Abstract
Reduced gene dosage of ribosomal protein subunits has been implicated in 5q− myelodysplastic syndrome and Diamond Blackfan anemia, but the cellular and pathophysiologic defects associated with these conditions are enigmatic. Using conditional inactivation of the ribosomal protein S6 gene in laboratory mice, we found that reduced ribosomal protein gene dosage recapitulates cardinal features of the 5q− syndrome, including macrocytic anemia, erythroid hypoplasia, and megakaryocytic dysplasia with thrombocytosis, and that p53 plays a critical role in manifestation of these phenotypes. The blood cell abnormalities are accompanied by a reduction in the number of HSCs, a specific defect in late erythrocyte development, and suggest a disease-specific ontogenetic pathway for megakaryocyte development. Further studies of highly purified HSCs from healthy patients and from those with myelodysplastic syndrome link reduced expression of ribosomal protein genes to decreased RBC maturation and suggest an underlying and common pathophysiologic pathway for additional subtypes of myelodysplastic syndrome.
Introduction
Myelodysplastic syndrome (MDS) is a heterogeneous group of blood cell disorders characterized by defective hematopoiesis and increased susceptibility to leukemia; it is thought to involve abnormalities in HSCs. Approximately 50% of affected patients have blood cell cytogenetic abnormalities, of which deletions of chromosome 5q are the most common and portend a favorable prognosis.1 Identification of causal genes in 5q− and other MDS subtypes has been challenging, but recent advances in genetics and genomics have enhanced our understanding of how specific chromosomal alterations and their molecular consequences contribute to the pathogenesis of MDS.2,3
On the basis of a large-scale RNAi screen, Ebert et al identified RPS14 as a critical gene on 5q whose hemizygosity in BM cells recapitulates many of the features in 5q− MDS.2 Intriguingly, erythroid abnormalities in 5q− MDS are similar to those in Diamond Blackfan anemia (DBA), a dominantly inherited disorder in which germline mutations in one allele of either a 40S (encoded by RPS genes) or a 60S (encoded by the RPL genes) ribosomal protein gene have been identified.4,5 However, there are also important differences between the blood cell phenotypes of the 2 conditions; thrombocytosis and megakaryocytic dysplasia are cardinal features of 5q− MDS but not of DBA. Furthermore, recent work from Starczynowski et al suggests that 2 microRNAs are critical mediators of the 5q− phenotype because knockdown of these genes in immature hematopoietic cells leads to megakaryocytic dysplasia and thrombocytosis after transplantation in mice.6 Increased understanding of how ribosomal protein mutations cause disease might provide additional insight into the pathogenesis of MDS and DBA.
Spontaneous and induced ribosomal protein mutations have been studied in many model organisms, including yeast, flies, plants, fish, and mice.7-13 Studies designed on the basis of on these models indicate that reduced dosage of ribosomal protein genes compromises ribosome biogenesis, protein synthesis, cell proliferation, and cell survival.11,12,14 Although these processes are likely to account for impaired organismal and cellular growth, the mechanisms responsible for tissue-specific phenotypes, including anemia and thrombocytosis, remain unresolved.
Additional insight into the pathogenesis of ribosomal protein-mediated BM failure has come from recent work in our laboratory: mice with ribosomal protein mutations develop epidermal melanocytosis and dark skin in which activation of the transcription factor p53 is a critical event.15,16 Here, we ask whether these observations can be applied to understand the pathogenesis of MDS on the basis of conditional and BM-specific ablation of a ribosomal protein subunit, Rps6.12 We found that activation of p53 is necessary to elicit a BM failure phenotype in mice whose features recapitulate essential aspects of 5q− MDS and that suggests specific roles for p53 activation in both HSC dysfunction and defects in red cell maturation. In addition, we found that ribosomal protein dysregulation occurs in non-5q− MDS in human patients, suggesting a shared pathogenic mechanism among different MDS subtypes.
Methods
Generation of ribosomal protein mutant mice and animal experiments
We obtained mice carrying Tg.MxCre from The Jackson Laboratory, Rps6lox from S. Volarevic and G. Thomas (University of Cincinnati),12 Tg.K5Cre from S. Artandi (Standard Unviersity), and J. Jorcano (Epithelial Biomedicine Division, CIEMAT),17 and Trp53ko from T. Jacks (Massachusetts Institute of Technology).18 Rps19Dsk3/+ (C3HeB/FeJ), Tg.K5Cre (C3HeB/FeJ),19 Trp53ko (129/SvJ), and Tg.MxCre (C57BL/6J) were maintained on an isogenic background. Mdm2puro/Δ7-9 and +/+ controls were (129/SvJ × C57BL/6J)F1 littermates.
Rps6lox/+;Tg.MxCre/+ were generated by crossing Rps6lox/+ (129/SvJ × C57BL/6J F1) with Tg.MxCre/+. Then, 7- to 9-week-old animals were given 250 μg of polyinosinic:polycytidylic acid (polyI:C) intraperitoneally (Invivogen) on days 1, 3, and 5. All experiments in which we used Rps6lox/+;Tg.MxCre/+ animals were performed at least 16 weeks after polyI:C treatment.
Erythrocyte adenosine deaminase (eADA) was evaluated as previously described.20 All experiments were performed under a protocol approved by the Stanford Administrative Panel on Laboratory Animal Care.
Immunofluorescence, histology, and cytology
Adult skin or femurs were fixed in 4% paraformaldehyde. Femurs were decalcified in 500mM EDTA, pH7.2. Immunofluorescence was carried out with p53 (Novocastra) or von Willebrand factor (VWF; Dako) antisera after antigen retrieval with 0.01M citrate buffer, pH 6, in a pressure cooker. Skin sections were incubated with goat antirabbit Cy3 antisera (Jackson ImmunoResearch Laboratories) and ProLong antifade reagent with DAPI (Invitrogen). Femur sections were incubated with goat antirabbit biotinylated antisera (Jackson ImmunoResearch Laboratories), Vectastain Elite Avidin-Biotin complex reagent (Vector Labs), and Tyramide-Cy3 amplification reagent (PerkinElmer). Ter119 antisera (BD Pharmingen), mouse antirat biotinylated antisera (BD Pharmingen), avidin-biotin complex reagent, and Tyramide-Fluorescein amplification reagent (PerkinElmer) were used for double immunofluorescence staining on femur sections after a second treatment with 0.01M citrate buffer, pH 6, in a pressure cooker. Histologic sections were stained with hematoxylin and eosin. BM cytospins, and peripheral blood smears were stained with Wright Giemsa reagents.
Immunophenotyping, purification of, and assay for Rsp6 protein from mouse BM
Total BM cells were c-kit enriched with the use of magnetic beads (Miltenyi Biotec) and stained with fluorochrome-conjugated antibodies (supplemental Table 3, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and propidium iodide (Molecular Probes). Cell populations were fractionated on the basis of Pronk et al21 or Socolovsky et al22 (supplemental Table 4). Quantitative measurements presented in Figure 5A, C, and D are based on 1 million cells from each of 2-3 animals per genotype, with values from Rps6lox/+;Tg.MxCre/+ animals divided by a factor of 2.13 (or 1.18 for Rps6lox/+;Tg.MxCre/+;Trp53ko/ko) to account for the reduction in BM cellularity (Table 2); comparisons between genotypes therefore correspond to absolute differences on a per-animal basis. Cells were sorted or immunophenotyped with FACS Aria II cell sorters (Becton Dickinson), and data were analyzed with FlowJo Version 7 software (TreeStar).
Rps6 protein levels were evaluated from BM cells after staining for surface antigens, fixation with 1.6% paraformaldehyde, permeabilization with cold methanol, and staining with fluorescently conjugated mouse Rps6 antisera (Cell Signaling Technology). Mean fluorescence intensity for a population of cells is a surrogate for Rps6 protein levels.
Results
Rps6 hemizygosity causes robust p53 activation in skin and BM
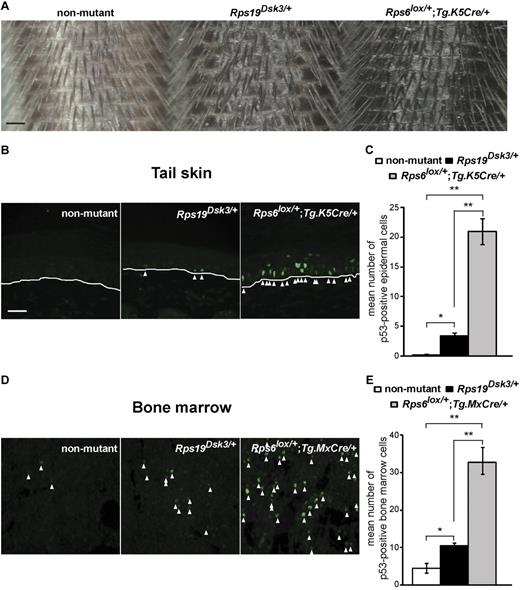
We previously reported mutations of Rps19 and Rps2016 as the cause of 2 dominantly inherited dark skin mutants, Dark skin 3 (Dsk3) and Dark skin 4 (Dsk4), respectively, that were identified during the course of a forward genetic screen for new pigmentary abnormalities.15,19 These animals exhibit increased accumulation of epidermal melanocytes in the footpads, ears, and tail that becomes apparent shortly after weaning and persists throughout adult life. We observed a similar phenotype in animals with keratinocyte-specific hemizygosity for a third ribosomal protein, Rps6 (Rps6lox/+;Tg.K5Cre/+).16 In additional studies, we showed that activation of p53 is both necessary and sufficient for the pigmentation phenotype. Thus, mutations in 3 different components of the 40S ribosome trigger a common pathophysiologic pathway that causes accumulation of p53, epidermal melanocytosis and dark skin.
Rps19Dsk3/+ animals develop a mild blood cell phenotype (a 5%-10% reduction in RBC counts), and Rps20Dsk4/+ animals have no blood cell abnormalities.16 However, we also noticed that Rps19Dsk3/+ animals exhibited skin color darkening that was intermediate between nonmutant and Rps6lox/+;Tg.K5Cre/+ animals (Figure 1A), which suggested that the level of skin darkening might serve as a proxy for the extent of p53 activation. We investigated this observation more carefully using tail skin sections processed and quantified in parallel. Immunofluorescence for p53 is undetectable in the skin of nonmutant animals, whereas Rps19Dsk3/+ and Rps6lox/+;Tg.K5Cre/+ animals have a 15-fold and 94-fold increase in staining, respectively, compared with nonmutant animals (Figure 1B-C). Thus, hemizygosity for Rps6 has a stronger effect on p53 activation than the Rps19Dsk3 mutation.
Effects of different ribosomal protein mutations on skin darkness and p53 levels. (A) Tails from nonmutant, Rps19Dsk3/+, and Rps6lox/+;Tg.K5Cre/+ animals. (B,D) Immunofluorescence for p53 in tail (B) and BM (D) sections from the ribosomal protein mutants. White lines (B) mark the dermal-epidermal junction; arrowheads mark p53-positive nuclei. (C,E) Quantitation of p53 immunostaining in the skin (C) and BM (E) of animals of the indicated genotype (n = 3-4 mice per group). Values are number of cells per high-power field (hpf) ± SEM (6 hpf per animal). P values are based on a 2-tailed t test, *P < .05, **P < .01; NS indicates not significant. Scale bars: 0.5 mm (A); 50μM (B,D).
Effects of different ribosomal protein mutations on skin darkness and p53 levels. (A) Tails from nonmutant, Rps19Dsk3/+, and Rps6lox/+;Tg.K5Cre/+ animals. (B,D) Immunofluorescence for p53 in tail (B) and BM (D) sections from the ribosomal protein mutants. White lines (B) mark the dermal-epidermal junction; arrowheads mark p53-positive nuclei. (C,E) Quantitation of p53 immunostaining in the skin (C) and BM (E) of animals of the indicated genotype (n = 3-4 mice per group). Values are number of cells per high-power field (hpf) ± SEM (6 hpf per animal). P values are based on a 2-tailed t test, *P < .05, **P < .01; NS indicates not significant. Scale bars: 0.5 mm (A); 50μM (B,D).
To investigate how the aforementioned difference would manifest itself in the BM, we crossed Rps6lox/+ mice to animals carrying an interferon-inducible Cre driver that has been used previously to modify genes in the adult BM, Tg.MxCre.23 At 7-9 weeks of age, nonmutant (+/+, Rps6lox/+ or Tg.MxCre/+) and mutant (Rps6lox/+;Tg.MxCre/+) animals were injected with polyI:C to activate the Mx promoter and were evaluated 16-75 weeks later.
The effect of Cre-mediated recombination on Rps6 mRNA levels was assessed by the use of quantitative RT-PCR on peripheral blood and sorted BM cells from nonmutant and Rps6lox/+;Tg.MxCre/+ animals. Overall, and in all cell populations examined (peripheral blood, CD71+ erythrocytes [supplemental Table 4], and megakaryocyte-erythrocyte progenitors), Rps6 expression was significantly reduced (P = .001, .042, and .006, respectively) in Rps6lox/+;Tg.MxCre/+ animals compared with nonmutant controls (supplemental Figure 1A-E). The levels of reduction varied, presumably because of differences in Cre-mediated excision (supplemental Figure 1A-E); yet, the overall decrement in Rps6 expression persisted over time (supplemental Figure 1D-E). Notably, the degree of Rps6 mRNA reduction correlated with alterations in peripheral blood cell counts (supplemental Figure 1C,E). We also measured Rps6 protein expression in mutant and nonmutant BM cells using a FACS-based approach and observed an ∼ 35% reduction (supplemental Figure 1F-G).
To assess the effects of ribosomal protein mutations on p53 in the BM, we developed a protocol for immunohistochemistry of femur sections. Although different transgenic strategies were used for Rps6 in the skin (Tg.K5Cre) and BM (Tg.MxCre followed by polyI:C injection), the level of p53 immunostaining in the BM still correlated with the extent of skin darkening (Figure 1B-E), consistent with previous reports that hemizygosity for Rps6 is generally more severe than hemizygosity for Rps19.10,12
Hematologic characterization of Rps6 mutants
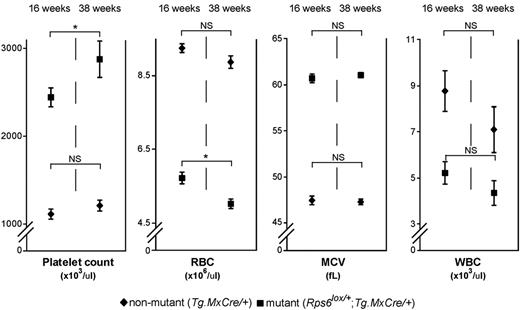
We first examined peripheral blood and BM from nonmutant (+/+, Rps6lox/+, and Tg.MxCre/+), and Rps6lox/+;Tg.MxCre/+ mutant animals. The MxCre transgene by itself had no effect on blood cell counts (supplemental Table 2); however, Rps6lox/+;Tg.MxCre/+ animals exhibited a robust macrocytic anemia that worsened over time (Figure 2, Table 1, supplemental Figure 2 and Table 2). At 16 weeks after induction of Cre recombinase, mutant animals had significant alterations in RBC count (36% reduction, P < .001) and mean corpuscular volume (25% increase, P < .001), and a modest reduction in reticulocyte count (supplemental Table 2). Mutant mice also exhibited marked leukopenia (and associated lymphopenia) and a progressive thrombocytosis (Figure 2, Table 1, supplemental Figure 2 and supplemental Table 2). In addition, a spectrophotometric assay for eADA function in peripheral blood (a biomarker for disease activity in DBA patients)20 demonstrated a 2-fold increase in enzyme activity in mutant compared with nonmutant control animals (Table 1).
Effects of BM-specific Rps6 hemizygosity on peripheral blood cell counts. Adult (7- to 9-week-old) Tg.MxCre/+ (n = 12) and Rps6lox/+;Tg.MxCre/+ (n = 16) animals were treated with polyI:C and blood cell counts (± SEM) determined 16 weeks and 38 weeks later. P values refer to a 2-tailed paired t test comparing the 16- and 38-week time points within a genotype, *P < .05; NS indicates not significant; and WBC, white blood cell count.
Effects of BM-specific Rps6 hemizygosity on peripheral blood cell counts. Adult (7- to 9-week-old) Tg.MxCre/+ (n = 12) and Rps6lox/+;Tg.MxCre/+ (n = 16) animals were treated with polyI:C and blood cell counts (± SEM) determined 16 weeks and 38 weeks later. P values refer to a 2-tailed paired t test comparing the 16- and 38-week time points within a genotype, *P < .05; NS indicates not significant; and WBC, white blood cell count.
Effect of Rps6 hemizygosity and Trp53 on blood counts and eADA activity*
| . | Nonmutant† . | Rps6lox/+; Tg.MxCre/+ . | Rps6lox/+; Tg.MxCre/+; Trp53ko/+ . | Rps6lox/+; Tg.MxCre/+; Trp53ko/ko . |
|---|---|---|---|---|
| No. of animals | 17 | 20 | 12 | 8 |
| WBC, × 103/μL | 7.58 ± 0.47 | 4.47 ± 0.35a‡ | 4.12 ± 0.53 | 6.39 ± 0.75b‖, NS¶ |
| RBC, × 106/μL | 9.72 ± 0.13 | 5.85 ± 0.19a‡ | 7.03 ± 0.11a§ | 9.12 ± 0.83a‖, NS¶ |
| MCV, fL | 47.0 ± 0.4 | 59.8 ± 0.7a‡ | 55.7 ± 0.7a§ | 48.3 ± 2.2a‖, NS¶ |
| Platelet count, × 103/μL | 930 ± 17 | 1490 ± 89a‡ | 1181 ± 65b§ | 911 ± 89b‖, NS¶ |
| No. of animals | 6 | 6 | 4 | 5 |
| eADA, IU/g hemoglobin | 1.06 ± 0.11 | 2.54 ± 0.45c‡ | 2.09 ± 0.15 | 1.13 ± 0.19c‖, NS¶ |
| . | Nonmutant† . | Rps6lox/+; Tg.MxCre/+ . | Rps6lox/+; Tg.MxCre/+; Trp53ko/+ . | Rps6lox/+; Tg.MxCre/+; Trp53ko/ko . |
|---|---|---|---|---|
| No. of animals | 17 | 20 | 12 | 8 |
| WBC, × 103/μL | 7.58 ± 0.47 | 4.47 ± 0.35a‡ | 4.12 ± 0.53 | 6.39 ± 0.75b‖, NS¶ |
| RBC, × 106/μL | 9.72 ± 0.13 | 5.85 ± 0.19a‡ | 7.03 ± 0.11a§ | 9.12 ± 0.83a‖, NS¶ |
| MCV, fL | 47.0 ± 0.4 | 59.8 ± 0.7a‡ | 55.7 ± 0.7a§ | 48.3 ± 2.2a‖, NS¶ |
| Platelet count, × 103/μL | 930 ± 17 | 1490 ± 89a‡ | 1181 ± 65b§ | 911 ± 89b‖, NS¶ |
| No. of animals | 6 | 6 | 4 | 5 |
| eADA, IU/g hemoglobin | 1.06 ± 0.11 | 2.54 ± 0.45c‡ | 2.09 ± 0.15 | 1.13 ± 0.19c‖, NS¶ |
eADA indicates erythrocyte adenosine deaminase activity; MCV, mean corpuscular volume; NS, not significant; polyI:C, polyinosinic:polycytidylic acid; RBC, red blood cell count; and WBC, white blood cell count.
Blood counts were evaluated 16 weeks after treatment with polyI:C compound. Blood counts given as mean ± SEM with P values based on multiple regression where sex and genotype are factors. A subset of animals was evaluated for eADA activity and values are mean ± SEM with P values based on a 2-tailed Student t test, aP < .001, bP < .01, cP < .05.
+/+ or Rps6lox/+.
Nonmutant vs Rps6lox/+;Tg.MxCre/+.
Rps6lox/+;Tg.MxCre/+ vs Rps6lox/+;Tg.MxCre/+;Trp53ko/+.
Rps6lox/+;Tg.MxCre/+ vs Rps6lox/+;Tg.MxCre/+;Trp53ko/ko.
Nonmutant vs Rps6lox/+; Tg.MxCre/+;Trp53ko/ko.
We next asked whether Trp53 was required for the hematologic changes caused by BM-specific reduction of Rps6 gene dosage. Rps6lox/+;Tg.MxCre/+ animals were crossed with those carrying a Trp53 knockout allele, Trp53ko,18 and assessed for peripheral blood cell counts and eADA levels. In Rps6lox/+;Tg.MxCre/+ animals, deficiency for p53 (Trp53ko/ko) completely mitigated the alterations in white blood cell count, RBC count, mean corpuscular volume, platelet count, and eADA activity (Table 1). Heterozygosity for Trp53 yielded an intermediate phenotype with partial rescue of the blood cell counts and parameters.
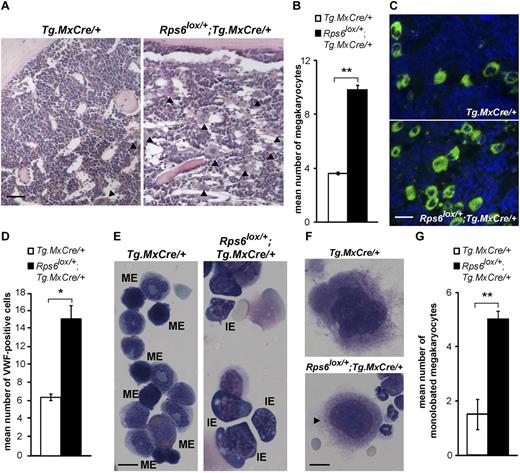
To investigate the origins of the blood cell abnormalities, we examined BM sections stained with hematoxylin and eosin and observed a prominent megakaryocytosis (a 2.7-fold increase in megakaryocyte number) and marked hypocellularity in Rps6lox/+;Tg.MxCre/+ compared with control animals (Figure 3A-B). Immunofluorescence for VWF, an immunohistochemical marker for megakaryocytes in normal BM and MDS,24 revealed a 2.3-fold increase in megakaryocytes in ribosomal protein mutant marrow (Figure 3C-D). To evaluate the degree of hypocellularity and to assess the role of p53 in this process, we flushed marrow from the femurs and tibias of nonmutant, Rps6lox/+;Tg.MxCre/+ and Rps6lox/+;Tg.MxCre/+;Trp53ko/ko animals and counted the number of cells. Mutant animals (Rps6lox/+;Tg.MxCre/+) exhibited a 50% reduction in cellularity that was abrogated completely by loss of p53 (Rps6lox/+;Tg.MxCre/+;Trp53ko/ko; Table 2).
Effects of BM-specific Rps6 hemizygosity on BM histology and cytology. (A) Representative histologic sections from animals of the indicated genotype; arrowheads indicate megakaryocytes, quantitated in panel B (n = 3 animals per genotype, mean number of megakaryocyte per hpf is shown ± SEM, 8 hpf per animal). (C) Immunofluorescence staining for VWF (green) in BM sections counterstained with DAPI (blue) from nonmutant and ribosomal protein mutant animals and quantitated in panel D (n = 3 animals per genotype, mean number of VWF-positive cells with large nuclei per hpf is shown ± SEM, 10 hpf per animal). (E) Cytospin preparations show increased numbers of immature erythroid (IE) cells and decreased numbers of mature erythroid (ME) cells in mutant compared with control samples. (F) Example of normal and abnormal (a monolobated micromegakaryocyte, arrowhead) megakaryocyte morphology in cytospin preparations from nonmutant and mutant BM, quantitated (G) as mean number of monolobated cells among 10 megakaryocytes evaluated per animal ± SEM (n = 4-5 animals per genotype). P values are based on a 2-tailed t test, *P < .05, **P < .01; NS indicates not significant. Scale bars: 50μM (A); 75μM (C); 20μM (E); 10μM (F).
Effects of BM-specific Rps6 hemizygosity on BM histology and cytology. (A) Representative histologic sections from animals of the indicated genotype; arrowheads indicate megakaryocytes, quantitated in panel B (n = 3 animals per genotype, mean number of megakaryocyte per hpf is shown ± SEM, 8 hpf per animal). (C) Immunofluorescence staining for VWF (green) in BM sections counterstained with DAPI (blue) from nonmutant and ribosomal protein mutant animals and quantitated in panel D (n = 3 animals per genotype, mean number of VWF-positive cells with large nuclei per hpf is shown ± SEM, 10 hpf per animal). (E) Cytospin preparations show increased numbers of immature erythroid (IE) cells and decreased numbers of mature erythroid (ME) cells in mutant compared with control samples. (F) Example of normal and abnormal (a monolobated micromegakaryocyte, arrowhead) megakaryocyte morphology in cytospin preparations from nonmutant and mutant BM, quantitated (G) as mean number of monolobated cells among 10 megakaryocytes evaluated per animal ± SEM (n = 4-5 animals per genotype). P values are based on a 2-tailed t test, *P < .05, **P < .01; NS indicates not significant. Scale bars: 50μM (A); 75μM (C); 20μM (E); 10μM (F).
Effect of Rps6 hemizygosity and Trp53 mutations on BM cell counts*
| . | Tg.MxCre/+ . | Rps6lox/+; Tg.MxCre/+ . | Rps6lox/+;Tg.MxCre/+; Trp53ko/ko . |
|---|---|---|---|
| Immature erythroid* | 13.50 ± 1.66 | 40.00 ± 3.24a† | 13.67 ± 3.71a‡, NS§ |
| Mature erythroid* | 37.5 ± 1.26 | 21.0 ± 3.72b† | 39.0 ± 4.16b‡, NS§ |
| Lymphocyte | 70.75 ± 6.02 | 29.75 ± 5.02a† | 52.00 ± 1.73a‡b, § |
| Myeloblast | 2.5 ± 0.65 | 3.25 ± 0.48 | 4.67 ± 1.67 |
| Immature myeloid* | 19.75 ± 0.75 | 21.00 ± 1.96 | 19.67 ± 3.53 |
| Mature myeloid* | 93.75 ± 1.89 | 121.75 ± 7.22 | 108.00 ± 6.93 |
| Monocyte/macrophage | 20 ± 3.87 | 22.25 ± 3.25 | 20.33 ± 3.53 |
| Cellularity (× 107)‖ | 7.59 ± 0.42 | 3.56 ± 0.28c† | 6.39 ± 0.61b‡, NS§ |
| . | Tg.MxCre/+ . | Rps6lox/+; Tg.MxCre/+ . | Rps6lox/+;Tg.MxCre/+; Trp53ko/ko . |
|---|---|---|---|
| Immature erythroid* | 13.50 ± 1.66 | 40.00 ± 3.24a† | 13.67 ± 3.71a‡, NS§ |
| Mature erythroid* | 37.5 ± 1.26 | 21.0 ± 3.72b† | 39.0 ± 4.16b‡, NS§ |
| Lymphocyte | 70.75 ± 6.02 | 29.75 ± 5.02a† | 52.00 ± 1.73a‡b, § |
| Myeloblast | 2.5 ± 0.65 | 3.25 ± 0.48 | 4.67 ± 1.67 |
| Immature myeloid* | 19.75 ± 0.75 | 21.00 ± 1.96 | 19.67 ± 3.53 |
| Mature myeloid* | 93.75 ± 1.89 | 121.75 ± 7.22 | 108.00 ± 6.93 |
| Monocyte/macrophage | 20 ± 3.87 | 22.25 ± 3.25 | 20.33 ± 3.53 |
| Cellularity (× 107)‖ | 7.59 ± 0.42 | 3.56 ± 0.28c† | 6.39 ± 0.61b‡, NS§ |
NS indicates not significant.
BM cytology was evaluated from cytospins from 4 nonmutant, 4 Rps6lox/+;Tg.MxCre/+, and 3 Rps6lox/+;Tg.MxCre/+;Trp53ko/ko animals. A total of 260 cells were evaluated per animal. Immature erythroid cells are Pre CFU-E or CFU-E cells as defined by Pronk et al21 (supplemental Table 4); mature erythroid cells are either basophilic, polychromatic or orthochromatic erythroblasts22 (supplemental Table 4); immature myeloid cells are promyelocytes or myelocytes; and mature myeloid cells are band or segmented neutrophils. The number of cells within each population is given as mean ± SEM with P values on the basis of 2-tailed Student t test, aP < .01, bP < .05, cP < .001.
Nonmutant versus Rps6lox/+;Tg.MxCre/+.
Rps6lox/+;Tg.MxCre/+ vs Rps6lox/+;Tg.MxCre/+;Trp53ko/ko.
Nonmutant vs Rps6lox/+;Tg.MxCre/+;Trp53ko/ko.
BM cellularity determined by flushing 2 femurs and 2 tibias from 7 nonmutant, 7 Rps6lox/+;Tg.MxCre/+, and 3 Rps6lox/+;Tg.MxCre/+;Trp53ko/ko animals. Values are given as mean ± SEM with P values based on 2-tailed Student t test, aP < .01, bP < .05, cP < .001.
We evaluated lineage specific blood cell precursors on the basis of BM cytology. From cytospin preparations of BM samples flushed from the lower extremities, we observed a marked reduction in the number of lymphoid cells in Rps6 mutant animals, whereas immature and mature granulocytic and monocytic populations were unaffected (Table 2). The number of lymphocytes in Rps6lox/+;Tg.MxCre/+ BM was significantly reduced (by 56%, P = .006), and partially rescued in the absence of p53 (Rps6lox/+;Tg.MxCre/+;Trp53ko/ko).
We examined the cytology of RBC precursors from Tg.MxCre/+ and Rps6lox/+;Tg.MxCre/+ and Rps6lox/+; Tg.MxCre/+; Trp53ko/ko BM on the basis of recent work by Pronk21 and Socolovsky22 in which the morphology of specific, sorted cell populations from the mouse myeloerythroid hierarchy was carefully assessed. We scored cells as either immature (PreCFU-E and CFU-E; supplemental Table 4) or mature erythroid precursors (basophilic, polychromatophilic, and orthochromatic erythroblasts; supplemental Table 4) and observed a significant relative increase in the number of immature erythroid precursors (P = .001, Figure 3E, Table 2, supplemental Table 4) and a significant decrease in mature erythroid cells (P = .016, Figure 3E, Table 2, supplemental Table 4). Both of these changes were dependent on Trp53 (Table 2).
As a complementary means of evaluating the erythroid lineage in Rps6 mutant animals, we performed immunofluorescence staining on sections from nonmutant and Rps6lox/+;Tg.MxCre/+ BM using Ter119 (a marker for mature RBCs)22 and p53. Akin to our previous findings (Figure 1D-E), we observed a significant (P = .013) increase in p53-positive cells in the BM of mutant animals (supplemental Figure 3A-B). Ter119 staining of nucleated cells (dapi-positive), however, was significantly reduced (P = .016) in the Rps6 mutant compared with nonmutant animal (supplemental Figure 3A,C). We also found that although a fraction of the nucleated, Ter119-positive cells also stained with p53, most p53-positive cells were not mature RBCs (Ter119-positive), suggesting that p53 acts at multiple stages during hematopoiesis.
Megakaryocytes were significantly increased and showed distinct cytologic changes, with frequent, small dysplastic, monolobated megakaryocytes in BM from Rps6lox/+;Tg.MxCre/+ mice (Figure 3F-G). Thus, from multiple perspectives, BM-specific hemizygosity for Rps6 causes defects that recapitulate key clinical features of 5q− MDS,25 including defects in red cell maturation and megakaryocyte dysplasia accompanied by thrombocytosis.
p53 activation mimics ribosomal protein deficiency in the BM
Except for the lymphoid abnormalities, the hematologic alterations we observed depended completely on the presence of Trp53. To ask whether activation of p53 was sufficient to trigger these abnormalities, we took 2 approaches to elicit moderate levels of p53 activation because robust activation of p53 in developing BM causes a failure to initiate primitive erythropoiesis and embryonic death.26
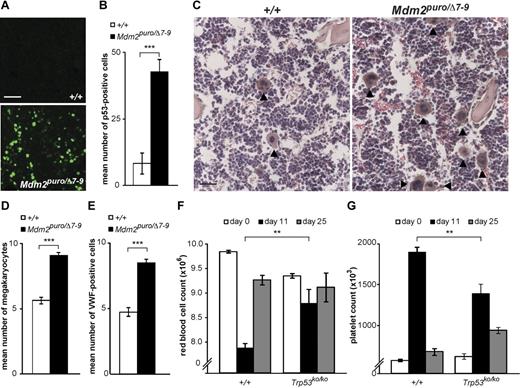
In one approach, we took advantage of Mdm2puro/Δ7-9 mice in which expression of Mdm2, a negative regulator of p53 stability and activity, is reduced ∼ 70% relative to wild-type mice.26 We measured p53 expression in the BM of Mdm2puro/Δ7-9 and nonmutant (+/+) controls. Although p53 expression in nonmutant marrow was negligible, immunofluorescent staining in Mdm2puro/Δ7-9 animals was increased by 5.2-fold compared with control animals (Figure 4A-B). To extend the initial results on the lymphocyte and red cell lineages described by Mendrysa et al,26 we counted megakaryocytes in BM sections and observed a significant megakaryocytosis (P < .001) in Mdm2puro/Δ7-9 compared with nonmutant (+/+) controls (Figure 4C-D). In addition, immunofluorescence for VWF was significantly increased in BM sections from Mdm2 mutant animals (Figure 4E).
Effects of Mdm2 gene dosage and 5-fluorouracil on p53 levels and hematologic phenotypes. Example (A) and quantitation (B) of p53 immunostaining in BM sections from animals with reduced Mdm2 activity (Mdm2puro/Δ7-9) compared with control (+/+) animals. For panel B, n = 5 mice per group, mean number of p53-positive cells per hpf ± SEM is shown, 6 hpf per animal. Example (C) and quantitation (D) of megakaryocyte abundance in BM sections from Mdm2puro/Δ7-9 and +/+ animals. For panel D, n = 3 animals per genotype, mean number of megakaryocytes per hpf ± SEM is shown, 10 hpf per animal. (E) Quantitation of VWF-positive cells in BM sections from nonmutant (+/+) and Mdm2 mutant (Mdm2puro/Δ7-9) animals (n = 4 animals per genotype, mean number of VWF-positive cells with large nuclei per hpf is shown ± SEM, 10 hpf per animal). (F-G) RBC and platelet counts (per μL) after administration of 5-fluorouracil to control (+/+, n = 6) and p53-deficient (Trp53ko/ko, n = 4) animals. Values before 5-fluorouracil treatment (white bars), 11 days after treatment (black bars), and 25 days after treatment (gray bars) are shown ± SEM. P values are based on a 2-tailed t test (B,D,E) or multiple regression where sex and genotype are factors (F-G), **P < .01, ***P < .001; NS indicates not significant. Scale bars: 75μM (A); 50μM (C).
Effects of Mdm2 gene dosage and 5-fluorouracil on p53 levels and hematologic phenotypes. Example (A) and quantitation (B) of p53 immunostaining in BM sections from animals with reduced Mdm2 activity (Mdm2puro/Δ7-9) compared with control (+/+) animals. For panel B, n = 5 mice per group, mean number of p53-positive cells per hpf ± SEM is shown, 6 hpf per animal. Example (C) and quantitation (D) of megakaryocyte abundance in BM sections from Mdm2puro/Δ7-9 and +/+ animals. For panel D, n = 3 animals per genotype, mean number of megakaryocytes per hpf ± SEM is shown, 10 hpf per animal. (E) Quantitation of VWF-positive cells in BM sections from nonmutant (+/+) and Mdm2 mutant (Mdm2puro/Δ7-9) animals (n = 4 animals per genotype, mean number of VWF-positive cells with large nuclei per hpf is shown ± SEM, 10 hpf per animal). (F-G) RBC and platelet counts (per μL) after administration of 5-fluorouracil to control (+/+, n = 6) and p53-deficient (Trp53ko/ko, n = 4) animals. Values before 5-fluorouracil treatment (white bars), 11 days after treatment (black bars), and 25 days after treatment (gray bars) are shown ± SEM. P values are based on a 2-tailed t test (B,D,E) or multiple regression where sex and genotype are factors (F-G), **P < .01, ***P < .001; NS indicates not significant. Scale bars: 75μM (A); 50μM (C).
As a complementary approach, we administered the chemotherapeutic agent, 5-fluorouracil, whose ability to cause apoptosis and growth arrest in cells has been attributed to its ability to induce p53,27 and which has been widely used in mice to investigate the biology of stress hematopoiesis. On the basis of a regimen from the work of Agosti et al,28 we administered a single dose of 150 mg/kg to wild-type control mice and observed an ∼ 20% decrease in RBC count and an ∼ 60% increase in platelet count 11 days later (Figure 4F-G). These values returned to near pretreatment levels by day 25. In contrast, the response to 5-fluorouracil in Trp53ko/ko animals was markedly attenuated; at day 11, both RBC and platelet counts were significantly different (P = .007 and P = .0075, respectively) from nonmutant animals. Thus, both genetic and pharmacologic approaches in vivo suggest that p53 activation induces erythrocyte and megakaryocyte abnormalities similar to those caused by Rps6 hemizygosity.
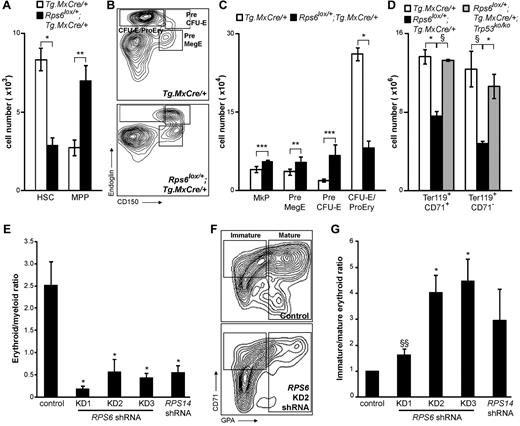
Effects of Rps6 hemizygosity on hematopoiesis
Recent work by Pronk et al demonstrated that specific cell populations in the mouse myeloerythroid hierarchy can be identified and prospectively separated via differential expression of the cell-surface markers endoglin (Eng/CD105), CD150 (Slamf1), and CD41 (Itga2b).21 Multipotent progenitors (MPPs) can be distinguished from HSCs, granulocyte-monocyte populations distinguished from megakaryocyte (MkP) and erythrocyte precursors, and erythroid progenitors can be divided into a maturation hierarchy: PreMegE (megakaryocyte-erythroid precursor), PreCFU-E, and CFU-E/ProEry populations.
To explore the ontogeny of the erythroid and megakaryocytic defects caused by BM-specific Rps6 hemizygosity in the mouse, we analyzed specific precursor populations from the BM as described in Pronk et al21 (HSC and MPP populations were identified on the basis of differential expression of CD34, MPP − CD34+, and HSC − CD34−.) We measured and compared 6 separate types of cells between Rps6lox/+;Tg.MxCre/+ mutant and nonmutant (Tg.MxCre/+) controls and observed 2 salient features. First, mutant animals exhibited a significant paucity in the absolute number of HSCs (P = .027), accompanied by significant increases of most downstream populations (MPP, P = .002; MkP, P < .001; PreMegE, P = .004; Pre CFU-E, P < .001; Figure 5A-C). Second, mutant animals also exhibited a significant (P = .017) reduction of more mature erythroid precursors (CFU-E/ProEry; Figure 5B-C).
Effects of BM-specific Rps6 hemizygosity on selected hematopoietic lineage subpopulations. (A) Number of HSC and MPP cells from Rps6 mutant and nonmutant BM. (B) Flow cytometric profiles of the erythroid lineage from Tg.MxCre/+ and Rps6lox/+;Tg.MxCre/+ animals. Immature erythroid precursors (Pre CFU-E) express greater levels of CD150 than mature erythroid precursors (CFU-E/ProEry); mutant animals exhibit a specific block in this step of erythroid maturation. (C) Number of megakaryocytic (MkP) and erythroid (PreMegE, Pre CFU-E, CFU-E/ProEry) cells from Rps6 mutant and nonmutant BM. (D) Number of mature erythroid cells based on expression of Ter119 and CD71 from Tg.MxCre/+, Rps6lox/+;Tg.MxCre/+, and Rps6lox/+;Tg.MxCre/+;Trp53ko/ko animals. Ter119+CD71+ cells are the sum of the Ter119+CD71high and Ter119+CD71int populations (supplemental Figure 4). For panels A, C, and D, open bars represent mean number observed per 1 million BM cells analyzed from nonmutant animals (± SEM, n = 3), black bars represent the corresponding numbers from Rps6lox/+;Tg.MxCre/+ animals (± SEM, n = 3) after dividing by a factor of 2.13 to account for the reduced number of total BM cells in mutant animals, and gray bars (D) represent the mean number of cells from Rps6lox/+;Tg.MxCre/+;Trp53ko/ko animals (± SEM, n = 2) after dividing by 1.18 (Table 2). (E-G) shRNA constructs targeting RPS6 (KD1, KD2, and KD3) or RPS14 decrease the ratio of mature erythroid (GPA+) to myeloid (CD11b+) cells (E) and also decrease the ratio of mature erythroid to immature erythroid (CD71+GPA−) cells (F-G). Each bar in panels E and G represents mean ± SEM, n = 3. P values are based on a 2-tailed t test, *P < .05, **P < .01, ***P < .001, §P = 0.07, and §§P = 0.08.
Effects of BM-specific Rps6 hemizygosity on selected hematopoietic lineage subpopulations. (A) Number of HSC and MPP cells from Rps6 mutant and nonmutant BM. (B) Flow cytometric profiles of the erythroid lineage from Tg.MxCre/+ and Rps6lox/+;Tg.MxCre/+ animals. Immature erythroid precursors (Pre CFU-E) express greater levels of CD150 than mature erythroid precursors (CFU-E/ProEry); mutant animals exhibit a specific block in this step of erythroid maturation. (C) Number of megakaryocytic (MkP) and erythroid (PreMegE, Pre CFU-E, CFU-E/ProEry) cells from Rps6 mutant and nonmutant BM. (D) Number of mature erythroid cells based on expression of Ter119 and CD71 from Tg.MxCre/+, Rps6lox/+;Tg.MxCre/+, and Rps6lox/+;Tg.MxCre/+;Trp53ko/ko animals. Ter119+CD71+ cells are the sum of the Ter119+CD71high and Ter119+CD71int populations (supplemental Figure 4). For panels A, C, and D, open bars represent mean number observed per 1 million BM cells analyzed from nonmutant animals (± SEM, n = 3), black bars represent the corresponding numbers from Rps6lox/+;Tg.MxCre/+ animals (± SEM, n = 3) after dividing by a factor of 2.13 to account for the reduced number of total BM cells in mutant animals, and gray bars (D) represent the mean number of cells from Rps6lox/+;Tg.MxCre/+;Trp53ko/ko animals (± SEM, n = 2) after dividing by 1.18 (Table 2). (E-G) shRNA constructs targeting RPS6 (KD1, KD2, and KD3) or RPS14 decrease the ratio of mature erythroid (GPA+) to myeloid (CD11b+) cells (E) and also decrease the ratio of mature erythroid to immature erythroid (CD71+GPA−) cells (F-G). Each bar in panels E and G represents mean ± SEM, n = 3. P values are based on a 2-tailed t test, *P < .05, **P < .01, ***P < .001, §P = 0.07, and §§P = 0.08.
Additional fractionation of the erythroid lineage on the basis of expression of CD71 and Ter11922 supports the aforementioned observations: the number of mature erythroid cells (Ter119+CD71+ and Ter119+CD71−; Figure 5D, supplemental Figure 4) was reduced in mutant (Rps6lox/+;Tg.MxCre/+) compared with nonmutant (Tg.MxCre/+) BM. This abnormality is also dependent on Trp53 (Figure 5D, supplemental Figure 4).
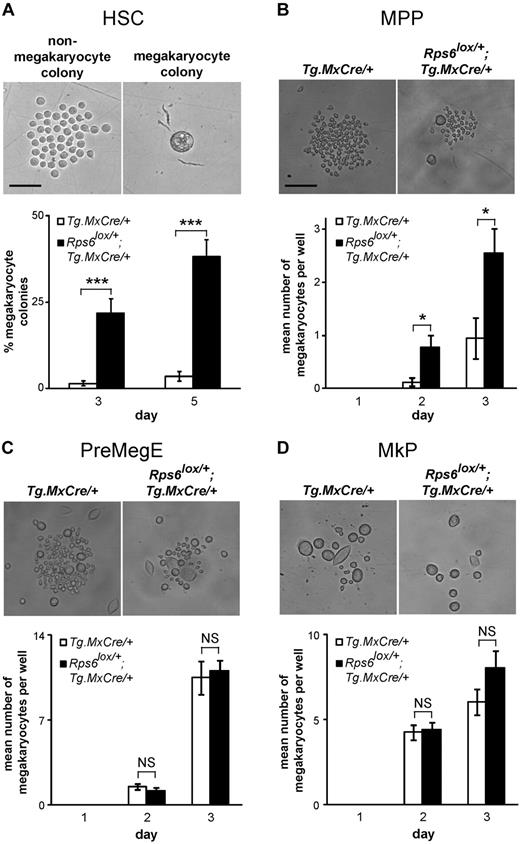
To further investigate how Rps6 hemizygosity (and p53 activation) might affect lineage commitment during blood cell development, we performed a clonal analysis for HSC differentiation. Single HSCs from Rps6lox/+;Tg.MxCre/+ mutant and nonmutant (Tg.MxCre/+) control BM were sorted into individual liquid culture wells of a 96-well plate, and the extent of megakaryocyte versus nonmegakaryocyte differentiation in each well was assessed visually after 3 and 5 days of culture. (The nature of this assay does not allow nonmegakaryocyte cell types to be subdivided by light microscopy, but megakaryocyte-like cells can be easily distinguished on the basis of their size and morphology, Figure 6A.) In nonmutant animals, the proportion of wells that give rise to megakaryocyte-only colonies was very low (generally 1-2 wells per 96-well plate). However, in mutant animals, there was a 20- to 30-fold increase in the proportion of wells that gave rise to megakaryocyte-only colonies (Figure 6A).
In vitro phenotypes of sorted hematopoietic populations from Rps6 mutant animals. (A) Photomicrographs (top) of a representative nonmegakaryocyte (left) and megakaryocyte (right) colony derived from a single HSC after 5 days in liquid culture. The percentage of wells that differentiate into megakaryocyte colonies at days 3 and 5 is shown for Tg.MxCre/+ and Rps6lox/+;Tg.MxCre/+ animals (bottom, n = 5 animals per genotype, at least 24 wells were evaluated per animal). (B-D) Photomicrographs (top) and the mean number of megakaryocytes (bottom) derived from 10 MPP (B), PreMegE (C), and MkP (D) cells sorted into a single well. Colonies were evaluated on days 1, 2, and 3. (n = 24 wells per animal with 2 animals of each genotype examined). P values are based on a 2-tailed t test, *P < .05, ***P < .001; NS indicates not significant. Scale bars: 50μM (A), 150μM (B-D).
In vitro phenotypes of sorted hematopoietic populations from Rps6 mutant animals. (A) Photomicrographs (top) of a representative nonmegakaryocyte (left) and megakaryocyte (right) colony derived from a single HSC after 5 days in liquid culture. The percentage of wells that differentiate into megakaryocyte colonies at days 3 and 5 is shown for Tg.MxCre/+ and Rps6lox/+;Tg.MxCre/+ animals (bottom, n = 5 animals per genotype, at least 24 wells were evaluated per animal). (B-D) Photomicrographs (top) and the mean number of megakaryocytes (bottom) derived from 10 MPP (B), PreMegE (C), and MkP (D) cells sorted into a single well. Colonies were evaluated on days 1, 2, and 3. (n = 24 wells per animal with 2 animals of each genotype examined). P values are based on a 2-tailed t test, *P < .05, ***P < .001; NS indicates not significant. Scale bars: 50μM (A), 150μM (B-D).
We performed a similar experiment sorting for MPP, PreMegE, or MkP populations, placing 10 cells per well in a 96-well plate, and then counting the number of megakaryocytes on each of the following 3 days. Similar to results observed with the single-cell assay for HSCs (Figure 6A), the number of megakaryocytes recovered from MPP cells (measured as mean number of megakaryocytes per well of a 96-well plate) was significantly greater (P = .01) in mutant compared with nonmutant animals (Figure 6B). However, no differences in megakaryocyte number were detected between mutant and control cultures established from PreMegE and MkP populations (Figure 6C-D), which is surprising given the conventional view of the megakaryocyte lineage in which there is a stepwise progression from HSC to MPP to PreMegE to MkP (Figure 7B).
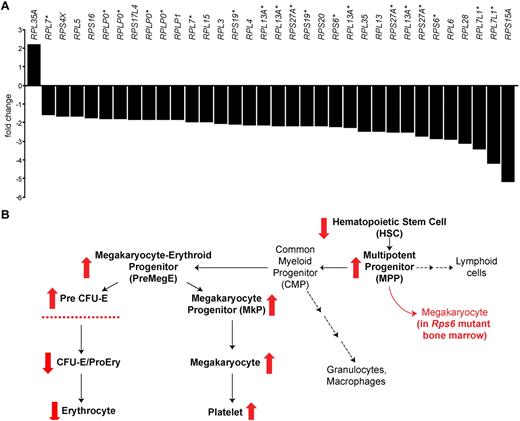
Ribosomal protein gene dysregulation in human MDS and summary of a mouse model. (A) Fold-change of mRNA for 8 patients with low-risk, non-5q− MDS compared with 11 healthy, age-matched control patients; each bar represents a different array probe for ribosomal protein genes that are differentially expressed. As described in the text, of 74 ribosomal protein genes represented on the array, 21 genes exhibited reduced mRNA levels and 1 gene exhibited increased mRNA levels (represented by a total of 34 probes). The figure shows all ribosomal protein gene probes that exhibit significant (< 0.01% false-discovery rate) differences; genes with multiple probes are indicated with an asterisk (*). (B) Diagram of the hematopoietic lineage and differences observed in BM-specific Rps6 mutant mice. Red arrows summarize alterations in Rps6 mutant animals on the basis of Figure 5A, C, and D, including a reduction in the size of the HSC population, a concomitant increase in the size of most downstream progenitor populations, and a block in erythroid maturation.
Ribosomal protein gene dysregulation in human MDS and summary of a mouse model. (A) Fold-change of mRNA for 8 patients with low-risk, non-5q− MDS compared with 11 healthy, age-matched control patients; each bar represents a different array probe for ribosomal protein genes that are differentially expressed. As described in the text, of 74 ribosomal protein genes represented on the array, 21 genes exhibited reduced mRNA levels and 1 gene exhibited increased mRNA levels (represented by a total of 34 probes). The figure shows all ribosomal protein gene probes that exhibit significant (< 0.01% false-discovery rate) differences; genes with multiple probes are indicated with an asterisk (*). (B) Diagram of the hematopoietic lineage and differences observed in BM-specific Rps6 mutant mice. Red arrows summarize alterations in Rps6 mutant animals on the basis of Figure 5A, C, and D, including a reduction in the size of the HSC population, a concomitant increase in the size of most downstream progenitor populations, and a block in erythroid maturation.
We considered whether this apparent paradox—increased numbers of MPP-derived megakaryocytes without a concomitant increase in PreMegE- or MkP-derived megakaryocytes—might be explained by differences in plating efficiency or cell survival. We measured plating efficiency by counting total cell number (without regard to morphology) after 1 day of culture and observed no differences between mutant and nonmutant HSC, MPP, PreMegE, or MkP cultures (supplemental Figure 5). We did, however, observe that cell growth over the next several days was reduced in all mutant cultures, consistent with a cell-autonomous process that causes general growth impairment.
In summary, hemizygosity for Rps6 in the BM causes not only a profound reduction in overall cellularity but also affects the relative distribution of hematopoietic subpopulations, with a relative decrease in the number of HSCs, CFU-E/ProEry, CD71+, and CD71− cells, accompanied by a relative increase in all other subpopulations. In culture, growth of each subpopulation is impaired, except for MPP-derived megakaryocytes, which are increased in mutant compared with nonmutant cultures.
Effect of reduced ribosomal protein gene dosage in human BM cells
To investigate how hemizygosity for RPS6 would affect human blood cell development, we studied the effects of RPS6 shRNAs in culture. Cord blood samples were sorted (Lin−CD34+CD38−CD90+CD45RA−) to enrich for HSCs,29 infected with a lentivirus containing short hairpin RNA sequences targeting RPS6 (or RPS14 for control), and then cultured under conditions to allow myeloid and erythroid differentiation.2 After 10 days, cultures were analyzed for cell-surface markers of myeloid (CD11b+), mature erythroid (GPA+), and immature erythroid (CD71+GPA−) differentiation.
In all cases, shRNA constructs targeting RPS6 or RPS14 caused a decrease in the erythroid/myeloid ratio (Figure 5E), the extent which correlated with the efficiency of knockdown as assessed by quantitative RT-PCR (supplemental Figure 6A). In addition, RPS6 knockdown led to a block in erythroid differentiation as assessed by presence of the erythroid lineage cell surface marker GPA (Figure 5F-G), similar to observations in BM-specific Rps6 mutant mice (Figure 5B-D), and consistent with the 5q− MDS phenotype.
A role for reduced ribosomal protein gene dosage in both DBA and 5q− MDS suggested to us that other forms of MDS might also involve altered expression of ribosomal protein genes. To explore this idea, we interrogated the results of Affymetrix microarray measurements carried out on 8 MDS patient HSC samples (7 low-risk and 1 intermediate-risk, none of whom carried 5q−; supplemental Table 1),30 compared with 11 age-matched healthy control samples. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE30201 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30201).
Among 54 676 probes on the array, mRNA levels for 3672 were decreased and 1159 were increased in the MDS patient samples at a false discovery rate of < 0.01%. However, ribosomal protein genes are disproportionately represented among the differentially expressed genes: 0.7% of the differentially expressed probes represented ribosomal protein genes, whereas 0.4% of all the probes on the array represented ribosomal protein genes (P = .005, 2-tailed z-statistic, 95% confidence interval, z = 2.821). More strikingly, of 74 ribosomal protein genes represented on the array, mRNA levels for 21 were decreased, and only 1 was increased (Figure 7A). Thus, ribosomal protein gene expression is dysregulated—and largely repressed— in HSCs from a group of non-5q− MDS patients.
Discussion
5q− MDS is characterized by abnormal erythrocyte maturation with macrocytic anemia, thrombocytosis, and a predisposition to leukemia.25 Intriguingly, DBA, a congenital BM failure syndrome, exhibits erythroid defects similar to 5q− syndrome, and in recent studies researchers suggest that both conditions are caused by reduced dosage of one or more ribosomal protein genes.2,5 To date, the underlying cellular pathophysiology and potential approaches to treatment for these diseases have been difficult to unravel.
During earlier studies of pigmentary mutations in mice, we realized that reduced dosage of the ribosomal protein genes, Rps6, Rps19, and Rps20, led to activation of p53 in which the differential and specific response of individual cell types in the skin and blood gave rise to a pleiotropic syndrome that was likely relevant to 5q− MDS and DBA.16 We have now tested this hypothesis more directly and found that hemizygosity for Rps6 in mouse BM and reduced dosage of ribosomal protein genes in human cells recapitulate cardinal features of the human condition. In addition, studies designed on the basis of genome-wide expression analysis reveal that abnormal expression—and specifically down-regulation—of ribosomal protein genes characterizes BM stem cells from patients with low-risk, non-5q− MDS. These results connect genotype to phenotype for a relevant animal model of MDS and suggest that ribosomal protein gene dysregulation may play a role in the pathogenesis of a broad group of BM-failure syndromes.
Potential involvement of ribosomal protein genes in 5q− MDS is based on work of Ebert and colleagues in which large-scale siRNA knockdown of candidate genes in cultured CD34+ cells implicates RPS14.2 Recent work from Starczynowski et al, however, suggests that 2 microRNAs, miR-145 and miR-146a, are critical mediators of the 5q− phenotype.6 Our work does not exclude a potential role for microRNAs as an alternative and/or secondary component of 5q− MDS but demonstrates that reduced dosage of a ribosomal protein gene within BM cells is sufficient to accurately model many aspects of the human condition. From this perspective, it is interesting to compare our results with those of Barlow et al, who engineered mice with a HSC-specific ∼ 500-kb deletion that removed 8 genes, including Rps14 (although not miR-145 or miR-146a).31 Hematologic abnormalities in the mouse model described by Barlow et al31 share many features with the BM-specific Rps6 mice described here but exhibit thrombocytopenia rather than thrombocytosis and a reduction rather than increase in megakaryocyte-erythroid progenitor subpopulations. Additional studies of miR-145 and miR146a expression and/or manipulation in vivo are likely to resolve the relative contributions of ribosomal protein and microRNA genes to 5q− MDS.
Our work uses Rps6 as a tool to model 5q− MDS (and to inform our understanding of DBA), yet human mutations of RPS6 have not been identified in either condition. However, germline mutations for RPS6 may be lethal (as they are in mice).32 Our results confirm that loss-of-function for Rps6 is more severe than loss-of-function for Rps19 or Rps20 because keratinocyte-specific hemizygosity for Rps6 has a more dramatic effect on p53 induction compared with germline mutations in Rps19 or Rps20 and because germline mutations in Rps19 or Rps20 have a very mild effect on BM.16 We note that Rps6 phosphorylation serves as an indicator of physiologic stress and is downstream of the mammalian target of rapamycin pathway, and from that perspective, Rps6 is not “just another ribosomal protein,” which may help to explain why lymphopenia develops in our BM-specific Rps6 mutants but not in other models of 5q− MDS (or DBA). However, our results also suggest that the effect of Rps6 mutation-induced BM disease in other BM lineages is similar to that caused by mutations in other ribosomal protein genes, at least with regard to dependence on p53.31,33 We also note that extensive ribosomal protein gene resequencing has not yet been reported for MDS; indeed, our results suggest that some patients with low-risk MDS and normal cytogenetics may have somatic mutations of ribosomal protein genes other than RPS14, including RPS6.
Similar to the work of Barlow et al,31 we find that many of the BM phenotypes in Rps6 mutant animals depend on the presence of p53. We find that activation of p53 is necessary for peripheral blood cell abnormalities (leukopenia, macrocytic anemia, thrombocytosis, and elevated eADA activity), BM hypocellularity, and erythrocyte maturation in mutant BM. We note that parallel mechanisms are likely to exist in other contexts. Activation of p53 during ischemia contributes to cell death, tissue injury, and organ failure,34,35 and recent studies of animal models of Fanconi anemia and Treacher Collins syndrome suggest that stabilization of p53 during embryogenesis induces characteristic developmental abnormalities.36,37 These observations support an emerging paradigm in which activation of p53 function can give rise to a variety of developmental or disease-related phenotypes depending on the amount and tissue-specific context in which activation occurs.38
Our studies of hematopoietic subpopulations suggest that reduced ribosomal protein dosage and consequent p53 activation can perturb multiple and independent steps in blood cell development. Impairment of HSC growth is consistent with both a general role for ribosome function and/or a proapoptotic effect of p53 activation on HSCs.3,39,40 In this context, the relative increase of downstream progenitors (MPP, PreMegE, Pre CFU-E, MkP) we observed is likely to reflect homeostatic regulation within the BM.41 However, the reduction of more mature erythroid progenitors (CFU-E/ProEry, CD71+, and CD71−) points to a specific block in erythroid maturation and may therefore account for the observation that 5q− MDS and DBA frequently present with a selective reduction in RBC development. Finally, akin to observations by Adolfsson et al,42 our studies of megakaryocyte-like growth from cultured hematopoietic cell subpopulations suggest that reduced ribosomal protein gene dosage unmasks an alternative pathway for megakaryocyte development directly from HSC and/or MPP rather than via PreMegE and MkP subpopulations.
In previous studies of large-scale gene expression in MDS CD34+ cells, investigators have shown that reduced expression of ribosomal protein genes is a hallmark of 5q− MDS43,44 ; revealed that pathways related to interferon, thrombopoietin, and Wnt signaling are dysregulated in all MDS subtypes3 ; and have also identified specific sets of genes that correlate with disease subtype and/or prognosis.3,44 At first glance, some of the results reported by Sridhar et al44 appear opposite to what we find, with elevated levels of ribosomal protein gene expression observed in CD34+ cells in non-5q− MDS. However, a key difference between the work reported here and previous studies3,43-45 is that our expression profiling studies use highly purified HSC (Lin−CD34+CD38−CD90+CD45RA−) from MDS patients. This approach both enriches for HSCs (which constitute approximately 1% of total CD34+ cells) and diminishes the presence of multilineage and lineage committed progenitor cells that are also CD34+. This is particularly important for patients with high-risk MDS, in which the CD34+ population is likely to contain a large proportion of actively dividing cells that are metabolically active. In fact, in the work from Sridhar et al, expression of ribosomal protein genes in all CD34+ cells is elevated in patients with high-risk (transformed) MDS compared with low-risk (stable) MDS,44 whereas the patients studied here all represent low-risk MDS. Nonetheless, additional studies, likely designed on the basis of large-scale sequencing, will be required to investigate whether the reduction of ribosomal protein gene expression we observe in HSCs from non 5q− MDS represent a critical pathophysiologic event or a secondary consequence of altered growth rate.
Taken together with genetic studies of Diamond-Blackfan anemia, our work suggests that the hematopoietic system is especially sensitive to alterations in ribosomal protein gene dosage. This intersection—of forward genetic approaches in human disease with reverse genetic approaches in mouse models—is reminiscent of an analogous connection between components of the primary cilium and a spectrum of human diseases represented by Bardet-Biedl syndrome, polycystic kidney disease, and heterotaxy.46 Additional studies in mouse models will illuminate the extent to which non 5q−MDS and other forms of BM failure represent ribosomopathies.
The online version of this article contains a data supplement
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Martin Hrabe de Angelis for Rps19Dsk3/+ mice, G. Thomas and S. Volarevic for Rps6lox mice, S. Artandi and J. Jorcano for Tg.K5Cre mice, and T. Jacks for Trp53ko mice. They also thank H. Manuel for technical support.
K.A.M. and C.Y.P were supported by Mentored Clinical Scientist Development Investigator Awards from the National Institutes of Health (NIH). C.Y.P. was also supported by an ARRA award from the NIH. W.W.P. is supported by the Stanford Medical Scientist Training Program. I.L.W. is supported by the Ludwig Center at Stanford University, NIH Grant 5U01HL099999-03, and the Leukemia & Lymphoma SCORE grant.
National Institutes of Health
Authorship
Contribution: K.A.M. generated the Rps6 mutant mice, analyzed the skin, peripheral blood, and BM phenotypes, performed experiments on Mdm2 mutant animals and analyzed the genome wide expression analysis from human bone marrow cells, with help from M.G.P. and C.Y.P.; W.W.P. carried out the siRNA knockdown and genome-wide expression analysis in human HSCs, with help from J.V.P. and C.Y.P.; R.B. evaluated Rps6 protein levels in the BM of mutant mice; B.E.G. measured eADA activity in the blood of mutant mice and provided experimental and intellectual advice about DBA; R.M. and S.M.M. designed experiments and generated the Mdm2 mutant mice and provided expertise about the role of p53 in the BM; and I.L.W. helped design experiments and provided intellectual insight into the role of hematopoietic stem cells in MDS. All experiments were performed under the guidance and leadership of C.Y.P. and G.S.B., who coordinated the project and wrote the manuscript with input from K.A.M.
The current affiliation for C.Y.P. is Department of Pathology and Clinical Laboratories, Memorial Sloan-Kettering Cancer Center, New York, NY.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gregory S. Barsh, Beckman Center B271A, Stanford University School of Medicine, Stanford, CA 94305; e-mail: gbarsh@stanford.edu; or Christopher Y. Park, Memorial Sloan-Kettering Cancer Center, Departments of Pathology and Clinical Labs and Human Oncology and Pathogenesis Program, New York, NY 10065; e-mail: parkc@mskcc.org.
References
Author notes
K.A.M. and W.W.P. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal