Abstract
Philadelphia chromosome–negative myeloproliferative neoplasms (MPNs) are clonal myeloid disorders with increased production of terminally differentiated cells. The disease course is generally chronic, but some patients show disease progression (secondary myelofibrosis or accelerated phase) and/or leukemic transformation. We investigated chromosomal aberrations in 408 MPN samples using high-resolution single-nucleotide polymorphism microarrays to identify disease-associated somatic lesions. Of 408 samples, 37.5% had a wild-type karyotype and 62.5% harbored at least 1 chromosomal aberration. We identified 25 recurrent aberrations that were found in 3 or more samples. An increased number of chromosomal lesions was significantly associated with patient age, as well as with disease progression and leukemic transformation, but no association was observed with MPN subtypes, Janus kinase 2 (JAK2) mutational status, or disease duration. Aberrations of chromosomes 1q and 9p were positively associated with disease progression to secondary myelofibrosis or accelerated phase. Changes of chromosomes 1q, 7q, 5q, 6p, 7p, 19q, 22q, and 3q were positively associated with post-MPN acute myeloid leukemia. We mapped commonly affected regions to single target genes on chromosomes 3p (forkhead box P1 [FOXP1]), 4q (tet oncogene family member 2 [TET2]), 7p (IKAROS family zinc finger 1 [IKZF1]), 7q (cut-like homeobox 1 [CUX1]), 12p (ets variant 6 [ETV6]), and 21q (runt-related transcription factor 1 [RUNX1]). Our data provide insight into the genetic complexity of MPNs and implicate new genes involved in disease progression.
Introduction
The 3 classic BCR-ABL–negative myeloproliferative neoplasms (MPNs) are polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF).1 The 3 forms share several clinical characteristics, such as clonal hematopoiesis, bone marrow hypercellularity, and a propensity to thrombosis and hemorrhage. Distinct hallmarks include elevated erythrocyte counts in PV, elevated platelet levels in ET, and bone marrow fibrosis in PMF.1 In general, MPNs are chronic diseases, but some patients exhibit clinical signs of disease progression. Progression stages include post-PV or post-ET secondary myelofibrosis (sMF) associated with an increase in reticulin fibrosis in the bone marrow and extramedullary hematopoiesis.1 Another progression stage often referred to as the “accelerated phase” (AP) is characterized by variable degrees of pancytopenia and the presence of blasts in up to 20% in bone marrow.2 In some of the patients, the disease transforms to post-MPN acute myeloid leukemia (AML), which is a major complication with a poor prognosis and a mean survival of around 5 months. The clinical hallmark is a blast count of > 20% in the bone marrow. The yearly risk for transformation is 0.38%, 0.37%, and 1.09% for PV, ET, and PMF, respectively.3
The genetic causes of MPN initiation and progression have been studied extensively in the last decades. Numerous reports have been published investigating chromosomal abnormalities using conventional cytogenetic technologies.4-7 Abnormal cytogenetics were found to be frequent in PMF but less prevalent in PV and ET.8 Some aberrations have been observed recurrently, most notably, deletions of chromosomes 20q, 13q, and 12p; trisomy 8 and 9; gains of 9p; and various translocations.4-7 The karyotyping methods used usually detect large-scale genomic changes, but these studies did not succeed in the identification of target genes. Microsatellite studies on chromosome 9 identified acquired uniparental disomy (UPD) of the short arm (9pUPD) as a common defect in MPN.9 In-depth investigation of this region resulted in the identification of the V617F mutation in the Janus kinase 2 gene (JAK2),10-13 which is present in ∼ 95% of PV and > 50% of ET and PMF patients. Studies on other members of the JAK2 pathway led to the identification of different mutations of the thrombopoietin receptor MPL, which were often associated with 1pUPDs.14-16
After high-resolution DNA microarrays became available, it became possible to study cytogenetic changes in great detail. Consequently, new target genes have been identified, such as the tet oncogene family member 2 (TET2), which is associated with deletions and UPDs of chromosome 4q,17,18 and the Cas-Br-M (murine) ecotropic retroviral transforming sequence (CBL), which is associated with aberrations of chromosome 11q.19,20 Different mutations have a variety of roles in MPN pathogenesis; some, such as JAK2 or MPL mutations, define the MPN phenotype, whereas mutations in additional sex combs like 1 (ASXL1)21 and isocitrate dehydrogenase 1/2 (IDH1/2)22 or deletions of IKAROS family zinc finger 1 (IKZF1)23 are associated with transformation to post-MPN AML. The exact influence of mutations in TET2 and CBL on the disease course is still not well understood. A few recent studies have assessed the cytogenetic profiles of MPN patients in the chronic phase and in transformation to AML; however, the number of studied samples has been rather limited.24,25 In the present study, we present the results of a study on a large MPN cohort using high-resolution single-nucleotide polymorphism (SNP) arrays to detect chromosomal aberrations. We investigated chromosomal changes, together with clinical parameters such as post-MPN AML transformation, and describe genes so far unknown for their role in MPN pathogenesis.
Methods
Patient samples
Peripheral blood samples were collected from MPN patients after written informed consent at the Medical University of Vienna, Austria, the University of Pavia, Italy, and the University of Florence, Italy, according to the regulations of the ethics committees of each participating institution. Genomic DNA was isolated from either granulocyte or mononuclear cell fractions of these blood samples according to standard procedures.
Microarray analysis
Patient DNA was processed and hybridized to Genome-Wide Human SNP 6.0 arrays (Affymetrix) according to the manufacturer's instructions. The raw data of 444 samples were processed using Genotyping Console Version 3.0.2 software (Affymetrix). The data were first analyzed for quality, and samples showing high background signal were excluded. In addition, several mononuclear cell samples showing deletions at the TCR locus on chromosome 14 in > 50% of the cells, which is suggestive of a significant number of T cells in the sample, were excluded from further analysis. In total, we excluded 36 samples and used the remaining 408 (representing 398 patients) for further analysis. For 10 patients, we had 2 samples each from 2 different disease stages (supplemental Table 2, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Chromosomal aberrations such as deletions, gains, and UPDs were annotated after loss of heterozygosity and copy number regions were identified (as implemented in the Genotyping Console software). Our criteria for UPDs were a terminal location (at the ends of chromosomal arms) and a size of at least 1 Mb. Terminal UPDs in patients with numerous extensive (> 10Mb) interstitial runs of homozygosity were not included. In general, we cannot exclude that some chromosomal aberrations were germline, because we did not test constitutional DNA for all samples. We also did not annotate aberrations that mapped to known copy number variation loci according to the Database of Genomic Variants (DGV version 5, human reference genome assembly hg18).
The microarray raw data (CEL files) and the processed data (CNCHP files) are available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) under the accession number E-MTAB-608.
Single-gene mutational analysis
All primer sequences and PCR conditions are provided in supplemental Table 1. JAK2 mutational status was determined using allele-specific PCR for the JAK2-V617F mutation, as described previously.26 Exon sequencing of runt-related transcription factor 1 (RUNX1), tumor protein p53 (TP53), IDH1, IDH2, cut-like homeobox 1 (CUX1), and SH2B adaptor protein 2 (SH2B2) was performed using the BigDye Terminator version 3.1 cycle-sequencing kit and the 3130xl Genetic Analyzer (Applied Biosystems). Sequence analysis was performed using Sequencher Version 4.9 software (Gene Codes). Nucleophosmin (NPM1) exon 12 was analyzed for duplications. The exon was amplified with dye-labeled primers, and PCR product length differences were detected on the Genetic Analyzer. The data were analyzed using Gene Mapper Version 4.0 software (Applied Biosystems). We also screened for fms-related tyrosine kinase 3 internal tandem duplications (FLT3-ITD) and FLT3-D835 mutations, as described previously.27
Statistical analysis
Differences in the distributions of chromosomal aberrations between different sample groups were tested statistically using the Kruskal-Wallis test (in the case of multiple comparisons) or the Mann-Whitney test (in the case of 2-sample comparisons). Differences in frequencies of individual chromosomal aberrations between chronic-phase MPN and sMF/AP or post-MPN AML, as well as between JAK2-V617F–positive and -negative samples were tested for significance using the Fisher exact test.
Results
Whole-genome analysis of MPN patients
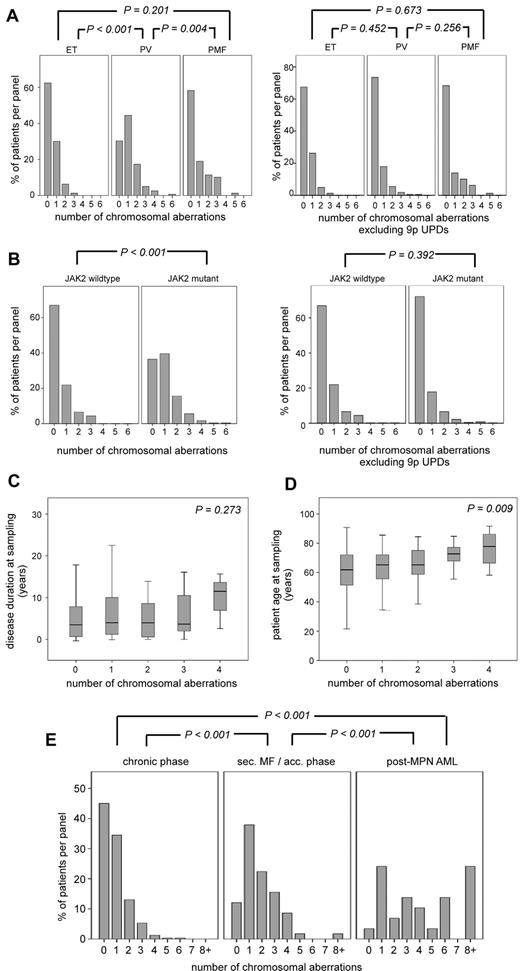
We investigated 408 MPN samples for chromosomal aberrations using high-resolution SNP microarrays that deliver 1.8 million copy number measurements and 0.9 million SNP genotypes per genome. The patients were diagnosed as chronic-phase PV (n = 162), chronic-phase ET (n = 80), chronic-phase PMF (n = 79), or post-MPN AML (n = 29) at the time of sampling (Figure 1A). The remaining 58 patients showed clear signs of disease progression (they were not in chronic phase), but did not fulfill the clinical criteria for post-MPN AML diagnosis. Diagnoses for these patients included post-PV or post-ET (secondary) myelofibrosis, as well as the clinical stage of AP, which was defined by variable degrees of pancytopenia and blasts in bone marrow up to 20%. This group of 58 patients will be referred to as sMF/AP (Figure 1A). The whole cohort included 199 male and 209 female samples. Of the total 408 samples, 297 (72.8%) harbored mutations in the JAK2 gene (Figure 1B). The analysis for chromosomal aberrations revealed 153 samples (37.5%) with a normal karyotype, whereas 255 (62.5%) harbored at least 1 chromosomal aberration (Figure 1C). All samples with UPD on chromosome 9p were positive for the JAK2-V617F mutation, whereas 6 of 7 samples with UPD on chromosome 1p were positive for the MPL-W515L mutation. All detected chromosomal aberrations are provided in supplemental Table 2.
Cohort descriptives. (A) Diagnosis distribution. (B) Fraction of the samples with mutant or wild-type JAK2. (C) Distribution of chromosomal aberrations in the entire MPN cohort.
Cohort descriptives. (A) Diagnosis distribution. (B) Fraction of the samples with mutant or wild-type JAK2. (C) Distribution of chromosomal aberrations in the entire MPN cohort.
Correlation of genomic aberrations with clinical parameters
To investigate whether chromosomal aberrations cluster in a certain patient group defined by clinical criteria, we first focused on the patients in the chronic phase of the disease (n = 321). We detected a total of 50 gains, 76 deletions, and 147 UPDs, with sizes ranging from 0.1-146.3 Mb distributed across the genome (Figure 2). We compared the distribution of chromosomal aberrations between the 3 MPN disease entities (Figure 3A) and found that PV patients harbored significantly more chromosomal aberrations than ET (P < .001) or PMF (P = .004) patients. Because 92% of the PV patients were positive for the JAK2 mutation and 68% of these patients harbored the associated 9pUPD, we examined whether the observed difference was mainly based on the high prevalence of 9pUPDs in PV patients. We reanalyzed the data excluding 9pUPDs and could no longer observe a significant difference in chromosomal aberrations between the disease entities (Figure 3A).
Karyoview of all chromosomal aberrations detected by Affymetrix SNP array analysis in 321 chronic-phase MPN patients. Bars depict the physical position and the size of the aberration; green indicates gains; red, deletions; and blue, UPDs. Thin black lines that connect 2 bars indicate multiple aberrations in the same patient.
Karyoview of all chromosomal aberrations detected by Affymetrix SNP array analysis in 321 chronic-phase MPN patients. Bars depict the physical position and the size of the aberration; green indicates gains; red, deletions; and blue, UPDs. Thin black lines that connect 2 bars indicate multiple aberrations in the same patient.
Correlation of chromosomal aberrations with clinical parameters.P values refer to differences between the indicated distribution patterns. Shown are the distributions of chromosomal aberrations in the 3 MPN entities PV, ET, and PMF (A) and in patients positive or negative for JAK2 mutations (B). Also shown are the associations of the number of chromosomal aberrations with disease duration (C) and patient age (D); outliers are not shown. (E) Distribution of chromosomal aberrations comparing patients in the chronic phase of the disease, patients with sMF/AP, and patients who transformed to post-MPN AML.
Correlation of chromosomal aberrations with clinical parameters.P values refer to differences between the indicated distribution patterns. Shown are the distributions of chromosomal aberrations in the 3 MPN entities PV, ET, and PMF (A) and in patients positive or negative for JAK2 mutations (B). Also shown are the associations of the number of chromosomal aberrations with disease duration (C) and patient age (D); outliers are not shown. (E) Distribution of chromosomal aberrations comparing patients in the chronic phase of the disease, patients with sMF/AP, and patients who transformed to post-MPN AML.
To investigate the relationship between JAK2 mutational status and chromosomal aberrations, we compared the frequency of aberrations in patients with and without JAK2 mutations (V617F and exon 12 mutations combined; Figure 3B). We observed a significant difference (P < .001), which was again lost after the exclusion of 9pUPDs from the analysis (P = .392). We also compared the distribution of individual recurrent chromosomal abnormalities (found in at least 3 patients) between JAK2 mutation–positive and -negative patients. The only significant difference was the clustering of 9pUPD in JAK2-positive MPN patients (Table 1). This result indicates that patients positive for JAK2 mutations do not carry more chromosomal aberrations compared with JAK2-negative patients. Furthermore, we did not detect an aberration specifically associated with JAK2-V617F–negative MPN.
Association of individual chromosomal aberrations with JAK2 mutational status
| Aberration type . | JAK2 mutant (n = 297) . | JAK2 wild-type (n = 111) . | P . | P* . | |
|---|---|---|---|---|---|
| UPD | 9p | 169 (56.90%) | 0 (0.00%) | 1.35 × 10−32 | 3.38 × 10−31 |
| UPD | 1p | 2 (0.67%) | 6 (5.41%) | .0062 | .1548 |
| deletion | 12p | 2 (0.67%) | 4 (3.60%) | .0492 | 1 |
| gain | 9p | 6 (2.02%) | 0 (0.00%) | .1959 | 1 |
| trisomy | 9 | 9 (3.03%) | 1 (0.90%) | .2983 | 1 |
| UPD | 7q | 2 (0.67%) | 2 (1.80%) | .2990 | 1 |
| deletion | 3p | 5 (1.68%) | 0 (0.00%) | .3293 | 1 |
| deletion | 4q | 11 (3.70%) | 2 (1.80%) | .5278 | 1 |
| deletion | 11q | 4 (1.35%) | 0 (0.00%) | .5783 | 1 |
| deletion | 5q | 3 (1.01%) | 2 (1.80%) | .6164 | 1 |
| deletion | 6p | 3 (1.01%) | 2 (1.80%) | .6164 | 1 |
| UPD | 22q | 3 (1.01%) | 2 (1.80%) | .6164 | 1 |
| deletion | 12q | 4 (1.35%) | 2 (1.80%) | .6653 | 1 |
| UPD | 14q | 7 (2.36%) | 1 (0.90%) | .6888 | 1 |
| gain | 1q | 11 (3.70%) | 3 (2.70%) | .7668 | 1 |
| deletion | 13q | 11 (3.70%) | 3 (2.70%) | .7668 | 1 |
| deletion | 20q | 11 (3.70%) | 5 (4.50%) | .7752 | 1 |
| gain | 3q | 3 (1.01%) | 1 (0.90%) | 1 | 1 |
| deletion | 7q | 7 (2.36%) | 3 (2.70%) | 1 | 1 |
| deletion | 7p | 3 (1.01%) | 1 (0.90%) | 1 | 1 |
| trisomy | 8 | 6 (2.02%) | 2 (1.80%) | 1 | 1 |
| UPD | 11q | 2 (0.67%) | 1 (0.90%) | 1 | 1 |
| deletion | 18p | 4 (1.35%) | 1 (0.90%) | 1 | 1 |
| UPD | 19q | 2 (0.67%) | 1 (0.90%) | 1 | 1 |
| trisomy | 21 | 2 (0.67%) | 1 (0.90%) | 1 | 1 |
| Aberration type . | JAK2 mutant (n = 297) . | JAK2 wild-type (n = 111) . | P . | P* . | |
|---|---|---|---|---|---|
| UPD | 9p | 169 (56.90%) | 0 (0.00%) | 1.35 × 10−32 | 3.38 × 10−31 |
| UPD | 1p | 2 (0.67%) | 6 (5.41%) | .0062 | .1548 |
| deletion | 12p | 2 (0.67%) | 4 (3.60%) | .0492 | 1 |
| gain | 9p | 6 (2.02%) | 0 (0.00%) | .1959 | 1 |
| trisomy | 9 | 9 (3.03%) | 1 (0.90%) | .2983 | 1 |
| UPD | 7q | 2 (0.67%) | 2 (1.80%) | .2990 | 1 |
| deletion | 3p | 5 (1.68%) | 0 (0.00%) | .3293 | 1 |
| deletion | 4q | 11 (3.70%) | 2 (1.80%) | .5278 | 1 |
| deletion | 11q | 4 (1.35%) | 0 (0.00%) | .5783 | 1 |
| deletion | 5q | 3 (1.01%) | 2 (1.80%) | .6164 | 1 |
| deletion | 6p | 3 (1.01%) | 2 (1.80%) | .6164 | 1 |
| UPD | 22q | 3 (1.01%) | 2 (1.80%) | .6164 | 1 |
| deletion | 12q | 4 (1.35%) | 2 (1.80%) | .6653 | 1 |
| UPD | 14q | 7 (2.36%) | 1 (0.90%) | .6888 | 1 |
| gain | 1q | 11 (3.70%) | 3 (2.70%) | .7668 | 1 |
| deletion | 13q | 11 (3.70%) | 3 (2.70%) | .7668 | 1 |
| deletion | 20q | 11 (3.70%) | 5 (4.50%) | .7752 | 1 |
| gain | 3q | 3 (1.01%) | 1 (0.90%) | 1 | 1 |
| deletion | 7q | 7 (2.36%) | 3 (2.70%) | 1 | 1 |
| deletion | 7p | 3 (1.01%) | 1 (0.90%) | 1 | 1 |
| trisomy | 8 | 6 (2.02%) | 2 (1.80%) | 1 | 1 |
| UPD | 11q | 2 (0.67%) | 1 (0.90%) | 1 | 1 |
| deletion | 18p | 4 (1.35%) | 1 (0.90%) | 1 | 1 |
| UPD | 19q | 2 (0.67%) | 1 (0.90%) | 1 | 1 |
| trisomy | 21 | 2 (0.67%) | 1 (0.90%) | 1 | 1 |
After Bonferroni correction for multiple testing; significant associations are bold.
We further examined the correlation between the cytogenetic complexity of patients and disease duration and age. The median disease duration of the chronic-phase patients was 4 years (range 0-26), and the median age at sample was 65 years (range 22-92). As shown in Figure 3C, patients without chromosomal lesions and patients with increasing numbers of lesions did not significantly differ in their disease duration (P = .273). Conversely, we observed a significant difference related to the patient age at sampling. Patients with normal karyotypes were younger than those with complex karyotypes (P = .009; Figure 3D).
As a next step, we compared the distributions of chromosomal aberrations in the 3 stages of disease progression (Figure 3E). Samples diagnosed with sMF/AP harbored significantly more chromosomal aberrations than samples in the chronic phase of the disease (P < .001) and significantly fewer aberrations compared with samples diagnosed with post-MPN AML (P < .001; Figure 3E).
Association of specific chromosomal aberrations with sMF/AP and post-MPN AML
To identify specific chromosomal lesions associated with disease progression, we compared the distribution of recurrent chromosomal abnormalities (found at least in 3 patients) between samples in chronic phase, sMF/AP, and post-MPN AML. Gains of chromosomes 1q and UPDs of 9p were significantly associated with sMF/AP compared with chronic phase (Table 2). Chromosome 1q and 3q amplifications; deletions of 7q, 5q, 6p, and 7p; and UPDs of 19q and 22q showed significant association with post-MPN AML compared with chronic phase (Table 3). We did not observe any associations of recurrent aberrations comparing sMF/AP with post-MPN AML. Common chromosomal aberrations in MPN, such as deletions of 4q (TET2 gene deletions), 13q, and 20q, were evenly distributed among the groups.
Association of individual chromosomal aberrations with progression to secondary myelofibrosis/accelerated phase
| Aberration type . | sMF/AP (n = 58) . | Chronic MPN (n = 321) . | P . | P* . | |
|---|---|---|---|---|---|
| gain | 1q | 7 (12.07%) | 2 (0.62%) | 3.98 × 10−5 | .0010 |
| UPD | 9p | 38 (65.52%) | 120 (37.38%) | 8.15 × 10−5 | .0020 |
| deletion | 12q | 3 (5.17%) | 1 (0.31%) | .0122 | .3053 |
| UPD | 14q | 4 (6.90%) | 4 (1.25%) | .0216 | .5393 |
| UPD | 11q | 2 (3.45%) | 0 (0.00%) | .0231 | .5770 |
| deletion | 20q | 5 (8.62%) | 7 (2.18%) | .0239 | .5963 |
| deletion | 7q | 3 (5.17%) | 2 (0.62%) | .0272 | .6798 |
| deletion | 4q | 4 (6.90%) | 5 (1.56%) | .0344 | .8605 |
| deletion | 12p | 2 (3.45%) | 2 (0.62%) | .1125 | 1 |
| deletion | 7p | 1 (1.72%) | 0 (0.00%) | .1530 | 1 |
| trisomy | 21 | 1 (1.72%) | 0 (0.00%) | .1530 | 1 |
| UPD | 1p | 2 (3.45%) | 4 (1.25%) | .2299 | 1 |
| UPD | 22q | 1 (1.72%) | 1 (0.31%) | .2830 | 1 |
| trisomy | 9 | 2 (3.45%) | 5 (1.56%) | .2916 | 1 |
| UPD | 7q | 1 (1.72%) | 2 (0.62%) | .3933 | 1 |
| deletion | 13q | 3 (5.17%) | 9 (2.80%) | .4051 | 1 |
| deletion | 3p | 1 (1.72%) | 3 (0.93%) | .4869 | 1 |
| gain | 9p | 1 (1.72%) | 4 (1.25%) | .5662 | 1 |
| gain | 3q | 0 (0.00%) | 1 (0.31%) | 1 | 1 |
| deletion | 5q | 0 (0.00%) | 1 (0.31%) | 1 | 1 |
| deletion | 6p | 0 (0.00%) | 1 (0.31%) | 1 | 1 |
| trisomy | 8 | 1 (1.72%) | 5 (1.56%) | 1 | 1 |
| deletion | 11q | 0 (0.00%) | 3 (0.93%) | 1 | 1 |
| deletion | 18p | 0 (0.00%) | 4 (1.25%) | 1 | 1 |
| UPD | 19q | 0 (0.00%) | 0 (0.00%) | 1 | 1 |
| Aberration type . | sMF/AP (n = 58) . | Chronic MPN (n = 321) . | P . | P* . | |
|---|---|---|---|---|---|
| gain | 1q | 7 (12.07%) | 2 (0.62%) | 3.98 × 10−5 | .0010 |
| UPD | 9p | 38 (65.52%) | 120 (37.38%) | 8.15 × 10−5 | .0020 |
| deletion | 12q | 3 (5.17%) | 1 (0.31%) | .0122 | .3053 |
| UPD | 14q | 4 (6.90%) | 4 (1.25%) | .0216 | .5393 |
| UPD | 11q | 2 (3.45%) | 0 (0.00%) | .0231 | .5770 |
| deletion | 20q | 5 (8.62%) | 7 (2.18%) | .0239 | .5963 |
| deletion | 7q | 3 (5.17%) | 2 (0.62%) | .0272 | .6798 |
| deletion | 4q | 4 (6.90%) | 5 (1.56%) | .0344 | .8605 |
| deletion | 12p | 2 (3.45%) | 2 (0.62%) | .1125 | 1 |
| deletion | 7p | 1 (1.72%) | 0 (0.00%) | .1530 | 1 |
| trisomy | 21 | 1 (1.72%) | 0 (0.00%) | .1530 | 1 |
| UPD | 1p | 2 (3.45%) | 4 (1.25%) | .2299 | 1 |
| UPD | 22q | 1 (1.72%) | 1 (0.31%) | .2830 | 1 |
| trisomy | 9 | 2 (3.45%) | 5 (1.56%) | .2916 | 1 |
| UPD | 7q | 1 (1.72%) | 2 (0.62%) | .3933 | 1 |
| deletion | 13q | 3 (5.17%) | 9 (2.80%) | .4051 | 1 |
| deletion | 3p | 1 (1.72%) | 3 (0.93%) | .4869 | 1 |
| gain | 9p | 1 (1.72%) | 4 (1.25%) | .5662 | 1 |
| gain | 3q | 0 (0.00%) | 1 (0.31%) | 1 | 1 |
| deletion | 5q | 0 (0.00%) | 1 (0.31%) | 1 | 1 |
| deletion | 6p | 0 (0.00%) | 1 (0.31%) | 1 | 1 |
| trisomy | 8 | 1 (1.72%) | 5 (1.56%) | 1 | 1 |
| deletion | 11q | 0 (0.00%) | 3 (0.93%) | 1 | 1 |
| deletion | 18p | 0 (0.00%) | 4 (1.25%) | 1 | 1 |
| UPD | 19q | 0 (0.00%) | 0 (0.00%) | 1 | 1 |
After Bonferroni correction for multiple testing; significant associations are bold.
Association of individual chromosomal aberrations with progression to post-MPN AML
| Aberration type . | Post-MPN AML (n = 29) . | Chronic MPN (n = 321) . | P . | P* . | |
|---|---|---|---|---|---|
| gain | 1q | 5 (17.24%) | 2 (0.62%) | 5.20 × 10−5 | .0013 |
| deletion | 7q | 5 (17.24%) | 2 (0.62%) | 5.20 × 10−5 | .0013 |
| deletion | 5q | 4 (13.79%) | 1 (0.31%) | 1.82 × 10−4 | .0046 |
| deletion | 6p | 4 (13.79%) | 1 (0.31%) | 1.82 × 10−4 | .0046 |
| deletion | 7p | 3 (10.34%) | 0 (0.00%) | 5.16 × 10−4 | .0129 |
| UPD | 19q | 3 (10.34%) | 0 (0.00%) | 5.16 × 10−4 | .0129 |
| gain | 3q | 3 (10.34%) | 1 (0.31%) | .0019 | .0487 |
| UPD | 22q | 3 (10.34%) | 1 (0.31%) | .0019 | .0487 |
| deletion | 4q | 4 (13.79%) | 5 (1.56%) | .0036 | .0904 |
| trisomy | 21 | 2 (6.90%) | 0 (0.00%) | .0066 | .1662 |
| deletion | 20q | 4 (13.79%) | 7 (2.18%) | .0084 | .2102 |
| deletion | 12q | 2 (6.90%) | 1 (0.31%) | .0189 | .4728 |
| trisomy | 9 | 3 (10.34%) | 5 (1.56%) | .0217 | .5413 |
| deletion | 12p | 2 (6.90%) | 2 (0.62%) | .0359 | .8970 |
| UPD | 1p | 2 (6.90%) | 4 (1.25%) | .0808 | 1 |
| UPD | 11q | 1 (3.45%) | 0 (0.00%) | .0829 | 1 |
| trisomy | 8 | 2 (6.90%) | 5 (1.56%) | .1073 | 1 |
| deletion | 13q | 2 (6.90%) | 9 (2.80%) | .2289 | 1 |
| UPD | 7q | 1 (3.45%) | 2 (0.62%) | .2291 | 1 |
| deletion | 3p | 1 (3.45%) | 3 (0.93%) | .2936 | 1 |
| deletion | 11q | 1 (3.45%) | 3 (0.93%) | .2936 | 1 |
| gain | 9p | 1 (3.45%) | 4 (1.25%) | .3528 | 1 |
| deletion | 18p | 1 (3.45%) | 4 (1.25%) | .3528 | 1 |
| UPD | 9p | 11 (37.93%) | 120 (37.38%) | 1 | 1 |
| UPD | 14q | 0 (0.00%) | 4 (1.25%) | 1 | 1 |
| Aberration type . | Post-MPN AML (n = 29) . | Chronic MPN (n = 321) . | P . | P* . | |
|---|---|---|---|---|---|
| gain | 1q | 5 (17.24%) | 2 (0.62%) | 5.20 × 10−5 | .0013 |
| deletion | 7q | 5 (17.24%) | 2 (0.62%) | 5.20 × 10−5 | .0013 |
| deletion | 5q | 4 (13.79%) | 1 (0.31%) | 1.82 × 10−4 | .0046 |
| deletion | 6p | 4 (13.79%) | 1 (0.31%) | 1.82 × 10−4 | .0046 |
| deletion | 7p | 3 (10.34%) | 0 (0.00%) | 5.16 × 10−4 | .0129 |
| UPD | 19q | 3 (10.34%) | 0 (0.00%) | 5.16 × 10−4 | .0129 |
| gain | 3q | 3 (10.34%) | 1 (0.31%) | .0019 | .0487 |
| UPD | 22q | 3 (10.34%) | 1 (0.31%) | .0019 | .0487 |
| deletion | 4q | 4 (13.79%) | 5 (1.56%) | .0036 | .0904 |
| trisomy | 21 | 2 (6.90%) | 0 (0.00%) | .0066 | .1662 |
| deletion | 20q | 4 (13.79%) | 7 (2.18%) | .0084 | .2102 |
| deletion | 12q | 2 (6.90%) | 1 (0.31%) | .0189 | .4728 |
| trisomy | 9 | 3 (10.34%) | 5 (1.56%) | .0217 | .5413 |
| deletion | 12p | 2 (6.90%) | 2 (0.62%) | .0359 | .8970 |
| UPD | 1p | 2 (6.90%) | 4 (1.25%) | .0808 | 1 |
| UPD | 11q | 1 (3.45%) | 0 (0.00%) | .0829 | 1 |
| trisomy | 8 | 2 (6.90%) | 5 (1.56%) | .1073 | 1 |
| deletion | 13q | 2 (6.90%) | 9 (2.80%) | .2289 | 1 |
| UPD | 7q | 1 (3.45%) | 2 (0.62%) | .2291 | 1 |
| deletion | 3p | 1 (3.45%) | 3 (0.93%) | .2936 | 1 |
| deletion | 11q | 1 (3.45%) | 3 (0.93%) | .2936 | 1 |
| gain | 9p | 1 (3.45%) | 4 (1.25%) | .3528 | 1 |
| deletion | 18p | 1 (3.45%) | 4 (1.25%) | .3528 | 1 |
| UPD | 9p | 11 (37.93%) | 120 (37.38%) | 1 | 1 |
| UPD | 14q | 0 (0.00%) | 4 (1.25%) | 1 | 1 |
After Bonferroni correction for multiple testing; significant associations are bold.
The chromosomal defects most significantly associated with sMF/AP and post-MPN AML transformation were gains of chromosome 1q (Tables 2 and 3). We recently reported that the minimal amplified region on chromosome 1q harbors the MDM4 gene, a potent inhibitor of p53. We also showed that p53 mutations are associated with post-MPN AML.28 In the present study, we extended our analysis on this finding and sequenced the TP53 gene in a total of 129 chronic-phase and the 29 post-MPN AML samples. We found 2 samples in the chronic phase (1.6%) and 6 samples with post-MPN AML (20%) carrying mutations in TP53. This result confirms the reported association of TP53 mutations with post-MPN AML (P < .001). The chromosome 9p UPDs, which were also associated with sMF/AP, amplified JAK2-V617F mutations.
The role of other de novo AML-specific mutations in post-MPN AML
To gain deeper insight into other pathways involved in the transformation to AML, we screened all post-MPN AML patients for point mutations common in de novo AML affecting the genes RUNX1, FLT3, NPM1, IDH1, and IDH2 (Table 4). Sample 393 carried 2 nonsense mutations in RUNX1 and also tested positive for somatic trisomy 21. Sample 396 had a hemizygous single-gene deletion of RUNX1. Sample 373 carried a L29S missense mutation and sample 381 harbored1 missense mutation and a 5-bp insertion. Three patients harbored mutations in FLT3 and 1 patient carried a NPM1 insertion. IDH1-R132G, IDH2-R140W, and IDH2-R140Q mutations were detected in samples 407, 304, and 381, respectively. All point mutations and chromosomal defects of post-MPN AML samples are summarized in Table 4.
Summary of point mutations and recurrent chromosomal aberrations in post-MPN AML patients
| Sample ID . | JAK2 . | RUNX1 . | FLT3 . | NPM1 . | IDH1 . | IDH2 . | TP53 . | gain 1q . | del 7q . | del 5q . | del 6p . | del 7p . | UPD 19q . | gain 3q . | UPD 22q . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sample_281 | V617F | wt | wt | wt | wt | wt | wt | Yes | |||||||
| sample_289 | V617F | wt | wt | wt | wt | wt | wt | Yes | |||||||
| sample_304 | V617F | wt | wt | wt | wt | R140W | wt | ||||||||
| sample_373 | V617F | L29S | wt | mut | wt | wt | wt | ||||||||
| sample_374 | wt | wt | wt | wt | wt | wt | wt | Yes | Yes | Yes | |||||
| sample_375 | wt | wt | wt | wt | wt | wt | wt | ||||||||
| sample_381 | wt | R135K/442insTGCCT_T150PfsX1 | ITD | wt | wt | R140Q | wt | ||||||||
| sample_382 | wt | wt | wt | wt | wt | wt | wt | ||||||||
| sample_383 | wt | wt | wt | wt | wt | wt | wt | Yes | Yes | Yes | Yes | ||||
| sample_389 | V617F | wt | wt | wt | wt | wt | wt | ||||||||
| sample_390 | V617F | wt | wt | wt | wt | wt | wt | Yes | |||||||
| sample_391 | V617F | wt | wt | wt | wt | wt | wt | ||||||||
| sample_392 | V617F | wt | D835 | wt | wt | wt | c.994–2 A > G (17pUPD) | Yes | Yes | ||||||
| sample_393 | V617F | Q235X/S373X | wt | wt | wt | wt | wt | Yes | Yes | ||||||
| sample_394 | V617F | wt | wt | wt | wt | wt | C135S / M246K | Yes | Yes | Yes | |||||
| sample_395 | V617F | wt | wt | wt | wt | wt | wt | ||||||||
| sample_396 | V617F | del 21q† | wt | wt | wt | wt | wt | Yes | Yes | Yes | |||||
| sample_397 | V617F | wt | wt | wt | wt | wt | wt | ||||||||
| sample_398 | V617F | wt | wt | wt | wt | wt | wt | ||||||||
| sample_399 | V617F | wt | wt | wt | wt | wt | N239D (17pUPD) | ||||||||
| sample_400 | V617F | wt | wt | wt | wt | wt | c.560–1 G > A / Y220H | Yes | Yes | ||||||
| sample_401 | V617F | wt | wt | wt | wt | wt | wt | ||||||||
| sample_402 | V617F | wt | wt | wt | wt | wt | K132E | Yes | |||||||
| sample_403 | V617F | wt | wt | wt | wt | wt | S261T / N239D | Yes | |||||||
| sample_404 | V617F | wt | D835 | wt | wt | wt | wt | Yes | |||||||
| sample_405 | V617F | wt | wt | wt | wt | wt | wt | ||||||||
| sample_406 | V617F | wt | wt | wt | wt | wt | wt | Yes | Yes | ||||||
| sample_407 | V617F | wt | wt | wt | R132G | wt | wt | Yes* | Yes* | ||||||
| sample_408 | wt | wt | wt | wt | wt | wt | wt | Yes |
| Sample ID . | JAK2 . | RUNX1 . | FLT3 . | NPM1 . | IDH1 . | IDH2 . | TP53 . | gain 1q . | del 7q . | del 5q . | del 6p . | del 7p . | UPD 19q . | gain 3q . | UPD 22q . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sample_281 | V617F | wt | wt | wt | wt | wt | wt | Yes | |||||||
| sample_289 | V617F | wt | wt | wt | wt | wt | wt | Yes | |||||||
| sample_304 | V617F | wt | wt | wt | wt | R140W | wt | ||||||||
| sample_373 | V617F | L29S | wt | mut | wt | wt | wt | ||||||||
| sample_374 | wt | wt | wt | wt | wt | wt | wt | Yes | Yes | Yes | |||||
| sample_375 | wt | wt | wt | wt | wt | wt | wt | ||||||||
| sample_381 | wt | R135K/442insTGCCT_T150PfsX1 | ITD | wt | wt | R140Q | wt | ||||||||
| sample_382 | wt | wt | wt | wt | wt | wt | wt | ||||||||
| sample_383 | wt | wt | wt | wt | wt | wt | wt | Yes | Yes | Yes | Yes | ||||
| sample_389 | V617F | wt | wt | wt | wt | wt | wt | ||||||||
| sample_390 | V617F | wt | wt | wt | wt | wt | wt | Yes | |||||||
| sample_391 | V617F | wt | wt | wt | wt | wt | wt | ||||||||
| sample_392 | V617F | wt | D835 | wt | wt | wt | c.994–2 A > G (17pUPD) | Yes | Yes | ||||||
| sample_393 | V617F | Q235X/S373X | wt | wt | wt | wt | wt | Yes | Yes | ||||||
| sample_394 | V617F | wt | wt | wt | wt | wt | C135S / M246K | Yes | Yes | Yes | |||||
| sample_395 | V617F | wt | wt | wt | wt | wt | wt | ||||||||
| sample_396 | V617F | del 21q† | wt | wt | wt | wt | wt | Yes | Yes | Yes | |||||
| sample_397 | V617F | wt | wt | wt | wt | wt | wt | ||||||||
| sample_398 | V617F | wt | wt | wt | wt | wt | wt | ||||||||
| sample_399 | V617F | wt | wt | wt | wt | wt | N239D (17pUPD) | ||||||||
| sample_400 | V617F | wt | wt | wt | wt | wt | c.560–1 G > A / Y220H | Yes | Yes | ||||||
| sample_401 | V617F | wt | wt | wt | wt | wt | wt | ||||||||
| sample_402 | V617F | wt | wt | wt | wt | wt | K132E | Yes | |||||||
| sample_403 | V617F | wt | wt | wt | wt | wt | S261T / N239D | Yes | |||||||
| sample_404 | V617F | wt | D835 | wt | wt | wt | wt | Yes | |||||||
| sample_405 | V617F | wt | wt | wt | wt | wt | wt | ||||||||
| sample_406 | V617F | wt | wt | wt | wt | wt | wt | Yes | Yes | ||||||
| sample_407 | V617F | wt | wt | wt | R132G | wt | wt | Yes* | Yes* | ||||||
| sample_408 | wt | wt | wt | wt | wt | wt | wt | Yes |
wt indicates wild-type; mut, mutant; ITD, internal tandem duplication; and del, deletion.
Monosomy 7.
Single-gene deletion of RUNX1.
CDRs that mapped to single genes
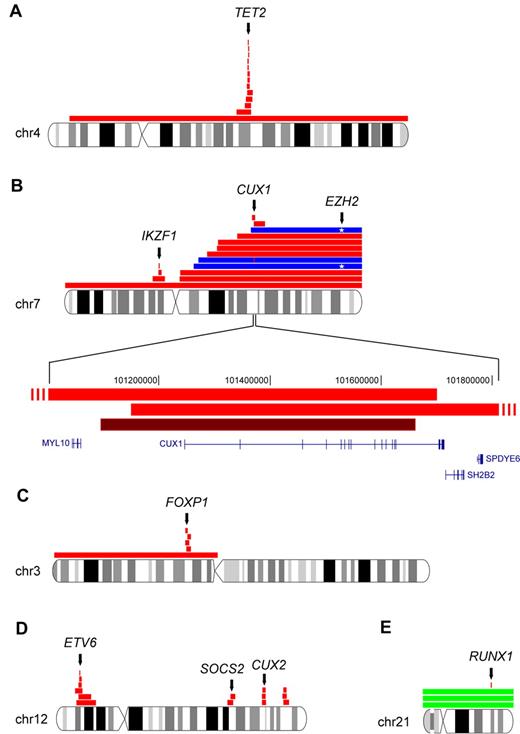
Common deleted regions (CDRs) were mapped as the minimum overlap of all deletions detected for a particular chromosome in our cohort. We found 6 such regions that mapped to single-target genes. The CDR of chromosome 4q harbors only the known target gene TET2 (Figure 4A), and the CDR of chromosome 7p contained only the IKZF1 gene (Figure 4B). Mutations of both genes have been reported previously.17,18,23 The deleted regions of chromosome 7q mapped to CUX1 (Figure 4B). Chromosome 7p and 7q deletions were among the lesions most significantly associated with post-MPN AML. Two patients tested positive for an acquired 7qUPD in the same chromosomal region. Sequence analysis of CUX1 in these 2 patients did not reveal any mutations. When we sequenced the enhancer of zeste homolog 2 gene (EZH2), a described target of 7qUPDs,29,30 we identified mutations in both patients. A third patient harboring a 7qUPD had wild-type EZH2 but harbored a homozygous single-gene deletion of CUX1 within the UPD region. A close neighboring gene of CUX1 is SH2B2, a homolog of the SH2B adaptor protein 3 gene (SH2B3 LNK). Aberrations in SH2B3 have been associated with myeloid malignancies.31,32 Because chromosome 7q aberrations were associated with post-MPN AML, we sequenced SH2B2 in the 29 leukemic patients of our cohort, and identified a single somatic missense mutation (G113C) in 1 patient.
Commonly deleted chromosomal regions in MPN that map to single-target genes. Red horizontal bars indicate deletions; dark red, homozygous deletion; and blue, UPDs. (A) Recurrent deletions on chromosome 4q target the TET2 gene. (B) Complexity of chromosome 7 aberrations in MPN. The CDR on 7p (IKZF1) and 7q (CUX1) are shown. Two events of 7qUPD associated with an EZH2 mutation (*) are shown in blue. Another 7qUPD is negative for EZH2 mutations but has homozygous CUX1 deletion. Two overlapping deletions in the CUX1 gene locus and the homozygous deletion of CUX1 are shown at higher magnification. (C) Five overlapping deletion events on chromosome 3p define the target gene as FOXP1. (D) Recurrent deletions on chromosome 12. The target of 12p deletions is ETV6, whereas 1 of the commonly deleted regions on 12q contains CUX2. (E) Aberrations on chromosome 21q affecting RUNX1: single-gene deletion of RUNX1 and trisomy 21 with 2 nonsense mutations in RUNX1. chr indicates chromosome and Mbp, mega base pairs.
Commonly deleted chromosomal regions in MPN that map to single-target genes. Red horizontal bars indicate deletions; dark red, homozygous deletion; and blue, UPDs. (A) Recurrent deletions on chromosome 4q target the TET2 gene. (B) Complexity of chromosome 7 aberrations in MPN. The CDR on 7p (IKZF1) and 7q (CUX1) are shown. Two events of 7qUPD associated with an EZH2 mutation (*) are shown in blue. Another 7qUPD is negative for EZH2 mutations but has homozygous CUX1 deletion. Two overlapping deletions in the CUX1 gene locus and the homozygous deletion of CUX1 are shown at higher magnification. (C) Five overlapping deletion events on chromosome 3p define the target gene as FOXP1. (D) Recurrent deletions on chromosome 12. The target of 12p deletions is ETV6, whereas 1 of the commonly deleted regions on 12q contains CUX2. (E) Aberrations on chromosome 21q affecting RUNX1: single-gene deletion of RUNX1 and trisomy 21 with 2 nonsense mutations in RUNX1. chr indicates chromosome and Mbp, mega base pairs.
The CDR on chromosome 3p (71.2-71.3 Mb) mapped to the forkhead box P1 (FOXP1) gene (Figure 4C). We found 6 patients with overlapping deletions on chromosome 12p. The smallest of these deletions mapped to the ets variant 6 gene (ETV6; Figure 4D). Three patients harbored chromosome 21 trisomies. In another patient with post-MPN AML, a deletion containing only the RUNX1 gene on chromosome 21q was detected (Figure 4E).
Discussion
Classic metaphase cytogenetics is widely used to detect chromosomal aberrations in hematologic malignancies and has emerged as a standard diagnostic tool.4,5,7 Recently, SNP array technology has been intensively discussed as a valuable complement to metaphase cytogenetics.8,33 We report here on the analysis of a large cohort of classic MPN patients at high resolution using Affymetrix 6.0 SNP arrays containing more than 1.8 million genomic markers. We found a highly diverse pattern of chromosomal aberrations among individual patients. Whereas studies of MPN using classic metaphase cytogenetics reported around 30% patients with aberrant karyotype,4,5,7 we found chromosomal changes in > 50% of the patients. This was clearly due to the fact that SNP arrays allow the detection of small aberrations as well as UPDs, both of which are undetectable by metaphase cytogenetics. It has been widely reported that mitotic recombination resulting in UPD often amplifies oncogene or tumor suppressor gene mutations. In MPN, chromosome 9pUPD amplifies the JAK2-V617F mutation,12 1pUPD amplifies MPL mutations,14-16 and chromosome 11qUPD has been shown to amplify various mutations of CBL.19 We detected chromosome 9pUPDs in 169 patients, all of whom tested positive for JAK2 mutations. Of the 7 1pUPD cases, only 6 were associated with mutations in the MPL gene. The remaining case raises the possibility that chromosome 1p harbors an unknown gene mutation relevant for MPN pathogenesis.
We did not observe any association of the number of chromosomal aberrations with any of the 3 MPN disease entities investigated in this study. This is in contrast to previous reports showing that PMF patients have a higher rate of chromosomal abnormalities.8 A recent publication suggested that the JAK2-V617F mutation induces genomic instability and might account for chromosomal lesions observed in MPN.34 Our results show that JAK2-V617F–positive patients do not have a higher frequency of chromosomal aberrations. We expected to observe accumulation of chromosomal aberrations over the course of the disease as the malignant clone evolved. Disease duration was not associated with the frequency of chromosomal aberrations in the studied patients. However, the age of the patients at the time of sample was positively correlated with the number of defects. In agreement with this finding, there have been reports showing that aging stem cells acquire chromosomal defects35 ; a study in PV showed a similar correlation.7
MPN patients diagnosed for sMF/AP had significantly more chromosomal aberrations than patients in the chronic phase of the disease. Patients with post-MPN AML harbored more aberrations than both chronic-phase and sMF/AP patients, which has been reported previously.5 Chromosome 9pUPDs were more closely associated with sMF/AP than with chronic-phase disease, indicating that a high burden of JAK2-V617F (located on chromosome 9p) predisposes for disease progression. JAK2-V617F homozygosity had already been associated with a higher risk for secondary myelofibrosis in PV and ET.36
Chromosome 1q amplifications were more closely associated with both sMF/AP and post-MPN AML compared with chronic phase, but no significant difference was observed between sMF/AP and post-MPN AML. All observed chromosome 1q gains amplified the MDM4 gene, which is a potent inhibitor of p53.37,38 TP53 itself has been shown to be involved in the leukemic transformation process in MPN,28,39 which was confirmed in the present study. It is also known that an impairment of the p53 pathway and altered cell-cycle regulation are associated with the accumulation of mutations. Whether chromosome 1q gains could set the stage for the acquisition of further aberrations that eventually induce leukemic transformation is a matter for further investigations. We also show herein that lesions of chromosomes 7, 5, 6, 19, 22, and 3 significantly cluster in post-MPN AML compared with chronic phase. It remains to be seen if the transformation-associated aberrations identified here will have prognostic value.
Six CDRs in our cohort mapped to single-target genes. We identified a new putative tumor suppressor gene on chromosome 7 that encodes CUX1, a DNA-binding protein that plays a role in gene transcription and cell-cycle regulation and is involved in hematopoiesis40,41 ; however, its precise role in MPN pathogenesis needs to be addressed in functional studies. Recently, Thoennissen et al reported an MPN patient harboring a 0.88-Mb deletion on chromosome 7q that only included the 2 genes CUX1 and SH2B2.24 Our data conclusively show that CUX1 is a major target of chromosome 7q deletions. It remains to be seen whether deletions of SH2B2 play a role in a smaller subset of patients. We also mapped 2 known tumor suppressor genes on chromosome 4 (TET2) and on chromosome 7p (IKZF1), which were reported previously.17,23 The CDR of chromosome 3p mapped to FOXP1, a member of the large family of forkhead transcription factors that are involved in various cellular processes. Differential expression of FOXP1 has been observed in several types of tumors, including both overexpression and loss of expression.42 Loss of heterozygosity of the region on chromosome 3p14 that harbors FOXP1 is common in cancer.43 FOXP1 has been shown to play important roles in B-cell development.44 Its role in myeloid malignancies is so far poorly understood. ETV6 is a transcription factor of the ets family and the only gene in the CDR of chromosome 12p. It was originally identified in a translocation between chromosomes 5 and 12 in a patient with chronic myelomonocytic leukemia,45 and was then shown to be involved in > 40 translocations in several different hematologic malignancies.46 Different fusion genes of ETV6 with other genes, including ABL1, JAK2, and RUNX1, have also been described.47 In addition to translocations, deletions of ETV6 are often reported, which may affect the remaining allele not involved in the translocations.48 Common deleted regions have been mapped to a small region including ETV6 and CDKN1B.47 To our knowledge, the present study is the first report of a single-gene deletion of ETV6 in MPN.
RUNX1 (AML1) has been implicated previously in leukemic transformation of MPNs.39,49 Both mutations and translocations of RUNX1/AML1 are frequently encountered in various leukemias. RUNX1 is a heterodimeric transcription factor that binds to the core element of many enhancers and promoters and is thought to be involved in the development of normal hematopoiesis.50 Inactivating mutations in RUNX1 result in differentiation arrest. Our observation of both point mutations and chromosomal aberrations at the RUNX1 locus in post-MPN AML samples confirms its role in leukemic transformation. With FOXP1, IKZF1, CUX1, ETV6, and RUNX1, 5 of the 6 target genes of CDRs are transcription factors, indicating that transcription factor networks may play crucial roles in MPN pathogenesis.
In this study, we have described several chromosomal regions with recurrent deletions in which the CDRs were small and contained only few genes. We identified a “hot spot” of complex deletions on chromosome 12q that contains CUX2, a homolog of CUX1. In a recent study, CUX2 was found in 1 MPN patient to be homozygously deleted,25 suggesting that both CUX family members play a role in MPN pathogenesis (Figure 4D). Our CDR on chromosome 17q includes the NF1 gene that has been mapped as the target of this chromosomal region.25 The pathogenesis of de novo AML and post-MPN AML involve different genetic mechanisms, because we found mutations of FLT3, IDH1, and IDH2 at lower frequencies than reported in de novo AML. Interestingly, 6 of 29 post-MPN AML patients carried no chromosomal aberrations except 9pUPDs, and were negative for all leukemia-associated mutations tested. This indicates that there are as-yet-unknown pathways of leukemic transformation in MPN.
The results of the present study suggest that the chromosomal aberrations detected in MPN can be separated into 2 groups: the first is compatible with the chronic phase of the disease and the second with disease progression. Understanding the modifications that lead to MPN disease progression has important implications for therapy. Prevention of the leukemic transformation and prolongation of the chronic phase of MPN could be associated with fewer clinical complications and a more favorable prognosis for MPN patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by the Austrian Academy of Sciences, the Austrian Science Fund (FWF, P20033-B11), and the MPN Research Foundation. Studies in Pavia (M.C.) and Florence (A.M.V.) were funded by the Associazione Italiana per la Ricerca sul Cancro (AIRC), Special Program Molecular Clinical Oncology 5 per mille, Milan, Italy; progetto AGIMM, http://www.progettoagimm.it). M.C. was also supported by Fondazione Cariplo (Milan, Italy).
Authorship
Contribution: T.K. and A.H. designed and performed research, analyzed and interpreted data, and wrote the manuscript; T.B., M.S., K.B., D.O., and R.J. performed research; B.G. and D.P. performed DNA extractions and biobanking; F.P. and E.R. managed patient accrual and collected clinical data; L.P. and P.G. managed patient accrual and analyzed data; I.I. and G.M. analyzed data; M.C. and H.G. collected samples and acquired and analyzed clinical data; A.M.V. designed the study, managed patient accrual, and analyzed data; and R.K. designed the research, interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robert Kralovics, PhD, Center for Molecular Medicine (CeMM) of the Austrian Academy of Sciences, Lazarettgasse 14, BT25.3, 1090 Vienna, Austria; e-mail: robert.kralovics@cemm.oeaw.ac.at.
References
Author notes
T.K. and A.H. contributed equally to this study.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal