Abstract
miRNAs have emerged as master regulators of cancer-related events. miRNA dysregulation also occurs in Kaposi sarcoma (KS). Exploring the roles of KS-associated miRNAs should help to identify novel angiogenesis and lymphangiogenesis pathways. In the present study, we show that Kaposi sarcoma-associated herpesvirus (KSHV), the etiological agent of KS, induces global miRNA changes in lymphatic endothelial cells (LECs). Specifically, the miR-221/miR-222 cluster is down-regulated, whereas miR-31 is up-regulated. Both latent nuclear antigen (LANA) and Kaposin B repress the expression of the miR-221/miR-222 cluster, which results in an increase of endothelial cell (EC) migration. In contrast, miR-31 stimulates EC migration, so depletion of miR-31 in KSHV-transformed ECs reduces cell motility. Analysis of the putative miRNA targets among KSHV-affected genes showed that ETS2 and ETS1 are the downstream targets of miR-221 and miR-222, respectively. FAT4 is one of the direct targets of miR-31. Overexpression of ETS1 or ETS2 alone is sufficient to induce EC migration, whereas a reduction in FAT4 enhances EC motility. Our results show that KSHV regulates multiple miRNA-mRNA networks to enhance EC motility, which eventually contributes to KS progression by promoting the spread of malignant KS progenitor cells. Targeting KSHV-regulated miRNAs or genes might allow the development of novel therapeutic strategies that induce angiogenesis or allow the treatment of pathogenic (lymph)angiogenesis.
Introduction
Kaposi sarcoma-associated herpesvirus (KSHV) was identified in 1994.1 KSHV is also associated with 2 other human B-cell leukemias, primary effusion lymphoma (PEL) and multicentric Castleman disease.2 Kaposi sarcoma (KS) is one of the few human cancers derived from a lymphatic endothelial cell (LEC) lineage,3,4 and typically appears as colored lesions or patches on the skin, although it can spread to internal organs.2 This disease has been recognized as a highly disseminated and angiogenic tumor.4 Analyzing KSHV clonality at different stages of KS has shown that KS begins as a polyclonal disease and then evolves into a mono/oligoclonal disease involving infected spindle cells.5 This model therefore implies that the spreading of a few malignant spindle cells in patients' bodies occurs during tumor progression. Moreover, distal metastasis, such as pulmonary KS, can be observed in AIDS-KS patients and causes diffused lung disease. Tumor cells of KSHV-associated B-cell neoplasias (PEL and multicentric Castleman disease) are also able to migrate into body cavities.6 The ability of KSHV to induce cell migration and invasion is therefore important to disease progression.
In vitro, KSHV infects both micro- and macrovascular endothelial cells (ECs), and these cells are useful when studying the role of KSHV in the pathogenesis of KS.7,8 KSHV has also become a powerful tool that aids in the study of EC biology.9 KSHV infects both LECs and blood vessel ECs (BECs) of microvascular origin in vitro.3,4 After infection, KSHV stimulates EC invasiveness by inducing extracellular matrix metalloproteinases (MMPs) such as MMP-1, MMP-2, and MMP-9.10 Because the majority of cells in KS lesions and in KSHV-infected ECs are latently infected by KSHV,4 viral latent proteins may, at least in part, contribute to the invasiveness of KSHV-infected cells. We have shown recently that K15M is a KSHV latent protein capable of inducing cell migration and invasion.11 Studies investigating whether other latent viral proteins also contribute to cell motility and how these genes achieve such an effect not only represent important research fields in KSHV biology, but also enhance our understanding of the regulation of (lymph)angiogenesis.
miRNAs are small RNAs 18-24 nucleotides in length that have emerged as master regulators of cell-lineage differentiation and also as key modulators of cancer.12 Such miRNAs have also been linked to angiogenesis, which is crucial to tumor formation and metastasis. For example, miR-221 and miR-222, which are encoded from the same miRNA cluster, can modulate the angiogenic properties of HUVECs by targeting c-Kit.13 Furthermore, miR-221 and miR-222 are able to reduce indirectly the expression of NOS3, another important factor associated with EC function.14 KSHV also manipulates the expression of cellular miRNAs.15,16 In KS and PEL, tumor-suppressor miRNAs such as miR-221, miR-222, and the let-7 family are underrepresented.16 By comparing primary patient biopsies with well-established culture and mouse tumor models, it was found that the pre-miRNA signatures delineate the various stages of HUVEC transformation.15 Loss of miR-221 and gain of miR-15 expression induce the transition of cells from merely immortalized to fully tumorigenic HUVECs.15 A recent study showed that the K13 vFLIP protein of KSHV is able to suppress CXCR4 expression by up-regulating miR-146a in HUVECs.17 We have contributed to this field by showing that the K15 protein of KSHV up-regulates miR-21 and miR-31 expression via its SH2-binding motif, which then induces cell migration and invasion.11
In the present study, we explored the genes and miRNAs involved in KSHV-induced cell motility and found that KSHV creates a tumor genotype in infected LECs and manipulates both mRNA and miRNA levels to stimulate cell migration. Furthermore, novel miRNA-target gene networks were identified by tandem array analysis and wetlab validation.
Methods
Plasmids
For details on plasmid construction, please refer to supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Cell culture and virus infection
Human primary LECs and BECs were purchased from Clonetics and were cultured as described previously.4 HMEC1 cells18 were cultured in EC growth medium MV (C-22020; PromoCell). LTCs were obtained from Dr Rolf Renne and maintained as described previously.19 A green fluorescent protein (GFP)–recombinant virus, rKSHV.219, which expresses the red fluorescent protein from the KSHV lytic PAN promoter, the GFP from the EF-1α promoter, and contains a gene for puromycin resistance as a selectable marker, was obtained from recombinant JSC-1 cells as described previously.20 All infections were performed at a multiplicity of infection of 10.
Immunofluorescence assay and immunoblotting
Immunofluorescence assays and immunoblots were performed as described previously.11 For details on these procedures, please refer to supplemental Methods.
miRNA microarray and data analysis
The Agilent Human miRNA Microarray Kit V2 (Agilent Technologies), which contains probes for 723 human miRNAs from the Sanger database v10.1, was used. For miRNA array data analysis, the raw dataset from the Agilent Feature Extraction software was normalized and analyzed using the “AgiMicroRna” package of the Bioconductor (http://www.bioconductor.org) suite of software for the R statistical programming language (http://www.r-project.org). The miRNA target prediction was performed using the miRTarG web tool (http://mirtar.mbc.nctu.edu.tw/) and TargetScan (http://www.targetscan.org/).
Array probes and computational analyses
Array data for KS, normal skin, BECs, LECs, KSHV-infected BECs, and KSHV-infected LECs were published previously and are deposited in the Gene Expression Omnibus database with an accession number of GSE16357,21 or were obtained from the ArrayExpress database (E-MEXP-66).4 For details on the analysis of the microarray data, please see supplemental Methods.
Transwell cell-migration assay and Matrigel invasion assay
Cell migration ability was evaluated using Costar Transwell Polycarbonate Permeable Supports (Corning) as described previously.11 The degree of cell invasiveness was examined by the Matrigel Basement Membrane Matrix invasion assay (BD Biosciences) according to the manufacturer's instructions. For details on the Transwell cell-migration assay and Matrigel invasion assay, please see supplemental Methods.
RNA extraction and real-time qRT-PCR
RNA extraction and real-time quantitative RT-PCR (qRT-PCR) were performed as described previously.11 For details, please see supplemental Methods.
miRNA expression
Expression of mature human miRNAs was determined by a stem-loop real-time qRT-PCR system22 using the appropriate primer pairs described in supplemental Table 2. The universal PCR reverse primer for the miRNAs was 5′-GTGCAGGGTCCGAGGT-3′. The miRNA expression data were normalized against U6 small nuclear RNA, which was amplified with the specific forward and reverse primers 5′-CTCGCTTCGGCAGCAC-3′ and 5′-AACGCTTCACGAATTTGCG-3′.
RNA interference-mediated knockdown of LANA and miRNA expression
To specifically knock down latent nuclear antigen (LANA) and miRNA expression, small interference RNA (siRNA) oligonucleotides were electroporated into cells using a Microporator MP-100 (NanoEnTek) according to the manufacturer's instructions. Briefly, 105 cells were resuspended in 10 μL of R buffer (provided in the microporation kit) and mixed with 1 μL of specific siRNA oligonucleotide (50μM) before microporation. After pulse application, the samples were transferred into fresh medium and then further incubated for 48 hours before being subjected to further assays. Anti-LANA siRNA was designed in-house and then purchased from Dharmacon. The sequences used for siLANA were as follows: sense, 5′-AAACAGGUCUCCGGAAAGAUG-3′; and antisense, 5′-CAUCUUUCCGGAGACCUGUUU-3′. For miRNA knockdown, we used commercially available hairpin antagomirs specific to human miR-31 (IH-300507-06-0005; Dharmacon), human miR-221 (IH-300578-07-0005; Dharmacon), and human miR-222 (IH-300579-08-0005; Dharmacon).
Luciferase assay
Luciferase assays were performed to determine the downstream targets of miRNA and the miR-221/miR-222 promoter-responsive regions targeted by KSHV viral proteins. For details on these procedures, please see supplemental Methods.
Results
KSHV introduces tumor traits and migration potential into lymphatic endothelium.
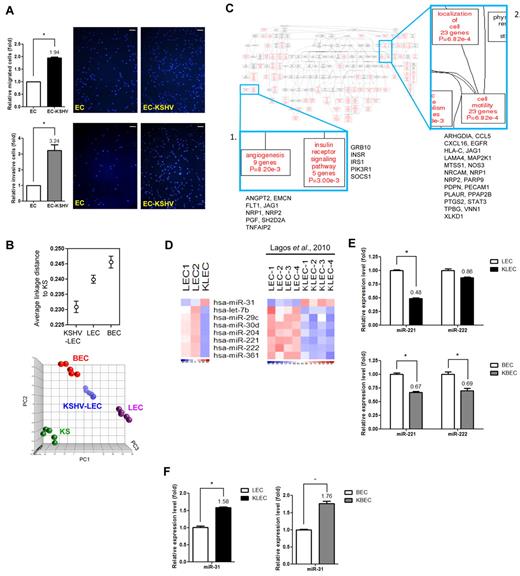
It has been shown that KSHV can induce cellular motility and invasiveness in infected ECs (Figure 1A).10,11 To identify the mechanisms involved, we analyzed KSHV-regulated genes in infected ECs. We used a recombinant KSHV expressing GFP20 to infect a pure population of LECs. At 48 hours after infection, uninfected LECs showed a characteristic contact-inhibited cobblestone appearance, but infected LECs (KSHV-LECs) became elongated and spindle shaped (supplemental Figure 1A). KSHV LANA was expressed in the infected cells and showed a typical nuclear stippling staining pattern (supplemental Figure 1B).
KSHV introduces tumor traits and motility potential and alters cellular miRNA expression in primary LECs. (A) KSHV induces EC motility. HMEC1 cells (EC) or KSHV-infected HMEC1 cells (EC-KSHV) were subjected to the Transwell cell-migration assay (top panel; n = 3) or the Matrigel invasion assay (bottom panel; n = 3). Migrated cells were stained (representative pictures are shown) and counted. Scale bar represents 200 μm. (B) Relationships between the infected and noninfected cell groups. Average linkage distance to KS were calculated using probe sets differentiating LECs, BECs, and KSHV-LECs (q < 0.001 plus > 1.5-fold changes; top panel). PCA using the LEC-BEC discriminatory gene signature (LECs vs BECs; n = 3878, q < 0.001) for LECs, BECs, infected LECs collected 2 days after infection, and KS (bottom panel). Each spot represents a single sample. (C) Altered functional modules in KSHV-LECs. KSHV-regulated LEC genes were subjected to a Gene Ontology database search via the WebGestalt interface. The numbers of genes, gene symbols, their percentages, and P values for each category that are significantly (P < .05) enriched are listed. (D) Heat maps show miRNAs commonly enriched in LECs or KSHV-LECs between our experiment and public data. Down-regulated miRNAs are shown in blue and up-regulated miRNAs in red. (E-F) Validation of the miRNA array data by qRT-PCR. Mean expression levels of the target miRNAs are compared with the U6 control.
KSHV introduces tumor traits and motility potential and alters cellular miRNA expression in primary LECs. (A) KSHV induces EC motility. HMEC1 cells (EC) or KSHV-infected HMEC1 cells (EC-KSHV) were subjected to the Transwell cell-migration assay (top panel; n = 3) or the Matrigel invasion assay (bottom panel; n = 3). Migrated cells were stained (representative pictures are shown) and counted. Scale bar represents 200 μm. (B) Relationships between the infected and noninfected cell groups. Average linkage distance to KS were calculated using probe sets differentiating LECs, BECs, and KSHV-LECs (q < 0.001 plus > 1.5-fold changes; top panel). PCA using the LEC-BEC discriminatory gene signature (LECs vs BECs; n = 3878, q < 0.001) for LECs, BECs, infected LECs collected 2 days after infection, and KS (bottom panel). Each spot represents a single sample. (C) Altered functional modules in KSHV-LECs. KSHV-regulated LEC genes were subjected to a Gene Ontology database search via the WebGestalt interface. The numbers of genes, gene symbols, their percentages, and P values for each category that are significantly (P < .05) enriched are listed. (D) Heat maps show miRNAs commonly enriched in LECs or KSHV-LECs between our experiment and public data. Down-regulated miRNAs are shown in blue and up-regulated miRNAs in red. (E-F) Validation of the miRNA array data by qRT-PCR. Mean expression levels of the target miRNAs are compared with the U6 control.
Total RNAs were extracted from infected and uninfected LECs, as well as from BECs and KS tissues, and their global mRNA expression patterns were determined by Affymetrix whole-genome microarray. To provide new insights into KSHV-induced transcriptome changes in LECs, the global mRNA expression pattern of KSHV-LECs was analyzed. It has been reported that KSHV-infected LECs show an up-regulation of genes associated with BECs.4,23 We examined the LEC-to-BEC transcriptome reprogramming present in our array data to confirm that the KSHV infection had been successful. By calculating the transcriptome distances between sample groups, we confirmed that KSHV-infected LECs were closer to BECs than parental LECs (supplemental Figure 1C). Furthermore, we found that the transcriptome of infected LECs was closer to that of KS than to uninfected ECs, suggesting that KSHV brought about oncogenic events early in the infected population (Figure 1B top panel). A principle component analysis (PCA) plot using genes differentiating LECs and BECs (n = 3878, pFDR q < 0.001) reconstructed the above sample group relationships (Figure 1B bottom panel).
To disclose motility-related genes changes in KSHV-LECs, we subjected the 1499 probe sets up-regulated in KSHV-LECs (with a false discovery rate q < 0.01 plus a fold change of > 1.5-fold; supplemental Table 1) to a Gene Ontology database search24 to identify statistically overrepresented functional groups within the gene lists. The WebGestalt web tool25 was applied to carry out the statistical analysis and visual presentation. The Gene Ontology categories of biologic processes being statistically overrepresented (P < .05) among the KSHV-LEC genes are shown in Figure 1C. Nine angiogenesis-related genes were induced by KSHV (Figure 1C panel 1); 23 genes involved in cell motility were also overexpressed after KSHV infection (Figure 1C panel 2). Other related predominant processes deregulated in the KSHV-LECs included those pertaining to cellular signaling pathways, such as integrin-mediated signaling, insulin receptor signaling, and the JAK-STAT pathway (supplemental Figure 2).
miRNA signature of KSHV-infected LECs
Another level of gene-expression regulation is through miRNAs.26,27 To provide a more comprehensive view of how transcriptome profiles might be correlated with KSHV biology and KS pathogenesis, we also determine the miRNA profiles of infected LECs. RNA was collected 2 days after KSHV infection, and miRNAs that were differentially expressed between infected and parental LECs were identified with a significance level of q < 0.05 plus > 1.5-fold change. Because there is another dataset describing KSHV-regulated miRNAs in LECs,28 we compared our data with this public dataset to aid the identification of the miRNAs commonly deregulated in both experiments. As shown in Figure 1D, both datasets suggested miR-31 is up-regulated in KSHV-LECs, whereas another 7 miRNAs are down-regulated (Figure 1D). Among these 7 miRNAs, miR-221 and miR-222 have also been found to be down-regulated in PEL and KS tissues.16
The differential expression of miR-221/miR-222, as well as miR-31, was examined by qRT-PCR, and their expression levels were also evaluated in KSHV-infected primary BECs. The expression of miR-221 was repressed in both infected LECs and BECs, whereas the level of miR-222 was only affected in infected BECs (Figure 1E). Conversely, miR-31 was up-regulated in both infected LECs and infected BECs (Figure 1F).
miR-221/miR-222 and miR-31 are involved in KSHV-induced EC migration
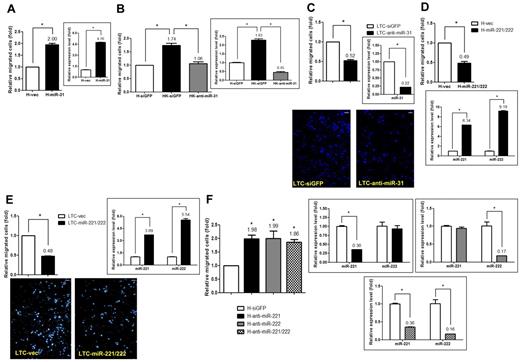
miR-31 is a novel angiogenic miRNA for ECs.11 Lagos et al showed recently that during KSHV infection, miR-31 is induced as early as at 6 hours after infection and then persists until at least 72 hours after infection.28 Conversely, miR-221 and miR-222, which are transcribed from the same primary microRNA (pri-miRNA) cluster, are known to inhibit HUVEC migration and endothelial nitric oxide synthase production.13,14 Whether these miRNAs directly contribute to whole virus–induced EC motility remains unexplored. We expressed miR-31 via a lentivirus infection system in an immortalized MVEC cell line, HMEC1.18 miR-31 alone was sufficient to increase EC migration (Figure 2A), and a similar result was observed in HUVECs (supplemental Figure 3A). The direct involvement of miR-31 in KSHV-induced cell motility was confirmed using a virus(+) background. Specifically, when endogenous miR-31 in KSHV-infected HMEC1 (HK) cells was knocked down with a siRNA antagomir specific to miR-31, KSHV-induced cell migration was reduced to a basal level even though KSHV was still present (Figure 2B). The direct role of miR-31 in KS pathogenesis was further examined in a KSHV-transformed, tumorigenic HUVEC line called LTC.19 When LTCs were transfected with an anti–miR-31 antagomir and subjected to the Transwell cell-migration assay, it was found that the reduction in miR-31 level reduced LTC motility (Figure 2C).
miR-221/miR-222 and miR-31 are involved in KSHV-induced endothelial migration. (A) miR-31 induces cell migration. HMEC1 cells stably transduced with miR-31 or the vector control by lentivirus were used to detect the expression level of miR-31 by qRT-PCR (right panel) and were also subjected to the Transwell cell-migration assay (left panel; n = 3). (B) Knockdown of miR-31 reduces KSHV-induced cell migration. KSHV-infected HMEC1 cells (HK cells) transfected with the siGFP control or the anti–miR-31 antagomir by electroporation were used to detect the expression of miR-31 (right panel) and were also subjected to the Transwell cell-migration assay (left panel; n = 3). (C) LTCs with miR-31 knockdown were subjected to the Transwell cell-migration assay (n = 3). Migrated cells were stained (representative pictures are shown in bottom panel) and counted. (D-F) Both miR-221 and miR-222 inhibit cell migration. HMEC1 cells (D) or LTCs (E) stably transduced with the miR-221/miR-222 cluster or HMEC1 cells knocked down with specific antagomirs to a single miRNA or to both by electroporation (F) were used to detect the expression levels of miR-221 and miR-222 and were also subjected to the Transwell cell-migration assay (n = 3). Scale bar represents 100 μm.
miR-221/miR-222 and miR-31 are involved in KSHV-induced endothelial migration. (A) miR-31 induces cell migration. HMEC1 cells stably transduced with miR-31 or the vector control by lentivirus were used to detect the expression level of miR-31 by qRT-PCR (right panel) and were also subjected to the Transwell cell-migration assay (left panel; n = 3). (B) Knockdown of miR-31 reduces KSHV-induced cell migration. KSHV-infected HMEC1 cells (HK cells) transfected with the siGFP control or the anti–miR-31 antagomir by electroporation were used to detect the expression of miR-31 (right panel) and were also subjected to the Transwell cell-migration assay (left panel; n = 3). (C) LTCs with miR-31 knockdown were subjected to the Transwell cell-migration assay (n = 3). Migrated cells were stained (representative pictures are shown in bottom panel) and counted. (D-F) Both miR-221 and miR-222 inhibit cell migration. HMEC1 cells (D) or LTCs (E) stably transduced with the miR-221/miR-222 cluster or HMEC1 cells knocked down with specific antagomirs to a single miRNA or to both by electroporation (F) were used to detect the expression levels of miR-221 and miR-222 and were also subjected to the Transwell cell-migration assay (n = 3). Scale bar represents 100 μm.
HMEC1 cells and LTCs were also used to link miR-221/miR-222 to KSHV-induced EC motility. When the miR-221/miR-222 pri-miRNA cluster was transduced into HMEC1 cells and LTCs by lentivirus, both transductants showed reduced cell motility (Figure 2D and E, respectively). In contrast, when the expression of endogenous miR-221 and/or miR-222 was knocked down with siRNA antagomirs specifically to miR-221, miR-222, or both, knocking down either of the miRNAs resulted in an increase in EC motility (Figure 2F). However, knocking down both miR-221 and miR-222 at the same time did not further increase cellular motility (Figure 2F).
KSHV viral gene library screening identified that LANA and Kaposin B down-regulate the levels of miR-221 and miR-222 and thus modulate EC motility
We have found previously that the K15 protein of KSHV is able to induce miR-31 expression in ECs11 ; however, which of the viral proteins may be involved in down-regulating the expression of miR-221/miR-222 remained unclear. We have previously shown that the K15 protein of KSHV is not able to regulate the level of either miR-221 or miR-222.11 A lentiviral library of selected KSHV latent proteins (including LANA, vCyclin, vFLIP, and K12/Kaposin B) was used in a series of screening experiments. The expression of all genes in HMEC1 cells was confirmed by RT-PCR (supplemental Figure 4A-B). The efficiency of lentiviral infection of HMEC1 cells was measured using a GFP-encoding lentivirus, and it was found that > 80% of cells were GFP positive when a multiplicity of infection of 10 was used (not shown). The expression of LANA and Kaposin B protein in transduced ECs was further verified by IFA, and a total of 80%-90% of the cells were found to be positive for LANA/Kaposin B staining (Figure 3A and B, respectively).
LANA and Kaposin B down-regulate miR-221 and miR-222 levels, resulting in modulation of EC motility. (A) Immunofluorescence staining for LANA proteins. HMEC1 cells with stable LANA expression or the vector control were fixed, and LANA protein was detected by anti-LANA mAb, followed by anti–rat IgG secondary antibody conjugated with rhodamine (red). An enlarged LANA staining pattern is shown in the bottom panel. (B) Immunofluorescence staining and immunoblotting for Kaposin B proteins. HMEC1 cells with stable Kaposin B expression or the vector control were fixed, and Kaposin B proteins were detected by anti-Flag mAb, followed by anti–mouse IgG secondary antibody conjugated with FITC (green). Note that Kaposin B is expressed in the nucleus. Scale bar represents 25 μm. Immunoblotting of Kaposin B proteins with anti-Flag mAb is shown. Actin was used as an internal control (bottom panel). (C) Both LANA and Kaposin B can repress the expression of miR-221 and miR-222. HMEC1 cells with stable LANA (top panel) or Kaposin B (bottom panel) expression were used to detect the expression levels of miR-221 and miR-222 by qRT-PCR. (D) Knocking down LANA expression in HK cells resulted in the restoration of the miR-221 level, but not the level of miR-222. HK cells electroporated with siLANA or siGFP control siRNA were subjected to the Transwell cell-migration assay (top panel; n = 2) and also used to detect the expression level of miR-221 and miR-222 (bottom panel). (E) LANA- or Kaposin B–induced cell migration is countered by miR-221/miR-222. HMEC1 cells with stable LANA (left panel) or Kaposin B (right panel) expression were transduced with vector or the miR-221/miR-222 cluster, and then the cells were subjected to the Transwell cell-migration assay (n = 3). (F) LANA and Kaposin B individually repress miR-221/miR-222 expression by targeting the promoter region. miR-221/miR-222 promoter reporter assays were performed by transfection with different promoter fragments inserted into luciferase reporter plasmids in stably LANA- or Kaposin B–expressing 293T cell lines. Note that LANA and Kaposin B repress miR-221/miR-222 promoter activity by targeting different regions of the promoter, namely the −150 to −200 bp and the −200 to −600 bp regions, respectively (n = 3).
LANA and Kaposin B down-regulate miR-221 and miR-222 levels, resulting in modulation of EC motility. (A) Immunofluorescence staining for LANA proteins. HMEC1 cells with stable LANA expression or the vector control were fixed, and LANA protein was detected by anti-LANA mAb, followed by anti–rat IgG secondary antibody conjugated with rhodamine (red). An enlarged LANA staining pattern is shown in the bottom panel. (B) Immunofluorescence staining and immunoblotting for Kaposin B proteins. HMEC1 cells with stable Kaposin B expression or the vector control were fixed, and Kaposin B proteins were detected by anti-Flag mAb, followed by anti–mouse IgG secondary antibody conjugated with FITC (green). Note that Kaposin B is expressed in the nucleus. Scale bar represents 25 μm. Immunoblotting of Kaposin B proteins with anti-Flag mAb is shown. Actin was used as an internal control (bottom panel). (C) Both LANA and Kaposin B can repress the expression of miR-221 and miR-222. HMEC1 cells with stable LANA (top panel) or Kaposin B (bottom panel) expression were used to detect the expression levels of miR-221 and miR-222 by qRT-PCR. (D) Knocking down LANA expression in HK cells resulted in the restoration of the miR-221 level, but not the level of miR-222. HK cells electroporated with siLANA or siGFP control siRNA were subjected to the Transwell cell-migration assay (top panel; n = 2) and also used to detect the expression level of miR-221 and miR-222 (bottom panel). (E) LANA- or Kaposin B–induced cell migration is countered by miR-221/miR-222. HMEC1 cells with stable LANA (left panel) or Kaposin B (right panel) expression were transduced with vector or the miR-221/miR-222 cluster, and then the cells were subjected to the Transwell cell-migration assay (n = 3). (F) LANA and Kaposin B individually repress miR-221/miR-222 expression by targeting the promoter region. miR-221/miR-222 promoter reporter assays were performed by transfection with different promoter fragments inserted into luciferase reporter plasmids in stably LANA- or Kaposin B–expressing 293T cell lines. Note that LANA and Kaposin B repress miR-221/miR-222 promoter activity by targeting different regions of the promoter, namely the −150 to −200 bp and the −200 to −600 bp regions, respectively (n = 3).
The screening for down-regulation of miR-221/miR-222 was done by monitoring miR-221 and miR-222 levels at 48 hours after infection using the different lentiviral preparations. Three independent screenings revealed that LANA and Kaposin B were able to decrease miR-221/miR-222 levels compared with empty vector (Figure 3C), whereas vCyclin and vFLIP did not repress miR-221/miR-222 levels significantly (supplemental Figure 4C-D). Knocking down LANA expression in HK cells resulted in the restoration of miR-221 expression, but not that of miR-222 (Figure 3D). We could not knock down the expression of endogenous Kaposin B in HK cells because the Kaposin B gene has many repeat sequences29 and is difficult to design effective siRNA oligos.
We hypothesized that LANA and Kaposin B by themselves are able to induce cell migration. HMEC1 cells overexpressing LANA or Kaposin B were subjected to a Transwell cell-migration assay, and LANA- or Kaposin B–transduced HMEC1 cells migrated faster than control cells transduced with vector only (Figure 3E). When the miR-221 and miR-222 expression levels were restored in LANA(+) or Kaposin B(+) HMEC1 cells by transducing the cells with the miR-221/miR-222 pri-miRNA cluster expression plasmid, LANA- and Kaposin B–induced cellular motility was suppressed (Figure 3E), implying a close link between miR-221/miR-222 and LANA- or Kaposin B–induced cell migration.
We next investigated how LANA and Kaposin B repress miR-221/miR-222 expression. LANA is known to regulate cellular gene expression by acting as a transcription cofactor.30,31 Because Kaposin B is also a nuclear protein, we hypothesize that Kaposin B may also be a novel transcription cofactor that can repress the promoter activity of the miR-221/miR-222 genes. We performed reporter assays to investigate whether the miR-221/miR-222 promoter can be repressed by LANA and Kaposin B, as well as to identify any critical binding sites for these viral proteins within the miR-221/miR-222 promoter. This was done by applying reporter constructs containing different lengths of the miR-221/miR-222 promoter (Figure 3F left panel).32 Repression of miR-221/miR-222 promoter activity was observed after transient transfection into stable LANA- or Kaposin B–expressing 293T cell lines (Figure 3F 1600-bp group). Deleting a region from −150 to −200 bp abolished the repression of the miR-221/miR-222 promoter by LANA (Figure 3F middle panel). Conversely, deleting a region from −200 to −600 bp abolished the repression of the miR-221/miR-222 promoter by Kaposin B (Figure 3F right panel). These results show that LANA and Kaposin B use different motifs within the miR-221/miR-222 promoter for regulation.
Identification of miRNA targets among KSHV-affected genes
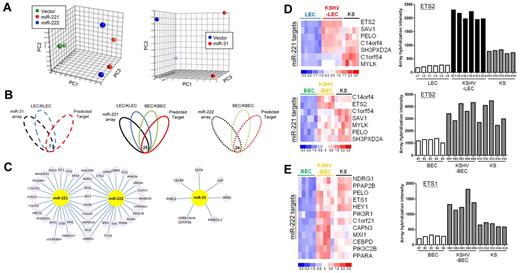
To gain insight into the mechanism underlying the functioning of these miRNAs, we used a combination of miRTarG and TargetScan33 algorithms together with gene-expression microarrays to obtain potential targets for miR-221, miR-222, and miR-31. Expression plasmids of these 3 miRNAs were transduced into HMEC1 cells by lentivirus, and the total RNAs were collected 3 days after infection. Microarray analysis revealed a total of 1872 and 1743 probe sets that were down-regulated by overexpression of miR-221 and miR-222, respectively. Overexpression of miR-31 resulted in the inhibition of 65 probe sets in ECs. The discrimination abilities of these probe sets were confirmed by PCA plot (Figure 4A).
Downstream target identification for KSHV-regulated miRNAs. (A) PCA using differentially expressed genes (P < .005 plus > 1.5-fold change). Each spot represents a single array. (B) Venn diagram showing the principle of selecting putative miRNA-regulated targets. Array data showing overexpression of the miRNA is shown in black, LECs versus KLECs are shown in blue, BECs versus KBECs are shown in green, and the computational prediction is shown in red. The number at the intersection indicates the overlapped genes across all groups. (C) Details of miRNA targets, with 9 common targets being found for miR-221 and miR-222. (D-E) Heat maps and histograms showing that our predicted miRNA targets are still deregulated in KS tissues.
Downstream target identification for KSHV-regulated miRNAs. (A) PCA using differentially expressed genes (P < .005 plus > 1.5-fold change). Each spot represents a single array. (B) Venn diagram showing the principle of selecting putative miRNA-regulated targets. Array data showing overexpression of the miRNA is shown in black, LECs versus KLECs are shown in blue, BECs versus KBECs are shown in green, and the computational prediction is shown in red. The number at the intersection indicates the overlapped genes across all groups. (C) Details of miRNA targets, with 9 common targets being found for miR-221 and miR-222. (D-E) Heat maps and histograms showing that our predicted miRNA targets are still deregulated in KS tissues.
To further narrow down the miRNA targets, we focused on KSHV-regulated genes among the filtered mRNA targets and the output data from the computational prediction programs. We focused on genes deregulated in LECs, and 7 putative miR-31 targets for down-regulation by KSHV in LECs were identified (Figure 4B left panel). In contrast, 161 candidate miR-221 targets were identified as being up-regulated in KSHV-LECs (not shown). To further narrow down candidate miR-221 targets, genes deregulated by KSHV in BECs (KSHV-BEC signature; q < 0.01, 2541 probe sets) were also taken into account. The result was that a total of 27 probe sets (25 genes) were identified (Figure 4B middle panel). For miR-222, because it was specifically diminished by KSHV only in BECs (Figure 1E), only the KSHV-BEC signature was used. A total of 26 probe sets (24 genes) for miR-222 were identified (Figure 4B right panel). Figure 4C illustrates the details of these miRNA targets. In total, 9 common targets were found for miR-221 and miR-222.
We further explored which of the above miRNA targets were also deregulated in KS tissues. Seven miR-221 targets (including ETS2 and PELO) were found to be up-regulated in KS tissues, as well as in KSHV-LECs and KSHV-BECs (Figure 4D). Conversely, 12 miR-222 targets (including ETS1, PELO, and HEY1) were up-regulated in KS tissue (Figure 4E).
ETS2 and ETS1 are novel downstream targets of miR-221 and miR-222, respectively
The transcription factors ETS1 and ETS2 are targets of the Ras/Raf/Mek/Erk pathway, which has been implicated in EC function and motility in vitro.34 The repression of both miR-221 and miR-222 by KSHV suggests that KSHV may exploit the ETS family pathway for increasing EC activities and (lymph)angiogenesis. The reduced expression level of ETS2 in miR-221 transfectants, and that of ETS1 in miR-222 transfectants, was verified by qRT-PCR and immunoblotting (Figure 5A-B), and the increase of ETS2 and ETS1 levels were observed in LECs transfected with anti–miR-221 or anti–miR-222 antagomir (supplemental Figure 5B and C, respectively). The abundant expression of both ETS1 and ETS2 in KSHV-infected LECs and BECs was also observed (Figure 5C-D). The protein levels of ETS1 and ETS2 were increased in KSHV-infected HMEC1 cells (Figure 5E). Overexpressing ETS2 or ETS1 in HMEC1 cells also clearly induced EC motility in a Transwell cell-migration assay (Figure 5F-G), and the same results were observed in HUVECs (supplemental Figure 3B).
ETS2 and ETS1, which are downstream targets of miR-221 and miR-222, respectively, induce EC migration. (A) ETS2 is repressed by miR-221. The expression level of ETS2 was detected by qRT-PCR (top panel) or immunoblotting (bottom panel) in HMEC1 cells with stably expressed miR-221. (B) ETS1 is repressed by miR-222. The mRNA and protein levels of ETS1 was detected in HMEC1 cells with stably expressed miR-222. (C-D) ETS2 and ETS1 are up-regulated in KSHV-infected LECs and BECs. The expression level of ETS2 (C) or ETS1 (D) was detected in KSHV-infected LECs or BECs. (E) ETS2 and ETS1 are up-regulated in KSHV-infected HMEC1 cells. The protein levels of ETS2 and ETS1 were detected by immunoblotting in HMEC1 cells or HK cells. Relative band intensities are shown. (F-G) Both ETS2 and ETS1 are able to induce EC migration. HMEC1 cells stably transduced with an ETS2-expressing plasmid (F) or an ETS1-expressing plasmid (G) were used to detect their protein expression level by immunoblotting (left panel) and were also subjected to the Transwell cell-migration assay (right panel; n = 3). (H) Structure of the ETS2 transcript and predicted seed region duplex formed between ETS2 and miR-221 (left panel). Reporter activity of the ETS2 3′UTR reporter construct after cotransfection with the miR-221–expressing or empty vector into 293T cells (right panel; n = 3). (I) Structure of the ETS1 transcript and 2 putative miR-222–binding sites (top panel). Reporter activity of the ETS1 3′UTR reporter constructs (UTR1 or UTR2) after cotransfection with the miR-222–expressing or empty vector into 293T cells (bottom panel; n = 3).
ETS2 and ETS1, which are downstream targets of miR-221 and miR-222, respectively, induce EC migration. (A) ETS2 is repressed by miR-221. The expression level of ETS2 was detected by qRT-PCR (top panel) or immunoblotting (bottom panel) in HMEC1 cells with stably expressed miR-221. (B) ETS1 is repressed by miR-222. The mRNA and protein levels of ETS1 was detected in HMEC1 cells with stably expressed miR-222. (C-D) ETS2 and ETS1 are up-regulated in KSHV-infected LECs and BECs. The expression level of ETS2 (C) or ETS1 (D) was detected in KSHV-infected LECs or BECs. (E) ETS2 and ETS1 are up-regulated in KSHV-infected HMEC1 cells. The protein levels of ETS2 and ETS1 were detected by immunoblotting in HMEC1 cells or HK cells. Relative band intensities are shown. (F-G) Both ETS2 and ETS1 are able to induce EC migration. HMEC1 cells stably transduced with an ETS2-expressing plasmid (F) or an ETS1-expressing plasmid (G) were used to detect their protein expression level by immunoblotting (left panel) and were also subjected to the Transwell cell-migration assay (right panel; n = 3). (H) Structure of the ETS2 transcript and predicted seed region duplex formed between ETS2 and miR-221 (left panel). Reporter activity of the ETS2 3′UTR reporter construct after cotransfection with the miR-221–expressing or empty vector into 293T cells (right panel; n = 3). (I) Structure of the ETS1 transcript and 2 putative miR-222–binding sites (top panel). Reporter activity of the ETS1 3′UTR reporter constructs (UTR1 or UTR2) after cotransfection with the miR-222–expressing or empty vector into 293T cells (bottom panel; n = 3).
The TargetScan program indicated a potentially favorable interaction between miR-221 and one 7-mer site in the 3′ untranslated region (UTR) of ETS2 (Figure 5H). Luciferase assays were used to confirm the predicted interaction of miR-221 with the 3′UTR of ETS2. miR-221 repressed a construct containing the ETS2 3′UTR downstream of the luciferase gene (Figure 5H left panel). This was reversed by mutating the predicted miR-221–binding site (Figure 5H UTR-mut group). In addition, 2 putative miR-222–binding sites were mapped in the 3′UTR of ETS1 (Figure 5I top panel). Reporter assays again showed that miR-222 could directly interact with both binding motifs and reduce the activity of a luciferase reporter gene fused to the ETS1 3′UTR (Figure 5I). Mutating either of the predicted miR-222–binding sites restored miR-222–mediated repression (Figure 5I).
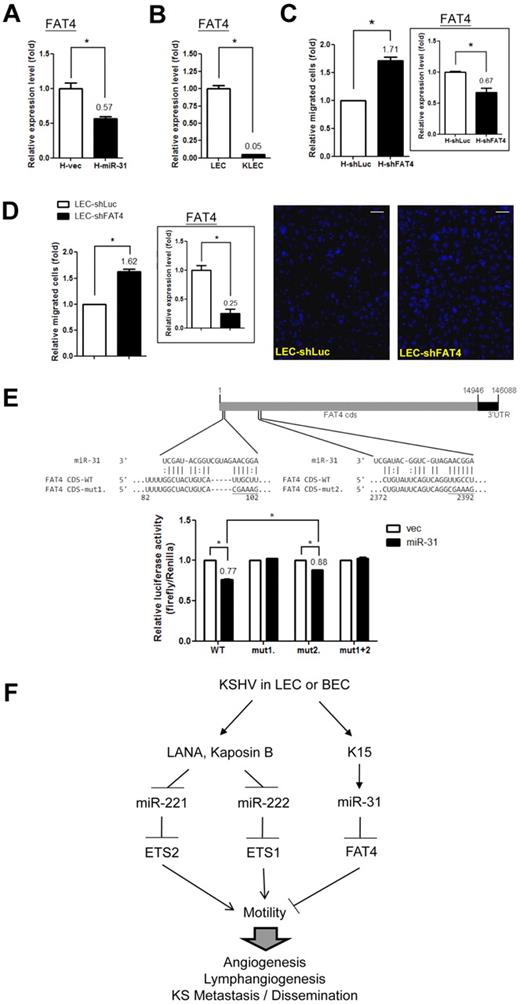
FAT4, a novel tumor suppressor and migration inhibitor, is a downstream target of miR-31 in LEC
Among the 7 identified putative miR-31 target genes, FAT4 is a novel tumor suppressor in breast cancer that can inhibit tumor growth in knockout mice.35 Overexpression of miR-31 in ECs reduced the level of FAT4 (Figure 6A for HMEC1 cells and supplemental Figure 5A for LECs), and the reduction in FAT4 expression in KSHV-LECs was confirmed by qRT-PCR (Figure 6B). Knocking down FAT4 in ECs resulted in an induction of cell motility (Figure 6C and supplemental Figure 3C). Knocking down FAT4 in LECs also enhanced cell migration ability (Figure 6D), suggesting a role for FAT4 in (lymph)angiogenesis. Bioinformatics analysis indicated 2 putative miR-31 target sites in the coding regions of FAT4 (Figure 6E top panel). Luciferase assays were used to confirm the predicted interaction of miR-31 with FAT4. miR-31 repressed constructs containing the FAT4-coding region fused downstream of the luciferase gene (Figure 6E). This repression was reversed by mutating either of the predicted miR-31–binding sites (Figure 6E). However, the binding site around nucleotide 100 in the FAT4 gene would seem to be more crucial than the other, because mutating the former binding site complete reversed miR-31–mediated repression (Figure 6E).
FAT4, a tumor suppressor and migration inhibitor, is a downstream target of miR-31. (A) FAT4 is down-regulated by miR-31. The expression level of FAT4 was detected by qRT-PCR in HMEC1 cells with stably expressed miR-31. (B) FAT4 is down-regulated in KSHV-infected LECs. The expression level of FAT4 was detected in KSHV-infected LECs. (C-D) Knockdown of FAT4 increases EC migration ability. HMEC1 cells (C) or LECs (D) that had undergone stable knockdown by shRNA lentivirus infection had their FAT4 expression level measured by qRT-PCR (right panel) and were also subjected to the Transwell cell-migration assay (left panel; n = 3). Migrated LECs were stained (representative photos are shown in the right panel) and counted. Scale bar represents 100 μm. (E) Structure of the FAT4 transcript and predicted seed region duplex formed between FAT4 and miR-31 (top panel). Reporter activity of the FAT4 CDS 1-2500 bp reporter construct after cotransfection with the miR-31–expressing or empty vector into 293T cells (bottom panel; n = 3). (F) Proposed model of EC motility regulation by KSHV.
FAT4, a tumor suppressor and migration inhibitor, is a downstream target of miR-31. (A) FAT4 is down-regulated by miR-31. The expression level of FAT4 was detected by qRT-PCR in HMEC1 cells with stably expressed miR-31. (B) FAT4 is down-regulated in KSHV-infected LECs. The expression level of FAT4 was detected in KSHV-infected LECs. (C-D) Knockdown of FAT4 increases EC migration ability. HMEC1 cells (C) or LECs (D) that had undergone stable knockdown by shRNA lentivirus infection had their FAT4 expression level measured by qRT-PCR (right panel) and were also subjected to the Transwell cell-migration assay (left panel; n = 3). Migrated LECs were stained (representative photos are shown in the right panel) and counted. Scale bar represents 100 μm. (E) Structure of the FAT4 transcript and predicted seed region duplex formed between FAT4 and miR-31 (top panel). Reporter activity of the FAT4 CDS 1-2500 bp reporter construct after cotransfection with the miR-31–expressing or empty vector into 293T cells (bottom panel; n = 3). (F) Proposed model of EC motility regulation by KSHV.
Discussion
ECs form the internal barrier of the vasculature and play fundamental roles in vascular development and disease.36 Aberrant endothelial motility and the resulting (lymph)angiogenesis cause a variety of diseases, such as ischemia, lymphedema, cancer, and metastasis. An increasing number of studies have shown that miRNAs, or angiomiRs, play a crucial role in regulating various aspects of cancer biology, including angiogenesis.36 In the present study, we found that KSHV exploits several cellular miRNA pathways to promote EC migration (Figure 6F), which may eventually benefit disease progression and KS/PEL cell spread in vivo. We have identified novel miRNA-target gene pairs for KSHV-infected primary ECs. The results also disclose for the first time to our knowledge that certain genes are also involved in (lymph)angiogenesis. For example, we have discovered a novel tumor suppressor, FAT4, which contributes directly to EC motility.
The involvement of cellular miRNA in viral pathogenesis has attracted much attention. For example, miR-132, a newly confirmed angiogenic miRNA,37 is also induced by KSHV to down-regulate host immunity.28 Whether KSHV exploits the miR-132 pathway to enhance EC motility has not been examined. In a study conducted to delineate the pre-miRNA signatures of KS tumor progression, the loss of miR-221 expression was shown to demark the transition from merely immortalized ECs to fully tumorigenic cells.15 Furthermore, the miR-221 and miR-222 tumor-suppressor miRNAs were found to be significantly down-regulated in PEL and KS.16 We found by array analysis that miR-221 and miR-222 target the ETS2 and ETS1 transcription factors, respectively. In vivo, the overlapping and redundant roles of ETS1 and ETS2 in the regulation of EC function and survival during embryonic angiogenesis has been discovered recently. Specifically, neither ETS1-null nor ETS2-null mice exhibited EC defects, yet the combined homozygous mutant alleles of these 2 genes result in defective blood vessel branching and embryonic lethality, which suggests that both proteins use a convergent mechanism for the remodeling of the various vascular systems.38 MMP9 and antiapoptotic genes (including Bcl-XL and cIAP2) are the common downstream targets of ETS1 and ETS2.38 The repression of the miR-221/miR-222 cluster and the induction of both ETS1 and ETS2 by KSHV therefore indicate that KSHV exploits this crucial and overlapping ETS1 and ETS2 pathway to ensure increased EC activities. Taking advantage of the structural similarities between the subfamilies in the DNA-binding domain of ETS1 and ETS2, it is possible to inhibit both oncogenes in KS or neoangiogenic blood vessels at the same time using a common therapeutic agent.39
We also found by gene profiling that miR-221 and miR-222 share redundant activities involving the targeting of common mRNAs. Overall, 39% of miR-222–regulated genes, including BCL6, C14orf4, CXCR4, HEY1, KIAA1946, LMBRD1, NDRG1, PELO, and SMPDL3A, are also the targets of miR-221 (Figure 4C). The shared targets between these 2 functionally close miRNA homologs suggests functional redundancy between these 2 close relatives, and also explains why knocking down both miRNAs together does not further induce EC motility (Figure 2F). From a therapeutic standpoint, recent studies support the important role of miR-221 and miR-222 as critical transcriptional repressors of tumor angiogenesis.36 Local administration of the miR-221/miR-222 pri-miRNA cluster may significantly block KS and PEL growth.
Because miR-221 and miR-222 are transcribed from the same pri-miRNA cluster, we anticipated a similar expression pattern for both miRNAs in tissues and cells. This is the case in PEL and KS, because both miR-221 and miR-222 are down-regulated.16 Although this is also true in infected BECs, we observed that there was asynchronous expression between miR-221 and miR-222 in KSHV-LECs. Furthermore, knocking down of LANA in KSHV-ECs restored only the expression level of miR-221 (Figure 3D). However, the pri-miR-221/miR-222 transcript level was still down-regulated in KSHV-LECs and up-regulated in HK-siLANA cells (not shown). These results suggest that there is a differential processing and degradation of the newly synthesized pre–miR-221 and pre–miR-222 transcripts. How miR-222 is controlled at the posttranscriptional stage in a cell-type–specific manner and whether and how other KSHV latent or lytic proteins may also be involved in miR-221/miR-222 biogenesis are exciting questions and will represent an important field of further study in KSHV as well as miRNA biology.
Kaposin B is one of the few viral genes expressed during latency, and is therefore expressed in all KSHV tumor cells.40 The Kaposin genes are expressed from a common locus in the viral genome: Kaposin A is a product of the K12 open reading frame, whereas Kaposin B and C are generated from the direct repeats upstream of K12 or from fused direct repeat–K12 sequences, respectively, with Kaposin B being the dominant form.29 Kaposin B has been reported to regulate cellular gene expression at the posttranscriptional level by increasing mRNA stability: Kaposin B activates the p38 MAPK–MK2 pathway to stabilize transcripts containing AU-rich elements.40 In the present study, we explored the mechanistic aspects of Kaposin B by showing that it, like another latent protein, LANA, is also a nuclear transcriptional cofactor that can regulate directly the promoter activities of cellular genes such as miR-221/miR-222. We further showed for the first time that Kaposin B and LANA can both induce EC motility (Figure 3E). Our study therefore provides novel mechanistic insights into how KSHV latent proteins induce cell motility and how Kaposin B regulates host and perhaps viral gene expression. Transcriptome analysis on ECs overexpressing Kaposin B revealed that Kaposin B regulates a significant group of genes involved in cellular motility and adhesion, and most of the deregulated mRNAs (including ETS1 and ETS2) do not contain an AU-rich element (data not shown). Kaposin B may regulate the promoter activities of these genes to enhance cell motility.
miR-31 is another angiogenic miRNA regulated by KSHV and is a commonly up-regulated miRNA as shown by our array data and another published dataset.28 It is known that miR-31 plays opposing roles in different tissues. For example, miR-31 is up-regulated in colon cancer tissue41 and can enhance colon cancer cell migration and metastasis.42 In contrast, miR-31 behaves as a tumor suppressor that inhibits metastasis in breast cancer.43 We recently found that the KSHV K15 protein can up-regulate miR-31 expression, thereby inducing EC migration and invasion.11 How miR-31 regulates either endothelial motility or angiogenesis remained unclear. A recent report showed that miR-31 is able to target the adhesion molecule E-selectin in HUVECs.44 Because E-selectin is involved in the inhibition of angiostatin-induced angiogenesis,45 inhibition of E-selectin by miR-31 will eventually benefit cell migration and angiogenesis. In the present study, we reveal a novel mechanism whereby miR-31 can induce EC motility by targeting FAT4, a novel tumor suppressor of breast cancer35 (Figure 6). FAT4 is a huge transmembrane protein (∼ 540 kDa) belonging to the cadherin family.46 The classic cadherins are Ca2+-dependent cell-cell adhesion proteins characterized by 5 repeated cadherin-specific motifs in their extracellular domain. FAT4 may decrease EC motility via increasing cell adhesiveness because it contains 34 cadherin motifs, 4 EGF-like repeats, a transmembrane domain, and a cytoplasmic region.35 FAT4 may also decrease EC motility via the transduction of inhibitory signals, because loss of FAT4 has been linked to the disruption of the noncanonical Wnt/PCP (planar cell polarity) pathway.47 How FAT4 prevents LEC migration and/or lymphangiogenesis remains to be explored in future studies.
Recently, miR-31 was defined as a negative regulator of lymphatic differentiation because of its direct repression of Prox1.48 We found that miR-31 is also up-regulated in KSHV-infected LECs (Figure 1F), and that KSHV induces LEC-to-BEC transcriptome reprogramming (supplemental Figure 1C). Therefore, direct targeting of master regulator Prox149 by miR-31 in LECs may be a mechanism involved in KSHV-induced LEC-to-BEC reprogramming.
In summary, our results reveal a series of differentially expressed miRNAs and protein-coding genes that have not previously been associated with Kaposi sarcomagenesis or with KSHV infection. Computational analyses and further wetlab validation has revealed that the miR221-ETS2, miR222-EST1, and miR31-FAT4 pairs are involved in manipulating EC motility. This study therefore provides direction for future mechanistic studies of KSHV-induced tumorigenesis and EC migration. Studies of KSHV-, LANA-, and Kaposin B–induced EC migration should help to improve our understanding of (lymph)angiogenesis, and will also benefit the development of new therapeutic approaches targeting the inhibition of pathogenic angiogenesis in tumors, the stimulation of angiogenesis in patients with stroke or diabetes, and the treatment of lymphatic disorders such as lymphedema.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Vieira at the University of Washington-Seattle for kindly providing the GFP-recombinant virus rKSHV.219 and Professor Ching-Hwa Tsai, Dr Su-Fang Lin, and Professor Chris Boshoff for inspiring discussions and critical reading of this manuscript. They also acknowledge the technical services provided by the Microarray & Gene Expression Analysis Core Facility, which is supported by the National Research Program for Genomic Medicine of the National Science Council at the Veterans General Hospital–Yang Ming Genome Research Center.
This work was supported by the National Science Council (NSC99-3111-B-010-003, NSC98-2320-B-010-020-MY3, and NSC 99-2911-I-009-101), the Yen Tjing Lin Medical Foundation (CI-98-11), the National Health Research Institutes (NHRI-EX99-9704BI), the Department of Health (CCMP99-RD-063), the Taipei Veterans General Hospital (V100E2-011 and Cancer Excellence Center Plan DOH100-TD-C-111-007), the MacKay Memorial Hospital (MMH-HB-100-01), and Yang-Ming University (Ministry of Education, Aim for the Top University Plan).
Authorship
Contribution: Y.-H.W. and H.-W.W. designed the analysis approach; Y.-H.W., T.-F.H., Y.-C.C., Y.-N.T., Y.-H.T., and C.-C.C. performed the research and analyzed the data; H.-W.W. provided biologic guidance during the analysis process; Y.-H.W. and H.-W.W. wrote the manuscript; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: H.-W. Wang, PhD, Institute of Microbiology and Immunology, National Yang-Ming University, No 155, Sec 2, Li-Nong Street, Taipei 112, Taiwan; e-mail: hwwang@ym.edu.tw.