody (Beckman Coulter), fixed in 2% ice-cold formaldehyde, and stored in 80% ethanol at −20°C overnight. Cells were then blocked with bovine serum albumin and treated with permeabilization reagent (Beckman Coulter). Anti-p53-phycoerythrin (Becton Dickinson) and anti-p21-fluorescein isothiocyanate (Calbiochem) or the appropriate isotype controls were incubated for 1 hour at 4°C.
Samples were measured using FACSCalibur (BD Biosciences) and analyzed with CellQuest Pro Version 5.1 software (BD Biosciences). The median of the fluorescent intensities of p53 and p21 was used for quantification.
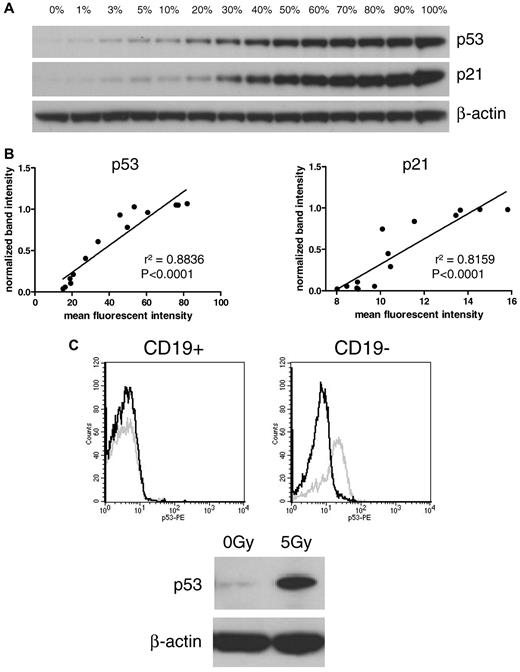
The reliability of the FACS assay was first tested by comparison with Western blot. For that purpose, a dilution series was made with cells of a CLL sample with high induction of both p53 and p21 after irradiation. Treated (positive) cells were mixed with untreated (negative) cells in different ratios, ranging from 0% to 100% positive cells. Samples were analyzed in parallel on Western blot and FACS for p53 and p21 (Figure 1A). Comparison of the median fluorescent intensities of the FACS analysis with the densitometric values of the Western blot showed linear correlations of both p53 and p21 (r2 = 0.8836, P < .0001 and r2 = 0.8159, P < .0001, respectively; Figure 1B).
Comparison of Western blot and FACS assay. (A) Dilution series of treated and untreated CLL cells on Western blot. Irradiated (5 Gy; 100%) cells were mixed with nonirradiated (0%) cells. (B) The intensity of the Western blot bands was quantified and compared with mean fluorescent intensities of the FACS assay. Linear regression shows a high correlation between both methods (left panel: p53, r2 = 0.8836; right panel: p21, r2 = 0.8159). (C) Discrimination between CD19-positive CLL cells and CD19-negative cells in the FACS assay is shown in an exemplary sample. The Western blot shows induction of p21 and p53. In the dissection of the contributing cellular component by FACS (CD19, p21, p53), it is clear that CLL cells showed impaired p53 up-regulation, whereas CD19-negative cells induced p53 after irradiation. Black line, p53 expression of nonirradiated sample; gray line, p53 expression of irradiated sample.
Comparison of Western blot and FACS assay. (A) Dilution series of treated and untreated CLL cells on Western blot. Irradiated (5 Gy; 100%) cells were mixed with nonirradiated (0%) cells. (B) The intensity of the Western blot bands was quantified and compared with mean fluorescent intensities of the FACS assay. Linear regression shows a high correlation between both methods (left panel: p53, r2 = 0.8836; right panel: p21, r2 = 0.8159). (C) Discrimination between CD19-positive CLL cells and CD19-negative cells in the FACS assay is shown in an exemplary sample. The Western blot shows induction of p21 and p53. In the dissection of the contributing cellular component by FACS (CD19, p21, p53), it is clear that CLL cells showed impaired p53 up-regulation, whereas CD19-negative cells induced p53 after irradiation. Black line, p53 expression of nonirradiated sample; gray line, p53 expression of irradiated sample.
To study the impact of T-cell/bystander cell contamination, we compared Western blot (all cells) with the results of the FACS analysis in CD19-positive and -negative fractions. Figure 1C shows an exemplary patient sample with a lymphocyte count of 79%. The FACS histograms show a strong induction of p53 in the CD19-negative cells compared with an absent induction in CLL cells. On the Western blot, a nondiscriminatory induction of p53 and p21 was seen (based on the induction in non-CLL cells).
Statistical analysis
Statistical analyses were performed with Prism Version 5.00 software (GraphPad). A 2-sided Mann-Whitney test was used to compare single parameters between 2 groups. An effect was considered statistically significant at P = .05.
Results
Establishment of FACS assay
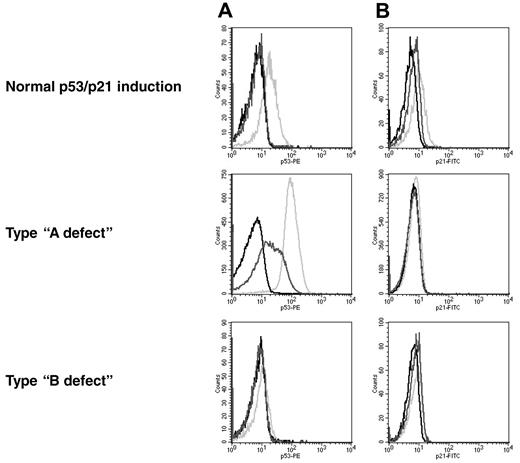
The p53/p21 induction was classified according to previous reports27 (Figure 2). Samples with a low basal p53 level and a high induction of p21 were referred to as a normal response. The type-A defect is characterized by a low p21 induction, along with a high basal p53 level. This defect is associated with a mutation of p53. Type-B defect cells show a low p21 induction in combination with a low basal p53 level and failure to up-regulate p53. The recently described type-C defect, with a low p21 induction combined with a normal p53 induction,31 was included in the group with the type-B defect.
Patterns of response to DNA damage in CLL. The histograms show the levels for p53 (A) and p21 (B). Black line, isotype control; dark gray line, nonirradiated sample; light gray line, irradiated sample. Normal response: low basal levels of p53 and p21 and induction of both proteins after irradiation. Type-A defect: high basal p53 level and impaired p21 induction. Type-B defect: low basal p53 level and impaired p21 induction.
Patterns of response to DNA damage in CLL. The histograms show the levels for p53 (A) and p21 (B). Black line, isotype control; dark gray line, nonirradiated sample; light gray line, irradiated sample. Normal response: low basal levels of p53 and p21 and induction of both proteins after irradiation. Type-A defect: high basal p53 level and impaired p21 induction. Type-B defect: low basal p53 level and impaired p21 induction.
In an effort to increase assay fidelity and in contrast to a previous report, we normalized the values of the CLL cells to the internal CD19-negative cells, similar to the FACS analysis of ZAP-70 described previously.34 The normalized values are displayed as a percentage of the levels in CD19-negative cells: ([CLL{0GyMedian}]/[CD19-negative{0GyMedian}] × 100% for p53 baseline levels and ([CLL{5GyMedian}/CLL{0GyMedian}]/[CD19-negative{5GyMedian}/CD19-negative{0GyMedian}]) × 100% for induction of p53 and p21. While germline polymorphisms of p53 pathway members may affect the response to radiation in normal (and malignant) cells, this approach offers the advantage of addressing the somatic changes with normalization to the individual's inherent variability of the p53 response because of host differences.38
To define cutoff levels, we used 9 healthy donor lymphocyte samples (data not shown). The cutoff levels for p21 induction and p53 induction were calculated as the mean of the healthy samples subtracted by 2 times the standard deviation. The cutoff level for the p53 basal level was determined accordingly, but because the basal levels of p53 were quite diverse in CLL cells (Figure 3), we used 3 times the standard deviation to the mean value.
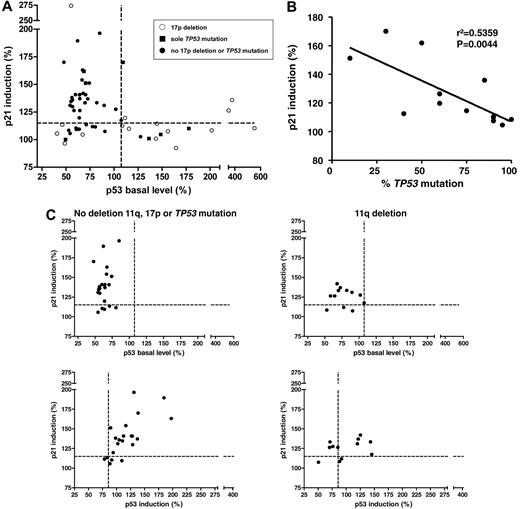
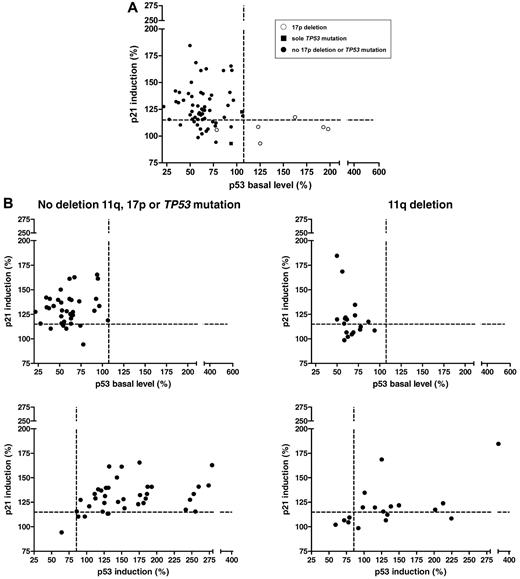
Baseline p53 and p21 induction in primary CLL cells shows distinct patterns. (A) The p53 response, defined as p21 induction versus baseline p53 levels (n = 59). Every dot represents 1 patient sample; the cutoffs are depicted by the dashed lines. A distinct pattern can be observed, with the normal response in the top left quadrant, the type-B defect in the bottom left quadrant, and the type-A defect in the bottom right quadrant. Whereas the type-A defect is strongly associated with 17p deletions and TP53 mutations, some samples with the p53 aberration can also be found among the cases with type-B defect and normal response. ○, 17p deletion; ■, sole TP53 mutation; •, no 17p deletion or TP53 mutation. (B) The clone size of the TP53 mutations (irrespective of 17p deletion) was negatively correlated with the level of p21 induction (r2 = 0.5359, P = .0044; cases with missense mutation only). (C) Top panel, p21 induction versus baseline p53 levels for patients with the 11q deletion (n = 12, right panel) and patients without the 17p deletion, the 11q deletion, or the TP53 mutation (n = 22, left panel). A minority of cases in both subgroups show impaired p21 induction with normal baseline p53 expression when based on the determined cutoff. Bottom panel, p21 induction versus p53 induction in patients with the 11q deletion (right) and in patients without the 11q deletion, 17p deletion, or TP53 mutation (left). p21 and p53 show a stronger induction after DNA damage in patients without the 11q deletion, the 17p deletion, or the TP53 mutation (median, 177.9) compared with samples with the 11q deletion (median: 151.7; P = .15).
Baseline p53 and p21 induction in primary CLL cells shows distinct patterns. (A) The p53 response, defined as p21 induction versus baseline p53 levels (n = 59). Every dot represents 1 patient sample; the cutoffs are depicted by the dashed lines. A distinct pattern can be observed, with the normal response in the top left quadrant, the type-B defect in the bottom left quadrant, and the type-A defect in the bottom right quadrant. Whereas the type-A defect is strongly associated with 17p deletions and TP53 mutations, some samples with the p53 aberration can also be found among the cases with type-B defect and normal response. ○, 17p deletion; ■, sole TP53 mutation; •, no 17p deletion or TP53 mutation. (B) The clone size of the TP53 mutations (irrespective of 17p deletion) was negatively correlated with the level of p21 induction (r2 = 0.5359, P = .0044; cases with missense mutation only). (C) Top panel, p21 induction versus baseline p53 levels for patients with the 11q deletion (n = 12, right panel) and patients without the 17p deletion, the 11q deletion, or the TP53 mutation (n = 22, left panel). A minority of cases in both subgroups show impaired p21 induction with normal baseline p53 expression when based on the determined cutoff. Bottom panel, p21 induction versus p53 induction in patients with the 11q deletion (right) and in patients without the 11q deletion, 17p deletion, or TP53 mutation (left). p21 and p53 show a stronger induction after DNA damage in patients without the 11q deletion, the 17p deletion, or the TP53 mutation (median, 177.9) compared with samples with the 11q deletion (median: 151.7; P = .15).
To test for reproducibility, we analyzed patients repeatedly. Two patients were analyzed 7 times, 3 patients 3 times, and 3 patients 2 times. For example, the mean coefficient of variation among all patients for p21 induction was 3%. Most importantly, none of the samples changed classification when analyzed several times.
Type-A defect is correlated with p53 aberrations
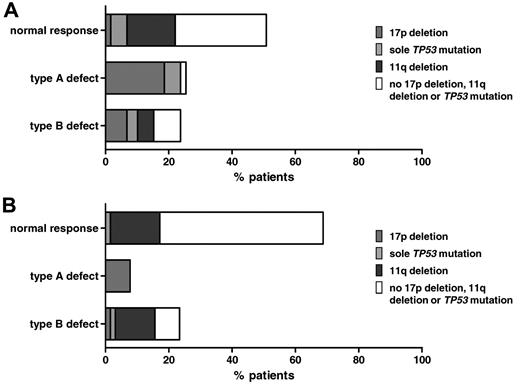
We analyzed a cohort of 59 patients and enriched the cohort for the poor prognostic subgroup's 17p deletion (n = 16), the sole TP53 mutation in the absence of 17p deletion (n = 8), and the 11q deletion (n = 12). Patient characteristics are summarized in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). We observed distinct patterns of p53/p21 response (Figure 3A). Thirty patients (51%) had a normal response (median p53 basal level, 62.52%; median p21 induction, 139.40%). The remaining patients showed a type-A defect (15 patients [25%]; median p53 basal level, 148.50%; median p21 induction, 109.90%) or a type-B defect (14 patients [24%]; median p53 basal level, 60.71%; median p21 induction, 109%; Table 1, Figure 4A).
Distribution of the genetic subgroups within the response groups in the original cohort
| . | Normal response . | Type-A defect . | Type-B defect . | Total . |
|---|---|---|---|---|
| Del 17p | 1 (3%) | 11 (73%) | 4 (29%) | 16 (27%) |
| Sole TP53 mutation | 3 (10%) | 3 (20%) | 2 (14%) | 8 (14%) |
| Del 11q | 9 (30%) | 0 (0%) | 3 (21%) | 12 (20%) |
| Absence of del 17p, del 11q, or TP53 mutation | 17 (57%) | 1 (7%) | 5 (36%) | 23 (39%) |
| Total | 30 (100%) | 15 (100%) | 14 (100%) | 59 (100%) |
| Median p53 basal level (95% CI) | 65.52 (63.78-75.35) | 148.50 (131.20-271.40) | 60.71 (53.43-70.08) | |
| Median p53 induction (95% CI) | 125.80 (112.00-135.30) | 84.26 (73.46-175.70) | 76.72 (52.75-82.70) | |
| Median p21 induction (95% CI) | 139.40 (136.80-159.50) | 109.90 (104.50-116.40) | 109.00 (104.90-110.60) |
| . | Normal response . | Type-A defect . | Type-B defect . | Total . |
|---|---|---|---|---|
| Del 17p | 1 (3%) | 11 (73%) | 4 (29%) | 16 (27%) |
| Sole TP53 mutation | 3 (10%) | 3 (20%) | 2 (14%) | 8 (14%) |
| Del 11q | 9 (30%) | 0 (0%) | 3 (21%) | 12 (20%) |
| Absence of del 17p, del 11q, or TP53 mutation | 17 (57%) | 1 (7%) | 5 (36%) | 23 (39%) |
| Total | 30 (100%) | 15 (100%) | 14 (100%) | 59 (100%) |
| Median p53 basal level (95% CI) | 65.52 (63.78-75.35) | 148.50 (131.20-271.40) | 60.71 (53.43-70.08) | |
| Median p53 induction (95% CI) | 125.80 (112.00-135.30) | 84.26 (73.46-175.70) | 76.72 (52.75-82.70) | |
| Median p21 induction (95% CI) | 139.40 (136.80-159.50) | 109.90 (104.50-116.40) | 109.00 (104.90-110.60) |
CI indicates confidence interval.
Functional classification of CLL samples in genetic subgroups. Percentages of patient samples with different p53/p21 FACS responses grouped by their genetic aberrations in (A) the original cohort and (B) the validation cohort. Dark gray, 17p deletion; light gray, sole TP53 mutation; black, 11q deletion; white, no 11q deletion, 17p deletion, or TP53 mutation.
Functional classification of CLL samples in genetic subgroups. Percentages of patient samples with different p53/p21 FACS responses grouped by their genetic aberrations in (A) the original cohort and (B) the validation cohort. Dark gray, 17p deletion; light gray, sole TP53 mutation; black, 11q deletion; white, no 11q deletion, 17p deletion, or TP53 mutation.
The type-A defect was strongly associated with 17p deletions and TP53 mutations. Eleven samples (73%) showed a deletion of 17p, whereas 3 cases (20%) exhibited sole TP53 mutations without a 17p deletion. In one case with a type-A defect, no deletion or mutation of TP53 was detectable (Figure 4A).
Three of the patients with a type-A defect showed some residual p21 induction (p21 induction median, 126.33; standard deviation, 8.126; patients 46, 54, and 55; supplemental Table 1).
Not all cases with TP53 mutation/17p deletion show distinct p53/p21 pattern
Most patients (14 of 24) with a 17p deletion or TP53 mutation were detected based on high basal p53 levels, in agreement with previous studies. As expected, the majority (n = 17) of patients with a normal response did not exhibit TP53 mutations or deletions of 17p or 11q (Figure 4A). These cases include samples with a 13q deletion, trisomy 12, and normal karyotype. However, 9 of the 12 samples with an 11q deletion were found in the group with a normal response when based purely on the determined cutoff. On the other hand, 5 patients with a normal karyotype exhibited a type-B defect.
Furthermore, 10 patients with a 17p deletion, a TP53 mutation, or both were identified in the groups with a normal response and a type-B defect. Three samples with the TP53 mutation showed a type-B defect. The TP53-mutated samples with a type-A defect mainly exhibited TP53 missense mutations, resulting in stabilized p53 protein. In contrast, cells with TP53 frameshift mutations showed the type-B defect (Figure 3A).
Three samples with the TP53 mutation had normal DNA-damage responses (patients 14, 16, and 28). This phenomenon is likely to be explained by the low clone size of the mutations that they exhibited (50%, 10%, and 20%, respectively) or the relatively high residual predicted activity toward transcriptional targets of the mutant p53 protein for patients 14 and 28 (41.7% and 15.7% of the wild-type p53 activity, respectively; Table 2).39 Comparison of the clone size of the TP53 mutation with the levels of p21 induction revealed a negative correlation (P = .0044), indicating a significant and relevant influence of the clone size on the degree of the damage response (Figure 3B). In contrast, the proportion of 17p deletions, which ranged from 44.5%-94.5% with a median of 87.75%, were not correlated with the p21 response or the p53 basal level (supplemental Figure 1A).
Overview of TP53 mutations in the first cohort
| UPN . | Del17p . | Mutation (aa) . | Type of mutation . | Clone size, % . | Predicted activity p21, %* . | Predicted activity Bax, %* . | P21 induction FACS, % . | P21 induction MLPA, -fold . | Bax induction MLPA, -fold . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | yes | splice site | splice site | 80 | N/A | N/A | 100.88 | 1.38 | 1.16 |
| 7 | no | p.W146_D148delinsC | frameshift | 5 | 0 | 0 | 100.96 | N/A | N/A |
| 11 | yes | p.F134C | missense | 75 | 15.6 | 10.2 | 114.52 | N/A | N/A |
| 12 | no | p.R209KfsX6 | frameshift | 75 | 0 | 0 | 109.47 | N/A | N/A |
| 14 | no | p.R283C | missense | 50 | 41.7 | 51.9 | 161.92 | 10.35 normal | 3.18 normal |
| 16 | no | p.R248Q | missense | 10 | 0 | 0 | 151.22 | N/A | N/A |
| 19 | yes | p.E180_R181delinsD | in frame deletion | 90 | N/A | N/A | 107.55 | 2.61 | 1.19 |
| 25 | yes | p.C238F | missense | 40 | 1.3 | 4.6 | 112.46 | N/A | N/A |
| 28 | no | p.P152L | missense | 20 | 15.7 | 16.4 | 170.04 | 5.09 normal | 2.17 normal |
| 31 | yes | p.Q167X | nonsense | 25 | 92.33 | 1.91 | 1.18 | ||
| p.C277F | missense | 60 | 0.5 | 0 | |||||
| 33 | yes | p.D281G | missense | 90 | 19.9 | 13.5 | 110.37 | N/A | N/A |
| 35 | no | p.D48VfsX1fs | frameshift | 20 | 110.03 | 1.80 | 1.41 | ||
| splice site | splice site | 20 | |||||||
| p.Y234H | missense | 10 | 0 | 2.3 | |||||
| p.I254N | missense | 5 | 10.2 | 16.8 | |||||
| p.E349X | nonsense | 10 | |||||||
| 40 | yes | p.P87fsX36 | frameshift | 70 | 0 | 0 | 105.55 | N/A | N/A |
| 41 | yes | p.G245D | missense | 90 | 3.2 | 11.9 | 109.91 | N/A | N/A |
| 43 | no | p.I162F | missense | 95 | 10.8 | 13.4 | 104.61 | 1.29 | 1.21 |
| 46 | yes | p.R110L | missense | 60 | 20.2 | 8.9 | 119.66 | N/A | N/A |
| 52 | yes | p.A138V | missense | 100 | 52.6 | 25.5 | 108.43 | N/A | N/A |
| 54 | yes | p.R248Q | missense | 60 | 0 | 0 | 126.33 | 1.84 | 1.56 |
| 55 | yes | p.R273H | missense | 85 | 1.7 | 2.4 | 135.83 | N/A | N/A |
| 58 | no | p.R209KfsX6 | frameshift | 80 | 0 | 0 | 100.07 | N/A | N/A |
| UPN . | Del17p . | Mutation (aa) . | Type of mutation . | Clone size, % . | Predicted activity p21, %* . | Predicted activity Bax, %* . | P21 induction FACS, % . | P21 induction MLPA, -fold . | Bax induction MLPA, -fold . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | yes | splice site | splice site | 80 | N/A | N/A | 100.88 | 1.38 | 1.16 |
| 7 | no | p.W146_D148delinsC | frameshift | 5 | 0 | 0 | 100.96 | N/A | N/A |
| 11 | yes | p.F134C | missense | 75 | 15.6 | 10.2 | 114.52 | N/A | N/A |
| 12 | no | p.R209KfsX6 | frameshift | 75 | 0 | 0 | 109.47 | N/A | N/A |
| 14 | no | p.R283C | missense | 50 | 41.7 | 51.9 | 161.92 | 10.35 normal | 3.18 normal |
| 16 | no | p.R248Q | missense | 10 | 0 | 0 | 151.22 | N/A | N/A |
| 19 | yes | p.E180_R181delinsD | in frame deletion | 90 | N/A | N/A | 107.55 | 2.61 | 1.19 |
| 25 | yes | p.C238F | missense | 40 | 1.3 | 4.6 | 112.46 | N/A | N/A |
| 28 | no | p.P152L | missense | 20 | 15.7 | 16.4 | 170.04 | 5.09 normal | 2.17 normal |
| 31 | yes | p.Q167X | nonsense | 25 | 92.33 | 1.91 | 1.18 | ||
| p.C277F | missense | 60 | 0.5 | 0 | |||||
| 33 | yes | p.D281G | missense | 90 | 19.9 | 13.5 | 110.37 | N/A | N/A |
| 35 | no | p.D48VfsX1fs | frameshift | 20 | 110.03 | 1.80 | 1.41 | ||
| splice site | splice site | 20 | |||||||
| p.Y234H | missense | 10 | 0 | 2.3 | |||||
| p.I254N | missense | 5 | 10.2 | 16.8 | |||||
| p.E349X | nonsense | 10 | |||||||
| 40 | yes | p.P87fsX36 | frameshift | 70 | 0 | 0 | 105.55 | N/A | N/A |
| 41 | yes | p.G245D | missense | 90 | 3.2 | 11.9 | 109.91 | N/A | N/A |
| 43 | no | p.I162F | missense | 95 | 10.8 | 13.4 | 104.61 | 1.29 | 1.21 |
| 46 | yes | p.R110L | missense | 60 | 20.2 | 8.9 | 119.66 | N/A | N/A |
| 52 | yes | p.A138V | missense | 100 | 52.6 | 25.5 | 108.43 | N/A | N/A |
| 54 | yes | p.R248Q | missense | 60 | 0 | 0 | 126.33 | 1.84 | 1.56 |
| 55 | yes | p.R273H | missense | 85 | 1.7 | 2.4 | 135.83 | N/A | N/A |
| 58 | no | p.R209KfsX6 | frameshift | 80 | 0 | 0 | 100.07 | N/A | N/A |
N/A indicates not applicable.
From IARC database.
In 4 samples with the 17p deletion, no TP53 mutation on the other allele could be identified. Of these samples, only one showed a normal response; the other 3 cases were classified as having a type-B defect.
Impaired p53/p21 response in cases with an 11q deletion
As indicated, the samples with an 11q deletion (n = 12) and the samples without aberration of 17p or 11q (n = 22) could not be clearly separated based on a cutoff, although the p21 levels differed (median of samples with the 11q deletion: 127.0; median of samples without the 17p deletion, 11q deletion, or TP53 mutation: 137.6; P = .054; Figure 3C). To further distinguish the samples, we also included the induction of p53 in the analysis (Figure 3C bottom panel). We observed a linear correlation between the induction of p53 and p21 in samples without the 17p deletion, the 11q deletion, or the TP53 mutation (r2 = 0.45, P = .0002).
The group with the 11q deletion lacked the marked dynamic shift of the whole sample population to the top right quadrant of the graph that is seen in the samples without the 17p deletion, the 11q deletion, or the TP53 mutation (Figure 3C). The induction dynamics were quantified using the length of the vectors of the data points (median of samples without the 17p deletion, 11q deletion, or TP53 mutation: 177.9; median of samples with the 11q deletion: 151.7; P = .15). These data suggest that a minority of cases with the 11q deletion will be distinguished based on the defined cutoff. At the same time, the overall dynamic p53/p21 induction appears to be impaired, which may be explained by a gene-dosage effect for ATM in the 11q-deleted samples. To further explore this phenomenon, we also performed quantitative real-time PCR for p21 in treated and untreated samples. Comparison of the levels of p21 induction in samples with the 11q deletion and samples without the 17p deletion, the 11q deletion, or the TP53 mutation again showed a reduced overall induction in cases with the 11q deletion (median p21 induction of samples with the 11q deletion: 6.825; median p21 induction of samples without the 17p deletion, 11q deletion, or TP53 mutation: 12.38; P = .06; data not shown).
We also compared data obtained by RT-MLPA for radiation-induced induction of p21 (median of samples with the 11q deletion: 4.867; median of samples without the 17p deletion, 11q deletion, or TP53 mutation: 11.42; P = .06), BAX (B-cell lymphoma 2–associated X protein; median of samples with the 11q deletion: 2.024; median of samples without the 17p deletion, 11q deletion, or TP53 mutation: 2.378; P = .19) and PUMA (p53-up–regulated modulator of apoptosis; median of sample with the 11q deletion: 2.069; median of samples without the 17p deletion, 11q deletion, or TP53 mutation: 4.096; P = .06; data not shown). This confirmed the trend seen by FACS and quantitative RT-PCR.
The clone size of the 11q deletion (by FISH) in the samples ranged from 30.5% to 91% (median, 76.25%). Correlation of the percentages of the 11q deletion with p21 induction levels measured by FACS showed borderline significance (P = .09), but no correlation with p53 induction was observed (supplemental Figure 1B).
Validation in an independent cohort
To verify our results in an independent dataset, we analyzed a cohort of patients prior to first-line treatment (n = 64). This cohort included samples with the 17p deletion (n = 6), sole TP53 mutation (n = 2), or 11q deletion (n = 18) and samples without the 17p deletion, 11q deletion, or TP53 mutation (n = 38). The distribution of the genetic subgroups more closely resembled the distribution that would be expected from a cohort with CLL and treatment indication.
The results of the FACS analysis are shown in Figure 5A and Table 3. Forty-four samples (69%) had a normal response (median p53 basal level: 60.71%; median p21 induction: 128.80%); 5 samples (8%) had a type-A defect (median p53 basal level: 162.60%; median p21 induction: 108.30%); and 15 (23%) samples had a type-B defect (median p53 basal level: 69.09%; median p21 induction: 106.60%; Figure 4B). Of the cases with the 17p deletion and the sole TP53 mutation (Figure 5A), 5 showed a type-A defect. As expected, they all exhibited stabilizing missense mutations. Two cases with a type-B defect had missense (Y126D) and splice site (673-2A > T) mutations, respectively. One case with a missense mutation (M246I) had a normal response and therefore would remain undetected based on the FACS assay.
p53/p21 induction in an independent cohort. (A) The DNA-damage response (p53 basal levels, p21 induction) in all patients of the validation cohort. ○, 17p deletion; ■, sole TP53 mutation; •, no 17p deletion or TP53 mutation. (B) Top panel, Patients without the 11q deletion, the 17p deletion, or the TP53 mutation (n = 38) compared with patients with the 11q deletion (n = 18). Similar to the observation in the initial cohort, based on the cutoff, 8 samples (44%) with the 11q deletion showed a type-B defect. In the cohort without the 11q deletion, 17p deletion, or TP53 mutation, 5 cases (13%) showed a type-B defect. Bottom panel, Impaired p53 and p21 induction in patients with the 11q deletion was confirmed in the validation cohort (median induction of samples without the 17p deletion, 11q deletion, or TP53 mutation: 196.9; median of samples with the 11q deletion: 170.5; P = .0505).
p53/p21 induction in an independent cohort. (A) The DNA-damage response (p53 basal levels, p21 induction) in all patients of the validation cohort. ○, 17p deletion; ■, sole TP53 mutation; •, no 17p deletion or TP53 mutation. (B) Top panel, Patients without the 11q deletion, the 17p deletion, or the TP53 mutation (n = 38) compared with patients with the 11q deletion (n = 18). Similar to the observation in the initial cohort, based on the cutoff, 8 samples (44%) with the 11q deletion showed a type-B defect. In the cohort without the 11q deletion, 17p deletion, or TP53 mutation, 5 cases (13%) showed a type-B defect. Bottom panel, Impaired p53 and p21 induction in patients with the 11q deletion was confirmed in the validation cohort (median induction of samples without the 17p deletion, 11q deletion, or TP53 mutation: 196.9; median of samples with the 11q deletion: 170.5; P = .0505).
Distribution of the genetic subgroups within the response groups in the independent cohort
| . | Normal response . | Type-A defect . | Type-B defect . | Total . |
|---|---|---|---|---|
| Del 17p | 0 (0%) | 5 (100%) | 1 (7%) | 6 (9%) |
| Sole TP53 mutation | 1 (2%) | 0 (0%) | 1 (7%) | 2 (3%) |
| Del 11q | 10 (23%) | 0 (0%) | 8 (53%) | 18 (28%) |
| Absence of del 17p, del 11q, or TP53 mutation | 33 (75%) | 0 (0%) | 5 (33%) | 38 (59%) |
| Total | 44 (100%) | 5 (100%) | 15 (100%) | 64 (100%) |
| Median p53 basal level (95% CI) | 60.71 (55.69-67.83) | 162.60 (115.90-204.70) | 69.09 (61.45-77.65) | |
| Median p53 induction (95% CI) | 149.90 (147.80-183.90) | 98.23 (64.63-147.60) | 91.86 (79.38-129.90) | |
| Median p21 induction (95% CI) | 128.8 (128.50-138.40) | 108.30 (95.87-117.80) | 106.60 (102.40-109.50) |
| . | Normal response . | Type-A defect . | Type-B defect . | Total . |
|---|---|---|---|---|
| Del 17p | 0 (0%) | 5 (100%) | 1 (7%) | 6 (9%) |
| Sole TP53 mutation | 1 (2%) | 0 (0%) | 1 (7%) | 2 (3%) |
| Del 11q | 10 (23%) | 0 (0%) | 8 (53%) | 18 (28%) |
| Absence of del 17p, del 11q, or TP53 mutation | 33 (75%) | 0 (0%) | 5 (33%) | 38 (59%) |
| Total | 44 (100%) | 5 (100%) | 15 (100%) | 64 (100%) |
| Median p53 basal level (95% CI) | 60.71 (55.69-67.83) | 162.60 (115.90-204.70) | 69.09 (61.45-77.65) | |
| Median p53 induction (95% CI) | 149.90 (147.80-183.90) | 98.23 (64.63-147.60) | 91.86 (79.38-129.90) | |
| Median p21 induction (95% CI) | 128.8 (128.50-138.40) | 108.30 (95.87-117.80) | 106.60 (102.40-109.50) |
CI indicates confidence interval.
As in the original cohort, subdivision of the remaining cases by their 11q deletion status revealed similar patterns of p21 induction versus p53 baseline (Figure 5B) and p53 induction (Figure 5B bottom panel). Ten of 18 cases with the 11q deletion had a normal p21/p53 response, whereas 8 showed a type-B defect. Again, the samples without the 17p deletion, 11q deletion, or TP53 mutation exhibited a higher induction of p53 and p21 than 11q-deleted samples (median of samples without 17p or 11q deletion: 196.9; median of samples with the 11q deletion: 170.5; P = .05).
When we combined both cohorts, the comparison of p53 and p21 induction between the 2 genetic subgroups was highly significant (median of samples with the 11q deletion: 168.2; median of samples without 17p or 11q deletion: 189.6; P = .006), suggesting that cases with the 11q deletion do show an impaired p53 response.
Comparison of FACS assay with other functional tests
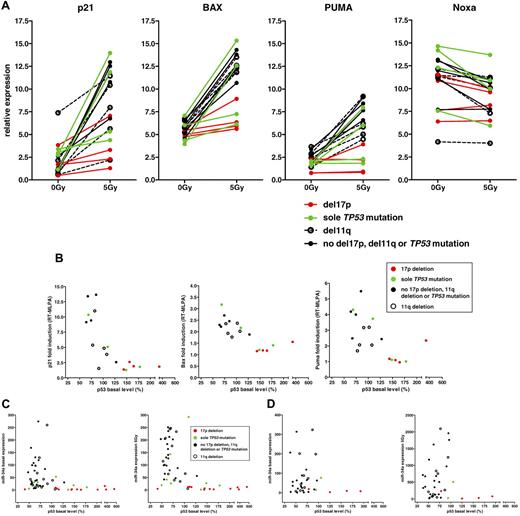
There is considerable interest in the development of functional p53 assays in CLL.40 To put the FACS assay into perspective, we compared the FACS data with other techniques. Figure 6A shows the levels of the p53 targets p21, BAX, PUMA, and NOXA in 18 patient samples. Whereas levels of NOXA did not change after irradiation, levels of p21, BAX, and PUMA were induced after DNA damage, with the highest induction level for p21. The samples with the 17p deletion and 2 samples with the TP53 mutation had impaired induction of all 3 genes (p21 median: 1.819 vs 7.269, P = .0043; BAX median: 1.198 vs 2.263, P = .0009; PUMA median: 1.109 vs 3.178, P = .0023). Two TP53-mutated cases (not detected by FACS) had a strong induction of p21, BAX, and PUMA, suggesting that these cases escaped detection irrespective of assay.
Comparison of the FACS assay with RT-PCR/MLPA. (A) RT-MLPA analysis of p53 target genes 16 hours after irradiation. P21, BAX, and PUMA were induced in samples with wild-type p53. Samples with the 17p deletion showed a reduced induction. NOXA, on the contrary, was not induced. (B) P21, BAX, and PUMA measured by RT-MLPA versus p53 basal levels determined by FACS. All genes identified the 17p-deleted cases and 2 of the samples with the TP53 mutation. The use of p21 induction measured by RT-MLPA seemed to provide the best separation among the 3 genes. (C) Analysis of miR-34a in 55 samples of the original cohort in untreated cells and 16 hours after 5-Gy irradiation. 17p-deleted samples could be detected by miR-34a expression at baseline and after 5-Gy irradiation and its absolute induction. (D) miR-34a analysis in 36 samples of the validation cohort in untreated cells and 24 hours after irradiation (5 Gy).
Comparison of the FACS assay with RT-PCR/MLPA. (A) RT-MLPA analysis of p53 target genes 16 hours after irradiation. P21, BAX, and PUMA were induced in samples with wild-type p53. Samples with the 17p deletion showed a reduced induction. NOXA, on the contrary, was not induced. (B) P21, BAX, and PUMA measured by RT-MLPA versus p53 basal levels determined by FACS. All genes identified the 17p-deleted cases and 2 of the samples with the TP53 mutation. The use of p21 induction measured by RT-MLPA seemed to provide the best separation among the 3 genes. (C) Analysis of miR-34a in 55 samples of the original cohort in untreated cells and 16 hours after 5-Gy irradiation. 17p-deleted samples could be detected by miR-34a expression at baseline and after 5-Gy irradiation and its absolute induction. (D) miR-34a analysis in 36 samples of the validation cohort in untreated cells and 24 hours after irradiation (5 Gy).
Because the range of the p21 induction in the FACS assay was not very pronounced and separation of the samples based on p21 induction was not distinct, we wanted to determine whether we could achieve a better separation using the RT-MLPA data for p21, BAX, and PUMA (Figure 6B) or the expression and induction of miR-34a in the cohorts (Figure 6C-D). All parameters were able to detect the samples with the 17p deletion. In contrast, the TP53-mutated cases were scattered, with 2 cases showing high residual induction of p21, BAX, and PUMA and high miR-34a expression. These cases corresponded to the samples with a low clone size. None of the assays showed a discernable cutoff between the 11q-deleted samples and samples without the 17p deletion, 11q deletion, or TP53 mutation, even if the overall level of induction was different.
The degree of up-regulation was most pronounced for p21 mRNA and miR-34a, suggesting that these p53 targets may deliver the greatest degree of distinction (Figure 6B-D).
The cases with a 17p deletion but not cases with 11q deletion could also be detected using baseline miR-34a expression or its induction irradiation.
In addition to p53 targets, the mRNA expression of genes involved in apoptosis were analyzed in the same experiment using RT-MLPA. Whereas p21, BAX, and PUMA showed a positive induction after treatment, various other genes were down-regulated after DNA damage to different extents. When we compared the samples with a type-A or type-B defect (n = 9) with the samples with a normal response (n = 9), we found a stronger relative down-regulation of the apoptosis pathway members in samples with a normal response (supplemental Figure 2). The differences in the apoptosis pathway between the response groups were correlated with the in vitro apoptosis of the CLL cells determined by forward scatter-side scatter in FACS. Samples with a normal response in the FACS assay showed the highest rate of apoptosis. The apoptosis in samples with a type-A or type-B defect was significantly reduced, with type-A defect samples having the lowest apoptosis rate (data not shown). The radiation-induced down-regulation of apoptosis-regulating genes in cases with functionally intact p53 could therefore be a consequence of preferential apoptosis in this group.
Discussion
The importance of the DNA-damage–response pathway in CLL is highlighted by the prognostic impact of mutations and deletions of the TP53 and ATM genes.6,8 While the prognostic impact of 17p deletions and TP53 mutations appears to be similar,10,11 the differential impact of ATM deletions and mutations is less well studied. In addition to the biological basis of drug resistance in CLL, practical issues concerning diagnostic workup and method of detection of p53/ATM defects remain unresolved.
In the current study, we sought to assess potential diagnostic approaches to CLL with p53 defects and to gain insight into the mechanisms underlying poor-risk CLL by a detailed analysis of the response of primary CLL cells to in vitro–induced DNA damage.
Encouraged by the pivotal studies of Pettitt et al,27,28,30 we used a FACS assay to quantitatively determine the levels of p53 and p21 after introducing DNA double-strand breaks by ionizing radiation. We increased the reproducibility by the use of internal CD19-negative control cells. Our aims were (1) to determine to what extent we could identify dysfunctional cases that could not be identified by other routinely used methods (FISH, TP53 mutation) and (2) to ascertain the fidelity for the detection of cases with a TP53 mutation or 17p deletion.
Of 123 samples tested, we observed only one patient without any abnormalities in TP53 FISH and TP53 mutation analysis with a type-A defect. This suggests that “functional assays” will only rarely detect additional type-A cases to mutational analysis of FISH (< 1% in our series). The clinical consequences of these abnormalities are unclear and are unlikely to be easily answered based on the rarity of the aberration.
In contrast, additional type-B cases were identified in samples with no 17p deletion, 11q deletion, or TP53 mutation (5 of 23), and are likely to represent cases with an ATM mutation. Analysis of phospho(Ser1981)–ATM in cases with a type-B defect (n = 7) revealed an impaired induction compared with samples with a normal response (n = 9; supplemental Figure 3 and Winkler et al41 ), supporting the assumption of a different ATM mutational status. The clinical consequences of type-B defects remain to be determined, but improved outcome for patients with the 11q deletion and FCR (fludarabine, cyclophosphamide, rituximab) treatment suggests that defects that phenocopy 17p deletion and TP53 mutation could be clinically more relevant.
Furthermore, the assay failed to identify 4 cases (17%) of the patients with a 17p deletion or TP53 mutation that showed a normal response. There was a tight correlation between the size of the TP53 mutant clone and the impaired p21/p53 response. Furthermore, 6 cases with a 17p deletion or TP53 mutation (25%) exhibited a type-B defect that was thought to be typical for 11q-deleted cases. This combination was also reported in previous studies.30 We found an association of the type-B defect with frameshift mutations, not giving rise to an accumulation of the mutant p53. These samples would have been misclassified if based only on the FACS assay. This is particularly relevant because TP53 frameshift mutations have been shown to account for about 20% of all TP53 mutations in CLL.42 This frequency is higher than in many other cancer types.43 A potential way to distinguish these cases from 11q-deleted samples would be the use of the MDM2 inhibitor Nutlin-3a, which restores the induction of p53 and p21 in 11q-deleted but not in TP53-mutated samples.29
We acknowledge that the case mix of the second cohort was different from that of the initial cohort, and the selection of cohort influences the performance of diagnostic tests. The second independent dataset is more representative of a cohort with first-line treatment indication.
One important observation from the comparison of different methods used to monitor the p53 response (Western blot, FACS, quantitative RT-PCR, RT-MLPA) was that the samples “misclassified” based on the FISH and TP53 mutation analysis showed similar results across techniques and targets, suggesting that the assays may have comparable potential. However, it was informative that the miR-34a and p21 mRNA levels showed the greatest amplitude of induction, suggesting that these 2 p53 targets may be most easily used. In addition, miR-34a (as previously reported by a number of groups) had suppressed baseline levels in the presence of the 17p deletion or TP53 mutation. This differentiated miR-34a from other p53 targets with baseline levels independent of 17p deletion/TP53 mutation, and suggests that miR-34a may be the most promising marker when unstressed cells are investigated.
Previous studies have achieved findings similar to those of our study, but have rarely compared different techniques or applied a validation cohort.25-30 Based on the present study, clonal heterogeneity is likely to be a limiting factor to the wider distribution of functional p53 assays, which may not be solved by the use of FACS. This is particularly important because there is convincing evidence that the TP53 mutation—even in subclones—is clinically relevant.37 While the current study was not designed as a technical comparison of all potential methods to assess “p53 function,” it does point to some potential pitfalls.
Some of the most interesting biological implications of our study are the demonstration of an impaired p53 response in all cases with an 11q deletion. Several CLL samples with the 11q deletion exhibited a normal response when based on predefined cutoffs (Figure 4). This suggests that the assays described in this study will not be well suited to identifying most cases with the 11q deletion (Figure 4). While there is some indication that the mutational status of ATM in the remaining allele is of prognostic significance, the clinical consequence of type-B defects remains to be determined. Our study suggests that there is impaired p53 pathway activity, potentially even in cases without ATM mutations. One potential explanation is a gene-dosage effect for ATM. This is in contrast to previous reports showing that mono-allelic deletions of 11q without mutation of the second ATM allele have a normal response to DNA-damaging agents by inducing p53 phosphorylation and p21 expression.9
We used a “functional” p53 assay (FACS) to determine its potential suitability for clinical applications. Even though the assay could identify most cases with defects, there was still a substantial heterogeneity. Therefore, 17p deletions and TP53 mutations with a low clone size are likely to be missed and frameshift mutants of p53 are likely to be misclassified.
Our findings demonstrate the current indispensability of TP53 mutational analysis and FISH in diagnostics.44,45 Further knowledge and understanding about the influence of different mutations of TP53 and ATM on the DNA-damage response and prognosis are needed to allow an application of functional assays in the future, which will then need to be tested in larger cohorts and ideally within clinical trials. One particularly interesting question in this regard is if we can relate the intensity of the p53 response to outcome variables even in cases with nondysfunctional p53.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by the Else-Kröner-Fresenius-Stifung (P20/07//A11/07), by the Deutsche Jose Carreras Leukämie-Stiftung (R06/28v, R08/26f), and the by the CLL Global Research Foundation.
Authorship
Contribution: J.M. designed research, performed experiments, and wrote the paper; H.H., M.F., A.B., D.W., M.V., and D.T. performed experiments and provided study reagents; E.E., A.P.K., and D.M. designed research and collected, analyzed, and interpreted the data; H.D. and S.S. collected, analyzed, and interpreted data; and T.Z. designed research, collected, analyzed, and interpreted data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thorsten Zenz, MD, Department of Internal Medicine III, University of Ulm, Albert Einstein Allee 23, 89081 Ulm, Germany; e-mail: thorsten.zenz@uniklinik-ulm.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal