Abstract
We evaluated concurrent gene mutations, clinical outcome, and gene expression signatures of CCAAT/enhancer binding protein alpha (CEBPA) double mutations (CEBPAdm) versus single mutations (CEBPAsm) in 1182 cytogenetically normal acute myeloid leukemia (AML) patients (16-60 years of age). We identified 151 (12.8%) patients with CEBPA mutations (91 CEBPAdm and 60 CEBPAsm). The incidence of germline mutations was 7% (5 of 71), including 3 C-terminal mutations. CEBPAdm patients had a lower frequency of concurrent mutations than CEBPAsm patients (P < .0001). Both, groups were associated with a favorable outcome compared with CEBPAwt (5-year overall survival [OS] 63% and 56% vs 39%; P < .0001 and P = .05, respectively). However, in multivariable analysis only CEBPAdm was a prognostic factor for favorable OS outcome (hazard ratio [HR] 0.36, P < .0001; event-free survival, HR 0.41, P < .0001; relapse-free survival, HR 0.55, P = .001). Outcome in CEBPAsm is dominated by concurrent NPM1 and/or FLT3 internal tandem duplication mutations. Unsupervised and supervised GEP analyses showed that CEBPAdm AML (n = 42), but not CEBPAsm AML (n = 18), expressed a unique gene signature. A 25-probe set prediction signature for CEBPAdm AML showed 100% sensitivity and specificity. Based on these findings, we propose that CEBPAdm should be clearly defined from CEBPAsm AML and considered as a separate entity in the classification of AML.
Introduction
In the current World Health Organization classification of acute myeloid leukemia (AML), AML with mutated CCAAT/enhancer binding protein alpha (AML with mutated CEBPA), has been designated as a provisional disease entity in the category “AML with recurrent genetic abnormalities.”1,2
CEBPA encodes a transcription factor that is essential for neutrophil development. Targeted disruption of Cebpa in mice results in a selective block in early granulocyte development, which is a hallmark of AML.3,4 Two proteins may be translated from the CEBPA transcripts, such as a 42-kDa (p42) and a shorter 30-kDa (p30) protein both translated from the same mRNA transcript. The p42 isoform contains 2 regulatory transactivation domains (TAD) in the N-terminus (TAD1 and TAD2), whereas the shorter p30 isoform only carries the TAD2 domain. Both isoforms contain the C-terminal basic DNA-binding domain and the leucine zipper (bZIP), involved in DNA binding and protein dimerization. In AML, CEBPA mutations mainly occur in cytogenetically normal AML (CN-AML) with an incidence of 5% to 14%.5-14 The 2 main types of mutations can be distinguished: N-terminal frame-shift mutations resulting in the translation of a 30-kDa protein only and the C-terminal in-frame mutations in the basic zipper region affecting DNA binding and homodimerization and heterodimerization.8,15 As a consequence, these mutations create an imbalance between proliferation and differentiation of hematopoietic progenitors.10,16
Patients who have AML with CEBPA mutations can be separated into 2 subgroups, namely, those with a single mutation CEBPA (CEBPAsm) and those with a double mutation CEBPA (CEBPAdm).17-21 In the majority of CEBPAdm AML, both alleles are mutated.19 These biallelic mutations frequently consist of an N-terminal mutation on one allele and a C-terminal bZIP mutation on the other. In CEBPAsm AML, mutations occur either in the N terminus or in the C terminus of the gene. In previous studies in which these 2 subgroups were not considered, AML patients with mutated CEBPA had a relatively good outcome.5,7,12,13 More recent data suggest that this favorable outcome is mainly observed in AML patients with CEBPAdm and not CEBPAsm.17-21 Moreover, it has been suggested that concurrent mutations may occur more frequently in CEBPAsm than in CEBPAdm AML. The impact of coexisting mutations remains elusive and needs to be validated in large cohorts.
By applying gene expression profiling (GEP), it was demonstrated that CEBPAdm AML can be distinguished from CEBPAsm and the majority of CEBPAwt AML based on a signature.18 However, a CEBPAdm GEP signature did not predict CEBPAdm AML with maximum accuracy because AML in which CEBPA was silenced (CEBPAsilenced) by promoter hypermethylation carried a highly similar signature.22,23
The objectives of this study were to evaluate the impact of CEBPAdm versus CEBPAsm on clinical outcome of CN-AML and to investigate the impact of concurrent mutations in nucleophosmin (NPM1mutant) and/or fms-like tyrosine kinase receptor-3 (FLT3) internal tandem duplication mutations (FLT3ITD). In addition, we searched for CEBPA-associated gene expression signatures and determined the frequency of predisposing CEBPA germline mutations. For these purposes, we combined datasets from the Dutch-Belgian Hemato-Oncology Cooperative Group (HOVON)-Swiss Group for Clinical Cancer Research (SAKK) and the German and Austrian AML Study Group (AMLSG).
Methods
Patients and molecular analyses
Diagnostic bone marrow or peripheral blood samples from 1182 younger adults (16 to 60 years of age) with CN-AML were analyzed; 193 patients were enrolled on HOVON-SAKK protocols -04, -04A, -29, and -42 (available at www.hovon.nl),24-27 and 989 patients on AMLSG protocols AML HD93 (n = 74),28 AML HD98A (n = 313),29 AMLSG 07-04 (n = 376), registered at www.clinicaltrials.gov as NCT00151242, AML SHG 02-95 (n = 94),30 and AML SHG 01-99 (n = 180), registered at www.clinicaltrials.gov as NCT00209833). All patients provided written informed consent in accordance with the Declaration of Helsinki. All trials were approved by the Institutional Review Board of Erasmus University Medical Center, University of Ulm, and Hannover Medical School.
Mutation analyses for the genes FLT3ITD and FLT3 tyrosine kinase domain (TKD) mutations (FLT3TKD) and the NPM1 were performed as described previously.31-33 CEBPAdm and CEBPAsm AML were identified by denaturing high-performance liquid chromatography or direct sequencing as described.18 Cases that carried an insertion polymorphism18,21 (www.ncbi.nlm.nih.gov/sites/snp; genome.ucsc.edu/cgi-bin/hgGateway; www.ensembl.org/Homo_sapiens/Gene/Variation_Gene) or variations that did not lead to amino acid changes were considered wild-type (wt). Cases were categorized as CEBPAdm when 2 different mutations or 1 homozygous mutation were present as determined by sequencing analysis; cases with only a single heterozygous mutation were designated as CEBPAsm. In 71 of the 151 patients with CEBPA mutations, DNA obtained from buccal swabs (n = 52), peripheral blood (n = 8), or bone marrow (n = 11) samples in complete remission (CR) were studied for the presence of CEBPA germline mutations. Patient demographics and molecular characteristics are summarized in Table 1. All CEBPA-mutated patients, except for patients treated within the AMLSG protocol 07-04, have been previously reported in different studies.7,13,18
Patient demographics and clinical and molecular characteristics of CEBPAwt, CEBPAsm, and CEBPAdm CN-AML cases
| Characteristic . | CEBPAwt (n = 1031) . | CEBPAsm (n = 60) . | P, CEBPAsm vs CEBPAwt . | CEBPAdm (n = 91) . | P, CEBPAdm vs CEBPAwt . | P, CEBPAsm vs CEBPAdm . |
|---|---|---|---|---|---|---|
| Median age, y (range) | 48 (16-60) | 46 (18-60) | .28 | 44 (16-60) | .04* | .66 |
| Sex, n (%) | .79 | .74 | .74 | |||
| Male | 500 (48) | 28 (47) | 46 (51) | |||
| Female | 531 (52) | 32 (53) | 45 (49) | |||
| WBC count, ×109/L | .23 | .062 | .86 | |||
| Median (range) | 28 (0.2-372) | 25 (1.1-345) | 28 (1.5-262) | |||
| Missing | 34 | 1 | 4 | |||
| Platelet count, ×109/L | .77 | < .0001* | < .0001* | |||
| Median (range) | 65 (5-746) | 62 (10-361) | 38 (4-265) | |||
| Missing | 40 | 3 | 4 | |||
| Bone marrow blasts | .83 | .53 | .76 | |||
| Median (range) | 80 (0-100) | 80 (0-97) | 78 (2-100) | |||
| Missing | 80 | 7 | 4 | |||
| Molecular abnormalities | ||||||
| FLT3ITD, n (%) | 347 (33.7) | 18 (30) | 1 | 7 (7.7) | < .0001* | .00015* |
| Missing | 69 | 9 | 5 | |||
| FLT3TKD, n (%) | 95 (9.2) | 4 (6.7) | .81 | 2 (2.2) | .018* | .2 |
| Missing | 48 | 6 | 3 | |||
| NPM1, n (%) | 560 (54.3) | 21 (35) | .018* | 3 (3.3) | 0 | < .0001* |
| Missing | 88 | 10 | 8 |
| Characteristic . | CEBPAwt (n = 1031) . | CEBPAsm (n = 60) . | P, CEBPAsm vs CEBPAwt . | CEBPAdm (n = 91) . | P, CEBPAdm vs CEBPAwt . | P, CEBPAsm vs CEBPAdm . |
|---|---|---|---|---|---|---|
| Median age, y (range) | 48 (16-60) | 46 (18-60) | .28 | 44 (16-60) | .04* | .66 |
| Sex, n (%) | .79 | .74 | .74 | |||
| Male | 500 (48) | 28 (47) | 46 (51) | |||
| Female | 531 (52) | 32 (53) | 45 (49) | |||
| WBC count, ×109/L | .23 | .062 | .86 | |||
| Median (range) | 28 (0.2-372) | 25 (1.1-345) | 28 (1.5-262) | |||
| Missing | 34 | 1 | 4 | |||
| Platelet count, ×109/L | .77 | < .0001* | < .0001* | |||
| Median (range) | 65 (5-746) | 62 (10-361) | 38 (4-265) | |||
| Missing | 40 | 3 | 4 | |||
| Bone marrow blasts | .83 | .53 | .76 | |||
| Median (range) | 80 (0-100) | 80 (0-97) | 78 (2-100) | |||
| Missing | 80 | 7 | 4 | |||
| Molecular abnormalities | ||||||
| FLT3ITD, n (%) | 347 (33.7) | 18 (30) | 1 | 7 (7.7) | < .0001* | .00015* |
| Missing | 69 | 9 | 5 | |||
| FLT3TKD, n (%) | 95 (9.2) | 4 (6.7) | .81 | 2 (2.2) | .018* | .2 |
| Missing | 48 | 6 | 3 | |||
| NPM1, n (%) | 560 (54.3) | 21 (35) | .018* | 3 (3.3) | 0 | < .0001* |
| Missing | 88 | 10 | 8 |
Number of cases (percentage), median, range, or missing values are indicated.
WBC indicates white blood cell.
P < .05 computed using the Mann-Whitney U test (continuous variables) and 2-sided Fisher exact test (categorical variables).
Gene expression profiling
Data from GEP analysis were available in 674 AML patients (53% CN-AML, HOVON-SAKK and AMLSG cohorts), generated using Affymetrix (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Sample processing and quality control were carried out as described previously.23,34 For both cohorts, normalization of raw data was processed with Affymetrix Microarray Suite 5 (MAS5) to target intensity values at 100. Intensity values were log2 transformed, and mean centered per probe set per cohort. GEP data are available at the Gene Expression Omnibus (National Center for Biotechnology Information; accession number GSE14468 [HOVON-SAKK cohort] and GSE22845 [AMLSG cohort]). There were 42 CEBPAdm and 18 CEBPAsm cases for which the GEP was determined (supplemental Table 1).
Statistical analyses
Statistical analyses were performed using Mathworks (Matlab R2009b) with the statistical, bioinformatics, and pattern recognition toolbox (Prtools). For clinical, molecular, univariate, and multivariate analyses, patients with CN-AML and age ≤ 60 years (supplemental Table 1) were included. Molecular and clinical variables of both patient cohorts (HOVON-SAKK and AMLSG) were comparable. Differences were assessed for CEBPAsm and CEBPAdm groups in comparison with CEBPAwt group (Table 1), using the Mann-Whitney U test for continuous variables and the 2-sided Fisher exact test for categorical variables.
Outcome measures of the HOVON-SAKK and AMLSG cohorts were comparable (log-rank test overall survival [OS], P = .08; event-free survival [EFS], P = .47; supplemental Figure 1A-B, for respective cohort). There were no statistical differences in outcome in patients receiving autologous or allogeneic hematopoietic stem cell transplantation between the HOVON-SAKK and AMLSG cohorts (log-rank test OS, P = .68; EFS, P = .89; supplemental Figure 2A-B, respectively).
For univariate analysis, significance was assessed using the stratified log-rank test and Kaplan-Meier estimates for OS, EFS and relapse-free survival (RFS). The recommended consensus criteria35 were used for the definition of CR and survival end points such as OS, EFS, and RFS. Multivariate analysis was performed using stratified Cox proportional hazard model. For all analyses, a value for P ≤ .05 was considered statistically significant and for survival analyses, values for P were computed using the full time span. Note that the closed testing procedure36 was applied, and a correction for multiple testing37 was only performed if the global log-rank test resulted in P > .05.
For gene expression–based classification of CEBPAdm cases, GEP of the HOVON-SAKK cohort was used to derive the 25-probe set predictive signature and the AMLSG cohort as validation set. To summarize, a logistic regression model with Lasso regularization (a continuous feature selection procedure) was used because it takes the correlation structure between the probe sets into account (see supplemental “creation and evaluation of the CEBPAdm predictive signature”).
Results
Frequency and type of acquired CEBPAdm and CEBPAsm mutations
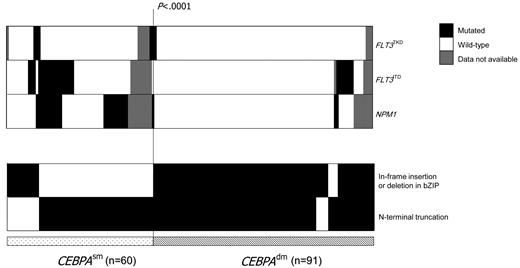
CEBPA mutations were detected in 151 of the 1182 (12.8%) CN-AML patients; 91 (60%) had CEBPAdm among which the combination of N- and C-terminal mutations was the predominant genotype (82 of the 91). CEBPAdm cases with only N- or C-terminal mutations were less frequently observed (4 of the 91 or 5 of the 91, respectively). A total of 60 of 151 (40%) CEBPA-mutated cases had CEBPAsm, which occurred most frequently in the N terminus (47 of the 60). Only 13 of the 60 CEBPAsm cases had in-frame insertion or deletion mutations affecting the bZIP domain (Figure 1).
Distribution of concurrent mutations in CEBPAdm and CEBPAsm patients. Columns represent patients with CEBPA mutations (91 CEBPAdm and 60 CEBPAsm) patients and rows represent the genotypes FLT3TKD, FLT3ITD, and NPM1mutant (filled black), wild-type (open), or missing (filled gray). The in-frame insertion or deletion in bZIP and N-terminal truncation mutations in CEBPA is shown and highlighted in black.
Distribution of concurrent mutations in CEBPAdm and CEBPAsm patients. Columns represent patients with CEBPA mutations (91 CEBPAdm and 60 CEBPAsm) patients and rows represent the genotypes FLT3TKD, FLT3ITD, and NPM1mutant (filled black), wild-type (open), or missing (filled gray). The in-frame insertion or deletion in bZIP and N-terminal truncation mutations in CEBPA is shown and highlighted in black.
CEBPA germline mutation analysis
Five of 71 (7%) CEBPA mutant AML patients analyzed carried CEBPA germline mutations: in 2 of the 5 patients, the germline mutation was localized in the N-terminus, and both acquired a C-terminal mutation. Both patients had a family history of AML and were diagnosed at a young age. In the remaining 3 patients, the germline mutation was in the C terminus; 1 of these patients gained an additional N-terminal mutation and the second patient an additional C-terminal mutation at the time of AML-diagnosis. None of these 3 patients had a family history of AML. Alignment to distinct Single Nucleotide Polymorphism databases (www.ncbi.nlm.nih.gov/sites/snp; genome.ucsc.edu; or www.ensembl.org/Homo_sapiens/Gene/Variation_Gene) did not identify one of these germline sequence variations as a polymorphism. Using the PolyPhen database (http://genetics.bwh.harvard.edu/pph/), all 3 C-terminal mutations were predicted to be damaging to the function and structure of the protein (Table 2).
Germline patient demographics and molecular characteristics
| Patient ID . | Age at diagnosis, y . | Germline mutation . | Acquired mutation* . | Additional mutation† . | Familial AML history . | CEBPA mutation . |
|---|---|---|---|---|---|---|
| 98A-751 | 28 | 338delC | 1080insGAA | None | Yes | CEBPAdm |
| 07/04-268 (ULM_10) | 25 | 307delG | 1122_1123ins1075_1225 | KRAS, WT1 | Yes | CEBPAdm |
| BioID 769 | 51 | 1096T>C | 478_485del | None | No | CEBPAdm |
| 98A-543 | 33 | 1164G>A | None | FLT3TKD, NPM1 | No | CEBPAsm |
| 07/04-48 (ULM_20) | 59 | 1036G>T | 1086insAAG | None | No | CEBPAdm |
| Patient ID . | Age at diagnosis, y . | Germline mutation . | Acquired mutation* . | Additional mutation† . | Familial AML history . | CEBPA mutation . |
|---|---|---|---|---|---|---|
| 98A-751 | 28 | 338delC | 1080insGAA | None | Yes | CEBPAdm |
| 07/04-268 (ULM_10) | 25 | 307delG | 1122_1123ins1075_1225 | KRAS, WT1 | Yes | CEBPAdm |
| BioID 769 | 51 | 1096T>C | 478_485del | None | No | CEBPAdm |
| 98A-543 | 33 | 1164G>A | None | FLT3TKD, NPM1 | No | CEBPAsm |
| 07/04-48 (ULM_20) | 59 | 1036G>T | 1086insAAG | None | No | CEBPAdm |
Characteristics of 5 of 71 (7%) CEBPA mutant AML patients who carried CEBPA germline mutations. CBL indicates Casitas B-lineage lymphoma; KRAS, Kirsten Rat sarcoma; NRAS, neuroblastoma Rat sarcoma; RUNX1, runt-related transcription factor 1; and WT1, Wilms tumor 1
Data according to GenBank accession no. Y11525.
Patients 98A-751, 07/04-268 (ULM_10), 98A-543, and 07/04-48 (ULM_20) were screened for FLT3ITD, FLT3TKD, NPM1, NRAS, KRAS, WT1, RUNX1, and CBL mutations; patient BioID 769 was analyzed for FLT3ITD, FLT3TKD, and NPM1 mutations.
Association of acquired CEBPAdm and CEBPAsm mutations with concurrent gene mutations and clinical characteristics
Concurrent mutations were seen less frequently in CEBPAdm than in CEBPAsm AML (22% vs 60%; P < .0001, Figure 1); frequency for NPM1mutant in CEBPAdm versus CEBPAsm AML was 3.3% and 35% (P < .0001), and for FLT3ITD was 7.7% and 30% (P < .001), respectively (Table 1). Comparing CEBPAsm and CEBPAwt AML, NPM1mutant were slightly less frequent in CEBPAsm AML (35% vs 54.3%; P = .018); the frequency for FLT3ITD was comparable between the 2 groups (30% vs 33.7%).
Regarding presenting clinical characteristics, CEBPAdm mutations were associated with younger age (median age 44 vs 48 years; P = .04) and lower platelet counts (median number 38 × 109/L vs 65 × 109/L; P < .0001) compared with CEBPAwt patients (Table 1).
Impact of CEBPAdm and CEBPAsm on response to induction therapy and clinical outcome
For correlation with clinical outcome, 1182 CN-AML were considered. CEBPAdm was associated with a higher CR rate compared with CEBPAsm (92% vs 78%, P = .02) and with CEBPAwt (92% vs 79%, P = .002). There was no difference to CR probability between CEBPAsm and CEBPAwt patients (78% vs 79%, P = .86).
The median follow-up time for survival in the 1182 CN-AML patients was 33 months (95% confidence interval [CI], 25.6-0.4); the estimated 5-year OS and RFS were 42% (95% CI, 39%-45%) and 34% (95% CI, 31%-38%), respectively.
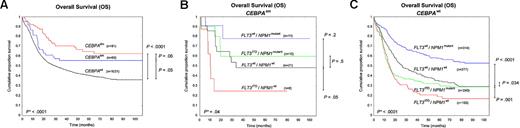
CEBPAdm AML was associated with a significantly superior outcome compared with CEBPAwt AML (5-year OS, 63% vs 39%, P < .0001; EFS, 45% vs 28%, P < .0001; RFS, 44% vs 32%, P = .05), as shown (Figure 2A and supplemental Figure 3A,D). A somewhat better outcome was also found for CEBPAsm AML compared with CEBPAwt AML (5-year OS, 55% vs 39%, P = .05; RFS, 49% vs 32%, P = .02; but not EFS, 37% vs 28%, P = .22). No significant difference was evident between CEBPAdm and CEBPAsm AML (5-year OS, P = .06; EFS, P = .16; RFS, P = .48). Of note, no differences in outcome were observed among CEBPAsm patients with either C-terminal (n = 13) or N-terminal (n = 47) mutations (5-year OS, 54% vs 56%, P = .58; supplemental Figure 4).
Kaplan-Meier survival curves for OS. (A) Kaplan-Meier survival curves for OS among the 3 groups CEBPAdm, CEBPAsm, and CEBPAwt. (B) Kaplan-Meier survival curves for OS of the 4 genotypes FLT3ITD/NPM1mutant, FLT3ITD/NPM1wt, FLT3wt/NPM1mutant, and FLT3wt/NPM1wt within the CEBPAsm group. (C) Kaplan-Meier survival curves for OS of the 4 genotypes within CEBPAwt. *P value by global log-rank test.
Kaplan-Meier survival curves for OS. (A) Kaplan-Meier survival curves for OS among the 3 groups CEBPAdm, CEBPAsm, and CEBPAwt. (B) Kaplan-Meier survival curves for OS of the 4 genotypes FLT3ITD/NPM1mutant, FLT3ITD/NPM1wt, FLT3wt/NPM1mutant, and FLT3wt/NPM1wt within the CEBPAsm group. (C) Kaplan-Meier survival curves for OS of the 4 genotypes within CEBPAwt. *P value by global log-rank test.
In multivariate analyses considering other prognostic indicators (Table 3), the presence of CEBPAdm was an independent prognostic factor for favorable OS (HR 0.36, P < .0001), EFS (HR 0.41, P < .0001), and RFS (HR 0.55, P = .001), whereas CEBPAsm did not impact these 3 end points (Table 3).
Multivariate analysis for overall, event-free, and relapse-free survival in CN-AML patients
| Variable* . | HR . | 95% CI . | P . |
|---|---|---|---|
| Overall survival | |||
| CEBPAsmα | 0.70 | 0.46-1.07 | .10 |
| CEBPAdmα | 0.36 | 0.23-0.55 | < .0001† |
| FLT3ITDβ | 1.78 | 1.49-2.14 | < .0001† |
| FLT3TKDβ | 0.84 | 0.61-1.15 | .28 |
| NPM1β | 0.56 | 0.46-0.67 | < .0001† |
| WBC countδ, ×109/L | 1.35 | 1.12-1.62 | < .0001† |
| Ageϵ | 1.02 | 1.01-1.03 | < .0001† |
| Event-free survival | |||
| CEBPAsmα | 0.86 | 0.60-1.22 | .40 |
| CEBPAdmα | 0.41 | 0.29-0.57 | < .0001† |
| FLT3ITDβ | 1.56 | 1.33-1.84 | < .0001† |
| FLT3TKDβ | 0.80 | 0.60-1.07 | .13 |
| NPM1β | 0.45 | 0.39-0.53 | < .0001† |
| WBC countδ, ×109/L | 1.27 | 1.08-1.50 | .003† |
| Ageϵ | 1.01 | 1.01-1.02 | .003† |
| Relapse-free survival | |||
| CEBPAsmα | 0.79 | 0.51-1.22 | .30 |
| CEBPAdmα | 0.55 | 0.38-0.79 | .001† |
| FLT3ITDβ | 1.75 | 1.45-2.12 | < .0001† |
| FLT3TKDβ | 0.82 | 0.59-1.13 | .22 |
| NPM1β | 0.56 | 0.46-0.68 | < .0001† |
| WBC countδ, ×109/L | 1.33 | 1.10-1.61 | .002† |
| Ageϵ | 1.01 | 1.00-1.02 | .001† |
| Variable* . | HR . | 95% CI . | P . |
|---|---|---|---|
| Overall survival | |||
| CEBPAsmα | 0.70 | 0.46-1.07 | .10 |
| CEBPAdmα | 0.36 | 0.23-0.55 | < .0001† |
| FLT3ITDβ | 1.78 | 1.49-2.14 | < .0001† |
| FLT3TKDβ | 0.84 | 0.61-1.15 | .28 |
| NPM1β | 0.56 | 0.46-0.67 | < .0001† |
| WBC countδ, ×109/L | 1.35 | 1.12-1.62 | < .0001† |
| Ageϵ | 1.02 | 1.01-1.03 | < .0001† |
| Event-free survival | |||
| CEBPAsmα | 0.86 | 0.60-1.22 | .40 |
| CEBPAdmα | 0.41 | 0.29-0.57 | < .0001† |
| FLT3ITDβ | 1.56 | 1.33-1.84 | < .0001† |
| FLT3TKDβ | 0.80 | 0.60-1.07 | .13 |
| NPM1β | 0.45 | 0.39-0.53 | < .0001† |
| WBC countδ, ×109/L | 1.27 | 1.08-1.50 | .003† |
| Ageϵ | 1.01 | 1.01-1.02 | .003† |
| Relapse-free survival | |||
| CEBPAsmα | 0.79 | 0.51-1.22 | .30 |
| CEBPAdmα | 0.55 | 0.38-0.79 | .001† |
| FLT3ITDβ | 1.75 | 1.45-2.12 | < .0001† |
| FLT3TKDβ | 0.82 | 0.59-1.13 | .22 |
| NPM1β | 0.56 | 0.46-0.68 | < .0001† |
| WBC countδ, ×109/L | 1.33 | 1.10-1.61 | .002† |
| Ageϵ | 1.01 | 1.00-1.02 | .001† |
Stratified Cox proportional hazard ratio (HR) model for multivariable analyses of CEBPAdm and CEBPAsm as prognostic marker for overall survival, event-free survival, and relapse-free survival. Analyses included 1182 CN-AML patients ≤ 60 years of age.
WBC indicates white blood cell.
Subgroup α CEBPA status vs CEBPAwt β FLT3ITD vs no FLT3ITD mutation β FLT3TKD vs no FLT3TKD mutation β NPM1 vs no NPM1 δ. WBC count > 20 × 109/L vs < 20 × 109/L ϵ. Age is used as a continuous variable.
P ≤ .05.
Treatment outcome of AML with CEBPAsm is dominated by FLT3/NPM1 genotypes
Finally, we performed explorative subgroup analyses in CEBPAsm and CEBPAwt AML to evaluate the impact of 4 FLT3/NPM1 genotype subgroups: FLT3ITD/NPM1mutant (n = 10); FLT3ITD/NPM1wt (n = 8); FLT3wt/NPM1mutant (n = 11); and FLT3wt/NPM1wt (n = 21). There were 10 cases from the CEBPAsm group excluded for which the genotypes were unknown.
Among patients with CEBPAsm AML, the FLT3ITD/NPM1wt genotype had an inferior OS compared with patients with the FLT3wt/NPM1wt genotype (5-year OS, 25% vs 49%, P = .05; Figure 2B); for EFS and RFS, there was a trend toward an inferior outcome (supplemental Figure 3B,E); in contrast, the FLT3wt/NPM1mutant genotype associated in trend with a favorable outcome compared with the FLT3wt/NPM1wt genotype (5-year OS, 78% vs 49%, P = .2; EFS, 59% vs 32%, P = .08; RFS, 66% vs 40%, P = .38; Figure 2B and supplemental Figure 3B,E). In analogy, in the CEBPAwt group, the FLT3ITD/NPM1wt genotype had a significantly inferior survival compared with the FLT3wt/NPM1wt genotype (5-year OS, 17% vs 34%, P = .001; EFS, 11% vs 14%, P = .04; RFS, 15% vs 24%, P = .002; Figure 2C and supplemental Figure 3C,F), whereas the FLT3wt/NPM1mutant genotype was associated with a favorable outcome (5-year OS, 57% vs 34%, P < .0001; EFS, 47% vs 14%, P < .0001; RFS, 50% vs 24%, P < .0001; Figure 2C and supplemental Figure 3C,F). Thus, we observed comparable trends for favorable (FLT3wt/NPM1mutant) and inferior (FLT3ITD/NPM1wt) outcome in the CEBPAsm and CEBPAwt subgroups. The outcome for all CEBPAsmFLT3/NPM1 genotypes was higher (not significantly, P > .05), compared with the CEBPAwt genotypes; however, the distinct groups were relatively small. For CEBPAdm AML, sample sizes of the composite genotypic subgroups were too small for analysis.
Unsupervised analyses of GEP showed homogeneity in CEBPAdm AML cases
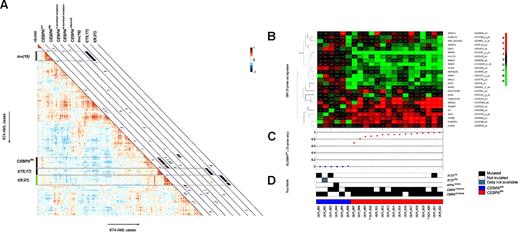
GEP was performed in a subset of the CN-AML patients and includes cytogenetically abnormal patients (n = 674) as shown (supplemental Table 1). Unsupervised analyses, by computing pairwise Pearson correlation coefficients of 674 AML cases, revealed distinct GEP clusters (Figure 3A), including the known clusters of AML with inv(16), t(15;17), or t(8;21), as shown previously.23 These subtypes revealed high correlation within the GEP cluster (average correlation, 0.42, 0.49, and 0.49, respectively) and differed significantly (P < .0001) among the AML cases without any of these aberrations, (supplemental Figure 5B-C and supplemental Figure 5E). We observed that the CEBPAdm AML cases were highly similar within the cluster (average correlation, 0.35) and differed significantly from cases without a CEBPAdm (P < .0001; supplemental Figure 5D). CEBPAsm AML cases showed reduced similarity (average correlation, 0.15) and did not differ from cases without CEBPAsm (P = .12; supplemental Figures 3A,5A).
Unsupervised analyses and classification results of candidate CEBPAdm cases with their GEP and their molecular characteristics. (A) Pairwise correlations between the 674 AML cases (supplemental Table 1). The cells in the visualization are colored by Pearson correlations values, depicting higher positive (red) or negative (blue) correlations, as indicated by the scale bar. CEBPAsm, CEBPAdm, CEBPAC-terminal mutation, CEBPAN-terminal mutation, CEBPAsilenced, together with inv(16), t(15;17), and t(8;21) cases are depicted on the diagonal with a red or blue colored bar. CEBPAC-terminal mutation and CEBPAN-terminal mutation indicates the presence of homozygous mutations. (B) Candidate CEBPAdm patients and the unambiguous CEBPAsm patients. The expression levels are defined by the 25-probe set signature. The colors of the hierarchical clustering are relative to the mean. (C) Computed posterior probabilities, indicating the prediction of a CEBPAdm case, given the 25 predictive probe set signature: P(CEBPAdm 25-probe sets). The ordering of patients is based on the classification probabilities. (D) True labels (molecular characteristics). Filled lanes indicate the mutation status in CEBPA (CEBPAdm or CEBPAsm), NPM1 (NPM1mutant), or FLT3 (TKD or ITD), open lanes represents no mutation in the particular patient, and a missing value is depicted in gray. *Germline CEBPAdm cases.
Unsupervised analyses and classification results of candidate CEBPAdm cases with their GEP and their molecular characteristics. (A) Pairwise correlations between the 674 AML cases (supplemental Table 1). The cells in the visualization are colored by Pearson correlations values, depicting higher positive (red) or negative (blue) correlations, as indicated by the scale bar. CEBPAsm, CEBPAdm, CEBPAC-terminal mutation, CEBPAN-terminal mutation, CEBPAsilenced, together with inv(16), t(15;17), and t(8;21) cases are depicted on the diagonal with a red or blue colored bar. CEBPAC-terminal mutation and CEBPAN-terminal mutation indicates the presence of homozygous mutations. (B) Candidate CEBPAdm patients and the unambiguous CEBPAsm patients. The expression levels are defined by the 25-probe set signature. The colors of the hierarchical clustering are relative to the mean. (C) Computed posterior probabilities, indicating the prediction of a CEBPAdm case, given the 25 predictive probe set signature: P(CEBPAdm 25-probe sets). The ordering of patients is based on the classification probabilities. (D) True labels (molecular characteristics). Filled lanes indicate the mutation status in CEBPA (CEBPAdm or CEBPAsm), NPM1 (NPM1mutant), or FLT3 (TKD or ITD), open lanes represents no mutation in the particular patient, and a missing value is depicted in gray. *Germline CEBPAdm cases.
CEBPAdm AML is accurately predicted based on GEP
The previously predictive CEBPAdm signature18 was hampered by the recently reported CEBPAsilenced AML cases that carry a similar GEP.22 The 2 independent AML cohorts were used to train and evaluate the predictive value of the CEBPAdm signature in terms of sensitivity and specificity. A predictive signature was created (Figure 3B and supplemental Table 2), containing 25-probe sets, using a logistic regression model with Lasso regularization38,39 that selects discriminative probe sets between the classes, CEBPAdm (n = 26) and all other AML cases, CEBPAwt and CEBPAsm (n = 494). Subsequently, a classifier was trained on the entire HOVON-SAKK cohort based on a 2-class approach; with 26 CEBPAdm versus 494 other (CEBPAwt and CEBPAsm) cases. This trained classifier subsequently classified 16 candidate CEBPAdm cases (supplemental Table 3) in the AMLSG cohort of 154 AML cases (16 CEBPAdm, 6 CEBPAsm, and 132 CEBPAwt; supplemental Table 1). Among the CEBPAdm cases were 5 with either homozygous N- or C-terminal CEBPAdm mutations, and a CEBPAdm patient with a germline C-terminal mutation. This approach showed perfect sensitivity and specificity (both 100%; Figure 3C). In addition, we performed a classification among CEBPAdm, CEBPAsm, and CEBPAwt to infer whether we were able to accurately classify CEBPAsm cases. We observed that all CEBPAsm cases were classified as CEBPAwt, thus CEBPAsm cases did not have a consistent gene expression pattern and were different from the CEBPAdm group.
Discussion
In this study, we established the value of CEBPAdm mutation in a large cohort of CN-AML patients from AMLSG and HOVON-SAKK treatment trials. Applying denaturing high-performance liquid chromatography and whole gene sequencing, we detected 91 (7.7%) CEBPAdm and 60 (5.1%) CEBPAsm mutations among 1182 patients. In multivariate analyses, we demonstrate that the presence of CEBPAdm but not CEBPAsm is an independent factor for favorable outcome in AML, which confirms previous findings reported in studies with relatively small cohorts.17-19,21
Concurrent mutations were significantly less frequent in CEBPAdm compared with CEBPAsm AML. This factor was true for FLT3ITD and in particular for NPM1mutant, which were virtually not present among CEBPAdm cases, a finding that is consistent with previously published data.20
Compared with previous studies17-21 and with the large number of cases, we were able to evaluate the prognostic impact of the CEBPA mutational status in the context of the FLT3/NPM1 genotypes. Among CEBPAsm AML, the 4 combined genotypes showed similar trend with regard to outcome compared with CEBPAwt AML (Figure 2B-C). Nevertheless, we observed a higher outcome (not significant) for all CEBPAsmFLT3/NPM1 genotypes compared with the CEBPAwt genotypes, but these groups are relatively small. These findings, supported by data from multivariable analysis, strongly suggest that not the existence of CEBPAsm per se but rather the combined effects of CEBPAsm and FLT3ITD and/or NPM1mutant determine outcome in these AML patients.
We have previously derived gene expression signatures that predict AML with inv(16), t(15;17), and t(8;21) with 100% accuracy. In this study, we generated a refined GEP signature of 25-probe sets that predict CEBPAdm AML cases (6 genes overlapped with the previous signature,18 as indicated in “Supplemental Materials”). This signature showed sensitivity and specificity of 100% and has a better predictive power than the CEBPAdm signature that we defined before.18 In fact, in contrast to the previous signature, the new signature also discriminates CEBPAdm from AML with hypermethylation of the proximal promoter region of CEBPA.22 Classification results were not affected by homozygous N- or C-terminal CEBPAdm mutations or because of germline mutation. Because this 25-probe set signature was optimized for classification it does not necessarily provide insight into the biologic meaning of CEBPAdm mutations.
Currently, nucleotide sequencing is used as the gold standard for the identification of CEBPA mutations. Because of the much higher effort required, the GEP technique should not be considered as a primary diagnostic tool in AML. However, GEP can be confirmatory, especially in cases in which the CEBPA gene appears difficult to sequence. More importantly, GEP provides relevant insights in the biology of the disease and the affected signaling pathways and therefore allows further classification/refinement of AML.
Finally, we evaluated the frequency of CEBPA germline mutations in this large cohort of CEBPA-mutated cases. Among 71 mutated patients, 5 revealed germline mutations. Of these cases, 4 developed CEBPAdm AML, that is, 4 cases acquired a mutation in the second allele. This finding is in line with previous data.40,41 Interestingly for the first time, we identified 3 C-terminal germline mutations of which 2 cases acquired a second CEBPA mutation at the time of AML diagnosis. In GEP analysis both cases clustered within the CEBPAdm group and were classified as a CEBPAdm, providing evidence that these C-terminal sequence variations are mutations rather than polymorphisms. In line with the GEP data, all 3 C-terminal germline mutations were predicted to be damaging for the function and the structure of the protein.
In the current World Health Organization classification of AML, AML with mutated CEBPA has been designated as a provisional disease entity in the category “AML with recurrent genetic abnormalities.” Based on our data obtained from a large patient cohort together with previous findings, we propose that CEBPAdm AML should be clearly distinguished from CEBPAsm AML and that only AML with CEBPAdm should be considered as an independent entity in the classification of the disease.
The online version of this article contains a data supplement.
Presented at the 15th Congress of the European Hematology Association, Barcelona, Spain, June 13, 2010.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Martin van Vliet and Jelle Goeman for the discussions.
This research was supported by the Center for Translational Molecular Medicine and by Else Kröner-Fresenius-Stiftung grant P38/05//A49/05//F03, the Network of Competence Acute and Chronic Leukemias grant 01GI9981, and the Bundesministerium für Bildung und Forschung, Germany, grant 01KG0605 (“IPD-meta-analysis: a model-based hierarchical prognostic system for adult patients with acute myeloid leukemia [AML]”).
Authorship
Contribution: E.T. performed research, data analysis, data interpretation, creation of the figures, and manuscript writing; L.B. and A.C. performed research, data analysis, and interpretation; M.A.S. performed data analysis, data interpretation, and manuscript writing; C.A.J.E., B.J.W., and S.C.v.d.P.-v.d.L. performed research; F.D. performed research and data interpretation; J.K. and A.G. provided study material; R.F.S. performed research, data interpretation, and manuscript writing; and B.L., R.D., H.D., P.J.M.V., and K.D. designed the study and performed data interpretation and manuscript writing.
Conflict-of-interest disclosure: B.L., R.D., and P.J.M.V. have declared ownership interests in Skyline, a spinoff company of Erasmus University Medical Center, held in a Special Purpose Foundation of Erasmus University Medical Center. The remaining authors declare no competing financial interests.
Correspondence: Konstanze Döhner, Department of Internal Medicine III, University of Ulm, Albert-Einstein-Allee 23, 89081 Ulm, Germany; e-mail: konstanze.doehner@uniklinik-ulm.de.
References
Author notes
P.J.M.V. and K.D. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal