Abstract
MicroRNAs (miRs) play an important role in cell differentiation and maintenance of cell identity, but relatively little is known of their functional role in modulating human hematopoietic lineage differentiation. Human embryonic stem cells (hESCs) provide a model system to study early human hematopoiesis. We differentiated hESCs by embryoid body (EB) formation and compared the miR expression profile of undifferentiated hESCs to CD34+ EB cells. miRs-126/126* were the most enriched of the 7 miRs that were up-regulated in CD34+ cells, and their expression paralleled the kinetics of hematopoietic transcription factors RUNX1, SCL, and PU.1. To define the role of miRs-126/126* in hematopoiesis, we created hESCs overexpressing doxycycline-regulated miRs-126/126* and analyzed their hematopoietic differentiation. Induction of miRs-126/126* during both EB differentiation and colony formation reduced the number of erythroid colonies, suggesting an inhibitory role of miRs-126/126* in erythropoiesis. Protein tyrosine phosphatase, nonreceptor type 9 (PTPN9), a protein tyrosine phosphatase that is required for growth and expansion of erythroid cells, is one target of miR-126. PTPN9 restoration partially relieved the suppressed erythropoiesis caused by miRs-126/126*. Our results define an important function of miRs-126/126* in negative regulation of erythropoiesis, providing the first evidence for a role of miR in hematopoietic differentiation of hESCs.
Introduction
The hematopoietic system is hierarchically organized, with a rare population of hematopoietic stem cells giving rise to specific progenitor cells and ultimately to diverse mature blood cell types. Lineage-specific fate decisions are intricately controlled by sets of master transcription factors.1 For example, erythroid versus myeloid lineage differentiation is based on transcriptional cross-antagonism between GATA-1 and PU.1.2 PU.1 and C/EBPα are required for the generation of both macrophages and neutrophils.3 In addition to transcription factors that are pivotal for hematopoietic cell lineage specification, several developmentally conserved signaling pathways, including Notch, Wnt, Sonic hedgehog, and TGF-β, have also been implicated in regulating lineage determination.4
MicroRNAs (miRs) are a recently discovered class of small (20-22 nt) noncoding RNAs that control gene expression post-transcriptionally by regulating mRNA translation or stability.5 The roles of miRs in regulating the hematopoietic system are still poorly understood, although a series of recent studies have provided some perspective on this important issue.6 There are several miRs, including miR-142, miR-181a, and miR-223, that are prominently expressed in hematopoietic cells.7,8 Ectopic expression of miR-181a in hematopoietic stem/progenitor cells promotes B-cell differentiation, whereas overexpression of miR-142 and miR-223 leads to an increase in the proportion of T-lineage cells.7 Some studies have not only delineated a role for miRs in hematopoiesis but also uncovered their molecular mechanisms of action involving regulation of lineage determining transcription factors. For example, the combination of gain-of-function and loss-of-function strategies has defined the requirement of miR-150 in B-cell differentiation by targeting the transcription factor c-Myb.9 During monocytopoiesis, down-regulation of miRs 17-5p-20a-106a relieves the suppression of AML1, thus facilitating monocytic differentiation and maturation.10
In addition to the role of specific miRs, the general requirement of miRs in hematopoiesis has been demonstrated, as deletion of Dicer in the thymus results in aberrant T-cell development in the murine system.11,12 There is expanding literature suggesting that miRs also play an important role in the pathology of hematologic malignancies as tumor suppressor genes or oncogenes. For example, both miR-15a and miR-16 are deleted or down-regulated in the majority of chronic lymphocytic leukemia cases,13 whereas C57BL6 mice transplanted with murine hematopoietic progenitor cells overexpressing miR-155 developed a myeloproliferative disorder.14
Given compelling evidence that miRs are essential in the hematopoietic system, we sought to elucidate the potential roles of miRs in early human hematopoietic development. Insights into hematopoietic development have largely relied on findings from gene knockout and transgenic mouse models as well as in other model organisms. Because of the inaccessibility of human embryos, human embryonic stem cells (hESCs) provide a unique tool for the study of gene function during the early stages of lineage specification and commitment. hESCs are pluripotent cells derived from the inner cell mass of embryos at the blastocyst stage of development.15 They have the ability to extensively proliferate and differentiate into all cell types in the human body, including a range of hematopoietic cell lineages.16 Hematopoietic differentiation of hESCs has been achieved by either formation of embryoid bodies (EBs) or coculture with stromal cells. Several studies indicate that hematopoietic commitment in differentiating hESCs parallels many aspects of embryonic hematopoiesis, making it a useful in vitro model to study early hematopoiesis.17-19
In this study, we identified miRs-126/126* as miRs highly enriched in CD34+ EB cells. We investigated their role in hematopoiesis with inducible miRs-126/126* overexpressing hESCs and revealed an inhibitory role of miRs-126/126 in erythropoiesis. We then identified protein tyrosine phosphatase, nonreceptor type 9 (PTPN9) as one target of miR-126 and showed that restored expression of PTPN9 partially rescued the phenotype. Our results underscore an important role for miRs in human hematopoietic lineage fate determination and demonstrate the value of hESCs in studying human hematopoietic development.
Methods
hESC culture and EB differentiation
Culture and EB differentiation of hESCs were performed as previously described.20 Briefly, hESC lines H1 (WiCell Research Institute) and HSF1 (University of California, San Francisco, CA) were maintained as undifferentiated cells by weekly passage with 1 mg/mL collagenase IV (Invitrogen) onto γ-irradiated mouse embryonic fibroblast feeder layers in Dulbecco modified Eagle medium/F-12 medium (Invitrogen) supplemented with 0.1mM nonessential amino acids (Invitrogen), 2mM l-glutamine (Invitrogen), 20% knockout serum replacement (Invitrogen), 0.1mM 2-mercaptoethanol (Sigma-Aldrich), and 10 ng/mL basic fibroblast growth factor (PeproTech). Medium was changed daily. To generate EBs, day 6 hESCs were incubated with 0.5 mg/mL dispase (Invitrogen) dissolved in Dulbecco modified Eagle medium/F-12 for 20 minutes at 37°C. Colonies were collected from the plates, washed once with EB differentiation medium consisting of Iscove minimal essential medium, 15% defined fetal bovine serum (HyClone Laboratories), 2mM l-glutamine, 0.1mM 2-mercaptoethanol, and plated in 6-well ultra low attachment plates (Corning Life Sciences) in EB differentiation medium. The next day, cultured EB cells were rinsed and given fresh medium. EB cells were cultured for up to 25 days with medium and doxycycline (Dox) treatment changes every other day.
Construction of lentiviral vectors and production of lentivirus
Vector pLTET1 and pLTRET-Luc were kindly provided by Dr MA Goodell (Baylor College of Medicine, Houston, TX) and were described previously.21 To generate vector pLTRET-hsa-miRs-126/126*, a fragment coding luciferase within pLTRET-Luc was released and replaced with genomic sequence coding hsa-miRs-126/126*. Primers for cloning miRs-126/126*: TAATGGATCCGGAATCTGGGCGGAAGGCGGTG; ATAAGAATGCGGCCGCATCGATAGAGCCAGAAGACTCAGGCCCAG; DNA fragment coding hsa-miRs-126/126* contains 185 bp upstream and 179 bp downstream from the miRs-126/126* precursor. A vector encoding the human PTPN9 cDNA was kindly provided by Dr GP Downey (University of Toronto, Toronto, ON). Amplified PTPN9 cDNA was digested with Sbf I and Age I and cloned into a lentiviral vector F-GFP-IRES-Neo (kindly provided by Dr J. A. Zack, University of California, Los Angeles, CA) to replace green fluorescent protein (GFP) and was called F-PTPN9-IRES-Neo. Production and titration of lentiviruses were performed in 293T cells as described previously.22
Transduction of hESCs
RtTA2SM2-expressing hESCs were derived by transduction of passage 40 H1 cells with lentiviruses pLTET1. Briefly, H1 cells were treated with 10μM ROCK inhibitor Y27632 (Tocris Bioscience) in hESC medium for 1 hour. Colonies were dissociated into single cells by incubation with 0.05% trypsin for 10 minutes at 37°C and passed through a 40-μm filter. Single hESCs and viral particles were then incubated together in an Eppendorf tube at multiplicity of infection 5 in hESC medium with 10 μg/mL polybrene and 10μM ROCK inhibitor Y27652 at 37°C for 4 hours, after which the cells were rinsed with hESC medium and plated onto fresh feeder plates. GFP+ colonies were manually picked and expanded to establish stable cell lines. Inducible miR-overexpressing cells were derived by transduction of H1/TET1 cells with lentiviruses pLTRET-hsa-miRs-126/126*. PTPN9 overexpressing cells were derived by transduction of inducible miRs-126/126* cells with lentiviruses F-PTPN9-IRES-Neo. Cells were then selected by culturing in medium supplemented with 100 μg/mL G418. After 2 weeks in culture, G418-resistant colonies were picked and expanded to establish stable cell lines.
Flow cytometric analysis and sorting
EB cells were dissociated into single-cell suspensions by incubation with 0.25% trypsin/2% chicken serum for 30 minutes at 37°C followed by passage through a 40-μm filter. Cells were then resuspended in phosphate-buffered saline/3% fetal bovine serum and stained for 15 minutes on ice with the following fluorochrome-conjugated monoclonal antibodies: CD34-allophycocyanin (APC), CD34-phycoerythrin-Cy5, CD45-APC, CD71-phycoerythrin-Cy5, CD235a-APC, annexin V-biotin (BD Biosciences). Cells were then washed with phosphate-buffered saline/3%fetal bovine serum and analyzed with a BD FACSCanto. For isolation of CD34+ and CD34− differentiated EB cells, dissociated single EB cells were stained with CD34-APC antibody and sorted on a BD FACSAria II.
CFU assays and Wright-Giemsa staining
Colony-forming unit (CFU) assays were performed by plating dissociated single EB cells harvested at various days of differentiation at a density of 5 × 104/plate or 2.5 × 104/plate into methylcellulose H4435 (StemCell Technologies). After 12 to 14 days of incubation at 37°C and 5% CO2 in a humidified atmosphere, colonies were scored. Some colonies were picked individually from methylcellulose culture, washed with phosphate-buffered saline, and spun onto slides with a cytospin apparatus. Cell were fixed and stained with Hema 3 stain set (Fisher Scientific)
miR profiling
The human mature miR expression profiling was performed using the TaqMan Low Density Array Human miRNA Panel containing 365 human mature miRs (Applied Biosystems) and an ABI PRISM 7900HT Real-Time PCR System. The analyzed samples include H1 hESCs, day 7 and 15 H1 EB cells, and a mixture of 3 adult bone marrow CD34+ cells (AllCells). The data were analyzed with SDS RQ Manager 1.2 (Applied Biosystems).
Real-time quantitative PCR
Total RNA was prepared from cells using the mirVana miRNA isolation kit (Ambion) according to the manufacturer's instructions. For mRNA quantitative polymerase chain reaction (PCR), RNA was subjected to reverse transcription with Superscript III (Invitrogen) and random primers according to protocols provided by the manufacturer. Oligonucleotide primer sequences for the subsequent PCR are shown in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Real-time PCR was performed using the iCycler IQ System and IQ SYBR Green Supermix (Bio-Rad). Human glyceraldehyde-3-phosphate dehydrogenase was used as the internal control. miR quantitative PCR was performed with TaqMan miR RT reagent and specific primers for each miR. The transcripts were amplified with TaqMan 2 times Universal PCR Master Mix (Applied Biosystems). RNU44 and 48b were used as the internal control. Each quantitative PCR reaction was done in triplicate, and relative expression was calculated using the comparative threshold cycle method.
Luciferase miR target reporter assay
For luciferase reporter experiments, a 550-bp fragment of the 3′-untranslated region (UTR) of the human PTPN9 predicted to interact with miR-126 was amplified by PCR from human genomic DNA. The PCR products were cloned into the sites of Sac I and Spe I in the luciferase reporter pMir-Report (Ambion). The predicted miR-126 seed regions were mutated or deleted using the Quickchange II XL Mutagenesis Kit (Stratagene). The 293T cells were cotransfected in 96-well plates with the reporter construct and miR-126 precursor or negative pre-miR control using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations. The β-galactosidase plasmid was used an internal control. Cells were lysed at 48 hours after transfection. Firefly luciferase and β-galactosidase activities were measured consecutively using a dual-light assay system. The results were expressed as relative activity.
Protein extraction, immunoprecipitation, and Western blot
Protein extraction, immunoprecipitation, and Western blot were performed as previously described.23 The antibodies used in this work were as follows: PTPN9 (sc-67049, Santa Cruz Biotechnology), PTPN9 (clone 291835, R&D Systems), and Erk2 (sc-154, Santa Cruz Biotechnology). Quantification of the Western blot data was performed using the National Institutes of Health ImageJ Version 1.43.
Statistical analysis
Values are mean plus or minus SD from the numbers of replicates described in legends to figures. Statistical significance was determined by Student t test with a significance threshold of P less than .05.
Results
Identifying miRs enriched in hESC-derived CD34+ cells associated with hematopoietic activity
hESC-derived hematopoietic cells have been identified by several cell surface markers or combinations of multiple cell surface markers.24-26 CD34 is considered the most inclusive marker for human hematopoietic stem and progenitor cells. hESC-derived CD34+ cells are highly enriched for hematopoietic colony-forming activity and give rise to both lymphoid and myeloid cells.25 However, the CD34+ population is heterogeneous. In addition to hematopoietic progenitors, it includes endothelial progenitors as well as hematoendothelial cells, which give rise to both hematopoietic and endothelial cells.27,28 We searched for miRs that were consistently up-regulated in CD34+ cells during EB differentiation.
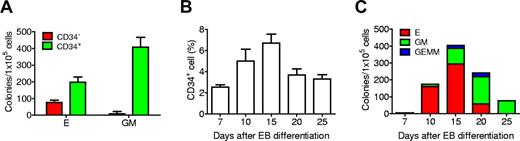
CD34+ and CD34− populations were isolated from day 15 EBs and subjected to CFU assay. Almost all the myeloid colonies arose from CD34+ cells. CD34+ selection also enriched for cells giving rise to erythroid colonies, although to a lesser extent (Figure 1A). This result is consistent with a previous report that CD34+ cells derived from hESC differentiation are highly enriched for cells with hematopoietic properties.25 To define the kinetics of hematopoiesis, EBs differentiated for up to 25 days were harvested and analyzed for expression of CD34 by fluorescence-activated cell sorter as well as colony formation capacity. Approximately 2.5% of EB cells were CD34+ at 7 days after differentiation, and levels increased to a peak on day 15 when this marker was expressed by 6.7% of cells (Figure 1B; supplemental Figure 1). Kinetic analysis of CFU emergence showed no colonies before day 7. On day 7, few erythroid colonies were detected. By day 10 of differentiation, the frequency of CFUs increased significantly, most of which were erythroid colonies. The total number of colonies peaked at day 15, after which the numbers of erythroid colonies as well as total hematopoietic colonies decreased. Myeloid colony numbers increased from day 10 to peak at day 20. On day 25, only myeloid colonies were found (Figure 1C).
Kinetic analysis of hematopoietic development from H1 EB differentiation. (A) CD34+ EB cells were highly enriched for hematopoietic colony-forming activity. (B) Expression kinetics of the cell surface marker CD34. (C) Hematopoietic colony-forming kinetics of differentiated EB cells. E indicates erythroid; GM, granulocyte-macrophage; and GEMM, granulocyte-erythroid-macrophage megakaryocyte. Data are mean ± SD (n = 3).
Kinetic analysis of hematopoietic development from H1 EB differentiation. (A) CD34+ EB cells were highly enriched for hematopoietic colony-forming activity. (B) Expression kinetics of the cell surface marker CD34. (C) Hematopoietic colony-forming kinetics of differentiated EB cells. E indicates erythroid; GM, granulocyte-macrophage; and GEMM, granulocyte-erythroid-macrophage megakaryocyte. Data are mean ± SD (n = 3).
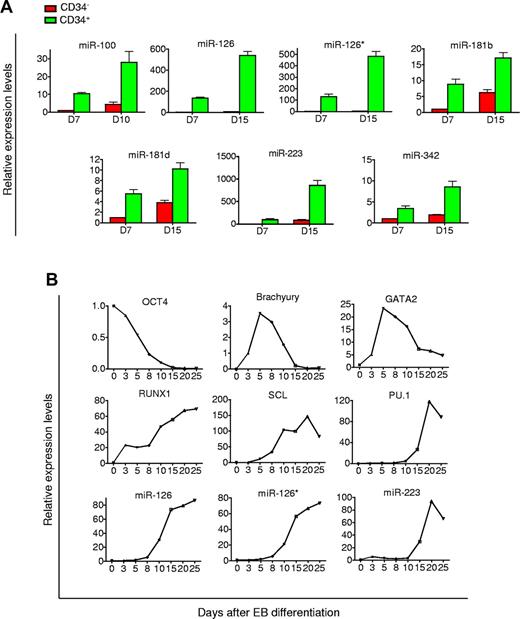
Based on these temporal patterns, we thus chose to harvest day 7 and 15 CD34+ EB cells and compared their miR profiles to H1 cells. Because of the heterogeneity of the CD34+ population, we included analysis of human bone marrow CD34+ cells to ensure that the identified miRs were truly characteristic of expression in hematopoietic cells. TaqMan miR array showed that 11 of 365 miRs were continuously up-regulated during 15 days of differentiation and also present in bone marrow CD34+ cells (supplemental Table 2). Real-time PCR analysis with specific primers verified the up-regulation. However, only 7 of these 11 miRs, including miR-100, 126, 126*,181b, 181d, 223, and 342 were enriched at least 2.5-fold in the CD34+ population compared with the CD34− population, whereas the other 4 were expressed at similar levels (Figure 2A). Interestingly, miR-126 and 126* are transcribed from the same miR precursor. Up-regulation and enrichment of these 7 miRs were also verified in CD34+ EB cells derived from HSF1 hESCs (data not shown). These results suggested that these 7 miRs might be associated with hematopoietic differentiation and play important roles in directing hematopoietic differentiation from hESCs.
Identification of miRs enriched in CD34+ EB cells. (A) miRs up-regulated and enriched in CD34+ cells derived from H1 EB differentiation. Levels of miRNA expression were determined by TaqMan real-time PCR analysis. Values of fold change in expression are relative to day 7 CD34− EB cells. (B) Kinetic analysis of pluripotency, mesodermal, hematopoietic genes, and miRs in differentiating EB cells. EBs were harvested on the indicated days, and levels of gene expression were determined by quantitative real-time PCR analysis. Values of fold change in expression are relative to undifferentiated H1 hESCs.
Identification of miRs enriched in CD34+ EB cells. (A) miRs up-regulated and enriched in CD34+ cells derived from H1 EB differentiation. Levels of miRNA expression were determined by TaqMan real-time PCR analysis. Values of fold change in expression are relative to day 7 CD34− EB cells. (B) Kinetic analysis of pluripotency, mesodermal, hematopoietic genes, and miRs in differentiating EB cells. EBs were harvested on the indicated days, and levels of gene expression were determined by quantitative real-time PCR analysis. Values of fold change in expression are relative to undifferentiated H1 hESCs.
Expression kinetics of miRs-126/126* and miR-223 during EB differentiation
miRs-126/126* and 223 were the most highly up-regulated miRs in CD34+ cells generated from EB differentiation. They have been implicated in the hematopoietic system, with miRs-126/126* being overexpressed in core binding factor acute myeloid leukemia and miR-223 playing an important in regulating granulopoiesis.29-31 We examined the dynamics of their expression along the 25-day EB differentiation regimen (Figure 2B). As hESCs differentiated, expression of the pluripotency marker OCT4 declined indicating a loss of pluripotency. BRACHYURY (T), a mesoderm marker, exhibited a significant increase 3 days after EB formation, and then quickly declined after day 5. Gene expression of GATA2, RUNX1, and SCL, which indicate commitment to hematopoietic fate, were up-regulated between days 3 to 5. Expression of PU.1, a transcription factor important for definitive hematopoietic cell differentiation, did not increase until after day 8. An increase in levels of miR-126, 126*, and 223 coincided with these hematopoietic transcriptional regulators. miR-126 and 126* were slowly induced in the first 5 days, after which there was an abrupt increase in their expression. The expression kinetics of miR-223 mirrored that of PU.1. The strong association between kinetics of hematopoietic transcription factors, and these miRs further suggested potential roles of these miRs in modulating hematopoietic differentiation from hESCs.
Generation of inducible miRs-126/126* overexpressing hESCs
Whereas miR-223 has been shown to regulate granulopoiesis,30,31 the roles of miRs-126/126* in hematopoiesis are less understood. To investigate the effects of miRs-126/126* on hematopoietic differentiation, we took advantage of a Dox-based conditional gene expression system.21 In this system, transduced hESCs retain pluripotentiality and demonstrate the ability to control the expression of a stable integrated transgene in a Dox-dependent manner. Conditional manipulation of gene expression enables us to analyze gene function at distinctive stages of hematopoietic differentiation from hESCs, which has not been shown previously, although it was reported by several groups in mouse embryonic stem cells.32,33
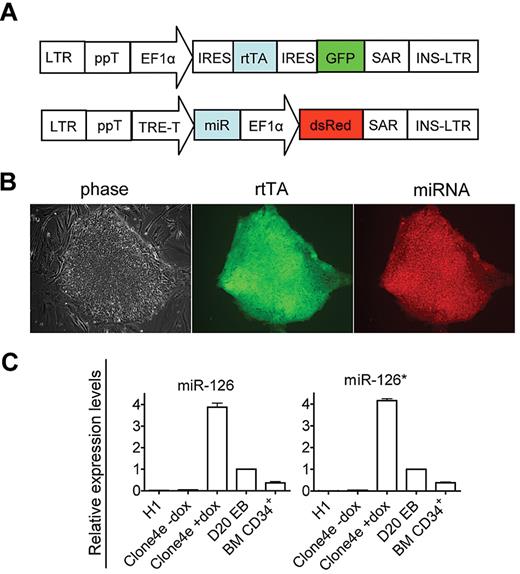
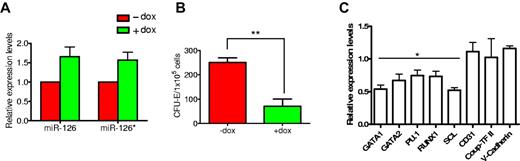
By sequentially infecting H1 cells with rtTA and inducible miRs-126/126* expressing lentiviral vectors (Figure 3A), we established several hESC lines conditionally expressing miRs-126/126* (I-miRs-126/126*). All experiments were performed on 2 independently derived lines, and they produced comparable results. Results from only one line are shown. In the undifferentiated state, I-miRs-126/126* cells displayed uniform GFP and dsRed expression and maintained normal hESC morphology (Figure 3B). They expressed robust levels of the stem cell markers Oct-4 and Tra1–81 and formed teratomas containing tissues of the 3 germ layers on injection into immunodeficient mice (supplemental Figure 2). These cells showed minimal “leaky” expression in the absence of Dox (Figure 3C). Addition of 1 μg/mL Dox induced robust expression of miRs-126/126* (Figure 3C). Levels of induced miR-126 and126* were approximately 4 fold of day 20 differentiated EB cells (Figure 3C). Induction of miRs-126/126* did not impair EB formation and the yields of EBs as well as the frequency of cells undergoing apoptosis were similar to control groups (supplemental Figure 3; and data not shown).
Generation of inducible miRs-126/126* overexpressing hESCs. (A) Schematic representation of pLTET1 and pLTRET1-miR lentiviral vectors used to derive inducible miR expressing hESCs (magnification 100×). Phase contrast and fluorescent micrographs of live cells in tissue culture media were acquired using a Zeiss Axiovert 200 model microscope with ECPlan Neofluor 10×/0.3 Ph1 objectives. Image was captured using an Optronics MagnaFIRE camera with MicroFIRE Version 070121-00X1 software. (B) Phase-contrast and immunofluorescence microscopy images of I-miRs-126/126* hESCs. (C) A total of 1 μg/mL Dox treatment induced miR-126 and 126* expression. miR expression is presented as fold induction relative to expression in day 20 EB cells.
Generation of inducible miRs-126/126* overexpressing hESCs. (A) Schematic representation of pLTET1 and pLTRET1-miR lentiviral vectors used to derive inducible miR expressing hESCs (magnification 100×). Phase contrast and fluorescent micrographs of live cells in tissue culture media were acquired using a Zeiss Axiovert 200 model microscope with ECPlan Neofluor 10×/0.3 Ph1 objectives. Image was captured using an Optronics MagnaFIRE camera with MicroFIRE Version 070121-00X1 software. (B) Phase-contrast and immunofluorescence microscopy images of I-miRs-126/126* hESCs. (C) A total of 1 μg/mL Dox treatment induced miR-126 and 126* expression. miR expression is presented as fold induction relative to expression in day 20 EB cells.
miRs-126/126* inhibit erythropoiesis
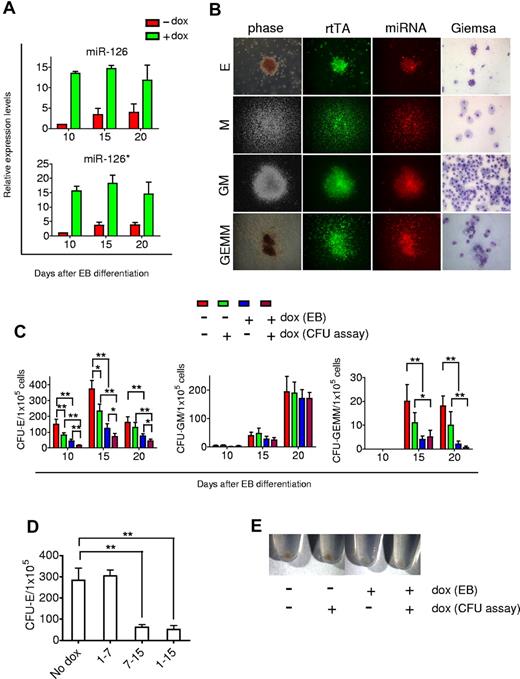
Having established the inducible miRs-126/126* expressing hESC lines, we examined how they regulated hematopoietic differentiation. I-miRs-126/126* cells were differentiated in the presence or absence of 1 μg/mL Dox over 20 days. Similar to H1 hESCs, levels of miRs-126/126* increased steadily in I-miRs-126/126* cells when no Dox was added. Dox treatment greatly induced miRs-126/126* expression. At day 10, levels were 12-fold higher than untreated control, and remained 3-fold higher at day 20 (Figure 4A).
Suppression of erythropoiesis by miRs-126/126*. (A) A total of 1 μg/mL Dox treatment induced expression of miRs-126/126* in differentiating EBs generated from I-miRs-126/126* hESCs. miR expression is presented as fold induction relative to expression in day 10 EB cells without Dox treatment. (B) Phase-contrast and fluorescence microscopic images taken from representative erythroid (E), macrophage (M), granulocyte-macrophage (GM), and mixed myelo-erythroid (GEMM) colonies derived from day 15 miRs-126/126* overexpressing EB cells. Images of E and GEMM colonies are 100× and of M and GM colonies, 50×. Wright-Geimsa images are 200×. Phase contrast and fluorescent micrographs of live cells in methylcellulose were acquired using a Zeiss Axiovert 200 model microscope with ECPlan Neofluor 5×/0.16 or 10×/0.3 Ph1 objectives. Wright-Geimsa–stained cytospins were imaged using a Nikon Eclipse E600 model microscope with PlanApo 20×/0.75 Ph2 DM objectives. Image was captured using an Optronics MagnaFIRE camera with MicroFIRE Version 070121-00X1 software. (C) Total number of hematopoietic CFU formed in methylcellulose. (D) CFU-E output of day 15 EB cells generated from different Dox treatments. (E) Appearance of the pellets of colonies grown in methylcellulose medium for 12 days. Data are mean ± SD (n = 4). *P < .05. **P < .01
Suppression of erythropoiesis by miRs-126/126*. (A) A total of 1 μg/mL Dox treatment induced expression of miRs-126/126* in differentiating EBs generated from I-miRs-126/126* hESCs. miR expression is presented as fold induction relative to expression in day 10 EB cells without Dox treatment. (B) Phase-contrast and fluorescence microscopic images taken from representative erythroid (E), macrophage (M), granulocyte-macrophage (GM), and mixed myelo-erythroid (GEMM) colonies derived from day 15 miRs-126/126* overexpressing EB cells. Images of E and GEMM colonies are 100× and of M and GM colonies, 50×. Wright-Geimsa images are 200×. Phase contrast and fluorescent micrographs of live cells in methylcellulose were acquired using a Zeiss Axiovert 200 model microscope with ECPlan Neofluor 5×/0.16 or 10×/0.3 Ph1 objectives. Wright-Geimsa–stained cytospins were imaged using a Nikon Eclipse E600 model microscope with PlanApo 20×/0.75 Ph2 DM objectives. Image was captured using an Optronics MagnaFIRE camera with MicroFIRE Version 070121-00X1 software. (C) Total number of hematopoietic CFU formed in methylcellulose. (D) CFU-E output of day 15 EB cells generated from different Dox treatments. (E) Appearance of the pellets of colonies grown in methylcellulose medium for 12 days. Data are mean ± SD (n = 4). *P < .05. **P < .01
EBs were then dissociated into single cells and subjected to hematopoietic cell surface marker analysis. Flow cytometry showed that overexpression of miRs-126/126* did not change the frequency of CD34+, CD45+, or CD34+CD45+ cells (supplemental Figure 4). Dissociated EB single cells, which contain hematopoietic progenitors, were then plated in methylcellulose medium to form colonies. A similar spectrum of colony types was observed with miRs-126/126* induction, and these colonies were uniformly double positive for GFP and dsRed (Figure 4B). Surprisingly, induction of miRs-126/126* during EB formation resulted in a 3- to 4-fold reduction in the erythroid colony numbers at all time points examined (Figure 4C). Numbers of granulocyte-erythroid-macrophage megakaryocyte colonies, which contained erythroid cells as well as other myeloid lineage cells, were also reduced (Figure 4C). In contrast, induction of miRs-126/126* did not affect the number of myeloid colonies (Figure 4C). No obvious changes were observed in gross morphology of cells composing colonies derived from miRs-126/126* overexpressing EB cells (Figure 4B).
As shown earlier, the emergence of hematopoietic progenitors occurred at day 7 of EB differentiation. To determine whether miRs-126/126* function before or after hematopoietic commitment, we induced miRs-126/126* expression from day 1 to day 7 or from day 7 to day 15 of EB differentiation and analyzed erythroid colony formation potential at the end of day 15. There was no change in the number of erythroid colonies compared with control noninduced EB cells when miRs-126/126* were induced from day 1 to day 7. In contrast, induction of miRs-126/126* from day 7 to day 15 fully recapitulated the extent of inhibition when miRs-126/126* was induced from day 1 to 15 (Figure 4D), suggesting an effect after initiation of hematopoietic commitment.
We then asked how miRs-126/126* influenced hESC-derived hematopoietic progenitors by adding Dox to the methylcellulose medium. Induction of miRs-126/126* further reduced erythroid and granulocyte-erythroid-macrophage megakaryocyte colony numbers by approximately 2-fold but had no effect on the number of myeloid colonies whether or not these EB cells had been exposed to Dox earlier (Figure 4C). Despite the further reduction, miRs-126/126* induction in the methylcellulose cultures did not change the morphology of cells composing colonies (data not shown). The inhibition of erythropoiesis by miRs-126/126* induction was made evident by the reduction of red color when cells grew in methylcellulose medium from 4 experimental conditions were spun down (Figure 4E). Despite a reduction in the numbers of erythroid colonies on miRs-126/126* induction, sizes of erythroid colonies, expression of various globins, and frequency of erythroid colony-forming cells undergoing apoptosis were similar under all conditions (data not shown). Importantly, addition of Dox to H1 or H1/pTET cells did not interfere with either hematopoietic commitment during EB formation or hematopoietic progenitor proliferation and differentiation (data not shown).
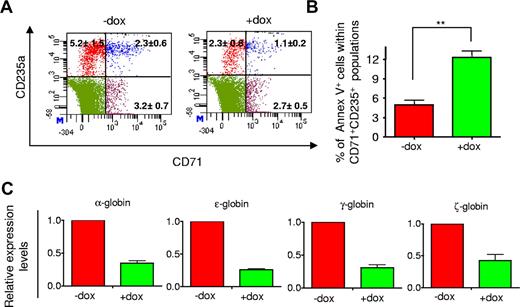
To further characterize the inhibited erythropoiesis, dissociated day 15 EB cells were analyzed by fluorescence-activated cell sorter for expression of erythoid cell-surface markers CD71 (transferrin receptor) and CD235a (glycophorin A). In response to Dox, we observed a significant decrease in the frequency of cells expressing these surface markers (Figure 5A). To determine whether the reduction was associated with increased cell death, we analyzed apoptosis levels of CD71+CD235a+ cells with annexin V staining. The frequency of CD71+CD235a+ annexin V+ cells was approximately 2.5-fold higher in the presence of Dox than the control (Figure 5B). Consistent with the inhibited erythropoiesis, miRs-126/126* induction also suppressed expression of embryonic and fetal globin genes by 3- to 5-fold (Figure 5C).
miRs-126/126* reduced the frequency of erythroid progenitors during EB differentiation. (A) Representative flow cytometric analysis of cell surface markers CD71 and CD235a of day 15 EB cells. Numbers indicate the percentage of cells within each category. (B) miRs-126/126* increased levels of apoptosis of CD71+CD235a+ cells of day 15 EBs. (C) miRs-126/126* induction suppressed expression of embryonic and fetal globins of day 15 EB cells. Values of fold change in expression are relative to cells without Dox treatment. Data are mean ± SD (n = 3). **P < .01.
miRs-126/126* reduced the frequency of erythroid progenitors during EB differentiation. (A) Representative flow cytometric analysis of cell surface markers CD71 and CD235a of day 15 EB cells. Numbers indicate the percentage of cells within each category. (B) miRs-126/126* increased levels of apoptosis of CD71+CD235a+ cells of day 15 EBs. (C) miRs-126/126* induction suppressed expression of embryonic and fetal globins of day 15 EB cells. Values of fold change in expression are relative to cells without Dox treatment. Data are mean ± SD (n = 3). **P < .01.
The inhibitory effect of miRs-126/126* is unexpected because initially our strategy was aimed to search for miRs that are enriched in hematopoietic cells and function as enhancers of hematopoietic development. We examined expression of miRs-126/126* in purified hematopoietic progenitors and erythroid cells from human first trimester placenta and second trimester fetal liver. miRs-126/126* were detected at significantly lower levels in both primitive and definitive erythroid cells compared with both endothelial and hematopoietic progenitors (supplemental Figure 5). Altogether, these data suggest that sustained expression of miRs-126/126* beyond their normal time frame inhibits erythroid differentiation.
miRs-126/126* inhibit erythropoiesis from CD34+ EB cells
Because cells composing the EBs are heterogeneous, to verify the induction of miRs-126/126* in CD34+ EB cells and exclude the possibility that effects of miRs-126/126* were solely the result of cell nonautonomous patterning, we fractionated dissociated day 15 EB cells based on expression of CD34. Dox treatment up-regulated miRs-126/126* in the CD34+ population (Figure 6A). Fractionated CD34+ EB cells were then subjected to CFU assays. Similar to the whole EB cells, induction of miRs-126/126* inhibited the capacity of CD34+ cells to form erythroid colonies (Figure 6B) but had no effect on myeloid colony formation (data not shown).
miRs-126/126* inhibit erythroid colony formation of CD34+ EB cells. (A) Dox treatment induced miRs-126/126* expression in day 15 CD34+ EB cells. (B) miRs-126/126* inhibit CFU-E capacity of CD34+ EB cells. Data are mean ± SD (n = 4). **P < .01. (C) Hematopoietic and endothelial gene expression in CD34+ EB cells. Expression values are relative to cells without Dox treatment. *P < .05.
miRs-126/126* inhibit erythroid colony formation of CD34+ EB cells. (A) Dox treatment induced miRs-126/126* expression in day 15 CD34+ EB cells. (B) miRs-126/126* inhibit CFU-E capacity of CD34+ EB cells. Data are mean ± SD (n = 4). **P < .01. (C) Hematopoietic and endothelial gene expression in CD34+ EB cells. Expression values are relative to cells without Dox treatment. *P < .05.
Expressions of transcripts important for hematopoietic differentiation were then compared between CD34+ cells with or without Dox treatment. miRs-126/126* induction led to approximately 50% reduction in the expression of GATA1 and SCL, known to be critical for erythroid differentiation. GATA2, PU.1, and RUNX1, which are not erythroid specific, were also down-regulated, although to a lesser extent (Figure 6C). In contrast, expression of endothelial genes CD31, COUP-TFII, and VE-CADHERIN was not affected by miRs-126/126* induction (Figure 6C). This is consistent with the finding in mouse ESCs, where overexpression of mouse miRs-126/126* does not promote differentiation toward an endothelial lineage.34
PTPN9 is a target of miR-126
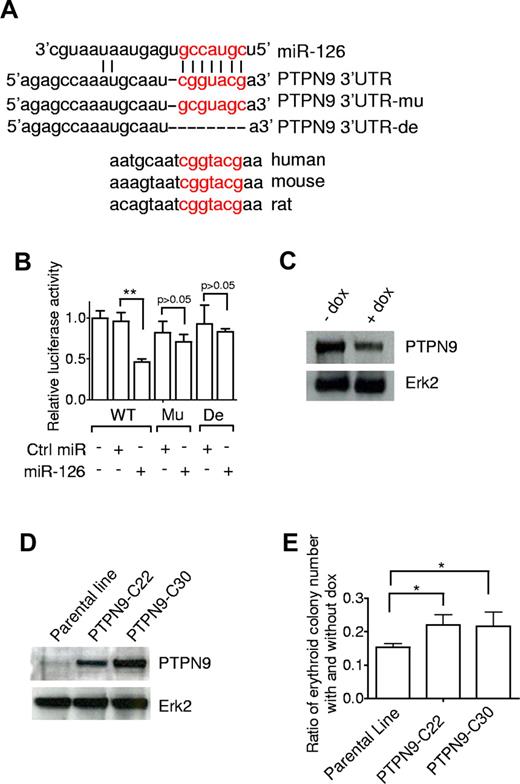
To elucidate the mechanism for inhibiting erythroid potential, we evaluated miRs-126/126* targets computationally predicted by publicly available programs. PTPN9 is one of the highest scored among the predicted miR-126 targets, and the predicted miR-126 binding site carries an identical sequence in human, mouse, and rat mRNA orthologs (Figure 7A). Interestingly, PTPN9 has been shown to play an important role in the development of erythroid cells.35 To test that PTPN9 is a miR-126 target, a standard luciferase assay was set up in which a fragment of the 3′-UTR of PTPN9 that contains the seed region was inserted downstream of a luciferase open reading frame. Constructs containing a mutated or deleted sequence of the seed region were produced as controls. Transfection of miR-126 precursor down-regulated the luciferase activity of the wild-type reporter constructs. This effect was dependent on the seed region in the PTPN9 mRNA 3′-UTR, as mutation or deletion of this site relieved the repressive effect of miR-126 (Figure 7B). To confirm miR-126-mediated modulation of endogenous PTPN9, day 15 EB cells with or without Dox treatment were lysed and subjected to immunoprecipitation and Western blot analysis. Induction of miRs-126/126* decreased PTPN9 protein expression by approximately 60% (Figure 7C).
PTPN9 is a target of miR-126 and can partially rescue the suppression of erythropoiesis. (A) Predicted binding site for the seed sequence of miR-126 in the 3′-UTR of PTPN9 mRNA. The seed region of miR-126 (top) matches the 3′-UTR of PTPN9. Mutated (mu) and deleted (de) forms of the 3′-UTR of PTPN9 used for creating the luciferase reporter construct are shown. (B) miR-126 inhibited wild-type, but not mutated or deleted PTPN9–3′-UTR reporter activity. The 293T cells were transfected with miR-126 (or control nontargeting oligonucleotide) and luciferase construct containing different forms of PTPN9–3′-UTR as indicated. After 36 hours of incubation, cells were subjected to luciferase assay. (C) Western blot analysis of PTPN9 expression in day 15 EB cells with or without Dox treatment; ERK2 levels are shown as an internal control. (D) Western blot analysis of PTPN9 expression in parental hESCs (I-miRs-126/126*) and 2 PTPN9 overexpressing hESC lines. (E) Restored expression of PTPN9 attenuated the suppressed erythropoiesis. I-miRs-126/126* and PTPN9 overexpressing hESCs were differentiated with or without Dox for 15 days. Dissociated single EB cells were then subjected to CFU assay. The number of erythroid colonies under both conditions was counted and the ratio was determined. Data are mean ± SD (n = 5). *P < .05. **P < .01.
PTPN9 is a target of miR-126 and can partially rescue the suppression of erythropoiesis. (A) Predicted binding site for the seed sequence of miR-126 in the 3′-UTR of PTPN9 mRNA. The seed region of miR-126 (top) matches the 3′-UTR of PTPN9. Mutated (mu) and deleted (de) forms of the 3′-UTR of PTPN9 used for creating the luciferase reporter construct are shown. (B) miR-126 inhibited wild-type, but not mutated or deleted PTPN9–3′-UTR reporter activity. The 293T cells were transfected with miR-126 (or control nontargeting oligonucleotide) and luciferase construct containing different forms of PTPN9–3′-UTR as indicated. After 36 hours of incubation, cells were subjected to luciferase assay. (C) Western blot analysis of PTPN9 expression in day 15 EB cells with or without Dox treatment; ERK2 levels are shown as an internal control. (D) Western blot analysis of PTPN9 expression in parental hESCs (I-miRs-126/126*) and 2 PTPN9 overexpressing hESC lines. (E) Restored expression of PTPN9 attenuated the suppressed erythropoiesis. I-miRs-126/126* and PTPN9 overexpressing hESCs were differentiated with or without Dox for 15 days. Dissociated single EB cells were then subjected to CFU assay. The number of erythroid colonies under both conditions was counted and the ratio was determined. Data are mean ± SD (n = 5). *P < .05. **P < .01.
Having confirmed that miR-126 negatively regulates PTPN9 protein levels, we asked whether PTPN9 is the key mediator of miRs-126/126*. PTPN9 lacking its 3′-UTR was introduced into I-miRs-126/126* hESCs and 2 lines with different levels of PTPN9 overexpression were established and subjected to the CFU assay (Figure 7D). Overexpression of PTPN9 resulted in a modest yet significant recovery in erythropoiesis in the presence of miRs-126/126* overexpression (Figure 7E). These results suggest that, although PTPN9 is a target of miR-126, it alone is not sufficient to rescue the effects that miRs-126/126* have on erythropoiesis. The function of miRs-126/126* in inhibition of erythropoiesis must involve other targets that act in concert with PTPN9.
Discussion
In this study, we explored the role of miRs in the regulation of human hematopoietic differentiation with a hESC EB system. We showed that a set of miRs, including miR-100, 126, 126*, 181b, 181d, 223, and 342 were consistently up-regulated in CD34+ EB cells, which indicates that these 7 miRs may represent a common feature of hematopoietic differentiation from hESCs. Among them, miRs-126/126*,29,36 181b,37 181d,38 22330,31 and 34239 have a well-characterized association with the hematopoietic system and miRs-126/126* are the most highly up-regulated. miR-126 and 126* are processed from the same primary miR and located in an intron of Egfl7, a gene that is highly expressed in endothelial cells.40 They are found to be highly enriched in endothelial cells derived from mouse embryonic stem cells.34 Although the role of miR-126* is less understood, miR-126 was shown to play a critical role in governing vascular integrity and angiogenesis.34,41 The identification of miR-126 in this study is consistent with the notion that CD34 is also a marker for endothelial cells. Indeed, in addition to miR-126 and 126*, miR-100, and 181b are also expressed in endothelial cells.42
Our results demonstrated an inhibitory role of miRs-126/126* in erythropoietic development. Although we searched for miRs that are constantly up-regulated in CD34+ cells and function as enhancers of hematopoietic development, we unexpectedly found an inhibitory role for the highly expressed miRs-126/126*. We further showed that miRs-126/126* are highly expressed in hematopoietic progenitors and down-regulated as differentiation progresses toward erythroid cells in vivo. The hematopoietic differentiation process requires temporal regulation of transcription factors and signaling pathways. For example, a recent study revealed the dynamic changes of requirements in Wnt, Activin, and BMP signaling during blood development using a mouse ESC differentiation model.43 The development of hematopoietic cells from ESCs involves sequential steps: mesoderm induction, hematoendothelial cell formation, and hematopoietic specification. We showed that induction of miRs-126/126* before the emergence of hematopoietic progenitors does not suppress erythropoiesis, which indicates that miRs-126/126* unlikely suppress commitment to mesoderm and/or hematoendothelial cells. In agreement with this conclusion, expression of the mesoderm marker BRACHYURY and the hematoendothelial cell marker KDR were not affected by miRs-126/126* induction (data not shown). Combined with the observation that induction of miRs-126/126* does not interfere with the commitment of GM progenitors, it implies that miRs-126/126* most likely have a direct effect on the commitment to erythroid progenitors and their survival. Taken together, our data provide an example of a dynamic requirement of miR in hematopoietic differentiation.
Erythropoiesis is a complex developmental process that is regulated by various factors and transcriptional networks.44 Several studies have recently described roles of miRs during this development process. miR-221 and 222 are down-regulated in erythropoietic culture of human cord blood CD34+ progenitor cells, which promotes erythropoiesis by unblocking expression of kit receptor expression.45 In contrast, miR-451 is significantly up-regulated during erythroid differentiation. It plays a crucial role in promoting erythroid maturation, in part via its target Gata-2.46 Our results have added miRs-126/126* to this expanding repertoire and strongly demonstrate that expression of miRs must be properly regulated to control erythroid differentiation.
We identified and experimentally verified PTPN9 as a bona fide target of miR-126. PTPN9 is a nonreceptor tyrosine phosphatase and is required for growth and expansion of normal erythroid cells. It is hyperactivated in the erythroid progenitor cells in patients with polycythemia vera (PV).35 However, restored expression of PTPN9 only modestly rescued the suppressed erythropoiesis. Our result suggests that in addition to PTPN9, other downstream target(s) must be suppressed by miRs-126/126* to inhibit erythropoiesis. Because PTPN9 plays an important role in PV, our finding suggests that it might be worthwhile to examine the potential role of miRs, especially miRs-126/126*, in this disease. Indeed, aberrant expression of miRs was found in peripheral blood cells from patients with PV.47 Our finding suggests that delivery of miRs-126/126* may provide a strategy to treat PV.
In our EB differentiation system, miRs-126/126* are induced in every EB cell. It is possible that overexpression of miRs-126/126* in nonhematopoietic cells might impact the differentiation of hematopoietic cells. By fractionating EB cells, we showed that miRs-126/126* are induced in CD34+ cells and their induction can inhibit the erythropoietic potential of CD34+ cells. Despite this, nonautonomous effects of miRs-126/126* still could not be ruled out. Further insight into this question will require the expression of miRs-126/126* specifically in erythroid progenitors.
Because the genomic fragment cloned into the lentiviral vector encoded both miR-126 and 126*, another unsolved issue raised by this study is to dissect the respective roles of miR-126 and 126* in erythropoiesis. It was assumed that, during the processing of miR, one strand of the RNA duplex preferentially binds to the silencing complex, whereas the other strand (miR* or passenger strand) is degraded, representing a functionally irrelevant carrier strand. However, miR* species are detected in increasing numbers with large-scale small RNA sequencing efforts. The selective or coaccumulation of both strands of miRs is highly tissue dependent and both strands of a miR pair can functionally suppress the expression of their target genes.48,49 We have found in this study that, in hESCs-derived CD34+ cells, miR-126* was present at significant levels at approximately 12.5% of miR-126 (both endogenous and ectopic expression; data not shown). Because restored expression of miR-126 target PTPN9 only modestly rescued the phenotype, one explanation is that miR-126* also plays a role.
In conclusion, our studies reveal a unique role for miRs-126/126* as a negative regulator of erythroid development. This observation suggests that miRs might provide a common mechanism in hematopoietic lineage fate determination.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Amander Clark and Dr Gay Crooks for critical reading of the manuscript, Dr Margaret Goodell for providing vector pLTET1 and pLTRE-Luc, Dr Gregory Downey for providing PTPN9 cDNA, Dr Jerome Zack for providing vector F-GFP-IRES-Neo, and Barbara Anderson for help with preparation of the manuscript.
This work was supported by CIRM RT1-01022 Tool/Technologies Award and CIRM New Faculty Award RN1-00557-1 (H.K.A.M). The Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at University of California, Los Angeles provided partial support for this study. B.V.H was supported by the National Institutes of Health (Ruth Kirschstein National Research Service Award GM007185). O.N.W. is an investigator of the Howard Hughes Medical Institute.
National Institutes of Health
Authorship
Contribution: X.H. designed and performed experiments and wrote the paper; E.G., B.V.H., and D.C. performed experiments; H.K.A.M. designed experiments; and O.N.W. designed experiments and wrote the paper.
Conflict-of-interest disclosure: B.V.H. is a founder of and owns a significant financial stake in Novogenix Laboratories LLC. The remaining authors declare no competing financial interests.
Correspondence: Owen N. Witte, Howard Hughes Medical Institute, 675 Charles E. Young Dr S, Los Angeles, CA 90095; e-mail: owenwitte@mednet.ucla.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal