Abstract
Genomic aberrations are of predominant importance to the biology and clinical outcome of patients with chronic lymphocytic leukemia (CLL), and FISH-based genomic risk classifications are routinely used in clinical decision making in CLL. One of the known limitations of CLL FISH is the inability to comprehensively interrogate the CLL genome for genomic changes. In an effort at overcoming the existing limitations in CLL genome analysis, we have analyzed high-purity DNA isolated from FACS-sorted CD19+ cells and paired CD3+ or buccal cells from 255 patients with CLL for acquired genomic copy number aberrations (aCNAs) with the use of ultra-high-density Affymetrix SNP 6.0 arrays. Overall, ≥ 2 subchromosomal aCNAs were found in 39% (100 of 255) of all cases analyzed, whereas ≥ 3 subchromosomal aCNAs were detected in 20% (50 of 255) of cases. Subsequently, we have correlated genomic lesion loads (genomic complexity) with the clinical outcome measures time to first therapy and overall survival. With the use of multivariate analyses incorporating the most important prognostic factors in CLL together with SNP 6.0 array–based genomic lesion loads at various thresholds, we identify elevated CLL genomic complexity as an independent and powerful marker for the identification of patients with aggressive CLL and short survival.
Introduction
Chronic lymphocytic leukemia (CLL) has a varied clinical course, and genomic aberrations are recognized as important to the diverse biologic and clinical phenotypes of CLL.1,2 In particular, the recurrent chromosomal deletions del17p and del11q are associated with aggressive CLL.1,3,4 Over the past few years, multiple additional chromosomal phenotypes, including recurrent translocations (mostly unbalanced), complex aberrant karyotypes, and single nucleotide polymorphism (SNP) array–defined complex karyotypes (elevated genomic complexity) have been correlated with clinical outcome measures.5-10 The overriding conclusion that can be drawn from these studies is that the inability to maintain genomic stability/integrity is associated with more aggressive disease.
More recently, it was shown that CLL cells with elevated apoptotic resistance to ex vivo external radiation often display elevated genomic complexity and, further, that the degree of radiation resistance was associated with short survival in univariate outcome analysis.11 This finding was true for CLL cohorts inclusive of TP53-mutated cases (TP53 mutations confer absolute radiation resistance to CLL cells ex vivo) as well as for cohorts from which TP53-mutated cases were excluded. It is thus clear that the inability to maintain genomic integrity is linked to apoptotic defects because of mutated genes (of which TP53 is dominant, ATM contributory, and other contributory genes not yet identified); this is possibly because of a permissive cellular context for the formation and persistence of DNA double-strand (ds)–breaks (without obligatory DNA ds-break–induced CLL cell apoptosis) and subsequent accumulation of acquired genomic copy number aberrations (aCNAs). In principle therefore, accurate and quantitative measurements of aCNAs should allow for the measurement of clinical risk that affects CLL through (1) impaired DNA ds-break repair and response pathways, which include defective DNA ds-break–induced apoptosis and associated resistance to genotoxic chemotherapy; (2) specific known gene defects (as exemplified by TP53 and del17p) or as-yet unidentified gene defects associated with individual recurrent genomic changes and therapy resistance; and (3) telomere-shortening–induced karyotypic instability and its postulated consequences.12,13
Various clinical observations suggest that the identification of high-risk CLL (CLL with short survival) with the use of currently available biomarkers or clinical criteria is incomplete. (1) CLL FISH does not identify all patients with aggressive clinical behavior and, conversely, even within del17p or del11q patient cohorts, some patients display relatively more indolent disease.14-17 (2) TP53 mutations do not identify all cases of aggressive CLL (and probably less than one-half of all such cases) and are not yet routinely clinically measured in a comprehensive manner.18-22 (3) Within all other marker-stratified CLL cohorts, persons with aggressive disease exist that are not readily identifiable with the use of conventional clinical or marker-based testing approaches.
Given prior observations of the value of SNP array–based genomic copy number analysis in CLL (albeit with the use of lower-resolution platforms or either analysis of tumor cells in the absence of paired normal DNA, which precludes accurate genomic complexity assessments) and other hematologic malignancies, we have for this study interrogated the genomes of 255 CLL cases for aCNAs with the use of ultra-high-density SNP 6.0 arrays.23-31 Subsequently, we have correlated the absolute aCNA load at various lesion thresholds with the survival of patients within this cohort. Through these efforts we have identified a high-risk CLL subgroup (≥ 2 aCNAs) comprising ∼ 40% of all CLL with short survival. Finally, with the use of comprehensive multivariate analysis, we have identified SNP array–based CLL genomic complexity as a powerful and independent prognostic factor of aggressive CLL. These data have clear implications for the development of novel CLL-directed therapeutic approaches for the subgroup of CLL patients with unstable genomes.
Methods
Patients
Between January 2005 and September 2009, 266 patients evaluated at the University of Michigan Comprehensive Cancer Center were enrolled onto this study. The trial was approved by the University of Michigan Institutional Review Board (IRBMED no. 2004-0962), and written informed consent was obtained from all patients before enrollment in accordance with the Declaration of Helsinki. Data from 255 of these 266 patients were included in this report (5 patients enrolled on the study were excluded because of a diagnosis that was not CLL, and 6 patients had insufficient cryopreserved cells available for the analyses described).
Regardless of whether the subjects' disease was originally diagnosed at our institution or another, we used the same CLL diagnostic criteria, based on the National Cancer Institute Working Group Guidelines for CLL.32 Eligible patients needed to have an absolute lymphocytosis (> 5000 mature lymphocytes/μL), and lymphocytes needed to express CD19, CD23, sIg (weak), and CD5 in the absence of other pan-T-cell markers.
Time to first therapy (TTFT) and overall survival (OS) were based on the CLL trial enrollment date (which is equal to the specimen procurement date) or alternatively the diagnosis date (see supplemental Figures 1-9, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) as previously defined.5 CLL treatment was defined as cytotoxic chemotherapy, monoclonal antibody therapy, or both for CLL. Clinical information, including Rai stage and all treatments given, was collected on all patients. Patient samples were characterized for selected CLL-associated chromosomal aberrations on the day of trial enrollment as a routine clinical test at the Mayo Clinic with the use of FISH (CLL-FISH). All outcome analyses described here are based on FISH-25 results (CLL-FISH findings detected in ≥ 25% of nuclei). Given the absence of published data that would model the effect of FISH results when tested as a continuous variable on outcome and the absence of data that compare the degree of FISH positivity for a lesion in paired samples (diagnostic and relapsed samples), we have standardized on FISH-25 as one operational way to categorize a small subset of CLL cases that have low clonal representation of the FISH finding. As can be seen in Table 1, this changes the FISH category for only a small subset of patients.
Cell isolation: flow cytometric sorting of CLL specimens
Cryopreserved PBMCs (frozen after Ficoll-gradient purification) from CLL blood specimens were prepared for FACS and sorted into CD19+ and CD3+ cells as previously described.5 The range, mean, and median of the absolute lymphocyte count on the day of enrollment/specimen procurement for this study were as follows: range of 5000-497 000 cells/μL, mean of 44 700 cells/μL, and median of 18 500 cells/μL.
Preparation of sample DNA
DNA used for SNP 6.0 profiling was extracted from CD19+ and CD3+ cells sorted with FACS as described.5 For 11 cases, paired buccal DNA was used instead of CD3+ cell–derived DNA.
Array data analysis
The DNA was prepared for hybridization to SNP 6.0 arrays according to the manufacturers' recommendations. Affymetrix CEL files for each cell sample were analyzed with Genotyping Console Version 2.0 software for initial quality control, followed by use of the Affymetrix “Birdseed” algorithm to generate tab-delimited SNP call files in text format. Call rates for the entire group of CD19+ samples included in this report were between 95.06% and 99.63%, with a mean call rate of 98.53%. Corresponding call rates for CD3+ or buccal DNA were between 93.14% and 99.66%, with a mean call rate of 98.49%.
Sample copy number heatmap displays were obtained from CEL files through the use of the freely available software dChip with a build date of February 25, 2009,33 adapted to run on a 64-bit computer environment. For genomic copy number analysis, we visually inspected parallel heatmap copy number images of CD19+ and paired CD3+/buccal DNA samples generated through dChipSNP and with the use of the median smoothening functionality. Only those copy number changes detected in CD19+ DNA that were not found at the same position in paired CD3+/buccal DNA were called somatic. This approach followed our previously externally referenced (FISH) SNP 6.0 genomic lesion calling method in acute myelogenous leukemia that resulted in 100% concordance between SNP 6.0 profiling and FISH results (for a total of 56 lesions analyzed).23 With the use of this approach, the 3 shortest identified acquired copy number (aCN) changes were 0.024, 0.042, and 0.052 Mb in length and were defined by 18, 27, and 19 consecutive SNP positions, respectively. Most aCN changes were defined by > 100 consecutive SNP positions (see supplemental Table 8).
SNP 6.0 array data files for all 510 patient samples analyzed have been deposited in the Gene Expression Omnibus public database (accession no. GSE30777).
Exon resequencing of TP53
Primers to amplify and sequence exons 2-10 of human TP53 and adjacent intronic sequences, including splice junctions, were designed with the primer 3 program (http://frodo.wi.mit.edu/primer3/), and sequence information was generated as described.34 Mutations were confirmed with paired patient CD3+/buccal DNA as templates.
Determination of ZAP70 and IgVH status
Determination of ζ-associated protein 70 (ZAP70), IgVH, and CD38 status was performed as described.5
Statistical methods
TTFT or OS was defined as the time (in months) between CLL trial enrollment date (or alternatively the diagnosis date; see supplemental Figures 1-9) and the date (in months) of first treatment received or the patient's death, respectively. Patient status was updated within the last week of October 2010; therefore, for patients still alive or without new treatment, the date of censoring was stipulated as November 1, 2010. Univariate and bivariate analyses were based on Kaplan-Meier estimates of survivor functions, proportional hazards modeling, and log-rank testing. Median survival times were estimated directly from the survivor function estimates. Statistical comparisons of survivor functions estimated from nonoverlapping subsets of patients were based on the log-rank test. Hazard ratios associated with group membership were estimated with Cox proportional hazards models, fit with the use of dummy variables indicating group membership. For bivariate analyses, subjects were stratified into 4 groups on the basis of the joint values of 2 factors, and Kaplan-Meier estimates of the survivor functions were used for visual display. Log-rank tests were then used to assess for differences between the survivor functions of any 2 of the 4 groups. Multivariate analyses were based on Cox proportional hazard models with additive effects for the factors as reported in the results. The reported significance levels assess whether the hazard for a given factor differs from 1 when the other factors in the model are held fixed.
Quantitative SNP 6.0 array–based aCNA analysis.
To understand how the quantitative SNP 6.0 genomic lesion count (aCNA) relates to risk, we considered univariate proportional hazards models with the use of various transformed versions of the aCNA value to understand how different quantitative aCNA levels compare in terms of risk. To do this, we considered 3 families of transformed aCNA levels (see supplemental Methods and Results).
Quantitative multivariate SNP 6.0 array–based aCNA analysis.
In univariate analysis, we determined the exponent p that maximized the hazard ratio (HR) of aCNAp for OS. For OS, the optimal exponent was approximately p = .5 (a square root transform), and for TTFT the optimal exponent was approximately p = 1 (no transformation). We thus used the square root of aCNA in multivariate analyses of OS in addition to aCNA untransformed.
Results
Patient characteristics
Characteristics of the 255 patients with CLL analyzed in this study are summarized in Table 1. Of these patients, 198 (78%) were untreated and 57 (22%) relapsed at study enrollment (median number of prior therapies, 1). The median time from diagnosis to enrollment and from enrollment to data analysis for previously untreated patients was 7 months and 51 months, respectively. All outcome analysis described next is based on biomarker measurements performed on samples procured at study enrollment.
Patient characteristics
| Patient characteristics . | Treatment naive at enrollment . | Relapsed at enrollment . |
|---|---|---|
| Sample size (N = 255 patients), n (%) | 198 (78) | 57 (22) |
| Age, y | ||
| Median | 61 | 63 |
| Range | 35-96 | 46-82 |
| Sex | ||
| Female, n (%) | 71 (36) | 14 (25) |
| Male, n (%) | 127 (64) | 43 (75) |
| Rai stage | ||
| Low (0), n (%) | 91 (46) | 10 (17) |
| Intermediate (I-II), n (%) | 98 (49) | 30 (53) |
| High (III-IV), n (%) | 9 (5) | 17 (30) |
| Median time from diagnosis to enrollment, mo | ||
| Median | 7 | 76 |
| Range | 0-306 | 5-244 |
| Median time from enrollment to analysis, mo | ||
| Median | 51 | 44 |
| Range | 14-69 | 14-69 |
| IgVH mutational status | ||
| Unmutated (≥ 98% homology to germ line), n (%) | 82 (41) | 34 (60) |
| Mutated (< 98% homology to germ line), n (%) | 108 (55) | 20 (35) |
| Not evaluable, n (%) | 8 (4) | 3 (5) |
| Prioritized interphase FISH | ||
| 17p deletion, n (%) | 17 (9) | 9 (16) |
| 11q deletion, n (%) | 18 (9) | 12 (21) |
| 12 trisomy, n (%) | 26 (13) | 10 (17) |
| Normal karyotype, n (%) | 43 (22) | 9 (16) |
| 13q deletion (sole abnormality), n (%) | 90 (45) | 16 (28) |
| FISH data not available, n (%) | 4 (2) | 1 (2) |
| Prioritized interphase FISH-25* | ||
| 17p deletion, n (%) | 16 (8) | 9 (16) |
| 11q deletion, n (%) | 14 (7) | 10 (17) |
| 12 trisomy, n (%) | 24 (12) | 9 (16) |
| Normal karyotype, n (%) | 61 (31) | 9 (16) |
| 13q deletion (sole abnormality), n (%) | 79 (40) | 19 (33) |
| FISH data not available, n (%) | 4 (2) | 1 (2) |
| ZAP70 expression | ||
| Negative (≤ 20%), n (%) | 116 (59) | 19 (33) |
| Positive (> 20%), n (%) | 82 (41) | 38 (67) |
| TP53 exon 2-10 mutations | ||
| Wild-type, n (%) | 174 (88) | 45 (79) |
| Mutated, n (%) | 24 (12) | 12 (21) |
| Number of prior therapies | ||
| Median | NA | 1 |
| Range | NA | 1-7 |
| Patient characteristics . | Treatment naive at enrollment . | Relapsed at enrollment . |
|---|---|---|
| Sample size (N = 255 patients), n (%) | 198 (78) | 57 (22) |
| Age, y | ||
| Median | 61 | 63 |
| Range | 35-96 | 46-82 |
| Sex | ||
| Female, n (%) | 71 (36) | 14 (25) |
| Male, n (%) | 127 (64) | 43 (75) |
| Rai stage | ||
| Low (0), n (%) | 91 (46) | 10 (17) |
| Intermediate (I-II), n (%) | 98 (49) | 30 (53) |
| High (III-IV), n (%) | 9 (5) | 17 (30) |
| Median time from diagnosis to enrollment, mo | ||
| Median | 7 | 76 |
| Range | 0-306 | 5-244 |
| Median time from enrollment to analysis, mo | ||
| Median | 51 | 44 |
| Range | 14-69 | 14-69 |
| IgVH mutational status | ||
| Unmutated (≥ 98% homology to germ line), n (%) | 82 (41) | 34 (60) |
| Mutated (< 98% homology to germ line), n (%) | 108 (55) | 20 (35) |
| Not evaluable, n (%) | 8 (4) | 3 (5) |
| Prioritized interphase FISH | ||
| 17p deletion, n (%) | 17 (9) | 9 (16) |
| 11q deletion, n (%) | 18 (9) | 12 (21) |
| 12 trisomy, n (%) | 26 (13) | 10 (17) |
| Normal karyotype, n (%) | 43 (22) | 9 (16) |
| 13q deletion (sole abnormality), n (%) | 90 (45) | 16 (28) |
| FISH data not available, n (%) | 4 (2) | 1 (2) |
| Prioritized interphase FISH-25* | ||
| 17p deletion, n (%) | 16 (8) | 9 (16) |
| 11q deletion, n (%) | 14 (7) | 10 (17) |
| 12 trisomy, n (%) | 24 (12) | 9 (16) |
| Normal karyotype, n (%) | 61 (31) | 9 (16) |
| 13q deletion (sole abnormality), n (%) | 79 (40) | 19 (33) |
| FISH data not available, n (%) | 4 (2) | 1 (2) |
| ZAP70 expression | ||
| Negative (≤ 20%), n (%) | 116 (59) | 19 (33) |
| Positive (> 20%), n (%) | 82 (41) | 38 (67) |
| TP53 exon 2-10 mutations | ||
| Wild-type, n (%) | 174 (88) | 45 (79) |
| Mutated, n (%) | 24 (12) | 12 (21) |
| Number of prior therapies | ||
| Median | NA | 1 |
| Range | NA | 1-7 |
FISH findings in ≥ 25% of nuclei.
Correlations of CLL FISH and SNP 6.0 array profiling results
To test our previously externally validated (see “Array data analysis”) analytical approach for analysis of SNP 6.0 array data in our CLL cohort, we calculated the sensitivity and specificity for detection of conventional 13q14 deletions by SNP 6.0 profiling (13q14 deletions were selected because many are < 1 Mb or ∼ 1 Mb in length) compared with CLL FISH and CLL FISH-25 (FISH findings present in ≥ 25% of nuclei) data as the gold standard. Sensitivity and specificity for SNP 6.0 profiling were 89% and 95% with the use of all CLL FISH results and 98% and 92% for CLL FISH-25 results, respectively. We also calculated data for del17p (sensitivity and specificity for SNP 6.0 profiling were 92% and 99% with the use of all CLL FISH results and 100% and 99% for CLL FISH-25 results, respectively) and del11q (sensitivity and specificity for SNP 6.0 profiling were 73% and 99% with all CLL FISH results and 92% and 99% for CLL FISH-25 results, respectively). These results confirm very high specificity of SNP 6.0 profiling-based aCNA calling compared with FISH in CLL and also confirm that SNP 6.0 profiling is less sensitive for lesions present in < 25% of the input DNA (range of FISH-positive nuclei for 11q lesions that were SNP 6.0 profiling negative was 9%-40%; mean, 19%). Finally, we detected relative (pseudo) gains at chromosome 7 at physical position 38.27-38.36 in all 255 CLL CD19+ samples compared with the averaged paired normal samples because of loss of chromosomal material at the T-cell receptor γ locus as a consequence of T-cell receptor gene rearrangements, thus providing additional high confidence in our analytical approach.
Pathologic anatomy of acquired subchromosomal and chromosomal genomic copy number changes in CLL defined through SNP 6.0 array profiling
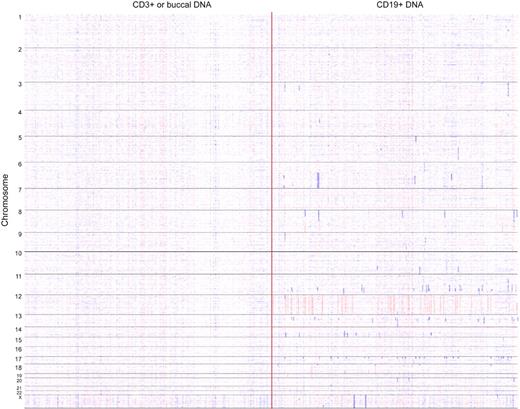
We catalogued all somatically aCNAs in our CLL cohort with the use of visual inspection of simultaneous displays of dChipSNP-based copy number estimates (heatmaps) for CD19+ cell–derived DNA and paired CD3+ cell–derived DNA (96% of all samples) or buccal DNA (4% of all samples). A whole-genome heatmap display of large CLL-associated aCNAs is displayed in Figure 1.
Whole genome copy number heatmap display of 255 CLL genomes. Copy number heatmap displays for paired DNA samples on the basis of SNP 6.0 array profiling were generated with dChipSNP. (Left) CD3+ or buccal DNA; (right) CLL CD19+ DNA. Samples are grouped by chromosome number (1-22 and X). Blue indicates copy loss, red indicates copy gain. Each column represents 1 patient.
Whole genome copy number heatmap display of 255 CLL genomes. Copy number heatmap displays for paired DNA samples on the basis of SNP 6.0 array profiling were generated with dChipSNP. (Left) CD3+ or buccal DNA; (right) CLL CD19+ DNA. Samples are grouped by chromosome number (1-22 and X). Blue indicates copy loss, red indicates copy gain. Each column represents 1 patient.
A total of 584 somatically aCN changes were detected in 255 CLL genomes (range, 0-22 aCN changes; see “Array data analysis”). We detected 3 losses (all monosomy X) and 45 gains of entire chromosomes (42 cases of trisomy 12), 474 subchromosomal losses (size range, 0.024-108.73 Mb) and 62 subchromosomal gains (size range, 0.114-94.626 Mb) for a total of 536 subchromosomal aCN changes (ratio of losses to gains of 7.6 to 1). In addition to well-described recurrent aCNAs (del17p, del11q, del13q14, and trisomy 12), recurrent albeit relatively infrequent aCNAs (with an incidence ≥ 2%) were identified on chromosomes 6 (N = 7 with multiple minimal deleted regions [MDRs]), 8p (N = 7; multiple MDRs), 8q gains (N = 5), 10q (N = 10), 14q (N = 9), 17q gains (N = 6), 18q (N = 8; multiple MDRs), and 18p gains (N = 6; see supplemental Table 8). Recurrent homozygous deletions other than biallelic 13q14 deletions were rare and identified only on chromosome 9 (N = 2 and spanning p16/CDKN2A) and the X chromosome (N = 2). Recurrent high-level genomic amplifications (> 3N) were not detected.
Two or more subchromosomal aCN changes were found in 100 of 255 (39%) of all cases analyzed, whereas ≥ 3 subchromosomal aCN changes were detected in 50 of 255 (20%) of cases. Within the group of previously untreated patients (N = 198), ≥ 2 subchromosomal aCN changes were found in 68 of 198 (34%) of all cases analyzed, whereas ≥ 3 subchromosomal aCN changes were detected in 31 of 198 (16%) of cases. Corresponding data for the group of relapsed patients (N = 57) were as follows: ≥ 2 subchromosomal aCN changes were found in 32 of 57 (56%) of all cases analyzed, whereas ≥ 3 subchromosomal aCN changes were detected in 19 of 57 (33%) of cases. Furthermore, 27% (144 of 536), 42% (227 of 536), and 60% (320 of 536) of subchromosomal deletions/gains were < 1 Mb, < 2 Mb, and < 5 Mb in length (see supplemental Table 8).
Next, we analyzed the TP53 gene status in the group of CLL with elevated SNP 6.0 array–based genomic complexity at various thresholds.35-38 The TP53 gene was mutated in 36 of 255 (14%) of the cases in this cohort, of which 25 also had del17p, 3 had acquired 17p-uniparental disomy, 7 did not have 17p abnormalities, and 1 case lacked FISH data but was SNP-A negative for 17p abnormalities. Of the group of CLL with ≥ 3 subchromosomal aCN changes (N = 50), 26 were TP53 exon 2-10 mutant (52%), whereas 33 of 100 (33%) of cases with ≥ 2 subchromosomal aCN changes were TP53 exon 2-10 mutant. These data therefore confirm a strong enrichment of TP53 mutants among the high-complexity CLL cases.11 Furthermore, the data indicate that a substantial fraction of CLL with acquired genomic complexity is TP53 wild-type and that genomic complexity in CLL is multifactorial in origin. Finally, patients with 11q (based on FISH 25) deletion and TP53 exon 2-10 wild-type status (N = 22) showed ≥ 2 or ≥ 3 subchromosomal aCNA counts in 77% and 41%, respectively, confirming prior observations of a strong association of del11q and elevated genomic complexity in CLL.11
Internal outcome validation of the CLL study cohort with the use of established prognostic factors
To validate our study cohort for the array-based genomic/clinical outcome analysis described below, we initially conducted univariate analyses of the most critical factors known to affect TTFT and OS of patients with CLL. These factors included the presence of del17p, del11q, TP53 exon 2-10 mutations, IgVH status unmutated,39,40 ZAP70 expression ≥ 20%,41 CD38 expression ≥ 30%, and Rai stage at enrollment. Kaplan-Meier plots for TTFT and OS for the group of previously untreated patients (N = 198), which were based on either the CLL trial enrollment date or alternatively the diagnosis date, are displayed in supplemental Figures 1 through 4. Data are summarized in supplemental Tables 1 and 2. Overall, the effects of these established prognostic factors were in line with published reports.
Results of univariate outcome analyses of SNP 6.0 array–detectable subchromosomal genomic lesions versus TTFT and OS in CLL
We subsequently determined the prognostic value of SNP 6.0 array–detectable aCNAs (combined total of subchromosomal losses and gains) on TTFT and OS in CLL with the use of univariate analysis; this showed a strong effect of increasing aCNA counts on short TTFT and OS. Kaplan-Meier plots for various aCNA thresholds are displayed in Figure 2A-D and E-H, and data are summarized in Table 2 (also see supplemental Figure 5, supplemental Table 3, and supplemental Results).
SNP 6.0 array–based lesion cutoffs and TTFT or OS in CLL (Kaplan-Meier plots). (A-D) Previously untreated patient group was analyzed from trial enrollment for TTFT. (E-H) Previously untreated patient group was analyzed from trial enrollment for OS.
SNP 6.0 array–based lesion cutoffs and TTFT or OS in CLL (Kaplan-Meier plots). (A-D) Previously untreated patient group was analyzed from trial enrollment for TTFT. (E-H) Previously untreated patient group was analyzed from trial enrollment for OS.
SNP 6.0 array–based lesion detection and TTFT and OS in CLL (univariate analysis)
| SNP 6.0 array genomic lesions . | HR . | CI . | P . |
|---|---|---|---|
| TTFT from enrollment date | |||
| Untreated patients (N = 198) | |||
| ≥ 1 vs 0 | 1.7 | 1-2.9 | .06 |
| ≥ 2 vs ≤ 1 | 1.9 | 1.2-3.1 | .01 |
| ≥ 3 vs ≤ 2 | 3.3 | 1.9-5.5 | < .01 |
| ≥ 4 vs ≤ 3 | 4.8 | 2.6-8.8 | < .01 |
| OS from enrollment date | |||
| Untreated patients (N = 198) | |||
| ≥ 1 vs 0 | 5.5 | 1.3-24 | .01 |
| ≥ 2 vs ≤ 1 | 4.7 | 2-11.1 | < .01 |
| ≥ 3 vs ≤ 2 | 6.2 | 2.7-14.2 | < .01 |
| ≥ 4 vs ≤ 3 | 14.5 | 6.3-33.5 | < .01 |
| OS from enrollment date | |||
| Relapsed patients (N = 57) | |||
| ≥ 1 vs 0 | 1 | 0.4-2.6 | .96 |
| ≥ 2 vs ≤ 1 | 1.9 | 0.9-4.0 | .1 |
| ≥ 3 vs ≤ 2 | 2.7 | 1.2-5.7 | .01 |
| ≥ 4 vs ≤ 3 | 3.5 | 1.6-7.6 | < .01 |
| SNP 6.0 array genomic lesions . | HR . | CI . | P . |
|---|---|---|---|
| TTFT from enrollment date | |||
| Untreated patients (N = 198) | |||
| ≥ 1 vs 0 | 1.7 | 1-2.9 | .06 |
| ≥ 2 vs ≤ 1 | 1.9 | 1.2-3.1 | .01 |
| ≥ 3 vs ≤ 2 | 3.3 | 1.9-5.5 | < .01 |
| ≥ 4 vs ≤ 3 | 4.8 | 2.6-8.8 | < .01 |
| OS from enrollment date | |||
| Untreated patients (N = 198) | |||
| ≥ 1 vs 0 | 5.5 | 1.3-24 | .01 |
| ≥ 2 vs ≤ 1 | 4.7 | 2-11.1 | < .01 |
| ≥ 3 vs ≤ 2 | 6.2 | 2.7-14.2 | < .01 |
| ≥ 4 vs ≤ 3 | 14.5 | 6.3-33.5 | < .01 |
| OS from enrollment date | |||
| Relapsed patients (N = 57) | |||
| ≥ 1 vs 0 | 1 | 0.4-2.6 | .96 |
| ≥ 2 vs ≤ 1 | 1.9 | 0.9-4.0 | .1 |
| ≥ 3 vs ≤ 2 | 2.7 | 1.2-5.7 | .01 |
| ≥ 4 vs ≤ 3 | 3.5 | 1.6-7.6 | < .01 |
OS data are grouped by disease status at trial enrollment (previously untreated group and relapsed group).
Next, we analyzed the prognostic significance of SNP 6.0 array–based aCNA thresholds on OS in the subgroup of CLL patients who were relapsed at trial enrollment (N = 57) and, again, noted elevated HRs for short OS for CLL cases with elevated genomic complexity at various thresholds (Kaplan-Meier plots for OS are displayed in supplemental Figure 6A-D and E-H, respectively).
Results of univariate analysis of SNP 6.0 array–based genomic complexity as a continuous variable in CLL outcome
Quantitative analysis of genomic lesions in CLL on the basis of SNP 6.0 array profiling described earlier can be used to determine the pattern of change in risk as the number of lesions increases. Of note, however, most CLL cases are characterized by lesion scores of 0 to 3; therefore, the aCNA incidence is not evenly distributed across the observed range of aCNAs (0-22) but heavily skewed. To use aCNA as a continuous variable in outcome analysis, we considered 3 families of transformed aCNA levels as detailed in supplemental Methods.
Results for the power transformation family I (Box-Cox transformation), which includes the linear transformation, along with transforms that give either increasing (exponent > 1) or decreasing (exponent < 1) incremental risks were as follows: for TTFT, the best fit to the data occurred for the linear transformation (exponent = 1), whereas for OS the best fit occurred with exponent < 1. That is, for OS it appears that the greatest increases in risk result from the first few lesions, with additional lesions incurring a lower (but non-zero) risk, whereas for TTFT it appears that the incremental risk is approximately constant throughout the observed range of lesion counts. Results are displayed in supplemental Figure 7.
Results of bivariate outcome analyses of traditional prognostic factors and SNP 6.0 array–detectable genomic lesions versus TTFT and OS in CLL
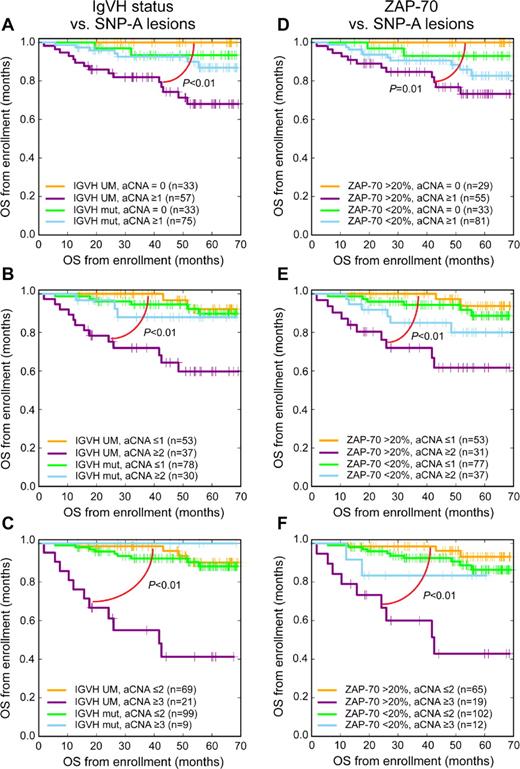
Next, we evaluated the prognostic value of SNP 6.0 array profiling-based lesion detection at various lesion thresholds in the setting of other important markers (del17p, del11q, TP53 mutations, IgVH status, ZAP70 status, and Rai stage) with the use of bivariate analyses in previously untreated patients. This analysis disclosed substantial survival differences within the CLL IgVH-unmutated group at all tested SNP 6.0 lesion thresholds, with the net effect that CLL with unmutated IgVH genes and low genomic lesion loads displayed survival similar to IgVH-mutated CLL (Figure 3A-C). Equally important, the negative prognostic effects of ZAP70-positive status in CLL were largely confined to cases with elevated genomic lesion loads (Figure 3D-F). The results for these bivariate analyses (aCNA and ZAP70 status and aCNA and IGVH status) could indicate the existence of interactions between these factors that may be quantifiable in future analyses involving patient cohorts that are many-fold larger (see “Discussion”).
SNP 6.0 array–based lesion cutoffs and IgVH status or ZAP70 status and OS in CLL (bivariate analysis; Kaplan-Meier plots). Previously untreated patient group was analyzed from trial enrollment (A-C) IgVH status and various SNP 6.0 array–based lesion cutoffs and (D-F) ZAP70 status and various SNP 6.0 array–based lesion cutoffs.
SNP 6.0 array–based lesion cutoffs and IgVH status or ZAP70 status and OS in CLL (bivariate analysis; Kaplan-Meier plots). Previously untreated patient group was analyzed from trial enrollment (A-C) IgVH status and various SNP 6.0 array–based lesion cutoffs and (D-F) ZAP70 status and various SNP 6.0 array–based lesion cutoffs.
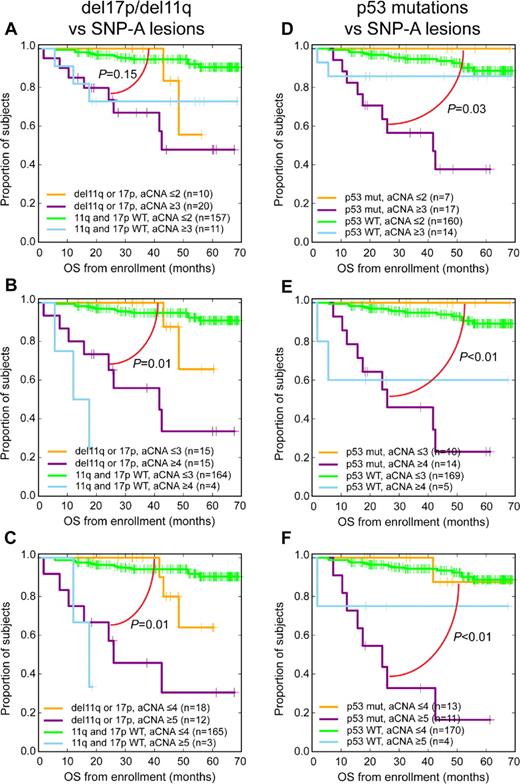
Bivariate analyses among the 2 CLL genomic subgroups del17p/del11q combined or TP53 exon 2-10 mutated together with SNP 6.0 array–based lesion thresholds disclosed that elevated genomic complexity at various SNP 6.0 array–based lesion thresholds separated the del17p/del11q and TP53 mutated subgroups into groups with significantly and substantially shorter and longer survival (Figure 4A-F).
SNP 6.0 array–based lesion cutoffs and del17p/del11q or TP53 exon 2-10 mutation status and OS in CLL (bivariate analysis; Kaplan-Meier plots). Previously untreated patient group analyzed from trial enrollment (A-C) del17p/del11q status and various SNP 6.0 array–based lesion cutoffs and (D-F) TP53 exon 2-10 mutation status and various SNP 6.0 array–based lesion cutoffs.
SNP 6.0 array–based lesion cutoffs and del17p/del11q or TP53 exon 2-10 mutation status and OS in CLL (bivariate analysis; Kaplan-Meier plots). Previously untreated patient group analyzed from trial enrollment (A-C) del17p/del11q status and various SNP 6.0 array–based lesion cutoffs and (D-F) TP53 exon 2-10 mutation status and various SNP 6.0 array–based lesion cutoffs.
In summary, bivariate analyses uncovered that elevated SNP 6.0 array–based genomic complexity identified high-risk persons in all established biomarker-based CLL subgroups (corresponding data on the basis of the CLL diagnosis date are displayed in supplemental Figures 8-9).
Bivariate analyses for TTFT and the various pairwise prognostic factor combinations are displayed in supplemental Figures 10 and 11, confirming the strong effects of elevated genomic complexity on short TTFT in CLL.
Results of multivariate outcome analyses of traditional prognostic factors and SNP 6.0 array–detectable genomic lesions at discrete thresholds versus OS in CLL
We proceeded with multivariate analyses for previously untreated patients (N = 187), initially incorporating base variables that are currently routinely available to physicians interested in CLL risk prognostication; these included IgVH status, ZAP70 status, CD38 status, del17p/del11q status (combined), Rai stage (coded as stages I-IV vs 0) together with SNP 6.0 array–based genomic lesion counts at various thresholds.
Importantly, for all models, elevated genomic complexity as measured through SNP 6.0 arrays emerged as the dominant and strongest predictor for short OS. For instance, for previously untreated patients analyzed from the date of trial enrollment the HR was 4.3 (P = .01) for a SNP 6.0 array–based lesion count of ≥ 3 (del17p/del11q combined: HR = 1.8, P = .38; ZAP70 ≥ 20% positive: HR = 0.7, P = .54; IgVH unmutated: HR = 1.8, P = .29; CD38 ≥ 30% positive: HR = 0.3, P = .06, and Rai stages 1-4: HR = 1.3, P = .59). Complete data for these analyses are summarized in Table 3.
Results of parallel multivariate analysis of ZAP70 status, CD38 status, IgVH status, FISH status for 17p and 11q combined or alternatively TP53 exon 2-10 mutations, Rai stage, and SNP 6.0 array–based lesion cutoffs on OS in previously untreated patients with CLL (n = 187)
| Variable . | n/N . | HR . | CI . | P . | Variable . | n/N . | HR . | CI . | P . |
|---|---|---|---|---|---|---|---|---|---|
| SNP 6.0 array genomic lesions ≥ 2 vs < 2 | 65/122 | 2.9 | 1.03-8.2 | .04 | SNP 6.0 array genomic lesions ≥ 2 vs < 2 | 65/122 | 2.9 | 1.06-7.9 | .03 |
| Del17p or del11q present vs not | 28/159 | 2.1 | 0.7-6.7 | .19 | TP53 exon 2-10 mutated vs not | 22/165 | 2.5 | 0.9-7.1 | .09 |
| Rai stage I-IV vs 0 | 90/97 | 1.3 | 0.6-3.2 | .53 | Rai stage I-IV vs 0 | 90/97 | 1.3 | 0.6-3.1 | .54 |
| ZAP70 ≥ 20% vs < 20% | 80/107 | 0.9 | 0.3-2.5 | .85 | ZAP70 ≥ 20% vs < 20% | 80/107 | 0.9 | 0.3-2.5 | .82 |
| IgVH UM vs M | 79/108 | 1.6 | 0.6-4.5 | .39 | IgVH UM vs M | 79/108 | 1.9 | 0.7-5.2 | .23 |
| CD38 ≥ 30% vs < 30% | 45/142 | 0.4 | 0.1-1.3 | .1 | CD38 ≥ 30% vs < 30% | 45/142 | 0.4 | 0.1-1.4 | .14 |
| SNP 6.0 array genomic lesions ≥ 3 vs < 3 | 28/159 | 4.3 | 1.4-13.4 | .01 | SNP 6.0 array genomic lesions ≥ 3 vs < 3 | 28/159 | 4.2 | 1.3-13.6 | < .01 |
| Del17p or del11q present vs not | 28/159 | 1.8 | 0.5-6.6 | .38 | TP53 exon 2-10 mutated vs not | 22/165 | 1.7 | 0.5-6 | .37 |
| Rai stage I-IV vs 0 | 90/97 | 1.3 | 0.5-3 | .59 | Rai stage I-IV vs 0 | 90/97 | 1.3 | 0.5-3 | .6 |
| ZAP70 ≥ 20% vs < 20% | 80/107 | 0.7 | 0.3-2 | .54 | ZAP70 ≥ 20% vs < 20% | 80/107 | 0.8 | 0.3-2.1 | .61 |
| IgVH UM vs M | 79/108 | 1.8 | 0.6-5.1 | .29 | IgVH UM vs M | 79/108 | 1.9 | 0.7-5.3 | .19 |
| CD38 ≥ 30% vs < 30% | 45/142 | 0.3 | 0.1-1.1 | .06 | CD38 ≥ 30% vs < 30% | 45/142 | 0.3 | 0.1-1.2 | .08 |
| SNP 6.0 array genomic lesions ≥ 4 vs < 4 | 17/170 | 19.5 | 3.9-97 | < .01 | SNP 6.0 array genomic lesions ≥ 4 vs < 4 | 17/170 | 18 | 4-81 | < .01 |
| Del17p or del11q present vs not | 28/159 | 0.8 | 0.2-3.7 | .78 | TP53 exon 2-10 mutated vs not | 22/165 | 0.9 | 0.3-3.3 | .89 |
| Rai stage I-IV vs 0 | 90/97 | 1.1 | 0.5-2.7 | .8 | Rai stage I-IV vs 0 | 90/97 | 1.1 | 0.5-2.7 | .8 |
| ZAP70 ≥ 20% vs < 20% | 80/107 | 0.6 | 0.2-1.7 | .3 | ZAP70 ≥ 20% vs < 20% | 80/107 | 0.6 | 0.2-1.6 | .26 |
| IgVH UM vs M | 79/108 | 1.2 | 0.4-3.8 | 0.78 | IgVH UM vs M | 79/108 | 1.2 | 0.4-3.8 | .8 |
| CD38 ≥ 30% vs < 30% | 45/142 | 0.4 | 0.1-1.6 | .2 | CD38 ≥ 30% vs < 30% | 45/142 | 0.4 | 0.1-1.5 | .2 |
| Variable . | n/N . | HR . | CI . | P . | Variable . | n/N . | HR . | CI . | P . |
|---|---|---|---|---|---|---|---|---|---|
| SNP 6.0 array genomic lesions ≥ 2 vs < 2 | 65/122 | 2.9 | 1.03-8.2 | .04 | SNP 6.0 array genomic lesions ≥ 2 vs < 2 | 65/122 | 2.9 | 1.06-7.9 | .03 |
| Del17p or del11q present vs not | 28/159 | 2.1 | 0.7-6.7 | .19 | TP53 exon 2-10 mutated vs not | 22/165 | 2.5 | 0.9-7.1 | .09 |
| Rai stage I-IV vs 0 | 90/97 | 1.3 | 0.6-3.2 | .53 | Rai stage I-IV vs 0 | 90/97 | 1.3 | 0.6-3.1 | .54 |
| ZAP70 ≥ 20% vs < 20% | 80/107 | 0.9 | 0.3-2.5 | .85 | ZAP70 ≥ 20% vs < 20% | 80/107 | 0.9 | 0.3-2.5 | .82 |
| IgVH UM vs M | 79/108 | 1.6 | 0.6-4.5 | .39 | IgVH UM vs M | 79/108 | 1.9 | 0.7-5.2 | .23 |
| CD38 ≥ 30% vs < 30% | 45/142 | 0.4 | 0.1-1.3 | .1 | CD38 ≥ 30% vs < 30% | 45/142 | 0.4 | 0.1-1.4 | .14 |
| SNP 6.0 array genomic lesions ≥ 3 vs < 3 | 28/159 | 4.3 | 1.4-13.4 | .01 | SNP 6.0 array genomic lesions ≥ 3 vs < 3 | 28/159 | 4.2 | 1.3-13.6 | < .01 |
| Del17p or del11q present vs not | 28/159 | 1.8 | 0.5-6.6 | .38 | TP53 exon 2-10 mutated vs not | 22/165 | 1.7 | 0.5-6 | .37 |
| Rai stage I-IV vs 0 | 90/97 | 1.3 | 0.5-3 | .59 | Rai stage I-IV vs 0 | 90/97 | 1.3 | 0.5-3 | .6 |
| ZAP70 ≥ 20% vs < 20% | 80/107 | 0.7 | 0.3-2 | .54 | ZAP70 ≥ 20% vs < 20% | 80/107 | 0.8 | 0.3-2.1 | .61 |
| IgVH UM vs M | 79/108 | 1.8 | 0.6-5.1 | .29 | IgVH UM vs M | 79/108 | 1.9 | 0.7-5.3 | .19 |
| CD38 ≥ 30% vs < 30% | 45/142 | 0.3 | 0.1-1.1 | .06 | CD38 ≥ 30% vs < 30% | 45/142 | 0.3 | 0.1-1.2 | .08 |
| SNP 6.0 array genomic lesions ≥ 4 vs < 4 | 17/170 | 19.5 | 3.9-97 | < .01 | SNP 6.0 array genomic lesions ≥ 4 vs < 4 | 17/170 | 18 | 4-81 | < .01 |
| Del17p or del11q present vs not | 28/159 | 0.8 | 0.2-3.7 | .78 | TP53 exon 2-10 mutated vs not | 22/165 | 0.9 | 0.3-3.3 | .89 |
| Rai stage I-IV vs 0 | 90/97 | 1.1 | 0.5-2.7 | .8 | Rai stage I-IV vs 0 | 90/97 | 1.1 | 0.5-2.7 | .8 |
| ZAP70 ≥ 20% vs < 20% | 80/107 | 0.6 | 0.2-1.7 | .3 | ZAP70 ≥ 20% vs < 20% | 80/107 | 0.6 | 0.2-1.6 | .26 |
| IgVH UM vs M | 79/108 | 1.2 | 0.4-3.8 | 0.78 | IgVH UM vs M | 79/108 | 1.2 | 0.4-3.8 | .8 |
| CD38 ≥ 30% vs < 30% | 45/142 | 0.4 | 0.1-1.6 | .2 | CD38 ≥ 30% vs < 30% | 45/142 | 0.4 | 0.1-1.5 | .2 |
Corresponding results of multivariate analyses inclusive of the TP53 exon 2-10 mutation status were HR of 4.2 (P < .01) for a SNP 6.0 array–based lesion count of ≥ 3 (TP53 exon 2-10 mutated status: HR = 1.7, P = .37; ZAP70 ≥ 20% positive, HR = 0.8, P = .61; IgVH unmutated: HR = 1.9, P = .19; CD38 ≥ 30% positive: HR = 0.3, P = .08; and Rai stages 1-4: HR = 1.3, P = .6). Complete data for these analyses are summarized in Table 3 and supplemental Table 4.
Next, we incorporated del17p/del11q combined and TP53 exon 2-10 mutations into models that also contained the above-referenced factors as well as SNP 6.0 array–based lesion counts. The results were HR of 3.7 (P = .03) for a SNP 6.0 array–based lesion count of ≥ 3 (TP53 exon 2-10 mutated status: HR = 1.5, P = .54; ZAP70 ≥ 20% positive: HR = 0.7, P = .53; IgVH unmutated: HR = 1.8, P = .25; CD38 ≥ 30% positive: HR = 0.3, P = .09; and del17p/del11q combined: HR = 1.5, P = .57). Data are summarized in supplemental Table 5.
Given the strong association of TP53 mutations and elevated genomic complexity in CLL, we wished to further clarify how much new information is gained when adding TP53 mutation status to SNP 6.0 array–based aCNA counts versus when adding SNP 6.0 array–based aCNA counts to TP53 mutation status. The likelihood ratio statistic is a way of comparing a model containing both aCNA counts and TP53 mutation status to models containing either of them separately. In supplemental Figure 12, we display the likelihood ratio statistic plotted for various values of the aCNA threshold. The gray area is the area where there is no significant difference. The most obvious interpretation of this is that for TTFT and OS, SNP 6.0 array–based aCNA adds to TP53 mutation status, but TP53 mutation status does not add to knowledge of SNP 6.0 array–based aCNA status.
Results of multivariate outcome analyses of traditional prognostic factors and SNP 6.0 array–detectable genomic lesions treated as a continuous variable versus OS in CLL
We also analyzed the SNP 6.0 array–based aCNA count as a continuous variable in comprehensive multivariate analysis (aCNA used either as a linear variable or as the square root of aCNA; see “Quantitative multivariate SNP 6.0 array–based aCNA analysis”). Results from theses analyses are summarized in Table 4 and supplemental Table 6, confirming SNP 6.0 array–based aCNA as a strong independent prognostic factor for short OS in CLL.
Results of parallel multivariate analysis of ZAP70 status, CD38 status, IgVH status, FISH status for 17p and 11q combined or alternatively TP53 exon 2-10 mutations, Rai stage, and SNP 6.0 array–based lesions treated as a continuous variable on OS in previously untreated patients with CLL (n = 187)
| Variable . | n/N . | HR . | CI . | P . | Variable . | n/N . | HR . | CI . | P . |
|---|---|---|---|---|---|---|---|---|---|
| SNP 6.0 array genomic lesions (continuous variable)* | N/A | 1.13 | 1.03-1.25 | .01 | SNP 6.0 array genomic lesions (continuous variable)* | N/A | 1.16 | 1.06-1.26 | < 0.01 |
| Del17p or del11q present vs not | 28/159 | 2 | 0.6-7.2 | .28 | TP53 exon 2-10 mutated vs not | 22/165 | 3 | 1.2-8.1 | .03 |
| Rai stage I-IV vs 0 | 90/97 | 0.9 | 0.4-2.4 | .85 | Rai stage I-IV vs 0 | 90/97 | 0.9 | 0.4-2.3 | .79 |
| ZAP70 ≥ 20% vs < 20% | 80/107 | 0.7 | 0.2-1.9 | .42 | ZAP70 ≥ 20% vs < 20% | 80/107 | 0.7 | 0.2-1.9 | .45 |
| IgVH UM vs M | 79/108 | 1.6 | 0.6-4.8 | .37 | IgVH UM vs M | 79/108 | 1.7 | 0.6-4.9 | .35 |
| CD38 ≥ 30% vs < 30% | 45/142 | 0.5 | 0.1-1.8 | .26 | CD38 ≥ 30% vs < 30% | 45/142 | 0.5 | 0.1-1.8 | .28 |
| Square root of SNP 6.0 array genomic lesions (continuous variable)† | N/A | 2.3 | 1.4-3.7 | < .01 | Square root of SNP 6.0 array genomic lesions (continuous variable)† | N/A | 2.3 | 1.5-3.5 | < .01 |
| Del17p or del11q present vs not | 28/159 | 1.2 | 0.3-4.7 | .77 | TP53 exon 2-10 mutated vs not | 22/165 | 2.1 | 0.8-5.9 | .13 |
| Rai stage I-IV vs 0 | 90/97 | 0.8 | 0.3-2.2 | .71 | Rai stage I-IV vs 0 | 90/97 | 0.9 | 0.3-2.2 | .76 |
| ZAP70 ≥ 20% vs < 20% | 80/107 | 0.7 | 0.2-2 | .5 | ZAP70 ≥ 20% vs < 20% | 80/107 | 0.7 | 0.3-2 | .48 |
| IgVH UM vs M | 79/108 | 1.6 | 0.5-4.7 | .4 | IgVH UM vs M | 79/108 | 1.5 | 0.5-4.6 | .43 |
| CD38 ≥ 30% vs < 30% | 45/142 | 0.5 | 0.1-1.8 | .28 | CD38 ≥ 30% vs < 30% | 45/142 | 0.5 | 0.1-1.7 | .24 |
| Variable . | n/N . | HR . | CI . | P . | Variable . | n/N . | HR . | CI . | P . |
|---|---|---|---|---|---|---|---|---|---|
| SNP 6.0 array genomic lesions (continuous variable)* | N/A | 1.13 | 1.03-1.25 | .01 | SNP 6.0 array genomic lesions (continuous variable)* | N/A | 1.16 | 1.06-1.26 | < 0.01 |
| Del17p or del11q present vs not | 28/159 | 2 | 0.6-7.2 | .28 | TP53 exon 2-10 mutated vs not | 22/165 | 3 | 1.2-8.1 | .03 |
| Rai stage I-IV vs 0 | 90/97 | 0.9 | 0.4-2.4 | .85 | Rai stage I-IV vs 0 | 90/97 | 0.9 | 0.4-2.3 | .79 |
| ZAP70 ≥ 20% vs < 20% | 80/107 | 0.7 | 0.2-1.9 | .42 | ZAP70 ≥ 20% vs < 20% | 80/107 | 0.7 | 0.2-1.9 | .45 |
| IgVH UM vs M | 79/108 | 1.6 | 0.6-4.8 | .37 | IgVH UM vs M | 79/108 | 1.7 | 0.6-4.9 | .35 |
| CD38 ≥ 30% vs < 30% | 45/142 | 0.5 | 0.1-1.8 | .26 | CD38 ≥ 30% vs < 30% | 45/142 | 0.5 | 0.1-1.8 | .28 |
| Square root of SNP 6.0 array genomic lesions (continuous variable)† | N/A | 2.3 | 1.4-3.7 | < .01 | Square root of SNP 6.0 array genomic lesions (continuous variable)† | N/A | 2.3 | 1.5-3.5 | < .01 |
| Del17p or del11q present vs not | 28/159 | 1.2 | 0.3-4.7 | .77 | TP53 exon 2-10 mutated vs not | 22/165 | 2.1 | 0.8-5.9 | .13 |
| Rai stage I-IV vs 0 | 90/97 | 0.8 | 0.3-2.2 | .71 | Rai stage I-IV vs 0 | 90/97 | 0.9 | 0.3-2.2 | .76 |
| ZAP70 ≥ 20% vs < 20% | 80/107 | 0.7 | 0.2-2 | .5 | ZAP70 ≥ 20% vs < 20% | 80/107 | 0.7 | 0.3-2 | .48 |
| IgVH UM vs M | 79/108 | 1.6 | 0.5-4.7 | .4 | IgVH UM vs M | 79/108 | 1.5 | 0.5-4.6 | .43 |
| CD38 ≥ 30% vs < 30% | 45/142 | 0.5 | 0.1-1.8 | .28 | CD38 ≥ 30% vs < 30% | 45/142 | 0.5 | 0.1-1.7 | .24 |
Continuous variable was displayed either as a linear variable or as the square root of the number of acquired lesions; see supplemental Methods. The hazard ratio for SNP 6.0 array–based lesions is multiplicative for any unit increase.
Multiplicative hazard for any unit increase.
Multiplicative hazard for any unit increase.
Results of multivariate outcome analyses of traditional prognostic factors and the total length of SNP 6.0 array–detectable subchromosomal genomic lesions treated as a continuous variable versus OS in CLL
Finally, we also analyzed the total combined length of all SNP 6.0 array–based subchromosomal genomic lesions (in Mb, exclusive of monosomies or trisomies) per patient as a continuous prognostic variable in multivariate analysis (total lesion length was used either as a linear variable or as a log-2 transformed variable). Results from these analyses are summarized in supplemental Table 7, indicating that greater subchromosomal genomic loss is an independent prognostic factor for short OS in CLL.
Discussion
In this study, we have used ultra-high-density SNP 6.0 arrays to interrogate the genomes of 255 CLL patients through comparing flow cytometry–sorted CD19+ cells with either paired sorted CD3+ cells or paired buccal DNA for acquired chromosomal copy number changes. CLL cases carried variable lesion loads (range, 0-22), with 28% of cases carrying no subchromosomal acquired lesions even at this gene-level resolution analysis. Overall, ≥ 2 subchromosomal aCN changes were found in 39% of all cases analyzed, whereas ≥ 3 subchromosomal aCN changes were detected in 20% of all cases. This compares with a TP53 exon 2-10 mutation frequency of 14% in this cohort (12% and 21% mutation frequency in previously untreated or relapsed patients, respectively). Through subsequent assessments of the prognostic effect of SNP 6.0 array–based genomic lesion identification on survival in CLL, we were able to determine that SNP array–based lesion detection (≥ 2 lesions and all higher cutoffs) identifies aggressive CLL with shortened OS. Importantly, in comprehensive multivariate analyses and incorporating FISH-based factors (del17p or del11q) as well as TP53 mutations, we were able to demonstrate that elevated SNP 6.0 array–based genomic complexity measurements are a dominant, comprehensive, and independent predictor for short OS in CLL.
Multivariate proportional hazards models establish that the risk captured by various biomarkers are complementary but do not address the potential for interactions in their effects. Specifically, in the proportional hazards context, lack of interaction indicates that the hazard associated with having 2 factors simultaneously is the product of the 2 hazards assessed in isolation. If, in fact, the hazard associated with having 2 risk factors is either greater than, or less than, the product of their individual risks, then an interaction is present. Our bivariate analysis for IGVH or ZAP70 status versus SNP 6.0 array–based aCNA thresholds visually suggest the possibility of interactions, whereas the parallel analysis for del17p/del11q combined or TP53 mutations versus SNP 6.0 array–based aCNA thresholds did not. However, our formal interaction analysis with the use of proportional hazards models proved to be underpowered to resolve any questions about interactions, and patient cohorts many-fold larger would be needed to address this question conclusively.
This study is characterized by methodologic strengths that, combined, are not found in any other relevant published report. These include paired SNP array analysis for all CLL samples, ultra-high-purity (sorted) cell sources as a source of DNA, and a rigorous and conservative genomic copy number data analysis schema that had previously been externally validated with FISH.23 The analysis of paired samples is of particular importance for the accurate determination of genomic complexity in cancer cell genomes in the setting of ultra-high-resolution SNP arrays because most germ line polymorphic copy number variants are < 1 Mb in length. Through comparison of CD19+ cell–derived and paired CD3+/buccal DNA, we were able to unequivocally identify small somatically acquired losses and gains with a high degree of certainty and to identify germ line polymorphic copy number variants directly for each patient. In all, our approach results in high specificity and sensitivity for aCNA, thus avoiding problems resulting from undercalling or overcalling of genomic lesions and the resulting confounding effects on the precision of genomic clinical correlative analysis.
This report is based on patients with CLL enrolled prospectively at a single center but treated sequentially with various treatment schemas. Given that most patients who required therapy at some point were treated with standard-of-care therapy, including potent chemoimmunotherapy programs,42-44 and given our focus on OS as the clinical end point, actual treatments given are not likely to substantially affect the results of this analysis. However, additional future analyses akin to the one presented here and with the use of patient cohorts treated on defined protocols may uncover differences in the prognostic value of elevated genomic complexity depending on treatment schemas. To minimize the effect of biases introduced through sample collections relative to prior treatments or actual dates of diagnosis, we have analyzed and presented data for the untreated and relapsed CLL cohort separately, using either the trial enrollment or the CLL diagnosis date as the reference dates. Importantly, these analyses in aggregate provided evidence for the main conclusion of this report that SNP 6.0 array–based lesion detection coincides with increased risk of short survival, and it identifies subgroups of patients with CLL for which current treatment approaches result in suboptimal outcomes.
Regarding potential mechanisms for the observed effects, 2 broad attractive hypotheses can be generated: (1) that CLL with elevated complexity is the result of multigene defects (in apoptosis and the DNA ds-break response and repair genes, which include defects in the TP53 gene) that allow for a permissive cellular context for the accumulation of genomic lesions,45 a phenotype that is also directly linked to defective apoptosis/cell death in response to genotoxic drugs11 ; and (2) that genomic complexity indicates the capacity for clonal plasticity/evolution and the possibility of the generation of successive clones with more aggressive clinical characteristics. This is particularly relevant after therapy. Clearly, both hypotheses are not mutually exclusive of one another and have implications for the current management strategies of such defined high-risk CLL.
In summary, our data show that a comprehensive assessment of genomic copy number aberrations as enabled through use of ultra-high-resolution SNP arrays adds independent information to CLL outcome prognostication and that SNP array–based genomic complexity assessments identify a large subpopulation of CLL with short survival for which alternative treatment approaches are indicated.46-50
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the microarray core of the University of Michigan Comprehensive Cancer Center for services provided.
This work was supported by the National Institutes of Health through grant 1R01 CA136537-01 (S.N.M.), by the Translational Research Program of the Leukemia & Lymphoma Society of America (S.N.M.), and in part by the National Institutes of Health through the University of Michigan's Cancer Center Support Grant (5 P30 CA46592).
National Institutes of Health
Authorship
Contribution: P.O., J.L., and S.N.M. performed the laboratory research; R.C., L.K., E.P., M.K., M.T., and S.N.M. enrolled patients and contributed and analyzed clinical data; K.S., S.S., and C.L. assisted with statistical analysis and software development for data analysis; S.N.M. conceived the study and supervised the work; and P.O., K.S., and S.N.M. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sami N. Malek, Department of Internal Medicine, Division of Hematology and Oncology, University of Michigan, 1500 E Medical Center Dr, Ann Arbor, MI 48109-0936; e-mail: smalek@med.umich.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal