Abstract
Mutations in the human erythroid Krüppel-like factor (EKLF) can lead to either anemia or the benign InLu phenotype. To elucidate the relationship between these mutations and the differing phenotypes, we prepared recombinant forms of wild-type and 5 mutant EKLF proteins and quantitated their binding affinity to a range of EKLF-regulated genes. Missense mutants (R328H, R328L, and R331G) from persons with InLu phenotype did not bind DNA. Hence, as with the heterozygous loss of function nonsense (L127X, S270X, and K292X) and frameshift (P190Lfs and R319Efs) EKLF mutations, monoallelic loss of EKLF does not result in haploinsufficiency at all loci. In contrast, K332Q has a slightly reduced DNA binding affinity (∼ 2-fold) for all promoters examined but exhibits a phenotype only in a compound heterozygote with a nonfunctional allele. E325K also has a reduced, but significant, binding affinity, particularly for the β-globin gene but results in a disease phenotype even with the wild-type allele expressed, although not as a classic dominant-negative mutant. E325K protein may therefore actively interfere with EKLF-dependent processes by destabilizing transcription complexes, providing a rational explanation for the severity of the disease phenotype. Our study highlights the critical role of residues within the second EKLF zinc finger domain.
Introduction
Erythroid Krüppel-like factor (EKLF; KLF1) is an erythroid specific transcription factor essential for β-globin expression, the switch from fetal to adult globin and definitive erythropoiesis.1,2 The role of EKLF in β-globin expression has been extensively studied.3 EKLF null mice die in utero around embryonic day 14 to 15 because of failure of β-globin expression during fetal liver erythropoiesis.4,5 Similarly, β-globin expression is absent in EKLF null mice containing a human β-locus transgene, whereas γ-globin is increased.6 Furthermore, accumulating evidence shows that EKLF also regulates many other erythroid genes and hence plays a critical and central role in erythropoiesis.2,7-10
Regulated by post-translational modifications, such as acetylation, phosphorylation, and sumoylation, EKLF modulates both chromatin remodeling and transcriptional activity via interaction with other proteins and complexes.11-14 EKLF is composed of 362 amino acids, which form a proline-rich amino terminal domain, thought to be the site of interaction with chromatin, and 3 highly conserved Krüppel-like zinc fingers (ZF1-ZF3) at the carboxy terminus, responsible for recognition of the consensus NCNCNCCCN promoter sequence.15,16 All of the mutations explored in this study fall within the second ZF domain, a key site of interaction with DNA promoter regions. Although the exact mechanism by which EKLF regulates gene expression is not yet fully elucidated, elegant data point toward EKLF as a coordinator of the 3-dimensional genomic architecture.17,18 In regard to the globin switch, EKLF is thought to play a central role in 3-dimensional chromatin looping and interaction of the locus control region with the proximal β-globin promoter, resulting in β-globin expression in adult erythroid cells.17 Consistent with this, EKLF has been shown to interact in vivo with HS2 and HS3 in the locus control region as well as with the β-globin proximal promoter.19
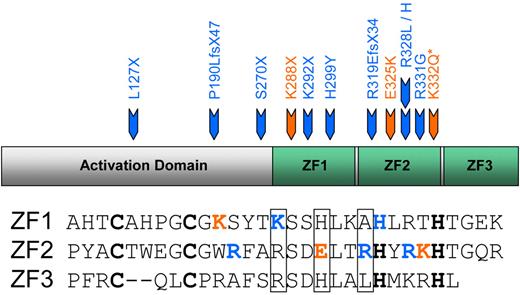
Recently, a series of diverse mutations in human EKLF have been described, which are associated with both benign and disease phenotypes (summarized in Figure 1). A variety of inactivating mutations on one EKLF allele have been identified in persons with the rare blood group phenotype InLu.10 The red cells of affected persons have a gross reduction in expression of the Lutheran blood group glycoprotein and have reduced expression of CD4410 and a slight (1%-3%) elevation in HbF20 but no associated anemia or overt disease. A single EKLF mutation (K288X) found in a large Maltese family with Hereditary Persistence of Fetal Hemoglobin (HPFH) has been linked with haploinsufficiency of EKLF, suggesting a relationship between EKLF and γ-globin expression.21 This mutation would also be predicted to result in the InLu phenotype but in this family gives a marked and variable increase in HbF (3.3%-19.5%). EKLF mutations have been associated with a disease phenotype as well as elevated HbF; E325K has been reported in 3 patients with severe congenital dyserythropoietic anemia (CDA)1,22 (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Red cells from the original patient had aberrant expression of several membrane proteins: reduced ICAM4, and loss of CD44 and the water transporter AQP1,23,24 had an HbF level of 40%-50%25 and persistent expression of ϵ- and ζ-embryonic globins.26 Mutation in the orthologous residue in mouse (E339D) has also been linked to disease: it has been found to cause the Nan (neonatal anemia) phenotype.27,28 Very recently, 2 more heterozygous mutations in human EKLF have been described.29 Interestingly, in this Sardinian family with mild anemia and HPFH, the K332Q mutation was only linked to high levels of HbF in compound heterozygotes (ie, in persons who also carried a second EKLF mutation, S270X). Persons with the S270X mutation alone had the InLu phenotype and normal levels of HbF.
Mutations in EKLF. Heterozygous mutations reported in human EKLF. Core DNA-binding residues in the ZF domains (ZF1-ZF3) are boxed. Blue represents mutations associated with the InLu blood group phenotype; and orange, mutations associated with elevated levels of HbF. *K332Q only when occurring as compound heterozygote with S270X. The “Introduction” contains references.
Mutations in EKLF. Heterozygous mutations reported in human EKLF. Core DNA-binding residues in the ZF domains (ZF1-ZF3) are boxed. Blue represents mutations associated with the InLu blood group phenotype; and orange, mutations associated with elevated levels of HbF. *K332Q only when occurring as compound heterozygote with S270X. The “Introduction” contains references.
In this paper, we compare the effect on DNA binding of missense mutations in the second ZF of human EKLF associated with a benign phenotype, such as InLu (R328L, R328H, and R331G) with those found in persons with mild anemia (K332Q) or severe CDA (E325K) to provide insight into the molecular mechanism(s) giving rise to these very different phenotypes. Investigation of these naturally occurring human models can help elucidate the relationship between the structure of EKLF and its function in erythropoiesis.
Methods
Human CD34+ cell isolation and culture
Blood from normal blood donors and from InLu persons was provided with written consent in accordance with the Declaration of Helsinki and approval from the local ethics committee at the University of Bristol. CD34+ cells were isolated and cultured as previously described30 using Stemspan serum-free medium supplemented with penicillin and streptomycin (Stem Cell Technologies), SCF (10 ng/mL; R&D Systems), IL-3 (1 ng/mL; R&D Systems), erythropoietin (3 U/mL), low-density lipoproteins (20 mg/mL; Merck Biosciences), and FK506 (0.1 ng/mL). At selected time points during culture, cells were harvested and frozen at −80°C.
Protein expression and purification
PCR was used to amplify the EKLF ZF domains (residues 278-362) from EKLF cDNA. Primers were designed for directional cloning using NdeI and XhoI into the pET24b vector (Novagen) with a C-terminal hexahistidine purification tag (supplemental Table 1). The resulting plasmid was transfected into the Rosetta strain of Escherichia coli (Novagen), protein expression induced with isopropyl β-D-1-thiogalactopyranoside, and the culture maintained at 30°C for 4 hours before harvesting. The cell pellet was resuspended in 25mM Tris buffer (pH 7.5), 300mM NaCl, 20mM imidazole, 10% (volume/volume) glycerol, 2mM βmercaptoethanol with EDTA-free protease inhibitor (Roche Diagnostics), and 5 μg/mL DNAse I (Sigma-Aldrich) and loaded onto a HiTrap Ni2+ affinity column (GE Healthcare). Nonspecifically bound DNA fragments were eluted by washing with buffer supplemented with 2M NaCl before elution of protein with a 0-500mM gradient of imidazole. Peak fractions containing protein, which was > 95% pure as judged by SDS-PAGE, were pooled for use in binding assays.
Generation of mutant proteins
QuikChange II mutagenesis kit (Stratagene) was used to generate all mutant EKLF constructs. Primer sequences are given in supplemental Table 1. All mutant proteins were expressed in the same manner as the wild-type (WT).
Stopped flow kinetic analysis
Stopped flow experiments were performed on a SX-17NV spectrophotometer (Applied Photophysics). A fixed concentration (50nM) of hexachlorofluorescein (Hex)–labeled DNA was rapidly mixed with 0.2μM WT or mutant EKLF ZF protein in reaction buffer (25mM HEPES, pH 7.5, 200mM NaCl, and 90μM ZnCl2). The stopped flow chamber was kept at 25°C, and the change in fluorescence was detected > 100 ms; reactions were excited at 536 nm and emission collected with a 550-nm cut-off filter. Approximately 10 datasets for each protein were obtained, averaged, and fitted to a single exponential rate equation: y = A1*(1 − e(−k1*t)) + c, where A1 is amplitude 1, K1 is the rate, and c is the offset, which was set to 4 V. Data were analyzed using Grafit 3 (Erithacus software). Background signal was determined for the reaction buffer and used to normalize each sample to start at zero.
ChIP assay
Day 10 ex vivo cultured human CD34+ cells were fixed in 1% formaldehyde for 10 minutes, lysed (50mM HEPES, pH 7.9, 140mM NaCl, 1mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1× Complete Protease inhibitor cocktail), and sonicated for 8 × 30 seconds at 50% power using a Sanyo Soniprep 150 MSE. Chromatin was collected and precleared using normal rabbit IgG (Santa Cruz Biotechnology) and protein G-Sepharose (GE Healthcare). An anti-EKLF antibody (Abcam ab2483) and protein G-Sepharose were used to isolate immune-specific complexes. ChIP was performed in parallel with an anti-GFP antibody as a nonspecific control. Samples were analyzed by PCR using primers flanking the EKLF CACCC recognition sequence of a number of gene promoters (supplemental Table 1). Primers to SURF1 were used to control for nonspecific DNA interactions.
fEMSA
Recombinant EKLF ZF domain proteins were incubated with 40nM Hex-labeled oligonucleotide (Sigma-Aldrich; supplemental Table 1) in TKM buffer (10mM Tris-HCl pH 8, 150mM KCl, 2mM MgCl2, 0.1% NP-40 and 150mM imidazole, 1mM dithiothreitol, Complete protease inhibitor [Roche Diagnostics], 0.1125 mg/mL poly-dI·dC [Sigma-Aldrich], and 0.1 mg/mL BSA) for 2 hours at 4°C. Free DNA and DNA/protein complexes were resolved on 5% native polyacrylamide gels and visualized on a Typhoon 9400 PhosphorImager with Image Quant analysis Version 5.2 software (Molecular Dynamics, Amersham Biosciences). Quantified bands were used to construct regression curves using GraphPad Prism Version 4.03 software.
Molecular modeling
Homology models were constructed of the EKLF ZF domains in complex with a duplex oligodeoxynucleotide (5′-CCACACCCT-3′) based on the crystal structure of a KLF4-DNA complex (PDB ID 2WBU)31 . Initial models of WT, E325K, and K332Q EKLF were created by manual mutation of amino acid and DNA residues, which differ from the template structure. Each model was then solvated with a 12 Å box of water and 0.3M NaCl added. Solvent was relaxed by 1000 steps of energy minimization using the CHARMM27 force-field with protein and DNA atoms restrained by a force constant of 999 kcal̇ mol−1Å−2. The macromolecular restraints were then relaxed for a further 2000 minimization steps. Molecular models were manually altered using the program COOT,32 solvated using the molecular visualization program VMD,33 and energy minimizations were performed using the molecular dynamics software NAMD.34 The geometry of the final models was analyzed with MolProbity,35 which ranked all models within the top 2% of crystallographic structures determined at approximately 2 Å resolution.
Results
We first checked the localization of the mutant EKLF proteins found in InLu. Whereas some mutations result in decreased overall levels of EKLF, consistent with the expected nonsense-mediated decay36 of nonsense and frameshift mutations (eg, R319Efs), missense mutations (eg, R331G) are expressed and translocate to the nucleus, resulting in both WT and mutant protein within the cell (supplemental Figure 2). Recombinant E325K22 is also stably expressed and translocates to the nucleus.
To understand why different EKLF mutations can result in such dramatically different phenotypes, we analyzed the binding affinity of recombinant forms of EKLF ZF domains (EKLF ZF) to a Hex-labeled oligonucleotide containing the β-globin promoter EKLF recognition sequence and surrounding residues.
Missense mutations in EKLF associated with the InLu phenotype eliminate DNA binding to the β-globin promoter
Stopped flow kinetic analyses were initially performed for WT and InLu mutant EKLF ZF proteins. Changes in fluorescence associated with protein/DNA complex formation were monitored to provide an indication of variations in binding affinity of WT and mutant EKLF for their target DNA sequence. A significant increase in the amplitude of fluorescence, indicating complex formation, was detected for WT EKLF but was much reduced for the R328H, R328L, and R331G mutants (Figure 2A). Observed rate constants for the binding of WT and E325K EKLF to the β-globin oligonucleotide did not increase with increasing protein concentration (data not shown), suggesting that initial complex formation was probably followed by rate-limiting conformational changes. This is consistent with the observed mode of binding of ZF proteins to DNA in which the ZF motifs bind and subsequently wrap around a promoter sequence (see “Structural basis for changes in binding affinity in mutant EKLFs”). This complex mode of recognition complicated the calculation of reliable association constants from these stopped flow data.
Binding affinity of WT and mutant EKLF for the β-globin promoter. (A) Stopped flow kinetic analyses. Increase in fluorescent emission intensity on binding of Hex-labeled double-stranded oligonucleotides composed of the β-globin EKLF binding site (50nM) to recombinant EKLF ZF domains (200nM). Each dataset is an average of 10 measurements. (B) Saturation fluorescent gel shift assays. Complex formation between recombinant WT, E325K, R331G, R328L, and R328H EKLF ZF protein (0-2500nM) with 40nM Hex-labeled double-stranded oligonucleotides composed of the β-globin EKLF binding site and surrounding residues (n = 6). Arrow indicates free Hex-labeled oligonucleotide. *Protein-DNA complexes. **The supershift band is considered nonspecific because at those protein concentrations EKLF binds both to its specific binding site (*) and to nonspecific DNA sequences. (C) Binding curve and Scatchard transformation of data from saturation fEMSAs (WT EKLF, 0-2500nM; E325K EKLF, 313-2500nM) and Hex-labeled oligonucleotide (40nM; n = 6). Quantified bands from fEMSA gels were used to construct regression curves using GraphPad Prism Version 4.03 software. [Bound] = [Bound]/[Bound] + [Free].
Binding affinity of WT and mutant EKLF for the β-globin promoter. (A) Stopped flow kinetic analyses. Increase in fluorescent emission intensity on binding of Hex-labeled double-stranded oligonucleotides composed of the β-globin EKLF binding site (50nM) to recombinant EKLF ZF domains (200nM). Each dataset is an average of 10 measurements. (B) Saturation fluorescent gel shift assays. Complex formation between recombinant WT, E325K, R331G, R328L, and R328H EKLF ZF protein (0-2500nM) with 40nM Hex-labeled double-stranded oligonucleotides composed of the β-globin EKLF binding site and surrounding residues (n = 6). Arrow indicates free Hex-labeled oligonucleotide. *Protein-DNA complexes. **The supershift band is considered nonspecific because at those protein concentrations EKLF binds both to its specific binding site (*) and to nonspecific DNA sequences. (C) Binding curve and Scatchard transformation of data from saturation fEMSAs (WT EKLF, 0-2500nM; E325K EKLF, 313-2500nM) and Hex-labeled oligonucleotide (40nM; n = 6). Quantified bands from fEMSA gels were used to construct regression curves using GraphPad Prism Version 4.03 software. [Bound] = [Bound]/[Bound] + [Free].
Quantitative analysis was therefore performed by saturation fluorescent-EMSA (fluorescent electrophoretic mobility shift assay [fEMSA]; Figure 2B). WT EKLF ZF interacted strongly with the β-globin oligonucleotide (n = 6) with a dissociation constant (Kd) determined from protein-DNA binding curves of 125.6 ± 14.7nM and Scatchard transformation of 120.4nM (Figure 2C). Binding of R331G, R328L, and R328H EKLF ZF (n = 3) was either absent or too low to calculate dissociation constants. Specificity of binding was confirmed using a scrambled β-globin oligonucleotide.
The missense mutation K332Q associated with mild anemia has near-normal binding to the β-globin promoter
Quantitative fEMSA was also used to assess binding of the mutant K332Q EKLF ZF to the β-globin oligonucleotide (Figure 3A). K332Q EKLF bound with a Kd of 156.1 ± 6.5nM (Figure 3B), indicating a 1.3-fold decrease in affinity compared with the WT EKLF ZF in this set of experiments (n = 3).
Binding affinity of WT and mutant K332Q EKLF for the β-globin promoter. (A) Saturation fluorescent gel shift assays using recombinant ZF proteins (0-2500nM, n = 3). (B) Binding curve and Scatchard transformation of data. Details are as in Figure 2.
Binding affinity of WT and mutant K332Q EKLF for the β-globin promoter. (A) Saturation fluorescent gel shift assays using recombinant ZF proteins (0-2500nM, n = 3). (B) Binding curve and Scatchard transformation of data. Details are as in Figure 2.
The missense mutation E325K associated with severe CDA retains binding for the β-globin promoter but with reduced affinity
Initial stopped flow kinetic analyses with the β-globin oligonucleotide showed a significant increase in the amplitude of fluorescence with the E325K EKLF ZF protein. Complex formation was slightly less than WT levels, although substantially more was seen with the InLu mutant proteins (Figure 2A). Quantitative fEMSA gave a Kd of 1286 ± 413.1nM and 1410nM from protein-DNA binding curves and Scatchard transformation, respectively (Figure 2C), indicating a 10-fold reduction in binding to the β-globin oligonucleotide compared with WT EKLF (Table 1).
Binding affinity of WT, E325K, and K332Q EKLF to a range of promoter EKLF recognition sequences
| Promoter . | EKLF recognition sequence . | Kd, nM . | Fold difference between WT and E325K . | Fold difference between WT and K332Q . | ||
|---|---|---|---|---|---|---|
| WT . | E325K . | K332Q . | ||||
| HBB | CCA CAC CCT | 122* | 1286 | 156 | 10.5 | 1.3 |
| LU | CCC CAC CCC | 332 | 1564 | 938 | 4.7 | 2.8 |
| CD44 | CCG CGC CCA | 1225 | — | — | — | — |
| ICAM4 | CTC CAC CCT | 187 | 788 | 426 | 4.2 | 2.3 |
| AQP1 | CCC CAC CCA | 245 | 870 | 550 | 3.6 | 2.2 |
| BCL11A | TCC CAC CCC | 461 | 979 | 1350 | 2.1 | 2.9 |
| p21 | CCC CGC CCG | 537 | — | 782 | — | 1.5 |
| Promoter . | EKLF recognition sequence . | Kd, nM . | Fold difference between WT and E325K . | Fold difference between WT and K332Q . | ||
|---|---|---|---|---|---|---|
| WT . | E325K . | K332Q . | ||||
| HBB | CCA CAC CCT | 122* | 1286 | 156 | 10.5 | 1.3 |
| LU | CCC CAC CCC | 332 | 1564 | 938 | 4.7 | 2.8 |
| CD44 | CCG CGC CCA | 1225 | — | — | — | — |
| ICAM4 | CTC CAC CCT | 187 | 788 | 426 | 4.2 | 2.3 |
| AQP1 | CCC CAC CCA | 245 | 870 | 550 | 3.6 | 2.2 |
| BCL11A | TCC CAC CCC | 461 | 979 | 1350 | 2.1 | 2.9 |
| p21 | CCC CGC CCG | 537 | — | 782 | — | 1.5 |
— indicates that the binding is too low to calculate the Kd value.
Structural basis for changes in binding affinity in mutant EKLFs
We examined the structural basis of the differential affinity for DNA of WT and the mutant EKLFs using molecular modeling. ZF motifs, common within transcription factors, such as the Krüppel-like factor, are highly conserved and have been previously studied in detail through a range of crystal structures.37-41 From these structures, a common mode of DNA recognition has emerged. Each ZF motif is composed of a pair of antiparallel strands packed against a single DNA recognition α-helix via a zinc atom coordinated by 2 histidine and 2 cysteine side chains. These motifs usually occur as tandem arrangements of 3 or more arrays that wrap around a double-helical DNA recognition sequence. Sequence analysis of EKLF has previously identified a 3-finger array (residues 278-362).16 As a very high level of homology (87% identity in amino acid sequence) exists between this region of EKLF and the existing crystal structure of murine KLF4 (PDB ID 2WBU), it was possible to readily assemble a reliable molecular model for the DNA-binding region of EKLF, including its probable complex with the β-globin promoter sequence (Figure 4). All 4 mutation sites are seen to lie within the DNA recognition helix of the second ZF. This molecular model represents a significant advance with respect to previous modeling studies based on the crystal structure of the Zif268 transcription factor.15,27,28 Although Zif268 does share a high level of homology with the ZF region of EKLF (44% identity in amino acid sequence), there are differences in the residues that contact DNA. However, the DNA-binding interface is completely conserved between murine KLF4 and human EKLF (supplemental Figure 3).
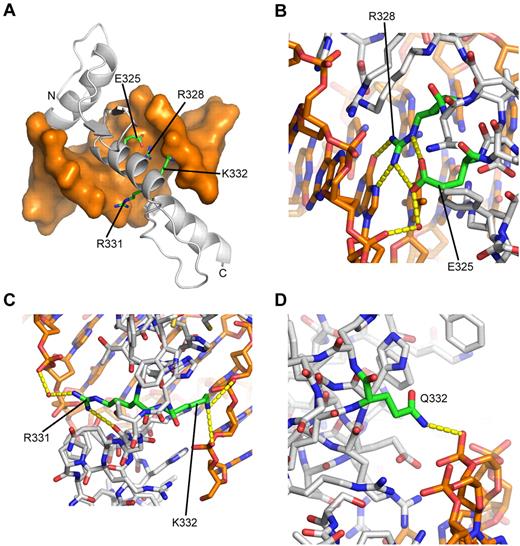
Molecular model of WT EKLF bound to the β-globin promoter. (A) Overall structure of the complex. DNA is shown as an orange molecular surface; protein is shown as a gray Cα-cartoon tracing the α-carbon (Cα) atoms. The amino terminus (A278) is labeled N; the carboxy terminus (L362) is labeled C. The side chains of mutated residues are labeled and highlighted as sticks with carbon atoms colored green. (B) Close-up showing the network of hydrogen bonds formed by E325 and R328, which cannot be replicated when the glutamate side chain is replaced by lysine. All atoms are colored by atom type, with carbon atoms colored as follows: orange represents DNA; green, E325 and R328; and gray, other protein residues. Red spheres represent selected water molecules. Dashed yellow lines indicate predicted hydrogen bonds. (C) Close-up showing interactions of R331 and K332. Displayed as in panel B, except that R331 and K332 carbon atoms are colored green. (D) Close-up of K332Q model showing predicted hydrogen bonding of Q332 to DNA backbone. Displayed as in panel B, except that Q332 carbon atoms are colored green.
Molecular model of WT EKLF bound to the β-globin promoter. (A) Overall structure of the complex. DNA is shown as an orange molecular surface; protein is shown as a gray Cα-cartoon tracing the α-carbon (Cα) atoms. The amino terminus (A278) is labeled N; the carboxy terminus (L362) is labeled C. The side chains of mutated residues are labeled and highlighted as sticks with carbon atoms colored green. (B) Close-up showing the network of hydrogen bonds formed by E325 and R328, which cannot be replicated when the glutamate side chain is replaced by lysine. All atoms are colored by atom type, with carbon atoms colored as follows: orange represents DNA; green, E325 and R328; and gray, other protein residues. Red spheres represent selected water molecules. Dashed yellow lines indicate predicted hydrogen bonds. (C) Close-up showing interactions of R331 and K332. Displayed as in panel B, except that R331 and K332 carbon atoms are colored green. (D) Close-up of K332Q model showing predicted hydrogen bonding of Q332 to DNA backbone. Displayed as in panel B, except that Q332 carbon atoms are colored green.
Residues 325 and 328.
In this model of EKLF, residue E325 is located in the +3 position of the α-helix of the second ZF, a key position where its charged side chain points toward the interface with DNA (Figure 4A). As in recent publications,22,27 we initially thought that the substitution of a positively charged lysine amino acid at this site would enhance binding to the predominantly negatively charged DNA. However, our model of WT EKLF bound to the β-globin promoter predicts that E325 does not project in the correct conformation to contact the DNA directly, except via water-mediated contacts (Figure 4B). Examination of the crystal structure of KLF4, in which the glutamate residue at this site is conserved, shows that the glutamate side chain consistently does not directly contact the DNA. Importantly, the dominant interaction of the glutamate side chain is to form a pair of hydrogen bonds with the side chain of R328, the (i+3) residue succeeding the glutamate. These bonds appear to orient the arginine side chain in the correct position to form 2 direct hydrogen bonds to a guanosine base (Figure 4B). Orientation of the equivalent arginine side-chain by the (i-3) glutamate residue has previously been described in a molecular model of murine EKLF (human R328, mouse R342)28 and is similar to the interaction formed between an arginine and an (i+2) aspartate residue, which is very common in ZF proteins, such as the second and third fingers of KLF4, all 3 fingers of Zif268 (1AAY),37 and the second, third, and fourth fingers of the Wilms tumor suppressor protein (2PRT).41 The extended length of a lysine side chain at position 325 prevents it from forming these interactions and is also too bulky to contact the base pairs directly as seen for histidine. The absence of either of these interactions, which are likely to stabilize complex formation, is consistent with the reduced affinity of the E325K mutant for all tested promoter sequences and would be predicted to result in a similar reduction in affinity for all other, as yet untested, promoter sequences. Binding is nonetheless retained as residue 325 is not in direct contact with the DNA, and this binding is largely independent of the DNA sequence. Substitution of the same glutamic acid residue (human E325; mouse E339) to aspartic acid has recently been reported to cause the Nan mouse phenotype.27,28 In the Nan case, the binding is sensitive to DNA sequence, with the β-carboxyl group of D339 causing a steric clash with the thymidine methyl group, strongly disfavoring thymidine in the central position of the promoter sequence. A lysine side chain, despite being larger overall than aspartate, is not branched at the γ-carbon position and therefore does not cause the same steric clash.
As discussed, the model of EKLF shows that the arginine side chain at position 328 makes direct hydrogen bonds with a guanosine from the promoter site (Figure 4B). Mutation of this residue to leucine or histidine, neither of which can replicate these bonds (the latter not having the extended length of arginine), would be expected to significantly diminish binding affinity for the DNA sequence, as observed in the binding assays.
Residues 331 and 332.
The role of arginine 331 is more complex. Arginine and lysine residues are commonly observed in this position at the end of the recognition helix in ZFs, where they often form hydrogen bonds to the main chain of the subsequent ZF motif, in addition to water-mediated contacts with the DNA phosphate backbone. This residue therefore probably forms an important contribution to aligning and correctly orienting an array of ZFs. These interactions are both observed in the EKLF/DNA model (Figure 4C), but neither contact is possible when no side chain (glycine) is at this position. It is therefore expected that a glycine at this location would have profound effects on the tandem ZF array structure; hence, it is unsurprising that this substitution proves deleterious for DNA binding.
The lysine residue at position 332 is predicted to make contacts with the DNA phosphate backbone but not form stabilizing intramolecular interactions with other amino acids (Figure 4C). Because of the lack of intramolecular interactions, we predict that the effect on the DNA-binding activity of the K332Q mutant is the result of alteration of protein-DNA interactions. The protein-DNA interactions are partially conserved in the model of the K332Q substitution (Figure 4D), which is commensurate with the mild effect on affinity for the β-globin promoter sequence.
In brief, the molecular model for EKLF fully explains the reduced affinity of E325K EKLF for the β-globin promoter sequence, the substantial reductions in affinity observed for the R328L, R328H, and R331G mutants, and the mild effect of the K332Q mutation.
Genes aberrantly expressed in InLu and CDA are bound by EKLF in vivo
Several red cell membrane proteins show aberrant expression in the different phenotypes associated with EKLF mutations. To determine whether the encoding genes are direct targets of EKLF in vivo, we performed ChIP on day 10 ex vivo cultured human CD34+ cells with an EKLF specific antibody (Figure 5). EKLF occupancy was demonstrated at the Lutheran, CD44, AQP1, and ICAM4 promoters. Regulation of these genes by EKLF is consistent with the reduced expression of these proteins in either the InLu and/or CDA phenotypes. These genes were not represented in the comprehensive genome-wide ChIP-seq for EKLF study,42 which may, at least in part, be attributable to differences in the regulation of erythroid genes between mouse and human. However, definitive demonstration of a direct functional effect of EKLF will require specific deletion of the various CACCC sites in these genes in vivo. Furthermore, EKLF may control expression of such genes by both direct and indirect mechanisms. We also demonstrated EKLF occupancy at the p21 cyclin-dependent kinase inhibitor 1A promoter, which is strongly bound in vitro by the Nan mutant EKLF (mouse E339D).28 The β-globin promoter served as a positive control, and the SURF1 promoter as a negative control, with EKLF binding detected at the former but not the latter promoter (Figure 5).
Genes aberrantly expressed in InLu and CDA are directly regulated by EKLF. EKLF ChIP analysis was performed using day 10 ex vivo cultured human erythroblasts. Lane 1 indicates molecular weight marker; lane 2, total genomic DNA; lane 3, positive input control DNA; lane 4, DNA precipitated with EKLF antibody; lane 5, DNA precipitated with GFP (control) antibody; and lane 6, water control. DNA was analyzed by polymerase chain reaction using primers flanking the EKLF recognition sequence of the β-globin, LU, CD44, AQP1, ICAM4, p21, and SURF1 (control) promoters. As some samples were run in a different order on gels, lanes were cut and aligned for consistency.
Genes aberrantly expressed in InLu and CDA are directly regulated by EKLF. EKLF ChIP analysis was performed using day 10 ex vivo cultured human erythroblasts. Lane 1 indicates molecular weight marker; lane 2, total genomic DNA; lane 3, positive input control DNA; lane 4, DNA precipitated with EKLF antibody; lane 5, DNA precipitated with GFP (control) antibody; and lane 6, water control. DNA was analyzed by polymerase chain reaction using primers flanking the EKLF recognition sequence of the β-globin, LU, CD44, AQP1, ICAM4, p21, and SURF1 (control) promoters. As some samples were run in a different order on gels, lanes were cut and aligned for consistency.
Mutant EKLF proteins show differential binding to a range of EKLF-regulated genes
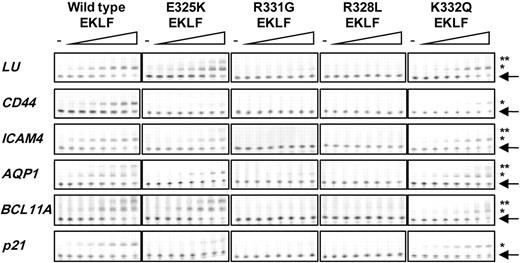
We extended our analyses to quantitate the binding of WT, E325K, R331G, R328L, and K332Q EKLF ZF proteins to the promoter region of other EKLF-regulated genes. Target genes were selected because of their involvement in the phenotypes of interest (“Genes aberrantly expressed in InLu and CDA are bound by EKLF in vivo,” and Figure 5). As for β-globin, we used fEMSA with Hex-labeled oligonucleotides containing the EKLF recognition sequence and surrounding residues (Figure 6; Table 1). The affinity of WT EKLF for the ICAM4 promoter was similar to that obtained for β-globin. Affinity for the Lutheran, AQP1, BCL11A, and p21 cyclin-dependent kinase inhibitor 1A promoters was lower, with least binding to the CD44 promoter. Little or no binding was detected for R331G and R328L EKLF to all oligonucleotides tested. K332Q EKLF showed a slight reduction in binding to all promoters compared with WT. Binding to the CD44 promoter was too low to calculate a Kd, but all other promoters were bound with a 1.5- to 3-fold reduction in affinity. E325K EKLF also bound all oligonucleotides but with a reduced affinity (2- to 4.7-fold down) compared with WT EKLF; affinity to CD44 and p21 was too low to calculate a Kd.
Binding affinity of WT and mutant EKLF for a number of EKLF-regulated genes. Saturation fluorescent gel shift assays were performed with recombinant WT, E325K, R331G, R328L, and K332Q EKLF ZF protein (0-2500nM) and 40nM Hex-labeled double-stranded oligonucleotides composed of EKLF recognition site and surrounding residues from the LU, CD44, ICAM4, AQP1, BCL11A, and p21 promoters (n = 3). Arrow indicates free Hex-labeled oligonucleotide. *Protein-DNA complexes. **Nonspecific binding or binding of EKLF to degenerate CACC sites.
Binding affinity of WT and mutant EKLF for a number of EKLF-regulated genes. Saturation fluorescent gel shift assays were performed with recombinant WT, E325K, R331G, R328L, and K332Q EKLF ZF protein (0-2500nM) and 40nM Hex-labeled double-stranded oligonucleotides composed of EKLF recognition site and surrounding residues from the LU, CD44, ICAM4, AQP1, BCL11A, and p21 promoters (n = 3). Arrow indicates free Hex-labeled oligonucleotide. *Protein-DNA complexes. **Nonspecific binding or binding of EKLF to degenerate CACC sites.
Discussion
It is evident that mutations in EKLF can have remarkably different functional effects. In EKLF with missense mutations associated with InLu, DNA binding is all but eliminated. In persons with InLu where heterozygous nonsense mutations in EKLF are found, such as the introduction of either stop codons or frameshifts, the production of these nonfunctional forms of EKLF is analogous to the missense forms in which DNA binding is disrupted. Hence, the main determinant of the benign InLu phenotype appears to be a reduction in the level of functional EKLF, suggesting a single functional EKLF allele is sufficient for erythropoiesis, a situation analogous to the EKLF+/− mouse, which is phenotypically normal.6
The mutant K332Q EKLF shows reduced binding to a number of DNA targets compared with WT EKLF. The reduction in binding to the β-globin and BCL11A promoters that we observed using purified recombinant ZF proteins (1.3-fold and 2.9-fold, respectively) was virtually identical to that reported by Satta et al using nuclear extracts (1.4-fold and 3-fold respectively).29 It would be interesting to determine the RBC phenotype of persons with the K332Q mutation (both heterozgyotes and compound heterozygotes). Our binding experiments suggest that red blood cells from K332Q compound heterozygotes may have a gross reduction in expression of CD44 protein and weakened expression of other membrane proteins, such as Lutheran, ICAM4, and Aquaporin 1. Importantly, in the presence of a WT allele, the K332Q mutation is benign (as in InLu) but when accompanied by the S270X InLu allele, the slightly reduced affinity of available EKLF is sufficient to cause a mild anemia and elevated HbF.29 This suggests that there is a threshold for EKLF activity that is less than, but probably close to, 50% of fully functional protein. It is possible that, in addition to the haploinsufficiency (because of the K288X mutation) reported by Borg et al,21 the Maltese family with HPFH have another genetic factor that either affects the expression of the WT EKLF allele or affects HbF levels in an alternative way.
In marked contrast, E325K EKLF has a reduced but significant binding affinity (0.8-1.5μM) for most promoters. The greatest reduction (10-fold) was seen at the β-globin promoter, consistent with the severe anemia in the CDA patient. The approximate 4-fold reduction in binding affinity at the ICAM4 and AQP1 promoters, and the low binding to the CD44 promoter, is also consistent with the reduced expression of these proteins on red cells from the patient. A direct relationship between EKLF levels and induction of the CD44 promoter is suggested by the CD44 promoter-reporter assay described by Arnaud et al.22 Hence, reduced expression of CD44 found in cells from both InLu and CDA persons may be attributable to haploinsufficiency at this promoter.
However, the approximately 5-fold reduced binding to the Lu promoter but normal expression of the Lutheran protein in the patient cannot currently be explained. Somewhat surprisingly, E325K EKLF only showed a 2-fold reduction in affinity to the BCL11A promoter. Perhaps the BCL11A promoter is particularly sensitive to the levels of bound EKLF, but some of the phenotypic effects are unlikely to be the result of reduced binding of EKLF alone. In the original patient with CDA, E325K EKLF occurs in a heterozygous background. As the level of fully functional (WT) EKLF in this phenotype must be similar to InLu, this suggests that the E325K protein actively interferes with EKLF-dependent processes. This hypothesis was corroborated by Arnaud et al,22 whereby reduced transcriptional activity of WT EKLF on cotransfection with E325K EKLF was observed. However, these authors predicted that E325K EKLF acts as a dominant-negative mutant with increased binding affinity compared with WT. Our modeling and experimental data do not support this. It is possible that the reduced affinity of the E325K EKLF for promoters destabilizes transcription complexes in a competitive manner that becomes evident when levels of WT complexes are limiting, such as in WT/E325K heterozygotes.
Substitution of the same glutamic acid residue (human E325; mouse E339) to aspartic acid has recently been reported to cause the Nan mouse phenotype,27,28 which presents with anemia and defects in erythrocyte membrane skeletal proteins. Using homology modeling, Heruth et al also postulated that this mutation would increase affinity of the mutant EKLF for DNA.27 However, Siatecka et al reported a much more complex picture with an altered gene expression profile, resulting from selective binding to different target sequences.28 They observed binding of E339D EKLF to sites that contained a “G” in the middle (which they termed a category I site) but no binding to sites with an “A” (a category II site). Interestingly, E339D EKLF interfered with expression of genes that contained a promoter category II site (ie, those the mutant EKLF did not bind). The authors proposed a novel unknown mechanism to account for this. In our quantitative fEMSA binding experiments, most of the target promoters are category II, but CD44 and p21 are category I sites. However, in contrast to the Nan mutant, we observed a reduction in binding affinity of E325K EKLF to all promoters examined. These studies highlight the critical role of this particular amino acid residue in the normal function of EKLF.
In this study, we have analyzed 5 mutations occurring within the second ZF domain of EKLF. Because these mutations lie within the DNA recognition helix, our assays have focused on the ability of the mutant EKLF proteins to bind different DNA targets. However, EKLF is also known to interact with other proteins (eg, BRG1-containing chromatin remodeling factor complexes).11,43,44 It is possible that such interactions could be impaired in the patient with CDA, which might in part explain the more severe phenotype. However, this seems unlikely, at least for BRG1, as binding of E325K and InLu mutant EKLFs (R328H and R331G) to the N-terminal of BRG1 was comparable to WT EKLF (supplemental Figure 4).
In considering the global effect of the E325K mutation, it is pertinent to also consider the essential role of EKLF in spatial genome organization. Binding of EKLF to gene promoters is essential for clustering and coassociation of EKLF-regulated genes at shared transcription factories where their expression is coregulated.18,45 Formation of reduced-affinity, and therefore destabilized, E325K promoter complexes may result in premature dissociation from the DNA with stalling of nascent transcription factory assembly or disruption of mature factories. In either case, the components (proteins and DNA) recruited thus far will be temporarily sequestered in nonfunctional or abnormally functional transcription factories, resulting in the skewed genetic output observed in CDA.
In conclusion, InLu, mild anemia with HPFH, and CDA are disparate human models for EKLF function and demonstrate that mutations resulting in differential affinity for DNA are physiologically significant. Whereas mutations that abolish or significantly impair protein function are more commonly associated with disease phenotypes, this study illustrates the importance of modulation in transcription factor binding for gene regulation. Suboptimal binding of E325K EKLF for its promoter sites is seen to lead to a more severe phenotype than nonfunctional mutants within a haploinsufficient environment. This has important implications for our understanding of the relationship between gene mutations and the development of disease. Further studies to investigate a wider range of mutations in EKLF are required to determine a comprehensive genotype-phenotype correlation.
Presented in part at the 51st annual meeting of the American Society of Hematology, New Orleans, LA, December 6, 2009.1
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Kate Heesom of the Bristol Proteomic facility for proteomic advice and 2D gel electrophoresis, Peter Martin of IBGRL for sequencing, Prof Beverley M. Emerson of the Salk Institute for Biologic Studies, San Diego, for kindly donating the GST-BRG1-N plasmid, and Dr Kevin Gaston of the University of Bristol for helpful advice on the manuscript.
This work was supported by the Biotechnology and Biological Sciences Research Council (CASE studentships; W.L. and V.S.S.F.) and Department of Health (England).
Authorship
Contribution: W.L., V.S.S.F., M.C.W., and B.K.S. designed and performed experiments; S.F.P., B.M.R., and K.T. gave practical assistance; N.M.B. constructed the molecular model; D.J.A., B.K.S., R.L.B., and J.F. conceived and supervised the study; J.F. wrote the paper; and D.J.A., R.L.B., B.K.S., and N.M.B. reviewed the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jan Frayne, School of Biochemistry, University of Bristol, Bristol, BS8 1TD, United Kingdom; e-mail: jan.frayne@bristol.ac.uk; David J. Anstee, Bristol Institute for Transfusion Sciences, National Health Service Blood and Transplant, Filton, Bristol BS34 7QG, United Kingdom; e-mail: david.anstee@nhsbt.nhs.uk; and R. Leo Brady, School of Biochemistry, University of Bristol, Bristol, BS8 1TD, United Kingdom; e-mail: l.brady@bristol.ac.uk.
References
Author notes
B.K.S., W.L., and V.S.S.F. contributed equally to this study.

![Figure 2. Binding affinity of WT and mutant EKLF for the β-globin promoter. (A) Stopped flow kinetic analyses. Increase in fluorescent emission intensity on binding of Hex-labeled double-stranded oligonucleotides composed of the β-globin EKLF binding site (50nM) to recombinant EKLF ZF domains (200nM). Each dataset is an average of 10 measurements. (B) Saturation fluorescent gel shift assays. Complex formation between recombinant WT, E325K, R331G, R328L, and R328H EKLF ZF protein (0-2500nM) with 40nM Hex-labeled double-stranded oligonucleotides composed of the β-globin EKLF binding site and surrounding residues (n = 6). Arrow indicates free Hex-labeled oligonucleotide. *Protein-DNA complexes. **The supershift band is considered nonspecific because at those protein concentrations EKLF binds both to its specific binding site (*) and to nonspecific DNA sequences. (C) Binding curve and Scatchard transformation of data from saturation fEMSAs (WT EKLF, 0-2500nM; E325K EKLF, 313-2500nM) and Hex-labeled oligonucleotide (40nM; n = 6). Quantified bands from fEMSA gels were used to construct regression curves using GraphPad Prism Version 4.03 software. [Bound] = [Bound]/[Bound] + [Free].](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/11/10.1182_blood-2011-04-349985/4/m_zh89991177870002.jpeg?Expires=1770983187&Signature=Lg0aJpG~y1zLez7y9iQQyqG38uQKYV12f-fefhVMu-Rk8FmWfeYrSAYl47LBLnMMHXaHflz3ukHi5pURifeA28zlOXuh0d1KwY4NYjvl12P94x8ZmcfqWYtqtGYS2dWVKnENIwwf~GgmXYyoKCmdxxn38nY3wGwKOzoJiRSFGPit~cFnmcbx2vQHl-ZEkUGjvT8zHgD8VT1D0fbhAlISsXlJWFqhrHHZuQ23GhR0p6Nb18Up8aCyeMfxc03BaB6fUXefY4Mo3jzUb7-nxQOni6bB-8t3MEacrAUefaMuZ4GyaXV4JnqFBP-~d5U8R0PSGLweF93x~bsxoC23E0k5Fg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)