Abstract
A role for specific human leukocyte antigen (HLA) variants in the etiology of childhood acute lymphoblastic leukemia (ALL) has been extensively studied over the last 30 years, but no unambiguous association has been identified. To comprehensively study the relationship between genetic variation within the 4.5 Mb major histocompatibility complex genomic region and precursor B-cell (BCP) ALL risk, we analyzed 1075 observed and 8176 imputed single nucleotide polymorphisms and their related haplotypes in 824 BCP-ALL cases and 4737 controls. Using these genotypes we also imputed both common and rare alleles at class I (HLA-A, HLA-B, and HLA-C) and class II (HLA-DRB1, HLA-DQA1, and HLA-DQB1) HLA loci. Overall, we found no statistically significant association between variants and BCP-ALL risk. We conclude that major histocompatibility complex-defined variation in immune-mediated response is unlikely to be a major risk factor for BCP-ALL.
Introduction
Acute lymphoblastic leukemia (ALL) is the major pediatric cancer in developed countries.1 B-cell precursor (BCP) ALL accounts for approximately 70% of childhood ALL and characteristically affects children between 3 and 5 years of age.
Although evidence linking most environmental exposures to risk of childhood ALL has largely been inconsistent, epidemiologic data for an infectious etiology are persuasive, albeit indirect.1 Implicit in a model of ALL development having an infectious etiologic basis is that ALL represents a rare sequela of infection. Because the major histocompatibility complex (MHC) plays a major role in regulating host response to infection, genetic variation in the MHC is an attractive basis for inherited genetic susceptibility to ALL.
The possible role of the human leukocyte antigen (HLA) system in the development of childhood ALL follows the demonstration of an MHC influence on the development of mouse leukemia. In 1964, Lilly et al showed that susceptibility to gross retroviral leukemia in the mouse was linked to the H-2k haplotype.2 Over the last 30 years, a large number of studies have sought to establish a relationship between MHC variation and risk of human ALL. Although numerous studies have been conducted suggesting associations with various class I (HLA-A2, A9, A11, B12, Cw4)3-5 and class II alleles (HLA-DR53, -DPB1*0201, -DQB1*0201/0501, -DQ, -DPw2/w5, -DPB1*0601),6-13 unambiguous statistical evidence to support a relationship is currently lacking.
Possible reasons for the inconsistencies in the associations include the existence of complex linkage disequilibrium patterns between the multiple risk loci mapping to the MHC region, supertypic associations, or failure to control for population stratification or disease heterogeneity. The studies of HLA and risk of ALL conducted so far have generally been based on small sample series (many involving < 100 cases), and there has been a failure to robustly address the issue of multiple testing in studies. In view of the inconsistencies and limitations of previously published studies on the relationship between genetic variation in HLA and ALL risk, we have been motivated to conduct a more rigorous analysis.
It has recently been demonstrated that it is possible to use single nucleotide polymorphism (SNP) variation within the 6p21 region to accurately predict alleles at key class I (HLA-A, HLA-B, and HLA-C) and class II (HLA-DRB1, HLA-DQA1, and HLA-DQB1) loci with better than 90% accuracy.14 Furthermore, the HLA alleles can be successfully predicted using SNPs included on commercial arrays used to conduct genome-wide association (GWA) studies. We have recently conducted a GWA study of childhood ALL using Illumina Infinium HD Human370 Duo BeadChips.15 Data from this study have thus allowed the interrogation of the role of genetic variation in the MHC region in the etiology of BCP-ALL.
Methods
Patients and DNA samples
Cases analyzed had been diagnosed with BCP-ALL and have been the subject of a GWA study of childhood ALL we have recently reported.15 Briefly, we analyzed the constitutional DNA of 824 pediatric ALL patients ascertained from the United Kingdom (464 male, 360 female; mean age at diagnosis, 5.4 years; SD, 3.6 years). Cases were derived from the United Kingdom Childhood Cancer study (UKCCS),16 the United Kingdom Medical Research Council ALL 97 (99) trial, and from the Northern Institute of Cancer Research. Diagnostic immunophenotyping and genotyping of patient samples were undertaken using standard diagnostic methodologies. To minimize population stratification, cases with documented non-Western European ancestry were excluded. Cytogenetic data were available on 632 persons with BCP-ALL with 289 persons having hyperdiploid ALL (≥ 50 chromosomes) and 126 with TEL/AML1 (alias ETV6/RUNX1) fusion.
Control series
We used publicly accessible data from the Wellcome Trust Case-Control Consortium 2 (WTCCC2) study for population SNP genotype frequencies. For the single SNP and haplotype analysis, data from 2420 persons from the 1958 Birth Cohort (58C, also known as the National Child Development Study)17 were used. For HLA imputation, we additionally made use of data on 2737 persons from the United Kingdom Blood Service Collection and also from the WTCCC2. Because the prevalence of childhood ALL survivors in adults is less than 1 in 10 000 in the United Kingdom, both control series can be considered representative of the non-ALL United Kingdom population.
Ethics
Collection of blood samples and clinicopathologic information from subjects was undertaken with informed consent and relevant ethical review board approval in accordance with the tenets of the Declaration of Helsinki.
Genotyping
As previously described,15 DNA was extracted and quantified from ethylenediaminetetraacetic acid-venous remission blood samples using conventional methodologies and a genome-wide scan of tagging SNPs conducted using Illumina Infinium HD Human370 Duo BeadChips according to the manufacturer's protocols (Illumina). We considered that a DNA sample had failed if it did not generate a genotype for more than 95% of loci. Similarly, an SNP was considered a failure if less than 95% of DNA samples generated a genotype at the locus. The genotyping call rate was 99.84%. To ensure quality of genotyping, a series of duplicate samples were genotyped on the same arrays.
Quality control
To identify samples showing relatedness, identity-by-state values were calculated for pairs of persons; and for any pair with more than 80% identical SNP genotypes, we removed the sample with the lower call rate from the analysis. We excluded SNPs on the basis of deviation from Hardy-Weinberg Equilibrium using a threshold of P less than 1.0 × 10−5 in controls. We also removed SNPs with minor allele frequency less than 0.05. To identify and exclude persons with non–Western European ancestry, case and control data were merged with persons of different ethnicities from the International HapMap Project, and dissimilarity measures were used to perform principal component analysis. For the WTCCC2 controls, we also followed the guidelines from the WTCCC and removed some additional controls. After imposing these stringent quality control measures, SNP genotypes were available on 824 cases and 4737 controls (2236 58C and 2501 United Kingdom Blood Service Collection).
Statistical and bioinformatics analysis
For single SNP and haplotype analysis, we considered the MHC to be defined by a 4.5-Mb region bordered by the RFP and MLN genes (rs209130 at 28 975 779 bps and rs1547668 at 33 883 424 bps, respectively) at the telomeric and centromeric ends of 6p21, respectively. For the HLA imputation, we made use of GWA study SNP data for an extended 6p21 region defined by rs1165196 and rs2772372 (25 921 129 bps and 33 535 328 bps, respectively).
Initially, we analyzed the association between each SNP and risk of ALL using the Cochran-Armitage trend test. Odds ratios and associated 95% confidence intervals were calculated by unconditional logistic regression.
In addition to single-SNP analysis, we performed haplotype analysis using HelixTree software, Version 6.4.2 (Golden Helix) through the use of a haplotype trend regression fitted to a model of additive effects of haplotypes. Analyses were performed with the use of sliding window sizes of 5 and 12 contiguous markers. Haplotype frequencies for each person were estimated with the use of an expectation-maximization algorithm. The minimum haplotype frequency was set at 0.01, and haplotypes with frequencies below this threshold were combined into a single group.
Prediction of the untyped SNPs within the 4.5-Mb region was carried out using IMPUTE, Version 0.3.2, based on HapMap Phase II haplotypes (HapMap Data Rel 21a/phase II Jan07 on NCBI B35 assembly, dbSNPb125). Imputed data were analyzed using SNPTEST, Version 2 to account for uncertainties in SNP prediction. Association between genotypes and BCP-ALL risk was performed using the Cochran-Armitage trend test.
From a reference database of SNP haplotypes carrying known HLA alleles, we imputed HLA genotypes for the 824 BCP-ALL cases as well as for the 1958 Birth Cohort for which classic HLA data are not available and United Kingdom Blood Service Collection controls as previously described.14 The reference database combines classic HLA data from the HapMap Project and the 1958 Birth Cohort. Imputation was performed for 3 class I and 3 class II loci: HLA-A (2474 reference samples), HLA-B (n = 3090), HLA-C (n = 2022), HLA-DQA1 (n = 175), HLA-DQB1 (n = 2629), and HLA-DRB1 (n = 2665). Imputation accuracy was assessed through a cross-validation analysis of the training data. Training was performed on two-thirds of the reference panel and tested on the remaining third at the SNPs chosen for imputation in the current study. Thresholding calls at a posterior probability of 0.74 provided call rates of between 0.90 (HLA-DRB1) and 0.99 (HLA-C) and accuracy of more than 0.95 for all loci at the 2-digit level and between 0.91 (HLA-DQA1) and 0.98 (HLA-DQB1) at the 4-digit level.
In all analyses, we considered an adjusted P value of .05 to be statistically significant. To address the issue of multiple testing, 100 000 permutations of case-control status were used to calculate adjusted P values. These analyses were undertaken using R, Version 2 software.
Results
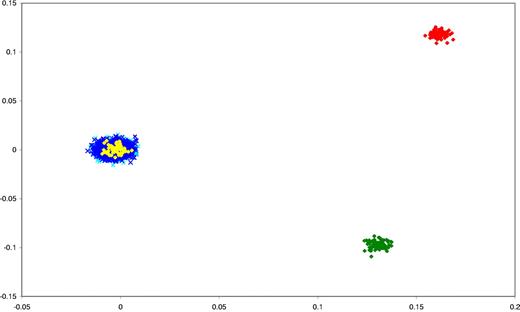
We have previously subjected cases and controls to rigorous quality control in terms of excluding samples and SNPs with poor call rates.15 Furthermore, we excluded SNPs showing significant departure from Hardy-Weinberg Equilibrium. Before pooling data from the 2 GWA case series and the WTCCC2 control series, we evaluated all of the datasets for ancestral differences by principal component analysis. Figure 1 shows that all sample series were ancestrally comparable and hence could be combined without introducing systematic bias.
Comparison of ethnicity in all of the sample series. The first 2 principal components of the analysis were plotted. Yellow plots represent HapMap CEU persons; green, CHB + JPT; red, YRI persons; light blue, BCP-ALL cases; and dark blue, WTCCC2 controls (58C and NBS).
Comparison of ethnicity in all of the sample series. The first 2 principal components of the analysis were plotted. Yellow plots represent HapMap CEU persons; green, CHB + JPT; red, YRI persons; light blue, BCP-ALL cases; and dark blue, WTCCC2 controls (58C and NBS).
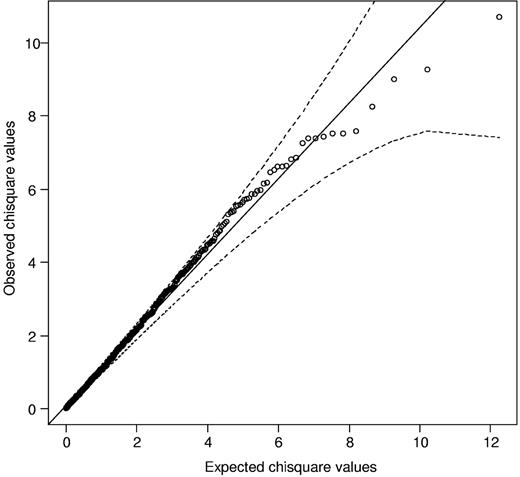
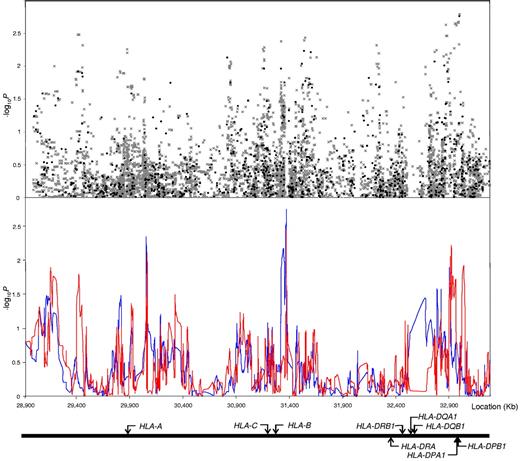
We initially considered 1075 SNPs mapping to the 4.5-Mb region encompassing the classic MHC region. Comparison of the observed and expected distributions from single SNP tests for complete GWA data showed little evidence for an inflation of the test statistics in the datasets,15 thereby excluding the possibility of significant hidden population substructure or differential genotype calling between cases and controls. Considering only those SNPs that mapped to the MHC region at the extreme end of the test statistic distribution, deviation from the null was not observed, thereby providing no evidence for a strong single SNP-based association (Figure 2). Figure 3 shows the strength of the associations across the 4.5-Mb region. The strongest singleSNP-based association was attained for rs3135034 mapping at 33 059 640 bps (P = .0017), not significant after adjustment for multiple testing.
Quantile-quantile (Q-Q) plot of Cochran-Armitage test for trend for SNPs mapping to the MHC region. The black line represents the null hypothesis of no true association; and dashed lines, 95% confidence intervals.
Quantile-quantile (Q-Q) plot of Cochran-Armitage test for trend for SNPs mapping to the MHC region. The black line represents the null hypothesis of no true association; and dashed lines, 95% confidence intervals.
Association between SNPs and haplotypes mapping to 6p21 and BCP-ALL risk. The x-axis represents the position of each SNP; and the y-axis, P values on a minus logarithmic scale. Cochran-Armitage trend test statistics are shown in black for directly genotyped SNPs and grey for imputed SNPs in the top panel. Lines in the bottom panel correspond to haplotype test statistics: blue defined by 5 SNPs and red by 12 SNPs. Relative positions of the major HLA genes are also shown. Chromosomal coordinates derived from the National Center for Biotechnology Information, build 36.
Association between SNPs and haplotypes mapping to 6p21 and BCP-ALL risk. The x-axis represents the position of each SNP; and the y-axis, P values on a minus logarithmic scale. Cochran-Armitage trend test statistics are shown in black for directly genotyped SNPs and grey for imputed SNPs in the top panel. Lines in the bottom panel correspond to haplotype test statistics: blue defined by 5 SNPs and red by 12 SNPs. Relative positions of the major HLA genes are also shown. Chromosomal coordinates derived from the National Center for Biotechnology Information, build 36.
To interrogate the MHC region more fully, SNPs were imputed using HapMap data. In total, 8176 SNPs were successfully imputed in the interval between 28 975 779 bps and 33 535 328 bps using data from the 1075 directly typed SNPs. This analysis provided no evidence for a relationship between genetic variation within the MHC region and BCP-ALL risk (Figure 3). Further evaluation of the data included conducting haplotype analysis. This analysis also provided little evidence for enigmatic disease alleles present on rare haplotypes that may have been missed by single SNP analyses (Figure 3).
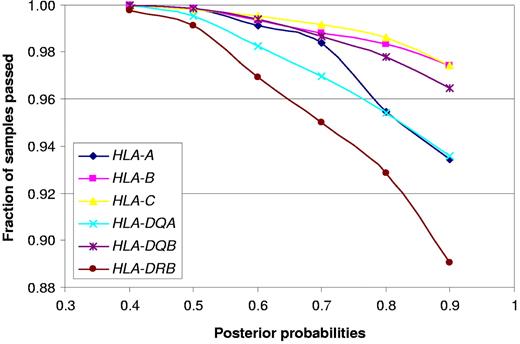
To survey the relationship between specific 2-digit and 4-digit HLA alleles and BCP-ALL, we predicted key class I and II HLA alleles. Comparison between observed and predicted HLA alleles for controls showed a high degree of concordance. To ensure accuracy of prediction, we restricted our case-control analysis to those alleles that were predicted with a posterior probability more than 90%, and this criterion was met by more than 88% of the data (Figure 4). Relaxing the posterior probability criterion made no substantial difference to the study findings (data not shown).
Fraction of samples passing posterior probability criteria for HLA predictions.
Four HLA class I alleles showed evidence of a relationship with BCP-ALL risk at the 0.05 threshold: A*26, A*29, C*02, and C*08 (Table 1). The strongest association was provided by C*08 (P = .01), albeit nonsignificant after adjustment for multiple tests. Only 2 of the 22 class II alleles showed marginal evidence for a relationship with ALL risk at the 0.05 threshold: DQB1*02 and DQB1*03 (Table 1). Both were nonsignificant after adjustment for multiple testing. Comparison of 4 digit-defined HLA subtypes provided no increased evidence for an association between a specific allele and ALL risk. The strongest associations were with A*2601, A*2902, B*1402, C*0202, and C*0802 class I alleles and DQB1*0201, DQB1*0302, DQB1*0303, and DRB1*1302 class II alleles, none attaining statistical significance after adjustment for multiple testing (Table 2).
Association results for 2-digit HLA alleles and BCP-ALL risk
| . | Cases, no. (%) . | Controls, no. (%) . | OR (95% CI) . | Pr (> t ) . |
|---|---|---|---|---|
| Class I | ||||
| HLA-A | ||||
| 01 | 331 (21.6) | 1824 (20.6) | 1.06 (0.93-1.21) | 0.38 |
| 11 | 103 (6.7) | 555 (6.3) | 1.08 (0.87-1.34) | 0.50 |
| 02 | 402 (26.2) | 2319 (26.1) | 1.00 (0.89-1.13) | 0.98 |
| 23 | 21 (1.4) | 119 (1.3) | 1.02 (0.64-1.63) | 0.94 |
| 24 | 133 (8.7) | 737 (8.3) | 1.05 (0.86-1.27) | 0.65 |
| 25 | 29 (1.9) | 182 (2.1) | 0.92 (0.62-1.36) | 0.67 |
| 26* | 16 (1.0) | 161 (1.8) | 0.57 (0.34-0.95) | 0.03 |
| 29* | 83 (5.4) | 370 (4.2) | 1.31 (1.03-1.68) | 0.03 |
| 03 | 221 (14.4) | 1348 (15.2) | 0.94 (0.80-1.09) | 0.41 |
| 30 | 26 (1.7) | 200 (2.3) | 0.75 (0.49-1.13) | 0.16 |
| 31 | 61 (4.0) | 269 (3.0) | 1.32 (1.00-1.76) | 0.05 |
| 32 | 56 (3.6) | 357 (4.0) | 0.90 (0.68-1.20) | 0.48 |
| 68 | 49 (3.2) | 359 (4.0) | 0.78 (0.58-1.06) | 0.11 |
| HLA-B | ||||
| 13 | 32 (2.0) | 182 (2.0) | 1.01 (0.69-1.48) | 0.95 |
| 14 | 48 (3.0) | 365 (4.0) | 0.75 (0.55-1.02) | 0.06 |
| 15 | 122 (7.7) | 670 (7.3) | 1.05 (0.86-1.29) | 0.62 |
| 18 | 69 (4.3) | 374 (4.1) | 1.07 (0.82-1.38) | 0.64 |
| 27 | 65 (4.1) | 421 (4.6) | 0.88 (0.68-1.15) | 0.37 |
| 35 | 106 (6.7) | 555 (6.1) | 1.11 (0.89-1.37) | 0.35 |
| 37 | 31 (1.9) | 130 (1.4) | 1.38 (0.93-2.05) | 0.11 |
| 38 | 17 (1.1) | 66 (0.7) | 1.49 (0.87-2.54) | 0.15 |
| 39 | 21 (1.3) | 107 (1.2) | 1.13 (0.71-1.81) | 0.61 |
| 40 | 108 (6.8) | 634 (6.9) | 0.98 (0.79-1.21) | 0.85 |
| 44 | 267 (16.8) | 1566 (17.1) | 0.98 (0.85-1.13) | 0.76 |
| 49 | 17 (1.1) | 103 (1.1) | 0.95 (0.57-1.59) | 0.84 |
| 51 | 57 (3.6) | 364 (4.0) | 0.90 (0.68-1.19) | 0.46 |
| 55 | 28 (1.8) | 178 (1.9) | 0.90 (0.60-1.35) | 0.62 |
| 57 | 73 (4.6) | 381 (4.2) | 1.11 (0.86-1.43) | 0.43 |
| 07 | 242 (15.2) | 1380 (15.0) | 1.01 (0.87-1.17) | 0.88 |
| 08 | 234 (14.7) | 1358 (14.8) | 0.99 (0.85-1.15) | 0.90 |
| HLA-C | ||||
| 01 | 46 (2.9) | 346 (3.8) | 0.77 (0.56-1.05) | 0.10 |
| 12 | 63 (4.0) | 313 (3.4) | 1.18 (0.89-1.55) | 0.24 |
| 15 | 22 (1.4) | 155 (1.7) | 0.82 (0.53-1.29) | 0.40 |
| 16 | 80 (5.1) | 400 (4.4) | 1.17 (0.92-1.50) | 0.21 |
| 02* | 74 (4.7) | 333 (3.6) | 1.31 (1.01-1.69) | 0.04 |
| 03 | 213 (13.6) | 1287 (14.1) | 0.96 (0.82-1.12) | 0.58 |
| 04 | 138 (8.8) | 768 (8.4) | 1.05 (0.87-1.27) | 0.62 |
| 05 | 156 (9.9) | 997 (10.9) | 0.90 (0.75-1.08) | 0.25 |
| 06 | 169 (10.8) | 874 (9.6) | 1.14 (0.96-1.36) | 0.14 |
| 07 | 552 (35.1) | 3183 (34.8) | 1.01 (0.91-1.13) | 0.81 |
| 08* | 41 (2.6) | 361 (3.9) | 0.65 (0.47-0.90) | 0.01 |
| Class II | ||||
| HLA-DQA | ||||
| 01 | 577 (37) | 3325 (37.5) | 0.98 (0.87-1.09) | 0.70 |
| 02 | 259 (16.6) | 1423 (16.0) | 1.04 (0.90-1.20) | 0.59 |
| 03 | 313 (20.1) | 1742 (19.6) | 1.03 (0.90-1.17) | 0.70 |
| 04 | 25 (1.6) | 192 (2.2) | 0.74 (0.48-1.12) | 0.15 |
| 05 | 387 (24.8) | 2189 (24.7) | 1.01 (0.89-1.14) | 0.92 |
| HLA-DQB | ||||
| 02* | 313 (20.6) | 2194 (23.8) | 0.83 (0.72-0.95) | 5.56 × 10−3 |
| 03* | 576 (37.9) | 3210 (34.9) | 1.14 (1.02-1.27) | 0.02 |
| 04 | 26 (1.7) | 197 (2.1) | 0.80 (0.53-1.20) | 0.28 |
| 05 | 246 (16.2) | 1392 (15.1) | 1.08 (0.93-1.26) | 0.29 |
| 06 | 359 (23.6) | 2208 (24.0) | 0.98 (0.86-1.11) | 0.75 |
| HLA-DRB | ||||
| 01 | 175 (12.4) | 926 (11.0) | 1.15 (0.96-1.36) | 0.12 |
| 11 | 49 (3.5) | 344 (4.1) | 0.84 (0.62-1.15) | 0.28 |
| 12 | 25 (1.8) | 146 (1.7) | 1.02 (0.67-1.57) | 0.92 |
| 13 | 142 (10.1) | 986 (11.7) | 0.84 (0.70-1.02) | 0.07 |
| 14 | 30 (2.1) | 219 (2.6) | 0.81 (0.55-1.20) | 0.30 |
| 15 | 233 (16.5) | 1285 (15.2) | 1.10 (0.94-1.28) | 0.23 |
| 03 | 238 (16.8) | 1386 (16.4) | 1.03 (0.89-1.20) | 0.70 |
| 04 | 210 (14.9) | 1362 (16.2) | 0.91 (0.77-1.06) | 0.22 |
| 07 | 244 (17.3) | 1375 (16.3) | 1.07 (0.92-1.24) | 0.36 |
| 08 | 25 (1.8) | 197 (2.3) | 0.75 (0.49-1.15) | 0.19 |
| 09 | 21 (1.5) | 119 (1.4) | 1.05 (0.66-1.68) | 0.82 |
| . | Cases, no. (%) . | Controls, no. (%) . | OR (95% CI) . | Pr (> t ) . |
|---|---|---|---|---|
| Class I | ||||
| HLA-A | ||||
| 01 | 331 (21.6) | 1824 (20.6) | 1.06 (0.93-1.21) | 0.38 |
| 11 | 103 (6.7) | 555 (6.3) | 1.08 (0.87-1.34) | 0.50 |
| 02 | 402 (26.2) | 2319 (26.1) | 1.00 (0.89-1.13) | 0.98 |
| 23 | 21 (1.4) | 119 (1.3) | 1.02 (0.64-1.63) | 0.94 |
| 24 | 133 (8.7) | 737 (8.3) | 1.05 (0.86-1.27) | 0.65 |
| 25 | 29 (1.9) | 182 (2.1) | 0.92 (0.62-1.36) | 0.67 |
| 26* | 16 (1.0) | 161 (1.8) | 0.57 (0.34-0.95) | 0.03 |
| 29* | 83 (5.4) | 370 (4.2) | 1.31 (1.03-1.68) | 0.03 |
| 03 | 221 (14.4) | 1348 (15.2) | 0.94 (0.80-1.09) | 0.41 |
| 30 | 26 (1.7) | 200 (2.3) | 0.75 (0.49-1.13) | 0.16 |
| 31 | 61 (4.0) | 269 (3.0) | 1.32 (1.00-1.76) | 0.05 |
| 32 | 56 (3.6) | 357 (4.0) | 0.90 (0.68-1.20) | 0.48 |
| 68 | 49 (3.2) | 359 (4.0) | 0.78 (0.58-1.06) | 0.11 |
| HLA-B | ||||
| 13 | 32 (2.0) | 182 (2.0) | 1.01 (0.69-1.48) | 0.95 |
| 14 | 48 (3.0) | 365 (4.0) | 0.75 (0.55-1.02) | 0.06 |
| 15 | 122 (7.7) | 670 (7.3) | 1.05 (0.86-1.29) | 0.62 |
| 18 | 69 (4.3) | 374 (4.1) | 1.07 (0.82-1.38) | 0.64 |
| 27 | 65 (4.1) | 421 (4.6) | 0.88 (0.68-1.15) | 0.37 |
| 35 | 106 (6.7) | 555 (6.1) | 1.11 (0.89-1.37) | 0.35 |
| 37 | 31 (1.9) | 130 (1.4) | 1.38 (0.93-2.05) | 0.11 |
| 38 | 17 (1.1) | 66 (0.7) | 1.49 (0.87-2.54) | 0.15 |
| 39 | 21 (1.3) | 107 (1.2) | 1.13 (0.71-1.81) | 0.61 |
| 40 | 108 (6.8) | 634 (6.9) | 0.98 (0.79-1.21) | 0.85 |
| 44 | 267 (16.8) | 1566 (17.1) | 0.98 (0.85-1.13) | 0.76 |
| 49 | 17 (1.1) | 103 (1.1) | 0.95 (0.57-1.59) | 0.84 |
| 51 | 57 (3.6) | 364 (4.0) | 0.90 (0.68-1.19) | 0.46 |
| 55 | 28 (1.8) | 178 (1.9) | 0.90 (0.60-1.35) | 0.62 |
| 57 | 73 (4.6) | 381 (4.2) | 1.11 (0.86-1.43) | 0.43 |
| 07 | 242 (15.2) | 1380 (15.0) | 1.01 (0.87-1.17) | 0.88 |
| 08 | 234 (14.7) | 1358 (14.8) | 0.99 (0.85-1.15) | 0.90 |
| HLA-C | ||||
| 01 | 46 (2.9) | 346 (3.8) | 0.77 (0.56-1.05) | 0.10 |
| 12 | 63 (4.0) | 313 (3.4) | 1.18 (0.89-1.55) | 0.24 |
| 15 | 22 (1.4) | 155 (1.7) | 0.82 (0.53-1.29) | 0.40 |
| 16 | 80 (5.1) | 400 (4.4) | 1.17 (0.92-1.50) | 0.21 |
| 02* | 74 (4.7) | 333 (3.6) | 1.31 (1.01-1.69) | 0.04 |
| 03 | 213 (13.6) | 1287 (14.1) | 0.96 (0.82-1.12) | 0.58 |
| 04 | 138 (8.8) | 768 (8.4) | 1.05 (0.87-1.27) | 0.62 |
| 05 | 156 (9.9) | 997 (10.9) | 0.90 (0.75-1.08) | 0.25 |
| 06 | 169 (10.8) | 874 (9.6) | 1.14 (0.96-1.36) | 0.14 |
| 07 | 552 (35.1) | 3183 (34.8) | 1.01 (0.91-1.13) | 0.81 |
| 08* | 41 (2.6) | 361 (3.9) | 0.65 (0.47-0.90) | 0.01 |
| Class II | ||||
| HLA-DQA | ||||
| 01 | 577 (37) | 3325 (37.5) | 0.98 (0.87-1.09) | 0.70 |
| 02 | 259 (16.6) | 1423 (16.0) | 1.04 (0.90-1.20) | 0.59 |
| 03 | 313 (20.1) | 1742 (19.6) | 1.03 (0.90-1.17) | 0.70 |
| 04 | 25 (1.6) | 192 (2.2) | 0.74 (0.48-1.12) | 0.15 |
| 05 | 387 (24.8) | 2189 (24.7) | 1.01 (0.89-1.14) | 0.92 |
| HLA-DQB | ||||
| 02* | 313 (20.6) | 2194 (23.8) | 0.83 (0.72-0.95) | 5.56 × 10−3 |
| 03* | 576 (37.9) | 3210 (34.9) | 1.14 (1.02-1.27) | 0.02 |
| 04 | 26 (1.7) | 197 (2.1) | 0.80 (0.53-1.20) | 0.28 |
| 05 | 246 (16.2) | 1392 (15.1) | 1.08 (0.93-1.26) | 0.29 |
| 06 | 359 (23.6) | 2208 (24.0) | 0.98 (0.86-1.11) | 0.75 |
| HLA-DRB | ||||
| 01 | 175 (12.4) | 926 (11.0) | 1.15 (0.96-1.36) | 0.12 |
| 11 | 49 (3.5) | 344 (4.1) | 0.84 (0.62-1.15) | 0.28 |
| 12 | 25 (1.8) | 146 (1.7) | 1.02 (0.67-1.57) | 0.92 |
| 13 | 142 (10.1) | 986 (11.7) | 0.84 (0.70-1.02) | 0.07 |
| 14 | 30 (2.1) | 219 (2.6) | 0.81 (0.55-1.20) | 0.30 |
| 15 | 233 (16.5) | 1285 (15.2) | 1.10 (0.94-1.28) | 0.23 |
| 03 | 238 (16.8) | 1386 (16.4) | 1.03 (0.89-1.20) | 0.70 |
| 04 | 210 (14.9) | 1362 (16.2) | 0.91 (0.77-1.06) | 0.22 |
| 07 | 244 (17.3) | 1375 (16.3) | 1.07 (0.92-1.24) | 0.36 |
| 08 | 25 (1.8) | 197 (2.3) | 0.75 (0.49-1.15) | 0.19 |
| 09 | 21 (1.5) | 119 (1.4) | 1.05 (0.66-1.68) | 0.82 |
Only alleles present in > 1% of cases are shown.
Alleles showing an association with ALL risk at P < .05.
Association results for 4-digit HLA alleles and BCP-ALL risk
| . | Cases, no. (%) . | Controls, no. (%) . | OR (95% CI) . | Pr (> t ) . |
|---|---|---|---|---|
| Class I | ||||
| HLA-A | ||||
| 0101 | 331 (21.6) | 1824 (20.6) | 1.06 (0.93-1.21) | 0.39 |
| 1101 | 103 (6.7) | 555 (6.3) | 1.08 (0.86-1.34) | 0.51 |
| 0201 | 391 (25.5) | 2243 (25.3) | 1.01 (0.89-1.14) | 0.91 |
| 2301 | 21 (1.4) | 119 (1.3) | 1.02 (0.64-1.62) | 0.94 |
| 2402 | 133 (8.7) | 732 (8.2) | 1.05 (0.87-1.28) | 0.60 |
| 2501 | 29 (1.9) | 182 (2.1) | 0.92 (0.62-1.36) | 0.67 |
| 2601* | 16 (1.0) | 156 (1.8) | 0.59 (0.35-0.98) | 0.04 |
| 2902* | 83 (5.4) | 370 (4.2) | 1.31 (1.03-1.67) | 0.03 |
| 0301 | 221 (14.4) | 1348 (15.2) | 0.94 (0.80-1.09) | 0.40 |
| 3101 | 61 (4.0) | 269 (3.0) | 1.32 (0.99-1.75) | 0.05 |
| 3201 | 56 (3.6) | 357 (4.0) | 0.90 (0.68-1.20) | 0.48 |
| 6801 | 45 (2.9) | 304 (3.4) | 0.85 (0.62-1.17) | 0.31 |
| HLA-B | ||||
| 1302 | 32 (2.0) | 182 (2.0) | 1.01 (0.69-1.48) | 0.95 |
| 1401 | 19 (1.2) | 114 (1.2) | 0.96 (0.59-1.56) | 0.86 |
| 1402* | 29 (1.8) | 251 (2.7) | 0.66 (0.45-0.97) | 0.03 |
| 1501 | 108 (6.8) | 616 (6.7) | 1.01 (0.82-1.25) | 0.93 |
| 1801 | 69 (4.3) | 374 (4.1) | 1.06 (0.82-1.38) | 0.64 |
| 2705 | 65 (4.1) | 419 (4.6) | 0.89 (0.68-1.16) | 0.38 |
| 3501 | 90 (5.7) | 466 (5.1) | 1.12 (0.89-1.41) | 0.35 |
| 3701 | 31 (1.9) | 130 (1.4) | 1.38 (0.93-2.05) | 0.11 |
| 3801 | 17 (1.1) | 66 (0.7) | 1.49 (0.87-2.54) | 0.15 |
| 4001 | 98 (6.2) | 551 (6.0) | 1.02 (0.82-1.28) | 0.83 |
| 4402 | 168 (10.6) | 1042 (11.4) | 0.92 (0.77-1.09) | 0.34 |
| 4403 | 95 (6.0) | 510 (5.6) | 1.08 (0.86-1.35) | 0.52 |
| 4901 | 17 (1.1) | 103 (1.1) | 0.95 (0.57-1.59) | 0.84 |
| 5101 | 57 (3.6) | 363 (4.0) | 0.90 (0.68-1.20) | 0.47 |
| 5501 | 28 (1.8) | 178 (1.9) | 0.90 (0.60-1.35) | 0.62 |
| 5701 | 73 (4.6) | 380 (4.1) | 1.11 (0.86-1.43) | 0.42 |
| 0702 | 242 (15.2) | 1380 (15.0) | 1.01 (0.87-1.17) | 0.89 |
| 0801 | 234 (14.7) | 1358 (14.8) | 0.99 (0.85-1.15) | 0.89 |
| HLA-C | ||||
| 0102 | 46 (2.9) | 346 (3.8) | 0.77 (0.56-1.05) | 0.10 |
| 1203 | 52 (3.3) | 274 (3.0) | 1.11 (0.82-1.50) | 0.51 |
| 1502 | 21 (1.3) | 146 (1.6) | 0.83 (0.53-1.32) | 0.44 |
| 1601 | 78 (5.0) | 389 (4.3) | 1.18 (0.92-1.51) | 0.20 |
| 0202* | 74 (4.7) | 333 (3.6) | 1.31 (1.01-1.69) | 0.04 |
| 0303 | 71 (4.5) | 525 (5.7) | 0.78 (0.60-1.00) | 0.05 |
| 0304 | 142 (9.0) | 761 (8.3) | 1.09 (0.91-1.32) | 0.35 |
| 0401 | 138 (8.8) | 768 (8.4) | 1.05 (0.87-1.27) | 0.62 |
| 0501 | 156 (9.9) | 997 (10.9) | 0.90 (0.75-1.08) | 0.25 |
| 0602 | 169 (10.8) | 874 (9.6) | 1.14 (0.96-1.36) | 0.14 |
| 0701 | 270 (17.2) | 1528 (16.7) | 1.03 (0.90-1.19) | 0.65 |
| 0702 | 256 (16.3) | 1501 (16.4) | 0.99 (0.86-1.15) | 0.90 |
| 0704 | 26 (1.7) | 154 (1.7) | 0.98 (0.65-1.49) | 0.93 |
| 0802* | 41 (2.6) | 361 (3.9) | 0.65 (0.47-0.90) | 0.01 |
| Class II | ||||
| HLA-DQA | ||||
| 0101 | 222 (14.2) | 1307 (14.7) | 0.96 (0.82-1.12) | 0.60 |
| 0102 | 287 (18.4) | 1560 (17.6) | 1.06 (0.92-1.21) | 0.45 |
| 0103 | 68 (4.4) | 458 (5.2) | 0.84 (0.64-1.09) | 0.18 |
| 0201 | 259 (16.6) | 1423 (16.0) | 1.04 (0.90-1.20) | 0.59 |
| 0301 | 313 (20.1) | 1742 (19.6) | 1.03 (0.90-1.17) | 0.70 |
| 0401 | 25 (1.6) | 192 (2.2) | 0.74 (0.48-1.12) | 0.15 |
| 0501 | 387 (24.8) | 2189 (24.7) | 1.01 (0.89-1.14) | 0.92 |
| Class II | ||||
| HLA-DQB | ||||
| 0201* | 189 (12.4) | 1322 (14.4) | 0.84 (0.72-0.99) | 0.04 |
| 0202 | 124 (8.2) | 871 (9.5) | 0.85 (0.70-1.03) | 0.10 |
| 0301 | 280 (18.4) | 1721 (18.7) | 0.98 (0.85-1.13) | 0.77 |
| 0302* | 192 (12.6) | 981 (10.7) | 1.21 (1.03-1.43) | 0.02 |
| 0303* | 104 (6.8) | 506 (5.5) | 1.26 (1.01-1.57) | 0.04 |
| 0402 | 26 (1.7) | 197 (2.1) | 0.79 (0.53-1.20) | 0.27 |
| 0501 | 206 (13.6) | 1093 (11.9) | 1.16 (0.99-1.36) | 0.07 |
| 0503 | 27 (1.8) | 231 (2.5) | 0.70 (0.47-1.05) | 0.08 |
| 0602 | 227 (14.9) | 1265 (13.7) | 1.10 (0.94-1.28) | 0.22 |
| 0603 | 77 (5.1) | 505 (5.5) | 0.92 (0.72-1.17) | 0.49 |
| 0604 | 37 (2.4) | 291 (3.2) | 0.76 (0.54-1.08) | 0.13 |
| HLA-DRB | ||||
| 0101 | 134 (9.5) | 689 (8.2) | 1.17 (0.96-1.42) | 0.11 |
| 0103 | 29 (2.1) | 149 (1.8) | 1.16 (0.78-1.73) | 0.47 |
| 1101 | 44 (3.1) | 240 (2.8) | 1.09 (0.79-1.51) | 0.60 |
| 1201 | 25 (1.8) | 144 (1.7) | 1.03 (0.67-1.58) | 0.89 |
| 1301 | 77 (5.4) | 489 (5.8) | 0.93 (0.73-1.19) | 0.57 |
| 1302* | 49 (3.5) | 410 (4.9) | 0.70 (0.52-0.95) | 0.02 |
| 1303 | 16 (1.1) | 85 (1.0) | 1.12 (0.65-1.91) | 0.68 |
| 1401 | 30 (2.1) | 217 (2.6) | 0.82 (0.56-1.20) | 0.30 |
| 1501 | 225 (15.9) | 1249 (14.8) | 1.08 (0.93-1.26) | 0.31 |
| 0301 | 238 (16.8) | 1386 (16.4) | 1.02 (0.88-1.19) | 0.76 |
| 0401 | 145 (10.3) | 884 (10.5) | 0.97 (0.81-1.17) | 0.76 |
| 0404 | 57 (4.0) | 349 (4.1) | 0.97 (0.73-1.29) | 0.83 |
| 0701 | 244 (17.3) | 1375 (16.3) | 1.06 (0.92-1.24) | 0.41 |
| 0801 | 23 (1.6) | 180 (2.1) | 0.75 (0.49-1.17) | 0.21 |
| 0901 | 21 (1.5) | 119 (1.4) | 1.05 (0.66-1.67) | 0.84 |
| . | Cases, no. (%) . | Controls, no. (%) . | OR (95% CI) . | Pr (> t ) . |
|---|---|---|---|---|
| Class I | ||||
| HLA-A | ||||
| 0101 | 331 (21.6) | 1824 (20.6) | 1.06 (0.93-1.21) | 0.39 |
| 1101 | 103 (6.7) | 555 (6.3) | 1.08 (0.86-1.34) | 0.51 |
| 0201 | 391 (25.5) | 2243 (25.3) | 1.01 (0.89-1.14) | 0.91 |
| 2301 | 21 (1.4) | 119 (1.3) | 1.02 (0.64-1.62) | 0.94 |
| 2402 | 133 (8.7) | 732 (8.2) | 1.05 (0.87-1.28) | 0.60 |
| 2501 | 29 (1.9) | 182 (2.1) | 0.92 (0.62-1.36) | 0.67 |
| 2601* | 16 (1.0) | 156 (1.8) | 0.59 (0.35-0.98) | 0.04 |
| 2902* | 83 (5.4) | 370 (4.2) | 1.31 (1.03-1.67) | 0.03 |
| 0301 | 221 (14.4) | 1348 (15.2) | 0.94 (0.80-1.09) | 0.40 |
| 3101 | 61 (4.0) | 269 (3.0) | 1.32 (0.99-1.75) | 0.05 |
| 3201 | 56 (3.6) | 357 (4.0) | 0.90 (0.68-1.20) | 0.48 |
| 6801 | 45 (2.9) | 304 (3.4) | 0.85 (0.62-1.17) | 0.31 |
| HLA-B | ||||
| 1302 | 32 (2.0) | 182 (2.0) | 1.01 (0.69-1.48) | 0.95 |
| 1401 | 19 (1.2) | 114 (1.2) | 0.96 (0.59-1.56) | 0.86 |
| 1402* | 29 (1.8) | 251 (2.7) | 0.66 (0.45-0.97) | 0.03 |
| 1501 | 108 (6.8) | 616 (6.7) | 1.01 (0.82-1.25) | 0.93 |
| 1801 | 69 (4.3) | 374 (4.1) | 1.06 (0.82-1.38) | 0.64 |
| 2705 | 65 (4.1) | 419 (4.6) | 0.89 (0.68-1.16) | 0.38 |
| 3501 | 90 (5.7) | 466 (5.1) | 1.12 (0.89-1.41) | 0.35 |
| 3701 | 31 (1.9) | 130 (1.4) | 1.38 (0.93-2.05) | 0.11 |
| 3801 | 17 (1.1) | 66 (0.7) | 1.49 (0.87-2.54) | 0.15 |
| 4001 | 98 (6.2) | 551 (6.0) | 1.02 (0.82-1.28) | 0.83 |
| 4402 | 168 (10.6) | 1042 (11.4) | 0.92 (0.77-1.09) | 0.34 |
| 4403 | 95 (6.0) | 510 (5.6) | 1.08 (0.86-1.35) | 0.52 |
| 4901 | 17 (1.1) | 103 (1.1) | 0.95 (0.57-1.59) | 0.84 |
| 5101 | 57 (3.6) | 363 (4.0) | 0.90 (0.68-1.20) | 0.47 |
| 5501 | 28 (1.8) | 178 (1.9) | 0.90 (0.60-1.35) | 0.62 |
| 5701 | 73 (4.6) | 380 (4.1) | 1.11 (0.86-1.43) | 0.42 |
| 0702 | 242 (15.2) | 1380 (15.0) | 1.01 (0.87-1.17) | 0.89 |
| 0801 | 234 (14.7) | 1358 (14.8) | 0.99 (0.85-1.15) | 0.89 |
| HLA-C | ||||
| 0102 | 46 (2.9) | 346 (3.8) | 0.77 (0.56-1.05) | 0.10 |
| 1203 | 52 (3.3) | 274 (3.0) | 1.11 (0.82-1.50) | 0.51 |
| 1502 | 21 (1.3) | 146 (1.6) | 0.83 (0.53-1.32) | 0.44 |
| 1601 | 78 (5.0) | 389 (4.3) | 1.18 (0.92-1.51) | 0.20 |
| 0202* | 74 (4.7) | 333 (3.6) | 1.31 (1.01-1.69) | 0.04 |
| 0303 | 71 (4.5) | 525 (5.7) | 0.78 (0.60-1.00) | 0.05 |
| 0304 | 142 (9.0) | 761 (8.3) | 1.09 (0.91-1.32) | 0.35 |
| 0401 | 138 (8.8) | 768 (8.4) | 1.05 (0.87-1.27) | 0.62 |
| 0501 | 156 (9.9) | 997 (10.9) | 0.90 (0.75-1.08) | 0.25 |
| 0602 | 169 (10.8) | 874 (9.6) | 1.14 (0.96-1.36) | 0.14 |
| 0701 | 270 (17.2) | 1528 (16.7) | 1.03 (0.90-1.19) | 0.65 |
| 0702 | 256 (16.3) | 1501 (16.4) | 0.99 (0.86-1.15) | 0.90 |
| 0704 | 26 (1.7) | 154 (1.7) | 0.98 (0.65-1.49) | 0.93 |
| 0802* | 41 (2.6) | 361 (3.9) | 0.65 (0.47-0.90) | 0.01 |
| Class II | ||||
| HLA-DQA | ||||
| 0101 | 222 (14.2) | 1307 (14.7) | 0.96 (0.82-1.12) | 0.60 |
| 0102 | 287 (18.4) | 1560 (17.6) | 1.06 (0.92-1.21) | 0.45 |
| 0103 | 68 (4.4) | 458 (5.2) | 0.84 (0.64-1.09) | 0.18 |
| 0201 | 259 (16.6) | 1423 (16.0) | 1.04 (0.90-1.20) | 0.59 |
| 0301 | 313 (20.1) | 1742 (19.6) | 1.03 (0.90-1.17) | 0.70 |
| 0401 | 25 (1.6) | 192 (2.2) | 0.74 (0.48-1.12) | 0.15 |
| 0501 | 387 (24.8) | 2189 (24.7) | 1.01 (0.89-1.14) | 0.92 |
| Class II | ||||
| HLA-DQB | ||||
| 0201* | 189 (12.4) | 1322 (14.4) | 0.84 (0.72-0.99) | 0.04 |
| 0202 | 124 (8.2) | 871 (9.5) | 0.85 (0.70-1.03) | 0.10 |
| 0301 | 280 (18.4) | 1721 (18.7) | 0.98 (0.85-1.13) | 0.77 |
| 0302* | 192 (12.6) | 981 (10.7) | 1.21 (1.03-1.43) | 0.02 |
| 0303* | 104 (6.8) | 506 (5.5) | 1.26 (1.01-1.57) | 0.04 |
| 0402 | 26 (1.7) | 197 (2.1) | 0.79 (0.53-1.20) | 0.27 |
| 0501 | 206 (13.6) | 1093 (11.9) | 1.16 (0.99-1.36) | 0.07 |
| 0503 | 27 (1.8) | 231 (2.5) | 0.70 (0.47-1.05) | 0.08 |
| 0602 | 227 (14.9) | 1265 (13.7) | 1.10 (0.94-1.28) | 0.22 |
| 0603 | 77 (5.1) | 505 (5.5) | 0.92 (0.72-1.17) | 0.49 |
| 0604 | 37 (2.4) | 291 (3.2) | 0.76 (0.54-1.08) | 0.13 |
| HLA-DRB | ||||
| 0101 | 134 (9.5) | 689 (8.2) | 1.17 (0.96-1.42) | 0.11 |
| 0103 | 29 (2.1) | 149 (1.8) | 1.16 (0.78-1.73) | 0.47 |
| 1101 | 44 (3.1) | 240 (2.8) | 1.09 (0.79-1.51) | 0.60 |
| 1201 | 25 (1.8) | 144 (1.7) | 1.03 (0.67-1.58) | 0.89 |
| 1301 | 77 (5.4) | 489 (5.8) | 0.93 (0.73-1.19) | 0.57 |
| 1302* | 49 (3.5) | 410 (4.9) | 0.70 (0.52-0.95) | 0.02 |
| 1303 | 16 (1.1) | 85 (1.0) | 1.12 (0.65-1.91) | 0.68 |
| 1401 | 30 (2.1) | 217 (2.6) | 0.82 (0.56-1.20) | 0.30 |
| 1501 | 225 (15.9) | 1249 (14.8) | 1.08 (0.93-1.26) | 0.31 |
| 0301 | 238 (16.8) | 1386 (16.4) | 1.02 (0.88-1.19) | 0.76 |
| 0401 | 145 (10.3) | 884 (10.5) | 0.97 (0.81-1.17) | 0.76 |
| 0404 | 57 (4.0) | 349 (4.1) | 0.97 (0.73-1.29) | 0.83 |
| 0701 | 244 (17.3) | 1375 (16.3) | 1.06 (0.92-1.24) | 0.41 |
| 0801 | 23 (1.6) | 180 (2.1) | 0.75 (0.49-1.17) | 0.21 |
| 0901 | 21 (1.5) | 119 (1.4) | 1.05 (0.66-1.67) | 0.84 |
Only alleles present in > 1% of cases are shown.
Alleles showing an association with ALL risk at P < .05.
In BCP-ALL, 2 chromosomal anomalies typify more than 50% of cases. Approximately 25% of patients carry a reciprocal translocation between chromosomes 12 and 21 [t(12;21)(12q13;21q22)], leading to the in-frame fusion of TEL(ETV6) at 12p13 and AML1(RUNX1) on 21q22. A further 30% of ALL patients are characterized by hyperdiploidy, whereby the modal chromosome number exceeds 50. In view of these molecular differences seen in BCP-ALL, we conducted a subgroup analysis for the TEL-AML and hyperdiploid cases separately. After correction for multiple testing, no statistically significant association between HLA alleles and specific BCP-ALL subtype was observed (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Discussion
The assertion that inherited genetic factors contribute to the development of childhood ALL has been vindicated by recent GWA studies of ALL. These analyses have identified several SNPs mapping to 7p12.2 (IKZF1), 9p21 (CDKN2A-CDKN2B), 10q21.2 (ARID5B), and 14q11.2 (CEBPE), which each confer a modest but highly significant increase in ALL risk.
There has been renewed interest in role of infection as an etiologic basis for ALL, centered on the hypothesis that the disease may be a consequence of common infection in nonimmune persons. It has been suggested that an HLA class association would provide a link between host genotype and response. However, as compelling as such an argument is, unambiguous evidence in the form of a statistically robust association has been lacking. To address this, we have conducted a large case-control study and have systematically examined the relationship between genetics variants and BCP-ALL risk for the entire 4.5-Mb region, which encompasses MHC. Moreover, prediction of HLA alleles from GWA SNP data has allowed us to systematically analyze the relationship between HLA supertypes and risk.
The increase in ALL risk conferred by any common polymorphic variant is almost certainly small (ie, typical relative risk < 1.6). The inherent statistical uncertainty of case-control studies involving just a few hundred cases and controls severely constrains study power to reliably identify genetic determinants, conferring modest, but potentially important, risks. A major strength of our study, therefore, is that we have based our analysis on a large sample set for a comparatively rare tumor. The issue of potential population stratification in previous association studies of HLA and resulting false positive results is of concern. If population subdivisions exist, it is possible that associations will be found between ALL and arbitrary markers that are unlinked to causative loci. This is especially an issue with studies of HLA alleles, which even among European populations can be highly variable in frequency.18 Using GWA SNP data has allowed us to minimize this potential problem as formal statistical analysis provided no significant evidence that population substructure is a confounding factor in our study. Survivorship is a potential source of bias if a variant influences prognosis as seen in the study of some cancers. This is unlikely to be of serious concern in the present study as survivorship from childhood ALL is high.
Through the analysis of single SNPs both observed and imputed haplotypes and alleles at HLA–A, -B, -C, -DRB1, -DQA1, and -DQB1, our study provides no evidence for a strong relationship between MHC variation and risk of BCP-ALL. Based on UKCCS data, associations between specific HLA-DPB1 alleles and ALL risk have been reported.8 Although we acknowledge that in our study we did not predict all of the HLA alleles, the assay of single SNPs and haplotypes we have conducted will have been sufficient to recover strong associations at untyped loci.
There are limitations to the interpretation of data obtained in this study in terms of generalizability to other forms of ALL. We have restricted our current study to the commonest form of childhood ALL, BCP-ALL, to have high power to detect associations. It is probable that the risk factors for BCP and T-cell ALL will be different given the differences in their biology and clinical response to treatment; hence, our findings do not preclude a role for the MHC in T-cell ALL.
The “delayed infection” model for the etiology of childhood BCP-ALL predicts some degree of inherited susceptibility operating via functional variants in genes regulating the immune response. However, because several or many different infections are anticipated to act as triggers for the development of ALL, proximal to diagnosis,1 a lack of specific HLA allele association is not inconsistent with an infectious or immunologic etiology.
In conclusion, our analysis does not provide support for the hypothesis that MHC variation is a major determinant of BCP-ALL risk. We cannot, however, exclude a role of the MHC in T-cell ALL or that HLA genotype plays a role in determining patient outcome.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Sue Richards and Julie Burrett (Clinical Trials Service Unit, Oxford), Christine Harrison, Lucy Chilton, and Anthony Moorman (Leukemia Research Cytogenetics Group, Northern Institute for Cancer Research, Newcastle University), Jill Simpson (University of York), Pamela Thomson and Adiba Hussain (Cancer Immunogenetics, School of Cancer Sciences, University of Manchester) for assistance with data harmonization, Irene Roberts and the Children's Cancer and Leukemia Group Biological Studies Steering Group for access to Medical Research Council ALL Trial samples, all the patients and persons for their participation, and the clinicians, other hospital staff, and study staff who contributed to the blood sample and data collection for this study. This study made use of control genotyping data generated by the Wellcome Trust Case-Control Consortium. The authors acknowledge use of genotype data from the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council (grant G0000934) and the Wellcome Trust (grant 068545/Z/02). A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk.
This work was supported by Leukaemia & Lymphoma Research (United Kingdom), the Kay Kendall Leukaemia Fund, Cancer Research UK (C1298/A8362 supported by the Bobby Moore Fund), and the Wellcome Trust (award 076113 and 085475).
Authorship
Contribution: R.S.H. and F.J.H. designed the study; R.S.H., G.M., and F.J.H. drafted the manuscript; F.J.H., Y.W., S.L., A.D., L.M., and S.E.D. performed statistical and bioinformatic analyses; E.P. oversaw laboratory analyses; E.S. and S.E.K. performed curation and sample preparation of Medical Research Council ALL 97 trial samples; T.L. and E.R. managed and maintained the UKCCS sample data; M.T. performed curation and sample preparation of UKCCS samples; J.M.A. and J.A.E.I. performed ascertainment, curation, and sample preparation of Northern Institute for Cancer Research case series; and R.S.H. and M.G. obtained funding and designed parent project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard S. Houlston, Section of Cancer Genetics, Institute of Cancer Research, Sutton, Surrey, SM2 5NG, United Kingdom; e-mail: Richard.houlston@icr.ac.uk.