Abstract
Oligo-array comparative genomic hybridization (CGH) and gene-expression profiling of natural killer (NK)–cell neoplasms were used in an effort to delineate the molecular pathogenesis involved. Oligo-array CGH identified two 6q21 regions that were most frequently deleted (14 of 39 or 36%). One of these regions included POPDC3, PREP, PRDM1, ATG5, and AIM1, whereas the other included LACE1 and FOXO3. All genes located in these regions, except for POPDC3 and AIM1, were down-regulated in neoplastic samples, as determined by gene-expression analysis, and were therefore considered to be candidate tumor-suppressor genes. A20 and HACE1, the well-known tumor-suppressor genes located on 6q21-23, were included as candidate genes because they also demonstrated frequent genomic deletions and down-regulated expression. The Tet-Off NK cell line NKL was subsequently established for functional analyses. Seven candidate genes were transduced into Tet-Off NKL and forced re-expression was induced. Re-expression of FOXO3 and PRDM1 suppressed NKL proliferation, but this was not the case after re-expression of the other genes. This effect was confirmed using another NK cell line, SNK10. Furthermore, genomic analyses detected nonsense mutations of PRDM1 that led to functional inactivation in one cell line and one clinical sample. PRDM1 and FOXO3 are considered to play an important role in the pathogenesis of NK-cell neoplasms.
Introduction
Natural killer (NK)–cell neoplasms are lymphoid malignancies with an aggressive clinical course; they are more prevalent in Asians and native Americans.1 The World Health Organization classification defines NK-cell neoplasms as 2 major disease entities: aggressive NK-cell leukemia (ANKL) and extranodal NK/T-cell lymphoma, nasal type (ENKTL).1,2 Neoplastic cells of both diseases express the NK-cell markers CD16 and CD56, cytotoxic molecules, granzyme B- and T-cell–restricted intracellular antigen (TiA1), and the EBV-associated molecules EBV-LMP1 and EBER.1,2
The molecular pathogenesis of NK-cell neoplasms remains unknown not just because of their rarity, but also because of the pathologic features of numerous inflammatory cell infiltrates and necrotic changes. Most studies describing chromosomal aberrations in NK-cell neoplasms have included fewer than 10 cases, and recurrent chromosomal translocations that are often recognized in B-cell neoplasms have not been found in NK-cell neoplasms.3-6 However, some gene-expression profiling and array comparative genomic hybridization (CGH) analyses of NK-cell neoplasms have been reported amidst the recent progress in molecular biotechnology.7-9 We analyzed genomic aberrations of 17 ANKL cases and 10 ENKTL cases using array CGH, and described 2q gain and deletions of 6q16-27 and 7p15-22 as frequent abnormalities.9 Iqbal et al reported on the array CGH analysis and gene-expression profiling of NK-cell neoplastic cell lines, and identified ATG5, PRDM1, and AIM1 as candidate tumor-suppressor genes.8 Huang et al also used gene-expression profiling and array CGH to investigate ENKTL and made a comparison to peripheral T-cell lymphoma.7 They described HACE1, a well-known tumor-suppressor gene located in 6q, as a candidate tumor-suppressor gene in ENKTL, although a functional analysis was not provided. They also reported that PDGFRA was up-regulated in ENKTL, and that its inhibitor imatinib suppressed proliferation in an NK cell line.
It has recently been accepted that, in addition to genomic aberrations and changes in expression level, functional effects are also important in the exploration of cancer-associated genes.10-12 Very few studies have investigated the functional effects associated with the molecular pathogenesis of NK-cell neoplasms by gene transduction.13 Therefore, identification of the most likely tumor-suppressor genes among the numerous candidate genes described in previous studies remains to be resolved.7-9
In the present study, we identified FOXO3 and PRDM1 as important tumor-suppressor genes of NK-cell neoplasms, as determined by a combination of array-CGH, gene-expression profiling, and functional analyses. We also detected nucleotide substitutions highly suspicious as somatic mutations in some clinical samples.
Methods
Samples and cell lines
We collected 35 cases of NK-cell neoplasms (9 ANKL and 26 ENKTL). The diagnoses were based on the World Health Organization classification.1,2 These samples were obtained from patients at Samsung Medical Center, Aichi Cancer Center, Fukuoka University, and other collaborating institutions. The analysis was performed at Aichi Cancer Center with the approval of the institutional review board. Patient characteristics are described in Table 1. Twenty-four cases (nos. 1-24) were cases analyzed in a previous report,9 and 11 cases (nos. 25-35) were newly collected cases in this study. DNA and RNA samples were extracted according to previously described methods.9,14 We examined rearrangement of the TCR gene (γ chain) in case numbers 26, 28, 29, 30, 31, 32, and 35 using PCR as described previously,15 and confirmed NK-cell lineage in all of the cases. Seven cell lines of NK-cell neoplasms (NK-YS,16 SNK6,17 NK92,18 NKL,19 HANK-1,20 KHYG-1,21 and SNK1022 ) were also analyzed (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Normal NK cells from 3 healthy volunteers were isolated and analyzed as control samples. The detailed method used for the isolation is described in supplemental Methods.
Patient characteristics
| No. . | Age . | Sex . | Site . | Diagnosis . |
|---|---|---|---|---|
| 1* | 40 | M | Heart | ANKL |
| 2* | 53 | F | Skin | ANKL |
| 3* | 63 | M | Nasal | ANKL |
| 4* | 12 | F | PB | ANKL |
| 5* | 55 | M | PB | ANKL |
| 6* | 52 | F | PB | ANKL |
| 7* | 23 | M | PB | ANKL |
| 8* | 34 | M | Spleen | ANKL |
| 9* | 18 | M | PB | ANKL |
| 10* | 31 | M | Nasal | ENKTL |
| 11* | 19 | F | Skin | ENKTL |
| 12* | 87 | F | Skin | ENKTL |
| 13* | 57 | F | Skin | ENKTL |
| 14* | 45 | F | Nasal | ENKTL |
| 15* | 71 | F | Nasal | ENKTL |
| 16* | 52 | F | Nasal | ENKTL |
| 17* | 63 | M | Skin | ENKTL |
| 18* | 53 | F | Nasal | ENKTL |
| 19* | 62 | M | Nasal | ENKTL |
| 20* | 72 | M | Nasal | ENKTL |
| 21* | 40 | F | Nasal | ENKTL |
| 22* | 46 | F | Nasal | ENKTL |
| 23* | 50 | F | Nasal | ENKTL |
| 24* | 38 | M | Nasal | ENKTL |
| 25† | 46 | F | Nasal | ENKTL |
| 26† | 31 | F | Nasal | ENKTL |
| 27† | 28 | M | Nasal | ENKTL |
| 28† | 63 | M | Nasal | ENKTL |
| 29† | 32 | M | Nasal | ENKTL |
| 30† | 60 | M | Nasal | ENKTL |
| 31† | 46 | F | Nasal | ENKTL |
| 32† | 34 | M | Nasal | ENKTL |
| 33† | 59 | M | Nasal | ENKTL |
| 34† | 54 | M | Skin | ENKTL |
| 35‡ | 43 | M | Nasal | ENKTL |
| No. . | Age . | Sex . | Site . | Diagnosis . |
|---|---|---|---|---|
| 1* | 40 | M | Heart | ANKL |
| 2* | 53 | F | Skin | ANKL |
| 3* | 63 | M | Nasal | ANKL |
| 4* | 12 | F | PB | ANKL |
| 5* | 55 | M | PB | ANKL |
| 6* | 52 | F | PB | ANKL |
| 7* | 23 | M | PB | ANKL |
| 8* | 34 | M | Spleen | ANKL |
| 9* | 18 | M | PB | ANKL |
| 10* | 31 | M | Nasal | ENKTL |
| 11* | 19 | F | Skin | ENKTL |
| 12* | 87 | F | Skin | ENKTL |
| 13* | 57 | F | Skin | ENKTL |
| 14* | 45 | F | Nasal | ENKTL |
| 15* | 71 | F | Nasal | ENKTL |
| 16* | 52 | F | Nasal | ENKTL |
| 17* | 63 | M | Skin | ENKTL |
| 18* | 53 | F | Nasal | ENKTL |
| 19* | 62 | M | Nasal | ENKTL |
| 20* | 72 | M | Nasal | ENKTL |
| 21* | 40 | F | Nasal | ENKTL |
| 22* | 46 | F | Nasal | ENKTL |
| 23* | 50 | F | Nasal | ENKTL |
| 24* | 38 | M | Nasal | ENKTL |
| 25† | 46 | F | Nasal | ENKTL |
| 26† | 31 | F | Nasal | ENKTL |
| 27† | 28 | M | Nasal | ENKTL |
| 28† | 63 | M | Nasal | ENKTL |
| 29† | 32 | M | Nasal | ENKTL |
| 30† | 60 | M | Nasal | ENKTL |
| 31† | 46 | F | Nasal | ENKTL |
| 32† | 34 | M | Nasal | ENKTL |
| 33† | 59 | M | Nasal | ENKTL |
| 34† | 54 | M | Skin | ENKTL |
| 35‡ | 43 | M | Nasal | ENKTL |
PB indicates peripheral blood.
Results of BAC array CGH reported previously.9
Both oligoarray CGH and gene-expression profiling were analyzed.
Only gene-expression profiling was analyzed.
Array CGH microarray and gene-expression profiling
We performed array CGH analysis using the Human Genome CGH 44A microarray (4426B; Agilent Technologies) comprising approximately 43 000 60-mer oligonucleotide probes. Normal human male genomic DNA from PBMCs was used in all experiments as a reference sample.23 Sample labeling and microarray processing were performed according to the manufacturer's protocol (www.agilent.com). Scanning analysis was performed using the Agilent Micro Array Scanner (Agilent Technologies), and acquired array images were processed using Feature Extractions Version 9.1 (Agilent Technologies). The analytical software DNA analytics Version 4.0 (Agilent Technologies) was then used for array CGH analysis. The ADM-1 algorithm (Threshold 6.0) was adopted to detect genomic aberrations.24 The probes on chromosomes X and Y were excluded from the analysis because each experiment was not sex-matched.
For analysis of gene-expression profiling, total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. The quality of the extracted RNA was ascertained using a Bioanalyzer (Agilent Technologies). The Low-RNA Input Linear Amplification and Gene Expression Hybridization kits (Agilent Technologies) were used for RNA labeling and amplification. The Whole Human Genome 4 × 44k Oligomicroarray Kit containing 43 377 60-mer probes and the Gene Expression Hybridization kit (Agilent Technologies) were used for the hybridization of labeled RNA. The experimental protocol used paralleled the manufacturer's protocol (www.agilent.com). Each oligomicroarray slide was scanned and converted into a dataset for array CGH analysis.
Statistical analysis of gene-expression profiles including Gene Set Enrichment Analysis (GSEA)25 and Gene Ontology Analysis were performed using normalized data. A detailed description of these analyses can be found in supplemental Methods.
FISH
Dual-color FISH was performed to confirm the deletion of 6q21 (FOXO3) detected by array CGH analysis according to a previously described method.26 For hybridization, a spectrum green–labeled probe at the centromeric region of chromosome 6 (CEP6; Abbott Laboratories) and a spectrum red–labeled RP11-118H13 probe were used for detection of the FOXO3 gene at chromosome band 6q21.
Semiquantitative RT-PCR
cDNA was synthesized from 1 μg of total RNA using SuperScript II Reverse Transcriptase (Life Technologies). Semiquantitative determination of FOXO3 and PRDM1 gene expression was assessed and compared, with β-actin gene as an internal control. The forward and reverse primers used for RT-PCR were as follows: β-actin; forward (5′-gactacctcatgaagatc-3′) and reverse (5′-gatccacatctgctggaa-3′), FOXO3; forward (5′-ggtgcgttgcgtgccctact-3′) and reverse (5′-ccgtggcagttccaccgtgc-3′), PRDM1; forward (5′-caaatgtcagacttgcaacaagggc-3′) and reverse (5′-ggccagaatttccttctccactaca-3′). The PCR conditions were: 95°C for 3 minutes and then 25 cycles (β-actin) or 35 cycles (FOXO3 and PRDM1) at 95°C for 45 seconds, 55°C (FOXO3 and β-actin) or 53°C (PRDM1) for 45 seconds, 72°C for 90 seconds, and a final extension at 72°C for 10 minutes. The expected fragment sizes were: 464 bp for β-actin, 543 bp for FOXO3, and 474 bp for PRDM1. The intensity of each band in the agarose gel image was calculated using ImageJ software (http://rsbweb.nih.gov/ij/). The Student t test was used to determine significant differences.
Mutation analysis of PRDM1 and FOXO3
The 7 coding exons of PRDM1 and the 2 coding exons of FOXO3 were amplified from genomic DNA using PCR. PCR primers were synthesized based on previous studies.27,28 Thirty-two clinical samples and 7 cell lines were analyzed. Because there was no remaining genomic DNA, case numbers 25, 33, and 34 were not analyzed. Direct sequencing of PCR products was performed through capillary electrophoresis using the ABI3100 sequencer (Applied Biosystems).
Gene transduction to the NK cell line
The pRetroX-Tet-Off Advanced retroviral vector and pRetroX-tight-Pur-multiple cloning site, included in the Retro-X Tet-Off Advanced Inducible Expression System (Clontech) were used in this study. A detailed description of plasmid construction can be found in supplemental Methods.
Stable Tet-Off cell lines were generated following the manufacturer's protocol. NKL and SNK10 were chosen because these possess genomic loss of 6q. Doxycycline was washed from Tet-Off NKL and 1 × 105 cells were resuspended in 1 mL of medium. The medium was changed and diluted 10-fold (NKL) or 5-fold (SNK10) at day 2. Cell numbers were determined using the trypan blue exclusion assay at days 2, 4, and 6. The Student t test was used to determine significant differences.
Western blotting
Western blot analysis was performed essentially as described previously.29 The transferred polyvinylidene fluoride membranes were incubated overnight with anti-FLAG M2 monoclonal antibody (Sigma-Aldrich), anti-actin antibody (AC-40; Sigma-Aldrich), or anti-A20 antibody (59A426; Abcam) in blocking buffer at 4°C. These were then washed extensively in PBS containing 0.05% Tween 20 T-PBS and incubated for 1 hour with HRP-conjugated anti–mouse IgG (Amersham) diluted at 1:1000 in blocking buffer without 0.05% NaN3. Antibody binding was visualized using an enhanced chemiluminescence detection kit (Amersham). We used FLAG-tagged genes for FOXO3, PRDM1, ATG5, HACE1, LACE1, and PREP (supplemental Table 2), so detection was conducted using anti-FLAG antibody. Because the A20 construct was made without a FLAG-tag, anti-A20 antibody was used. Anti-FLAG antibody or anti-A20 antibody was initially incubated to detect the target gene product. Membranes were subsequently stripped using 0.1M glycine-HCl buffer (pH 2.5) and then incubated with anti-actin antibody.
Apoptosis and cell-cycle assay
Apoptotic cell death was determined using the annexin V APC Apoptosis Detection Kit (eBioscience) according to the manufacturer's instructions. Briefly, cells were washed in serum-free medium and stained for 15 minutes at room temperature. For the cell-cycle assay, propidium iodide (PI) staining was performed according to a method described in a previous study with some modifications.30 Briefly, 5 × 105 cells were incubated in 1 mL of PBS containing 0.1% Triton X-100 (Sigma-Aldrich), 0.5% RNase A (QIAGEN), and 20 mg/mL PI (Wako). Analyses were performed using a FACSCalibur (BD Biosciences) and FlowJo Version 7.2.4 software (TreeStar).
Microarray data accession number
The microarray data obtained in this study have been submitted to ArrayExpress and assigned accession numbers E-MEXP-3002 (array CGH) and E-MEXP-2996 (gene expression).
Results
Gene-expression profile of NK-cell neoplasms
Comprehensive gene-expression profiles of 7 NK cell lines and 11 clinical samples were analyzed (Table 1 and supplemental Table 1). The gene-expression profiles of NK cells from normal donors were analyzed as control samples. Genes associated with the cell cycle, DNA replication, and angiogenesis were more highly expressed in neoplastic samples compared with normal NK cells (supplemental Tables 3, 5, and 7). We then compared the expression profiles of clinical samples and cell lines. Several pathways associated with inflammation (eg, the CCR5 pathway: Fos, Jun, CCL4, etc) and with immunosuppression (eg, the PD1 signal: CD4, PDCD1, CD274, etc) were highly expressed in clinical samples (supplemental Tables 4, 6, and 7).
Genomic profile of NK-cell neoplasms
Oligo-array CGH was performed on 32 clinical samples and 7 cell lines (Table 1 and supplemental Table 1). Twenty-four clinical samples included cases previously analyzed by bacterial artificial chromosome (BAC)–array CGH.9 High correlation and reproducibility were observed between the oligo-array CGH and the BAC-array CGH results (supplemental Figure 1A). Furthermore, oligo-array CGH could detect the narrow genomic aberrations that previous BAC-array CGH failed to detect (supplemental Figure 1B).
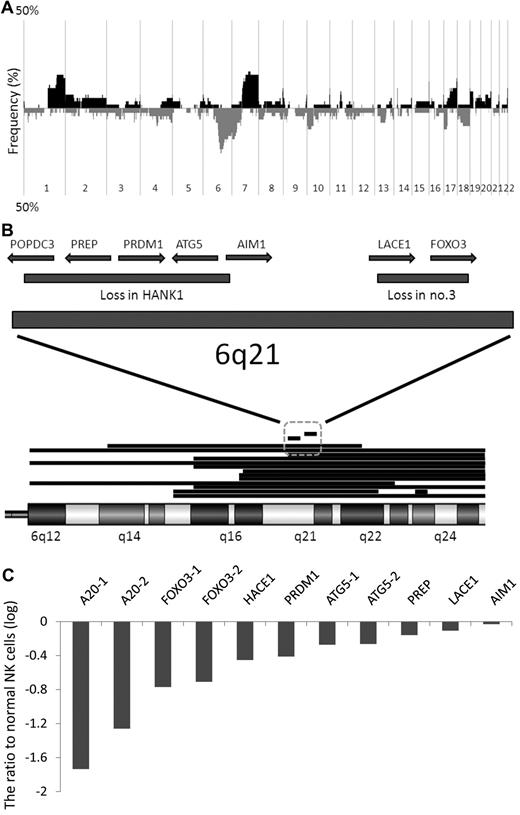
Figure 1A shows the results of oligo-array CGH for the 39 samples analyzed. The gains of 1q31.2-44, 7q11.22-36.3, 16p13.3 and 11p15.5, and the losses of 6q16.1-27 and 7p15.3-22.2, were found in more than 8 cases (20%). 6q21 loss was the most frequent aberration. The narrow deleted region of 6q21 found in HANK1 contained 5 known genes, POPDC3, PREP, PRDM1, ATG5, and AIM1, and that found in clinical sample no. 3 contained 2 known genes, LACE1 and FOXO3. These regions were considered as the most frequently deleted minimal common region (MCR; 14 of 39 or 36%; Figure 1B). FISH analysis showed heterozygous deletion of the genomic region covering FOXO3 in NKL, SNK6, and SNK10, whereas no aberration was found in KHYG1, NK92, HANK1, and NKYS. These results were in agreement with the results of the oligo-array CGH (Figure 4A).
Genomic and expression profiles of NK-cell neoplasms. (A) Frequency of genomic aberrations analyzed by oligo-array CGH. Thirty-two clinical samples and 7 cell lines were analyzed. Each probe is aligned from chromosome 1 to 22 and p to q. The vertical axis indicates penetrance of the genomic aberration among the analyzed cases. The black area shows gain/amplification and the gray area shows loss/deletion. (B) Genomic aberration of chromosome 6q15-25, the most frequently deleted region of the whole genome except for chromosomes X and Y. Fifteen cases with a 6q deletion are described. Each deleted region is indicated as a horizontal bar. Two narrow, independently deleted regions (∼ 1.5 Mb) are observed in HANK1 and clinical case number 3, and are the most frequently deleted MCRs. The deleted region in HANK1 includes POPDC3, PREP, PRDM1, ATG5, and AIM1, and the deleted region in clinical case number 3 includes LACE1 and FOXO3. (C) Gene-expression profiles were analyzed in 7 cell lines and 11 clinical samples using oligo-microarray. Candidate genes selected by genomic analysis are described. The probe intensity was converted to a numerical value, and the z-score was calculated as described in supplemental Methods. The vertical axis indicates the difference in average value between normal NK cells (n = 3) and neoplastic samples (n = 18). TNFAIP3 (A20), FOXO3, and ATG5 possess 2 probes. A20-1: A_24_P166527, A20-2: A_24_P157926, FOXO3-1: A_32_P102062, FOXO3-2: A_23_P345575, ATG5-1: A_24_P175059, ATG5-2: A_23_P111381. Most of the candidate genes, and in particular FOXO3 and A20, were down-regulated in neoplastic samples compared with normal NK cells, whereas a negligible difference was observed for AIM1.
Genomic and expression profiles of NK-cell neoplasms. (A) Frequency of genomic aberrations analyzed by oligo-array CGH. Thirty-two clinical samples and 7 cell lines were analyzed. Each probe is aligned from chromosome 1 to 22 and p to q. The vertical axis indicates penetrance of the genomic aberration among the analyzed cases. The black area shows gain/amplification and the gray area shows loss/deletion. (B) Genomic aberration of chromosome 6q15-25, the most frequently deleted region of the whole genome except for chromosomes X and Y. Fifteen cases with a 6q deletion are described. Each deleted region is indicated as a horizontal bar. Two narrow, independently deleted regions (∼ 1.5 Mb) are observed in HANK1 and clinical case number 3, and are the most frequently deleted MCRs. The deleted region in HANK1 includes POPDC3, PREP, PRDM1, ATG5, and AIM1, and the deleted region in clinical case number 3 includes LACE1 and FOXO3. (C) Gene-expression profiles were analyzed in 7 cell lines and 11 clinical samples using oligo-microarray. Candidate genes selected by genomic analysis are described. The probe intensity was converted to a numerical value, and the z-score was calculated as described in supplemental Methods. The vertical axis indicates the difference in average value between normal NK cells (n = 3) and neoplastic samples (n = 18). TNFAIP3 (A20), FOXO3, and ATG5 possess 2 probes. A20-1: A_24_P166527, A20-2: A_24_P157926, FOXO3-1: A_32_P102062, FOXO3-2: A_23_P345575, ATG5-1: A_24_P175059, ATG5-2: A_23_P111381. Most of the candidate genes, and in particular FOXO3 and A20, were down-regulated in neoplastic samples compared with normal NK cells, whereas a negligible difference was observed for AIM1.
We then analyzed the expression level of these 7 candidate tumor-suppressor genes located on the most frequently deleted MCR of NK-cell neoplasms. TNFAIP3 (A20) and HACE1 were included as candidate genes. The genomic regions including A20 and HACE1 were deleted in 28% and 31% of the analyzed cases, respectively. A20 has been reported as a tumor-suppressor gene in B-cell lymphoma,31,32 and HACE1 has been reported as a tumor-suppressor gene7,33 and as one of the candidate genes associated with ENKTL.7,33
Selection of candidate genes by a combination of genomic and expression profiles
The expression levels of candidate genes (POPDC3, PREP, PRDM1, ATG5, AIM1, LACE1, FOXO3, A20, and HACE1) within cell lines, clinical samples, and normal NK cells were analyzed by oligo-microarray. All genes had at least one probe on the microarray chip for the gene-expression profile, and a signal was detected for all genes except POPDC3 in normal NK cells. Because the signal intensity of the POPDC3 probe in normal NK cells was very low and we regarded it as “flagged data,” we could not compare the expression level of this gene and excluded it from analysis. Figure 1C shows the expression levels in neoplastic samples compared with those in normal NK cells. A20 and FOXO3 were the top 2 down-regulated genes in neoplastic samples, whereas AIM1 expression did not differ between normal NK cells and neoplastic samples.
Selection of candidate genes by functional analysis
POPDC3 and AIM1 were excluded from the candidate gene list because the former was not expressed in normal NK cells and expression of the latter was similar in normal NK cells and neoplastic samples. The remaining 7 genes were transduced into the NK cell line in an effort to investigate the functional effects.
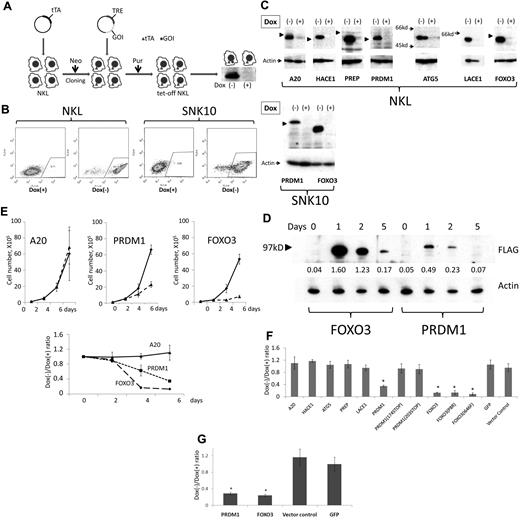
We chose NKL, SNK6, and SNK10 for gene transduction because these lines possess the 6q deletion. Gene transduction was initially attempted using lentivirus31 ; however, functional effect analysis was difficult using this method because the transduction efficiency was < 10% for each cell line used (data not shown). The generation of a Tet-Off cell line using drug selection was then attempted (Figure 2A). Although Tet-Off NKL and SNK10 lines were successfully established, the Tet-Off SNK6 line could not be established because of low transduction efficiency and difficulties associated with single-cell cloning. pRetroX-Tight-Pur-EGFP was transduced into Tet-Off NK cell lines to investigate the induction efficiency. As shown in Figure 2B, high enhanced green fluorescent protein (EGFP) expression was successfully induced in > 95% of total cells. Figure 2E through G shows that each candidate gene was transduced and induced in the Tet-Off NK cell lines. Tet-Off NKL with doxycycline-induced suppression of FOXO3 or PRDM1 expression proliferated logarithmically, whereas proliferation was suppressed with the induction of gene expression (Figure 2E top panel). Conversely, the induction of ATG5, A20, HACE1, PREP or LACE1 had no effect on cell proliferation, despite the large amount of protein expressed (Figure 2E-F). The suppression effect by re-expression of PRDM1 and FOXO3 was confirmed using Tet-Off SNK10 (Figure 2G). Although PRDM1 induction also suppressed Tet-Off NKL proliferation, the suppression efficiency was weaker compared with FOXO3 (Figure 2E-F). We then examined time-course changes in the expression of FOXO3 and PRDM1 at days 0, 1, 2, and 5 after removal of doxycycline. The expression of both genes decreased at day 2 after the initial increase at day 1, and the PRDM1 expression level was lower than that of FOXO3 (Figure 2D). From these findings, FOXO3 and PRDM1 were highly suspected to play more important roles as tumor-suppressor genes than the other candidate genes located on 6q21-23. Therefore, the status of FOXO3 and PRDM1 in NK-cell neoplasms was further analyzed in detail.
Re-expression of candidate genes into 6q-deleted NK cell lines. (A) Schema outlining gene transduction and induction using the Tet-Off system. The Retro-X Tet-Off Advanced inducible expression system (Clontech) was used. The Tet-controlled transactivator (tTA) was initially transduced by retroviral infection, and was followed by neomycin (G418) selection (Neo). Selected cells were cloned by limiting dilution, and the clone with the highest induction efficiency was isolated. cDNA of the gene of interest (GOI) was then inserted into pRetroX-Tight-Pur and transduced into the isolated clone. After puromycin selection (Pur), doxycycline was removed and cells were observed for 6 days. The Western blot shown is representative of the result obtained after incubation of anti-FLAG antibody with the cellular extract from the FLAG-PRDM1–transduced cell line. TRE indicates tetracycline response element. (B) Induction efficiency of the Tet-Off system was analyzed using pRetroX-Tight-Pur-GFP. pRetroX-Tight-Pur-GFP was transduced and selected as described in panel A. More than 95% of the cells showed induced GFP expression. (C) Western blot analysis. Gene expression was induced as described in panel A. A total of 1 × 106 cells was resuspended in 100 μL of 2× sample buffer (125mM Tris, pH 6.8, 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, and 0.02% bromophenol blue) and 10 μL was subjected to PAGE. After transfer of the separated proteins onto the membrane, the membrane was incubated with anti-FLAG antibody for FOXO3, PRDM1, ATG5, HACE1, LACE1, and PREP, and with anti-A20 antibody for A20 (top panel). Anti-actin antibody was then incubated with the membrane after stripping to measure cellular extract quantity (bottom panel). The molecular size marker 97 kDa is indicated by an arrowhead. (D) Time-course changes with FOXO3 and PRDM1 re-expression. Cell samples at days 0, 1, 2, and 5 were analyzed as described in panel C. Each band was converted to a numerical value using ImageJ software, and the ratio to actin was calculated and is shown below each band. (E) Effect of re-expressed PRDM1 or FOXO3 on NKL. A total of 1 × 105 cells was resuspended in medium with or without doxycycline (day 0). On day 2, cells were diluted by up to 10-fold and new medium was added because the cells had reached confluence. The horizontal axis indicates the time elapsed after doxycycline removal. The vertical axis indicates the average and standard deviation of the cell number ratio (Dox−/Dox+). Cell numbers were determined using the trypan blue dye-exclusion assay. Experiments were performed in triplicate. Cell proliferation was strictly suppressed on day 6 when FOXO3 and PRDM1 were re-expressed, unlike A20. (F) Each candidate gene, PRDM1 mutant (PRDM1174STOP and PRDM1203STOP), and FOXO3 mutant (FOXO3P8R and FOXO3I646F) was induced as described in panel D. Medium change and dilution were performed on day 2 and cell counts were determined on day 6. The vertical axis indicates the ratio of cell number with and without doxycycline. Experiments were performed in triplicate and standard deviations and average scores are described. Candidate genes, except in the case of FOXO3 and PRDM1, had no effect on cell proliferation, although these genes were induced and protein expression was confirmed. *Significantly lower than vector control (P < .05). (G) Effect of re-expressed PRDM1 or FOXO3 on SNK10. A total of 1 × 105 cells was resuspended in medium with or without doxycycline (day 0) and analyzed as described in panel D. On day 2, cells were diluted 5-fold. Cell proliferation was suppressed on day 6 when FOXO3 and PRDM1 were re-expressed, whereas the control genes (vector control and GFP) had no effect. *Significantly lower than vector control (P < .05).
Re-expression of candidate genes into 6q-deleted NK cell lines. (A) Schema outlining gene transduction and induction using the Tet-Off system. The Retro-X Tet-Off Advanced inducible expression system (Clontech) was used. The Tet-controlled transactivator (tTA) was initially transduced by retroviral infection, and was followed by neomycin (G418) selection (Neo). Selected cells were cloned by limiting dilution, and the clone with the highest induction efficiency was isolated. cDNA of the gene of interest (GOI) was then inserted into pRetroX-Tight-Pur and transduced into the isolated clone. After puromycin selection (Pur), doxycycline was removed and cells were observed for 6 days. The Western blot shown is representative of the result obtained after incubation of anti-FLAG antibody with the cellular extract from the FLAG-PRDM1–transduced cell line. TRE indicates tetracycline response element. (B) Induction efficiency of the Tet-Off system was analyzed using pRetroX-Tight-Pur-GFP. pRetroX-Tight-Pur-GFP was transduced and selected as described in panel A. More than 95% of the cells showed induced GFP expression. (C) Western blot analysis. Gene expression was induced as described in panel A. A total of 1 × 106 cells was resuspended in 100 μL of 2× sample buffer (125mM Tris, pH 6.8, 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, and 0.02% bromophenol blue) and 10 μL was subjected to PAGE. After transfer of the separated proteins onto the membrane, the membrane was incubated with anti-FLAG antibody for FOXO3, PRDM1, ATG5, HACE1, LACE1, and PREP, and with anti-A20 antibody for A20 (top panel). Anti-actin antibody was then incubated with the membrane after stripping to measure cellular extract quantity (bottom panel). The molecular size marker 97 kDa is indicated by an arrowhead. (D) Time-course changes with FOXO3 and PRDM1 re-expression. Cell samples at days 0, 1, 2, and 5 were analyzed as described in panel C. Each band was converted to a numerical value using ImageJ software, and the ratio to actin was calculated and is shown below each band. (E) Effect of re-expressed PRDM1 or FOXO3 on NKL. A total of 1 × 105 cells was resuspended in medium with or without doxycycline (day 0). On day 2, cells were diluted by up to 10-fold and new medium was added because the cells had reached confluence. The horizontal axis indicates the time elapsed after doxycycline removal. The vertical axis indicates the average and standard deviation of the cell number ratio (Dox−/Dox+). Cell numbers were determined using the trypan blue dye-exclusion assay. Experiments were performed in triplicate. Cell proliferation was strictly suppressed on day 6 when FOXO3 and PRDM1 were re-expressed, unlike A20. (F) Each candidate gene, PRDM1 mutant (PRDM1174STOP and PRDM1203STOP), and FOXO3 mutant (FOXO3P8R and FOXO3I646F) was induced as described in panel D. Medium change and dilution were performed on day 2 and cell counts were determined on day 6. The vertical axis indicates the ratio of cell number with and without doxycycline. Experiments were performed in triplicate and standard deviations and average scores are described. Candidate genes, except in the case of FOXO3 and PRDM1, had no effect on cell proliferation, although these genes were induced and protein expression was confirmed. *Significantly lower than vector control (P < .05). (G) Effect of re-expressed PRDM1 or FOXO3 on SNK10. A total of 1 × 105 cells was resuspended in medium with or without doxycycline (day 0) and analyzed as described in panel D. On day 2, cells were diluted 5-fold. Cell proliferation was suppressed on day 6 when FOXO3 and PRDM1 were re-expressed, whereas the control genes (vector control and GFP) had no effect. *Significantly lower than vector control (P < .05).
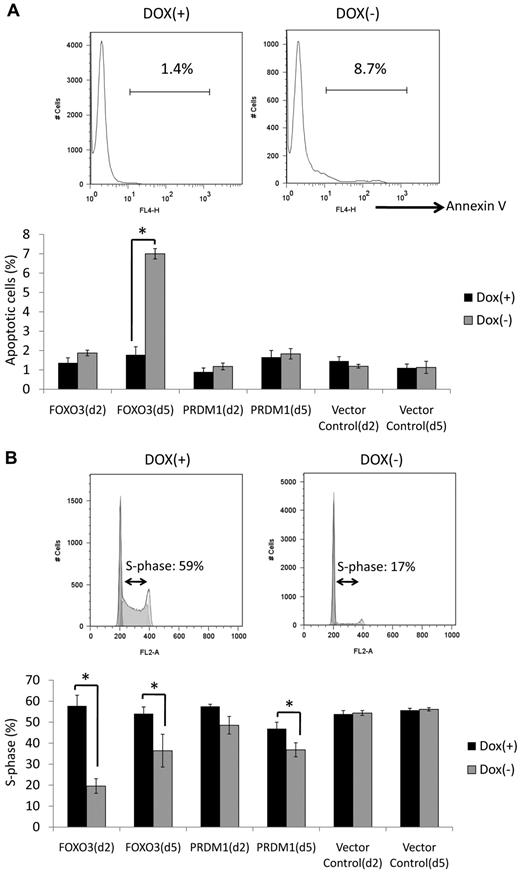
Apoptosis and cell-cycle arrest induced by re-expression of FOXO3 and PRDM1
Apoptosis and cell-cycle assays were performed in an effort to examine the effect of FOXO3 and PRDM1 re-expression (Figure 3A-B). Apoptosis assays using annexin V revealed that FOXO3 re-expression increased the number of apoptotic cells 5 days after removal of doxycycline (Figure 3A). No clear changes were observed in cells re-expressing PRDM1 at either day 2 or 5 after removal of doxycycline (Figure 3A). Cell-cycle assays using PI revealed that the proportion of cells in the S phase decreased in both FOXO3- and PRDM1-induced cells (Figure 3B). Interestingly, the suppression of S-phase cells was more obvious at day 2 than at day 5 in the FOXO3-induced cell line, whereas the suppression of S-phase cells was more obvious at day 5 than at day 2 in the PRDM1-induced cell line.
Apoptosis and cell-cycle assays. (A) Apoptosis assay. Annexin V–positive cells were quantified by flow cytometry at days 2 and 5 after removal of doxycycline. The top panel shows a representative result of FOXO3–re-expressing cells at day 5. More apoptotic cells were observed under conditions with doxycycline treatment than those without doxycycline treatment. The bottom panel indicates the proportion of annexin V–positive cells at days 2 and 5. FOXO3 re-expression increased the amount of apoptotic cells at day 5, whereas no clear changes were observed in PRDM1–re-expressing cells at days 2 or 5. Experiments were performed in triplicate and standard deviations and average scores are shown. Dox indicates doxycycline. *Significantly different Dox(−) and Dox(+) samples (P < .05). (B) Cell-cycle assay assays conducted using PI at days 2 and 5. The top panel shows a representative result of FOXO3 re-expressing cells at day 2. The proportion of S-phase cells was lower under conditions without doxycycline treatment than those with doxycycline treatment. The bottom panel shows the proportion of S-phase cells at days 2 and 5. FOXO3 re-expression decreased the proportion of S-phase cells at days 2 and 5, whereas a decrease in the proportion of S-phase cells only occurred at day 5 with PRDM1 re-expression. Experiments were performed in triplicate and standard deviations and average scores are shown. *Significantly different Dox(−) and Dox(+) samples (P < .05).
Apoptosis and cell-cycle assays. (A) Apoptosis assay. Annexin V–positive cells were quantified by flow cytometry at days 2 and 5 after removal of doxycycline. The top panel shows a representative result of FOXO3–re-expressing cells at day 5. More apoptotic cells were observed under conditions with doxycycline treatment than those without doxycycline treatment. The bottom panel indicates the proportion of annexin V–positive cells at days 2 and 5. FOXO3 re-expression increased the amount of apoptotic cells at day 5, whereas no clear changes were observed in PRDM1–re-expressing cells at days 2 or 5. Experiments were performed in triplicate and standard deviations and average scores are shown. Dox indicates doxycycline. *Significantly different Dox(−) and Dox(+) samples (P < .05). (B) Cell-cycle assay assays conducted using PI at days 2 and 5. The top panel shows a representative result of FOXO3 re-expressing cells at day 2. The proportion of S-phase cells was lower under conditions without doxycycline treatment than those with doxycycline treatment. The bottom panel shows the proportion of S-phase cells at days 2 and 5. FOXO3 re-expression decreased the proportion of S-phase cells at days 2 and 5, whereas a decrease in the proportion of S-phase cells only occurred at day 5 with PRDM1 re-expression. Experiments were performed in triplicate and standard deviations and average scores are shown. *Significantly different Dox(−) and Dox(+) samples (P < .05).
Down-regulated expression and genomic mutation of PRDM1 and FOXO3 genes in NK-cell neoplasms
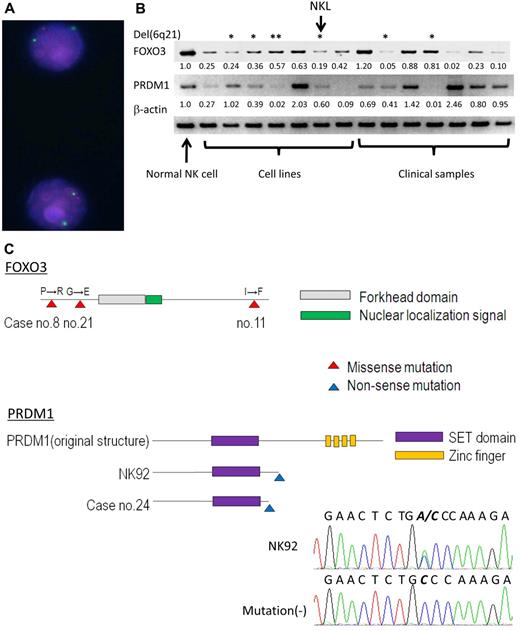
Semiquantitative RT-PCR of PRDM1 and FOXO3 was performed for 7 cell lines and 7 clinical samples to validate the gene-expression profile of oligo-microarray. Thirteen of 14 samples (FOXO3) and 10 of 14 samples (PRDM1) showed lower expression levels compared with normal NK cells (Figure 4B). The average expression levels of both genes in neoplastic samples were significantly lower than those in normal NK cells, reflecting the results from oligo-microarray. Although neoplastic samples with the 6q deletion tended to express both genes at a lower level than samples without the 6q deletion, some cases without the 6q deletion showed extremely low expression levels. Therefore, no significant difference was observed in the expression levels between neoplastic cases with the 6q deletion and those without the 6q deletion (for FOXO3, P = .44 and for PRDM1, P = .07).
Expression and mutation analyses of FOXO3 and PRDM1. (A) FISH analysis of NKL. The red signal indicates the BAC probe 65306RP11-118H13 encompassing FOXO3, and the green signal indicates the centromere probe for chromosome 6 (CEP6, D6Z1; Abbott). The FOXO3 region shows heterogeneous loss. (B) PRDM1 and FOXO3 expression in NK-cell neoplasms (cell line and clinical samples). Semiquantitative RT-PCR was performed to validate the results of oligo-microarray analysis in 7 NK cell lines including NKL (from left; NKYS, SNK6, SNK10, HANK1, NK92, NKL, and KHYG1) and 7 clinical samples (from left; sample nos. 25, 27, 28, 30, 26, 31, and 32). Normal NK cells from one representative donor were also analyzed. Each band was converted to a numerical value using ImageJ software, and the ratio to β-actin was calculated. The ratio of normal NK cells and each neoplastic sample is shown below each band. *Samples with a 6q21 deletion; **HANK1 accompanied the deletion of PRDM1, but not FOXO3. (C) Mutation analysis of FOXO3 and PRDM1. The schema outlines the functional domain and location of the mutation. The mutation is shown as triangle on the map. The electrofluorogram shows that the mutation is located on exon 5 in NK92 and is aligned with the case without mutation. Missense mutations of FOXO3 were found in 3 cases and nonsense mutations of PRDM1 were found in 2 cases.
Expression and mutation analyses of FOXO3 and PRDM1. (A) FISH analysis of NKL. The red signal indicates the BAC probe 65306RP11-118H13 encompassing FOXO3, and the green signal indicates the centromere probe for chromosome 6 (CEP6, D6Z1; Abbott). The FOXO3 region shows heterogeneous loss. (B) PRDM1 and FOXO3 expression in NK-cell neoplasms (cell line and clinical samples). Semiquantitative RT-PCR was performed to validate the results of oligo-microarray analysis in 7 NK cell lines including NKL (from left; NKYS, SNK6, SNK10, HANK1, NK92, NKL, and KHYG1) and 7 clinical samples (from left; sample nos. 25, 27, 28, 30, 26, 31, and 32). Normal NK cells from one representative donor were also analyzed. Each band was converted to a numerical value using ImageJ software, and the ratio to β-actin was calculated. The ratio of normal NK cells and each neoplastic sample is shown below each band. *Samples with a 6q21 deletion; **HANK1 accompanied the deletion of PRDM1, but not FOXO3. (C) Mutation analysis of FOXO3 and PRDM1. The schema outlines the functional domain and location of the mutation. The mutation is shown as triangle on the map. The electrofluorogram shows that the mutation is located on exon 5 in NK92 and is aligned with the case without mutation. Missense mutations of FOXO3 were found in 3 cases and nonsense mutations of PRDM1 were found in 2 cases.
Mutation analyses of FOXO3 and PRDM1 were then performed to detect genomic aberrations (Table 2). Genomic sequences of protein-coding regions and splice junctions were analyzed among the 7 cell lines and the available 33 clinical samples. The insertion and/or deletion of nucleotides was absent for both genes. One-nucleotide substitutions registered as single-nucleotide polymorphisms in the NCBI database (http://www.ncbi.nlm.nih.gov/gene/) were found in 27 cases for PRDM1 and in 4 cases for FOXO3 (data not shown). A synonymous one-nucleotide substitution not registered in the database was found in FOXO3 of HANK1. As shown in Figure 4C, a nonsense mutation of PRDM1 was found in one cell line (NK92) and in one clinical sample (no. 24). Re-expression of truncated PRDM1 mutants derived from case no. 24 (PRDM1174STOP) and NK92 (PRDM1203STOP) failed to show growth suppression, indicating that this is actually a loss-of-function mutation (Figure 2F). Nonsynonymous amino acid changes of FOXO3 were found in 3 clinical samples (nos. 8, 11, and 21), and these nucleotide substitutions were suspected as somatic missense mutations because the nucleotide substitutions were not registered in the single-nucleotide polymorphism database. FOXO3 mutants from case numbers 8 and 11 were successfully amplified using PCR. Re-expression of FOXO3 mutants suppressed cell growth as well as wild-type FOXO3 (Figure 2F). No mutations were detected in the splice junctions. Case 11 with a FOXO3 mutation and case 24 with a PRDM1 mutation possessed the 6q deletion, whereas the other samples did not.
Mutational analysis
| Gene/sample no. . | Mutation . | |||
|---|---|---|---|---|
| Location . | Nucleotide change* . | . | aa change† . | |
| FOXO3 | ||||
| 8 | Exon 1 | C366G | Missense | P8 → R |
| 11 | Exon 2 | A2279T | Missense | I646 → F |
| 21 | Exon 1 | G669A | Missense | G109 → E |
| PRDM1 | ||||
| 24 | Exon 4 | C864A | Nonsense | Y174 → STOP |
| NK92 | Exon 5 | C951A | Nonsense | C203 → STOP |
| Gene/sample no. . | Mutation . | |||
|---|---|---|---|---|
| Location . | Nucleotide change* . | . | aa change† . | |
| FOXO3 | ||||
| 8 | Exon 1 | C366G | Missense | P8 → R |
| 11 | Exon 2 | A2279T | Missense | I646 → F |
| 21 | Exon 1 | G669A | Missense | G109 → E |
| PRDM1 | ||||
| 24 | Exon 4 | C864A | Nonsense | Y174 → STOP |
| NK92 | Exon 5 | C951A | Nonsense | C203 → STOP |
Nucleotide position +1 equivalents to the first nucleotide of GenBank accession number NM_001198.3 (PRDM1) or GenBank accession number NM_001455.3 (FOXO3).
Amino acid location refers to GenBank accession number NP_001189 (PRDM1) and NP_001146.1 (FOXO3).
Discussion
The precise mechanisms associated with the molecular pathogenesis of NK-cell neoplasms remain to be delineated. The present study combined gene-expression analysis and genomic profiles to generate a list of candidate genes involved in NK-cell neoplasms. In an attempt to examine functional relevance, we established NK cell lines susceptible to induced gene expression. This approach of using a combination of different assays focused on fewer candidate genes more likely to be associated with the pathogenesis of NK-cell neoplasms.
The results of the gene-expression profiles reflected the neoplastic characters well. Genes associated with cell proliferation and the cell cycle were up-regulated in neoplastic samples compared with normal NK cells. Conversely, the expression levels of genes and gene sets associated with tumor microenvironments differed between clinical samples and cell lines. Huang et al analyzed gene-expression profiles of ENKTL and reported that gene sets associated with signal transduction, for example, the JAK/STAT and MAPK pathways, were up-regulated in neoplastic samples.34 The expression levels of these signal transduction-associated gene sets did not differ between neoplastic samples and normal NK cells in the present study. This may have been because our analyses were based on the MSigDB database (http://www.broadinstitute.org/gsea/msigdb/index.jsp) or Gene Ontology (http://bioinfo.vanderbilt.edu/webgestalt/analysis.php?gofile_organism = hsapiens). For example, the gene set of the JAK/STAT pathway in MSigDB (referred to as KEGG_JAK_STAT_SIGNALING_PATHWAY) contains no fewer than 155 genes, including genes up-regulated by JAK-STAT pathway activation in addition to those being down-regulated. If we had selected the same genes associated with the JAK/STAT pathway as used by Huang et al, this gene set would have been more highly expressed in neoplastic samples compared with normal NK cells (NES, −1.44; nom-P value, .095; data not shown).
We previously reported BAC-array CGH analysis of NK-cell neoplasms. In the present study, we performed oligo-array CGH analysis in an effort to identify more detailed and narrower genomic aberrations. Gain of 1q and 7q and loss of 6q and 7p were relatively frequent genomic aberrations. These abnormalities were consistent with previous studies.4,7-9,34 In particular, narrow regions of 6q21 that include POPDC3, PREP, PRDM1, ATG5, AIM1, LACE1, and FOXO3 were the most frequently deleted region of the whole genome except for the X and Y chromosomes. Iqbal et al also performed array CGH analysis of NK-cell neoplasms and reported PRDM1, ATG5, and AIM1 as strong candidate genes.8 We included TNFAIP3 (A20) and HACE1 in the candidate gene list because they have been described as tumor-suppressor genes in many previous studies.31-33,35,36 Although these were not located on the most frequently deleted MCR, 29% and 31% of cases showed deletion of these genes. The expression levels of POPDC3 and AIM1 did not differ between neoplastic samples and normal NK cells. Huang et al reported that AIM1 expression was down-regulated in ENKTL,7 although this down-regulated expression was not detected in other studies8 performed by Iqbal et al and by our group. In the present study, POPDC3 and AIM1 were excluded from our functional analysis. Further studies will be needed to confirm the expression level and detailed mechanism of AIM1. Forced re-expression of the other 7 genes was successfully achieved, and only FOXO3 and PRDM1 induction resulted in the suppression of cell proliferation.
A20 functions to suppress NF-κB signaling by targeting RIP1 and TRAF6.37,38 Frequent genomic aberrations of A20, comprising homogeneous deletions and biallelic inactivation with genomic mutation, have been described in B-cell lymphoma, including diffuse large B-cell lymphoma and MALT lymphoma.31,32,35,39 Kato et al32 and our group31 confirmed that A20 re-expression in the B-cell lymphoma cell line and the Hodgkin lymphoma cell line suppressed cell proliferation. However, this effect was not observed in the NK cell line even though NKL possessed a 6q deletion encompassing the A20 gene. In contrast to B-cell lymphoma, A20 was not located in the most frequently deleted MCR in NK-cell neoplasms and homogeneous deletion was not observed. This result indicates that A20 may not play as important a role in the pathogenesis of NK-cell neoplasms as it does in B-cell neoplasms. However, previous studies have shown NF-κB activation in NK-cell neoplasms as well as B-cell neoplasms.7,34,40,41 EBV, which is frequently detected in NK-cell neoplasms, was reported to induce NF-κB activation through LMP1.42 The action of the EB-virus may replace A20 inactivation in NF-κB activation of NK-cell neoplasms.
HACE1 is a gene encoding E3 ubiquitin ligase and was reported to be inactivated by DNA methylation in gastrointestinal tract cancer and Wilms tumor.33,36,43 Spontaneous multiorgan tumors frequently occur in HACE1−/− mice.33 From these findings, HACE1 is regarded as one of the tumor-suppressor genes in certain kinds of cancers as well as in NK-cell neoplasms.7,33,34,36,44 Huang et al described HACE1 as a candidate genes not only because it was located within the frequently deleted region, but also because its expression was down-regulated in tumor samples.7 In the present study, HACE1 expression in neoplastic samples was also down-regulated compared with normal NK cells (data not shown). However, an obvious tumor-suppressing effect was not observed in our functional analysis.
FOXO3 is a member of the forkhead family of proteins that possess the forkhead box domain, a DNA-binding domain that is highly conserved among various biologic species.45 Breast carcinoma cell proliferation was suppressed by re-expression of FOXO3.46 Spontaneous pituitary gland tumors occur in FOXO3−/− mice, and a progressive cancer prone condition was observed in broad FOXO (FOXO1, FOXO3, and FOXO4)–deleted mice.47 In hematologic malignancies, translocation involving the FOXO3 and MLL genes was detected in acute leukemia, indicating an association with pathogenesis.48 From these findings, FOXO3 is now recognized as a tumor-suppressor gene. However, there have been no studies describing the role of FOXO3 in malignant lymphoma, especially in the case of NK-cell neoplasms. In the present study, FOXO3 expression was down-regulated in most NK-cell neoplasms and re-expression of FOXO3 suppressed proliferation of the NK cell line. These findings indicate that FOXO3 plays a role as a tumor-suppressor gene in NK-cell neoplasms. It was shown that the inhibition of cell growth by FOXO3 re-expression was due to the induction of apoptosis and cell-cycle arrest (Figure 3A-B). These findings are consistent with previous studies.46,47 All cell lines possessing 6q deletions showed heterozygous loss, and mRNA expression was detected even though the expression was down-regulated. Furthermore, only one case showed a genomic mutation of FOXO3 with accompanied genomic loss. These findings tend to favor haploinsufficiency in lieu of biallelic inactivation as being the mechanism responsible for FOXO3 inactivation in NK-cell neoplasms.
PRDM1/BLIMP1 is a transcriptional repressor comprising Kruppel-type zinc fingers that combine DNA with the PR domain, and is a master regulator of B-cell differentiation.49 This gene product is considered to be a tumor-suppressor gene because approximately 25% of activated B-cell type diffuse large B-cell lymphoma cases showed genomic mutations that induced functional inactivation.28,50 Most of these cases with inactive mutations were accompanied by heterogeneous 6q deletions and DNA methylation, indicating that biallelic inactivation occurs frequently.28,50 Iqbal et al8 reported that PRDM1 expression was down-regulated in NK-cell neoplasms, whereas Huang et al7 reported no difference in expression levels between ENKTL and normal NK cells. In the present study, PRDM1 expression was down-regulated in NK-cell neoplasms compared with normal NK cells. However, the expression levels varied, with some neoplastic cases showing extremely up-regulated expression compared with normal NK cells (Figure 4B). This variation in expression level may account for previous studies showing the opposite effect. In the present study, the inhibition of cell growth by PRDM1 re-expression was less effective compared with FOXO3 re-expression (Figure 2E). The expression level of PRDM1 was less than that of FOXO3, even though the same Tet-Off system vector was used for the expression of both genes (Figure 2D). Differences in the expression level of each gene may have contributed to differences in the suppression effect observed. Genomic mutations in 2 cases represented nonsense mutations that resulted in inactive mutations (Figure 4C). One case was accompanied by a genomic deletion and biallelic inactivation was considered in this case. Although this indicates that PRDM1 may play a role as a classic tumor-suppressor gene based on the 2-hit theory, future studies using more cases are required to clarify this possibility.51
In the present study, we succeeded in identifying a small group of genes as candidate tumor-suppressor genes by combining array CGH, gene-expression profile, and functional analyses. The transduction of NK cell lines is generally difficult, even when using viral infection methods. This is the first study to establish a stable, inducible gene-expression system in the NK cell line, and the assay system developed should prove useful in delineating the molecular pathogenesis of NK-cell neoplasms. Interestingly, the well-known onco-suppressor genes A20 and HACE1 had no effect on NK-cell proliferation, whereas the suppression of proliferation was clearly demonstrated with PRDM1 and FOXO3 re-expression. Therefore, the down-regulation of these 2 genes is considered to play an important role in the pathogenesis of NK-cell neoplasms.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Hideki Murakami, Sivasundaram Karnan, Ken-ichi Suda, Tomoko Miyata, Harumi Kato, Akira Umino, Kiyoko Yamamoto, Dai Chihara, Katsuyoshi Takata, and Eisaku Kondo for discussions and encouragement throughout this study. The outstanding technical assistance of Kyoko Hirano, Seiko Sato, and Yumiko Kasugai is very much appreciated. Drs Ayako Demachi-Okamura and Yoshitoyo Kagami are acknowledged for their advice on NK-cell cultures.
This work was supported by a grant-in-aid from the Ministry of Health, Labor and Welfare of Japan; by the Ministry of Education, Culture, Sports, Science and Technology of Japan; by the Japan Society for the Promotion of Science (20890298, 22790368, and 20390277); and by a grant-in-aid for cancer research from the Ministry of Health, Labor and Welfare of Japan (21-6-3).
Authorship
Contribution: K.K. performed experiments, collected and analyzed the data, and wrote the manuscript; M.N., S.T., I.T., K.H., and Y.N. performed the experiments and analyzed the data; N.S. established and provided the cell lines; Y-H. K., Y.M., K.O., and S.N. collected the clinical samples and analyzed the data; and M.S. organized the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Masao Seto, MD, Division of Molecular Medicine, Aichi Cancer Center Research Institute, Chikusa-ku, Kanokoden 1-1, Nagoya 464-8681, Japan; e-mail: mseto@aichi-cc.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal