Abstract
Low MN1 expression bestows favorable prognosis in younger adults with cytogenetically normal acute myeloid leukemia (CN-AML), but its prognostic significance in older patients is unknown. We analyzed pretherapy MN1 expression in 140 older (≥ 60 years) de novo CN-AML patients treated on cytarabine/daunorubicin-based protocols. Low MN1 expressers had higher complete remission (CR) rates (P = .001), and longer overall survival (P = .03) and event-free survival (EFS; P = .004). In multivariable models, low MN1 expression was associated with better CR rates and EFS. The impact of MN1 expression on overall survival and EFS was predominantly in patients 70 years of age or older, with low MN1 expressers with mutated NPM1 having the best outcome. The impact of MN1 expression was also observed in the Intermediate-I, but not the Favorable group of the European LeukemiaNet classification, where low MN1 expressers had CR rates and EFS similar to those of Favorable group patients. MN1 expresser-status-associated gene- and microRNA-expression signatures revealed underexpression of drug resistance and adverse outcome predictors, and overexpression of HOX genes and HOX-gene–embedded microRNAs in low MN1 expressers. We conclude that low MN1 expression confers better prognosis in older CN-AML patients and may refine the European LeukemiaNet classification. Biologic features associated with MN1 expression may help identify new treatment targets.
Introduction
Over the past 3 decades, there has been relatively steady improvement of outcomes of patients with acute myeloid leukemia (AML) younger than 60 years. However, this has not occurred in older AML patients. Despite advances in our understanding of disease mechanisms and investigation of new therapies targeting distinct clinical, cytogenetic, and molecular subsets, the outcome of AML patients older than 60 years remains poor, with long-term survival rates of ∼ 7%-15%.1-3 The shorter survival of older AML patients compared with younger patients is probably related to clinical and biologic differences between them, including the failure to achieve a complete remission (CR) as a result of an increased intrinsic resistance of leukemic blasts to chemotherapy and the presence of specific cytogenetic and/or molecular alterations associated with worse outcome.4
As in younger patients, older patients with cytogenetically normal (CN) AML represent the largest AML subset.5 This group is molecularly heterogeneous.6,7 To date, however, the prognostic significance of molecular genetic alterations has been studied most extensively in younger (< 60 years) patients.6-8 Recently, some, but not all, of these markers have also been shown to impact on outcome of older (≥ 60 years) CN-AML patients. For example, NPM1 mutations,9 and lower expression levels of the BAALC and ERG10 genes have been associated with favorable outcome, whereas FLT3 internal tandem duplication (FLT3-ITD)11 and WT1 mutations12 have been shown to confer adverse prognosis in older patients, as they do in younger patients. However, to our knowledge, no study has investigated the prognostic impact of meningioma 1 (MN1) gene expression levels exclusively in CN-AML patients aged 60 years of age and older.
The MN1 gene is localized at human chromosome band 22q12 and encodes a transcriptional coregulator.13 MN1 is involved in myeloid malignancies as a fusion partner of the ETV6 gene in the recurrent translocation t(12;22)(p13;q11)14 and has been shown to be overexpressed in subsets of AML.15,16 We and others have shown that high MN1 expression levels are prognosticators for poor outcome in younger CN-AML patients.17,18
With the hope to better predict the course of the disease, adjust therapeutic approaches, and improve outcome, we explored herein the prognostic significance of MN1 expression in older de novo CN-AML patients. We have also analyzed genome-wide gene- and microRNA-expression profiles associated with MN1 expression in these patients, to gain insights into MN1-associated disease.
Methods
Patients and treatment
Pretreatment bone marrow (BM) samples of 140 adults 60 years of age or older with de novo CN-AML and material available were analyzed for MN1 expression. The patients were enrolled on Cancer and Leukemia Group B (CALGB) front-line intensive cytarabine/daunorubicin-based treatment protocols (for protocol details see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Institutional Review Board–approved, written informed consent for participation in these studies was obtained from all patients in accordance with the Declaration of Helsinki.
Cytogenetics and additional molecular markers
Pretreatment cytogenetic analyses of BM were performed by CALGB-approved institutional cytogenetic laboratories as part of CALGB 8461, a prospective cytogenetic companion study, and the results reviewed centrally.19,20 For a case to be considered CN, at least 20 metaphase cells had to be analyzed and the karyotype found to be normal.20
The presence or absence of FLT3-ITD mutations in the tyrosine kinase domain of the FLT3 gene (FLT3-TKD) and mutations in the CEBPA, IDH1, IDH2, NPM1, TET2, and WT1 genes was determined centrally in pretreatment samples as described previously.9,11,12,21-28 The expression levels of the BAALC and ERG genes in peripheral blood were also assessed centrally in pretreatment samples as previously described.10,29-32 miR-181a expression was evaluated as previously described.33
RNA extraction and real-time RT-PCR to measure MN1 expression levels
Preparation of pretreatment BM samples and the analysis of MN1 expression were performed as previously described.18 Briefly, total RNA was extracted using Trizol reagent, and complementary DNA was synthesized from total RNA. Quantitative real-time RT-PCR amplifications of MN1 and ABL1 were performed using standard curves. MN1 copy numbers were normalized to ABL1 copy numbers.
Gene- and microRNA-expression profiling
For gene- and microRNA-expression profiling, total RNA was extracted from pretreatment BM or blood mononuclear cells. Gene- and microRNA-expression profiling was performed using the Affymetrix U133 plus Version 2.0 array (Affymetrix) and The Ohio State University custom microRNA array (OSU_CCC Version 4.0), respectively, as previously reported9 and detailed in supplemental Methods.
Definition of clinical end points and statistical analysis
The main objective of this study was to evaluate the prognostic value of MN1 expression on clinical outcome in older de novo CN-AML patients. For these patients, the median MN1/ABL1 copy number value was chosen to define the low and high MN1 expressers. This cut-off was based on the trend in overall survival (OS) of patients divided into quartiles by MN1 level values; patients in the first 2 quartiles had a better outcome than patients in quartiles 3 and 4 (P = .04 test for trend).34
Definitions of clinical end points (ie, CR, disease-free survival [DFS], OS, and event-free survival [EFS]) are provided in supplemental Methods. Associations between patients with low or high expression of MN1 for baseline demographic, clinical, and molecular features were compared using the Fisher exact and Wilcoxon rank-sum tests for categorical and continuous variables, respectively. Estimated probabilities of DFS, OS, and EFS were calculated using the Kaplan-Meier method, and the log-rank test evaluated differences between survival distributions. Multivariable analyses are detailed in supplemental Methods. Briefly, multivariable logistic regression models were constructed to analyze factors related to the probability of achieving CR using a limited backward selection procedure. Multivariable proportional hazards models were constructed for OS and EFS to evaluate the impact of low or high expression of MN1 by adjusting for other variables using a limited backward selection procedure. For achievement of CR, estimated odds ratios, and for survival end points, hazard ratios with their corresponding 95% confidence intervals (CIs) were examined.
For the gene- and microRNA-expression profiling, summary measures of gene and microRNA expression were computed, normalized, and filtered (supplemental Methods). MN1-associated signatures were derived by comparing gene and microRNA expression between low and high MN1 expressers. Univariable significance levels of .001 for gene and .005 for microRNA expression profiling were used to determine, respectively, the probe sets and microRNA probes that constituted the signatures.
All analyses were performed by the CALGB Statistical Center.
Results
Associations of MN1 expression with clinical and molecular characteristics and clinical outcome in older CN-AML patients
At diagnosis, the low and high MN1 expresser groups did not differ significantly with regard to any of the clinical pretreatment characteristics. However, low MN1 expression was associated with mutated NPM1 (P < .001), lower BAALC expression levels (P < .001), and lower miR-181a expression levels (P = .04), as well as a trend for the presence of FLT3-TKD (P = .08; Table 1).
Clinical and molecular characteristics according to MN1 expression status in CN-AML patients 60 years of age or older
| Characteristic . | Low MN1 (n = 70) . | High MN1 (n = 70) . | P . |
|---|---|---|---|
| Age, y | .57 | ||
| Median | 66 | 69 | |
| Range | 60-81 | 60-81 | |
| Sex, no. (%) of males | 40 (57) | 32 (46) | .24 |
| Race, no. (%) | .49 | ||
| White | 66 (96) | 63 (93) | |
| Nonwhite | 3 (4) | 5 (7) | |
| Hemoglobin, g/dL | .16 | ||
| Median | 9.1 | 9.4 | |
| Range | 5.4-13.6 | 6.0-13.1 | |
| Platelets, × 109/L | .43 | ||
| Median | 63 | 72 | |
| Range | 20-271 | 11-850 | |
| WBC count, × 109/L | .15 | ||
| Median | 33.8 | 21.5 | |
| Range | 1.0-450.0 | 1.0-434.1 | |
| Blood blasts, % | 1.0 | ||
| Median | 45 | 49 | |
| Range | 0-96 | 0-99 | |
| BM blasts, % | .32 | ||
| Median | 71 | 64 | |
| Range | 15-97 | 7-96 | |
| Extramedullary involvement, no. (%) | 19 (28) | 15 (22) | .55 |
| FLT3-ITD, no. (%) | .59 | ||
| Present | 22 (31) | 26 (37) | |
| Absent | 48 (69) | 44 (63) | |
| FLT3-TKD, no. (%) | .08 | ||
| Present | 10 (14) | 3 (4) | |
| Absent | 60 (86) | 67 (96) | |
| CEBPA, no. (%) | .14 | ||
| Mutated | 6 (9) | 13 (19) | |
| Single mutated | 5 | 7 | |
| Double mutated | 1 | 6 | |
| Wild-type | 64 (91) | 57 (81) | |
| IDH1, no. (%) | .30 | ||
| Mutated | 10 (14) | 6 (9) | |
| Wild-type | 59 (86) | 63 (91) | |
| IDH2, no. (%) | .54 | ||
| IDH2-mutated | 13 (19) | 17 (24) | |
| R140-IDH2-mutated | 12 | 13 | |
| R172-IDH2-mutated | 1 | 4 | |
| Wild-type | 56 (81) | 53 (76) | |
| NPM1, no. (%) | < .001 | ||
| Mutated | 55 (79) | 26 (37) | |
| Wild-type | 15 (21) | 44 (63) | |
| TET2, no. (%) | .85 | ||
| Mutated | 19 (28) | 18 (26) | |
| Wild-type | 49 (72) | 51 (74) | |
| WT1, no. (%) | 1.0 | ||
| Mutated | 3 (4) | 3 (4) | |
| Wild-type | 67 (96) | 67 (96) | |
| BAALC expression,* no. (%) | < .001 | ||
| Low | 48 (73) | 24 (34) | |
| High | 18 (27) | 46 (66) | |
| ERG expression,* no. (%) | .23 | ||
| Low | 36 (55) | 30 (43) | |
| High | 30 (45) | 40 (57) | |
| miR-181a expression (continuous) | .04 | ||
| Median (log expression units) | 11.89 | 12.23 | |
| Range | 9.06-15.43 | 8.81-14.66 | |
| ELN risk group,† no. (%) | .03 | ||
| Favorable | 40 (57) | 26 (37) | |
| Intermediate-I | 30 (43) | 44 (63) |
| Characteristic . | Low MN1 (n = 70) . | High MN1 (n = 70) . | P . |
|---|---|---|---|
| Age, y | .57 | ||
| Median | 66 | 69 | |
| Range | 60-81 | 60-81 | |
| Sex, no. (%) of males | 40 (57) | 32 (46) | .24 |
| Race, no. (%) | .49 | ||
| White | 66 (96) | 63 (93) | |
| Nonwhite | 3 (4) | 5 (7) | |
| Hemoglobin, g/dL | .16 | ||
| Median | 9.1 | 9.4 | |
| Range | 5.4-13.6 | 6.0-13.1 | |
| Platelets, × 109/L | .43 | ||
| Median | 63 | 72 | |
| Range | 20-271 | 11-850 | |
| WBC count, × 109/L | .15 | ||
| Median | 33.8 | 21.5 | |
| Range | 1.0-450.0 | 1.0-434.1 | |
| Blood blasts, % | 1.0 | ||
| Median | 45 | 49 | |
| Range | 0-96 | 0-99 | |
| BM blasts, % | .32 | ||
| Median | 71 | 64 | |
| Range | 15-97 | 7-96 | |
| Extramedullary involvement, no. (%) | 19 (28) | 15 (22) | .55 |
| FLT3-ITD, no. (%) | .59 | ||
| Present | 22 (31) | 26 (37) | |
| Absent | 48 (69) | 44 (63) | |
| FLT3-TKD, no. (%) | .08 | ||
| Present | 10 (14) | 3 (4) | |
| Absent | 60 (86) | 67 (96) | |
| CEBPA, no. (%) | .14 | ||
| Mutated | 6 (9) | 13 (19) | |
| Single mutated | 5 | 7 | |
| Double mutated | 1 | 6 | |
| Wild-type | 64 (91) | 57 (81) | |
| IDH1, no. (%) | .30 | ||
| Mutated | 10 (14) | 6 (9) | |
| Wild-type | 59 (86) | 63 (91) | |
| IDH2, no. (%) | .54 | ||
| IDH2-mutated | 13 (19) | 17 (24) | |
| R140-IDH2-mutated | 12 | 13 | |
| R172-IDH2-mutated | 1 | 4 | |
| Wild-type | 56 (81) | 53 (76) | |
| NPM1, no. (%) | < .001 | ||
| Mutated | 55 (79) | 26 (37) | |
| Wild-type | 15 (21) | 44 (63) | |
| TET2, no. (%) | .85 | ||
| Mutated | 19 (28) | 18 (26) | |
| Wild-type | 49 (72) | 51 (74) | |
| WT1, no. (%) | 1.0 | ||
| Mutated | 3 (4) | 3 (4) | |
| Wild-type | 67 (96) | 67 (96) | |
| BAALC expression,* no. (%) | < .001 | ||
| Low | 48 (73) | 24 (34) | |
| High | 18 (27) | 46 (66) | |
| ERG expression,* no. (%) | .23 | ||
| Low | 36 (55) | 30 (43) | |
| High | 30 (45) | 40 (57) | |
| miR-181a expression (continuous) | .04 | ||
| Median (log expression units) | 11.89 | 12.23 | |
| Range | 9.06-15.43 | 8.81-14.66 | |
| ELN risk group,† no. (%) | .03 | ||
| Favorable | 40 (57) | 26 (37) | |
| Intermediate-I | 30 (43) | 44 (63) |
The median expression value was used as a cutpoint.
Favorable risk group consists of patients with CEBPA mutations or those who are FLT3-ITD-negative and harbor NPM1 mutations. Intermediate-I genetic group is composed of patients who are not in the Favorable group (ie, those with wild-type CEBPA and wild-type NPM1 with or without FLT3-ITD or mutated NPM1 with FLT3-ITD).
With a median follow-up for living patients of 4.0 years (range, 3-11.6 years) and for those who did not have an event of 4.2 years (range, 3.3-11.6 years), low MN1 expressers had a higher CR rate (80% vs 53%, P = .001) and longer OS (P = .03; Figure 1A) and EFS (P = .004; Figure 1B) than high MN1 expressers (Table 2). We did not observe a significant difference in DFS between high and low MN1 expressers (P = .29; Table 2)
Outcome of CN-AML patients 60 years of age or older with respect to MN1 expression. (A) OS. (B) EFS.
Outcome of CN-AML patients 60 years of age or older with respect to MN1 expression. (A) OS. (B) EFS.
Outcomes according to MN1 expression status in all CN-AML patients 60 years of age or older and, separately, in those 60-69 years of age and 70 years of age or older
| Outcome . | Low MN1 . | High MN1 . | P* . | OR/HR (95% CI) . |
|---|---|---|---|---|
| All patients | n = 70 | n = 70 | ||
| CR rate, no. (%) | 56 (80) | 37 (53) | .001 | 3.57 (1.68, 7.56) |
| DFS | .29 | 0.79 (0.51, 1.23) | ||
| Median, y | 0.9 | 0.6 | ||
| Disease-free at 3 y, % (95% CI) | 18 (9-29) | 11 (3-23) | ||
| OS | .03 | 0.68 (0.48, 0.96) | ||
| Median, y | 1.2 | 0.8 | ||
| Alive at 3 y, % (95% CI) | 23 (14-33) | 10 (4-18) | ||
| EFS | .004 | 0.50† (0.33, 0.76) | ||
| Median, y | 0.7 | 0.2 | ||
| Event-free at 3 y, % (95% CI) | 14 (7-23) | 6 (2-13) | ||
| Patients 60-69 y | n = 44 | n = 35 | ||
| CR rate, no. (%) | 35 (80) | 19 (54) | .03 | 3.28 (1.22, 8.81) |
| DFS | .82 | 0.94 (0.52, 1.69) | ||
| Median, y | 0.7 | 0.5 | ||
| Disease-free at 3 y, % (95% CI) | 9 (2-21) | 11 (2-28) | ||
| OS | .44 | 0.83 (0.52, 1.33) | ||
| Median, y | 1.1 | 0.8 | ||
| Alive at 3 y, % (95% CI) | 18 (9-31) | 11 (4-24) | ||
| EFS | .11 | 0.69 (0.44, 1.10) | ||
| Median, y | 0.6 | 0.2 | ||
| Event-free at 3 y, % (95% CI) | 7 (2-17) | 6 (1-17) | ||
| Patients 70 y or older | n = 26 | n = 35 | ||
| CR rate, no. (%) | 21 (81) | 18 (51) | .03 | 3.97 (1.22, 12.90) |
| DFS | .09 | 0.54 (0.27, 1.10) | ||
| Median, y | 1.3 | 0.7 | ||
| Disease-free at 3 y, % (95% CI) | 33 (15-53) | 11 (2-30) | ||
| OS | .006 | 0.46 (0.26, 0.81) | ||
| Median, y | 2.0 | 0.9 | ||
| Alive at 3 y, % (95% CI) | 31 (15-49) | 9 (2-21) | ||
| EFS | .007 | 0.48 (0.27, 0.84) | ||
| Median, y | 1.0 | 0.3 | ||
| Event-free at 3 y, % (95% CI) | 27 (12-44) | 6 (1-17) |
| Outcome . | Low MN1 . | High MN1 . | P* . | OR/HR (95% CI) . |
|---|---|---|---|---|
| All patients | n = 70 | n = 70 | ||
| CR rate, no. (%) | 56 (80) | 37 (53) | .001 | 3.57 (1.68, 7.56) |
| DFS | .29 | 0.79 (0.51, 1.23) | ||
| Median, y | 0.9 | 0.6 | ||
| Disease-free at 3 y, % (95% CI) | 18 (9-29) | 11 (3-23) | ||
| OS | .03 | 0.68 (0.48, 0.96) | ||
| Median, y | 1.2 | 0.8 | ||
| Alive at 3 y, % (95% CI) | 23 (14-33) | 10 (4-18) | ||
| EFS | .004 | 0.50† (0.33, 0.76) | ||
| Median, y | 0.7 | 0.2 | ||
| Event-free at 3 y, % (95% CI) | 14 (7-23) | 6 (2-13) | ||
| Patients 60-69 y | n = 44 | n = 35 | ||
| CR rate, no. (%) | 35 (80) | 19 (54) | .03 | 3.28 (1.22, 8.81) |
| DFS | .82 | 0.94 (0.52, 1.69) | ||
| Median, y | 0.7 | 0.5 | ||
| Disease-free at 3 y, % (95% CI) | 9 (2-21) | 11 (2-28) | ||
| OS | .44 | 0.83 (0.52, 1.33) | ||
| Median, y | 1.1 | 0.8 | ||
| Alive at 3 y, % (95% CI) | 18 (9-31) | 11 (4-24) | ||
| EFS | .11 | 0.69 (0.44, 1.10) | ||
| Median, y | 0.6 | 0.2 | ||
| Event-free at 3 y, % (95% CI) | 7 (2-17) | 6 (1-17) | ||
| Patients 70 y or older | n = 26 | n = 35 | ||
| CR rate, no. (%) | 21 (81) | 18 (51) | .03 | 3.97 (1.22, 12.90) |
| DFS | .09 | 0.54 (0.27, 1.10) | ||
| Median, y | 1.3 | 0.7 | ||
| Disease-free at 3 y, % (95% CI) | 33 (15-53) | 11 (2-30) | ||
| OS | .006 | 0.46 (0.26, 0.81) | ||
| Median, y | 2.0 | 0.9 | ||
| Alive at 3 y, % (95% CI) | 31 (15-49) | 9 (2-21) | ||
| EFS | .007 | 0.48 (0.27, 0.84) | ||
| Median, y | 1.0 | 0.3 | ||
| Event-free at 3 y, % (95% CI) | 27 (12-44) | 6 (1-17) |
OR indicates the odds of achieving a CR for low MN1 vs high MN1 expressers; HR, the hazard of having an event for low MN1 vs high MN1 expressers; and CI, confidence interval.
P values for categorical variables are from Fisher exact test. P values for time-to-event variables are from the log-rank test (OS, DFS, and EFS).
Does not meet the proportional hazards assumption, HR reported at 3 months.
In a multivariable model for CR, MN1 expression was a strong prognostic factor (P = .01), when controlling for BAALC expression (P < .001) and WBC (P = .01; Table 3). In a multivariable analysis for EFS, MN1 expression remained prognostic (P = .03), after adjustment for BAALC expression (P = .002), WBC (P < .001), and platelets (P = .002; Table 3). The risk of having an event (ie, induction failure, relapse, or death) for low MN1 expressers was half that for high expressers (hazard ratio [HR] = 0.54; 95% CI, 0.34-0.86). However, MN1 expression did not remain an important predictor in a multivariable model for OS.
Multivariable regression analysis for outcome according to the MN1 expression status in all older patients with de novo CN-AML, and in those 70 years of age or older
| End point . | Variables in final models . | OR/HR . | 95% CI . | P . |
|---|---|---|---|---|
| All patients | ||||
| CR* | MN1, low vs high | 3.16 | 1.29, 7.70 | .01 |
| BAALC, low vs high | 4.39 | 1.85, 10.44 | < .001 | |
| WBC, each 2-fold increase | 0.76 | 0.62, 0.93 | .01 | |
| EFS† | MN1, low vs high | 0.54 | 0.34, 0.86 | .03¶ |
| BAALC, low vs high | 0.43 | 0.27, 0.68 | .002¶ | |
| WBC, each 2-fold increase | 1.22 | 1.09, 1.35 | < .001¶ | |
| Platelets, each 50-unit increase | 1.15 | 1.05, 1.25 | .002 | |
| Patients 70 y or older | ||||
| CR‡ | MN1, low vs high | 3.97 | 1.22, 12.90 | .02 |
| OS§ | Interaction of MN1 and NPM1 | .29 | ||
| Mutated NPM1: | 0.41 | 0.17, 0.97 | .04 | |
| MN1, low vs high | ||||
| Wild-type NPM1: | .58 | |||
| MN1, low vs high | ||||
| FS‖ | Interaction of MN1 and NPM1 | .14 | ||
| Mutated NPM1: | 0.37 | 0.15, 0.88 | .02 | |
| MN1, low vs high | ||||
| Wild-type NPM1: | .87 | |||
| MN1, low vs high | ||||
| End point . | Variables in final models . | OR/HR . | 95% CI . | P . |
|---|---|---|---|---|
| All patients | ||||
| CR* | MN1, low vs high | 3.16 | 1.29, 7.70 | .01 |
| BAALC, low vs high | 4.39 | 1.85, 10.44 | < .001 | |
| WBC, each 2-fold increase | 0.76 | 0.62, 0.93 | .01 | |
| EFS† | MN1, low vs high | 0.54 | 0.34, 0.86 | .03¶ |
| BAALC, low vs high | 0.43 | 0.27, 0.68 | .002¶ | |
| WBC, each 2-fold increase | 1.22 | 1.09, 1.35 | < .001¶ | |
| Platelets, each 50-unit increase | 1.15 | 1.05, 1.25 | .002 | |
| Patients 70 y or older | ||||
| CR‡ | MN1, low vs high | 3.97 | 1.22, 12.90 | .02 |
| OS§ | Interaction of MN1 and NPM1 | .29 | ||
| Mutated NPM1: | 0.41 | 0.17, 0.97 | .04 | |
| MN1, low vs high | ||||
| Wild-type NPM1: | .58 | |||
| MN1, low vs high | ||||
| FS‖ | Interaction of MN1 and NPM1 | .14 | ||
| Mutated NPM1: | 0.37 | 0.15, 0.88 | .02 | |
| MN1, low vs high | ||||
| Wild-type NPM1: | .87 | |||
| MN1, low vs high | ||||
OR > 1 (< 1) indicates higher (lower) CR rate for the higher values of the continuous variables and the first category listed for the categorical variables. HR > 1 (< 1) indicates higher (lower) risk for an event for the higher values of the continuous variables and the first category listed for the categorical variables.
Variables considered in the model based on univariable analyses were MN1 expression (high vs low; median cut), BAALC expression (high vs low; median cut), FLT3-ITD (positive vs negative), IDH2 (mutated vs wild-type), NPM1 (mutated vs wild-type), WBC (continuous, log base 2), and platelets (continuous, 50-unit increase).
Variables considered in the model based on univariable analyses were MN1 expression (high vs low; median cut), BAALC expression (high vs low; median cut), ERG expression (high vs low; median cut), FLT3-ITD (positive vs negative), IDH2 (mutated vs wild-type), NPM1 (mutated vs wild-type), WT1 (mutated vs wild-type), WBC (continuous, log base 2), and platelets (continuous, 50-unit increase).
Variables considered in the model based on univariable analyses were MN1 expression (high vs low; median cut), BAALC, expression (high vs low; median cut), platelets (continuous, 50-unit increase), and NPM1 (mutated vs wild-type).
Variables considered in the model based on univariable analyses were MN1 expression (high vs low; median cut), BAALC, expression (high vs low; median cut), IDH1 (mutated vs wild-type), NPM1 (mutated vs wild-type), and platelets.
Variables considered in the model based on univariable analyses were MN1 expression (high vs low; median cut), BAALC, expression (high vs low; median cut), IDH2 (mutated vs wild-type), NPM1 (mutated vs wild-type), and platelets.
Does not meet the proportional hazards assumption. For EFS, the HR for BAALC, high vs low (median cut), MN1, high vs low (median cut), and WBC are reported at 3 months.
Prognostic impact of MN1 expression by 60 to 69 years of age and 70 years of age or older subgroups
We recently reported that the prognostic significance of FLT3-ITD and NPM1 mutations in older adults differed between patients 60-69 years of age and those 70 years of age or older, with the adverse impact of FLT3-ITD being found mostly in the former11 and the favorable impact of NPM1 mutations in the latter.9 Therefore, we analyzed the prognostic impact of MN1 expression in these 2 age subgroups (Table 2). Low MN1 expression was associated with higher CR rates both in patients 60-69 years of age (80% vs 54%, P = .03) and in those 70 years of age or older (81% vs 51%, P = .03). In contrast, a significantly longer OS (P = .006; 3-year OS rates, 31% vs 9%) and EFS (P = .007; 3-year EFS rates, 27% vs 6%) and a trend toward longer DFS (P = .09; 3-year DFS rates, 33% vs 11%) were observed only in patients 70 years of age and older (Table 2).
MN1 expression status remained independently associated with probability of achieving a CR for both age subgroups (60-69 years of age, P = .02, data not shown; 70 years of age or older, P = .02, Table 3), with no other variable remaining in the final model. Concerning patients 70 years of age or older (Table 3), low MN1 expressers had almost 4 times greater odds of attaining a CR (odds ratio [OR] = 3.97; 95% CI, 1.22-12.90; Table 3). When we considered OS and EFS in this age group, we found an interaction between MN1 expression and NPM1 mutation status. The favorable impact of low MN1 expression on OS and EFS appeared to be limited to patients who simultaneously carried an NPM1 mutation (P = .04 and P = .02, respectively; Table 3), whereas there was no significant difference in OS or EFS between low and high MN1 expressers with wild-type NPM1 (P = .58 and P = .87, respectively; Table 3).
Taking into account the aforementioned OS and EFS interaction and the fact that we previously reported that the impact of NPM1 mutations on outcome was much stronger in the 70 years of age or older subgroup,9 we examined the relationship between NPM1 mutation status and MN1 expression status within this patient subgroup more closely. Among CN-AML patients 70 years of age or older, those with NPM1 mutations who had low MN1 expression had a trend for better CR rates (P = .15) and significantly longer DFS (P = .003), OS (P = .002; Figure 2A), and EFS (P = .002; Figure 2B) compared with the 3 other molecular subsets combined (ie, low MN1 expressers with wild-type NPM1, high MN1 expressers with mutated NPM1, and high MN1 expressers with wild-type NPM1).
Outcome of CN-AML patients 70 years of age or older with respect to MN1 expression and NPM1 mutation status. (A) OS. (B) EFS.
Outcome of CN-AML patients 70 years of age or older with respect to MN1 expression and NPM1 mutation status. (A) OS. (B) EFS.
Prognostic impact of MN1 expression within the ELN classification
Recently, the European LeukemiaNet (ELN) guidelines classified CN-AML patients into Favorable or Intermediate-I genetic groups based on the mutational status of the CEBPA, NPM1, and FLT3 genes.8 The ELN Favorable genetic group is composed of CN-AML patients with CEBPA mutation and/or NPM1 mutation without FLT3-ITD, whereas the Intermediate-I genetic group encompasses all other CN-AML patients (ie, CN-AML patients with wild-type CEBPA and either NPM1 mutation with FLT3-ITD or wild-type NPM1 with or without FLT3-ITD). We thus investigated the prognostic impact of MN1 expression within these ELN genetic groups. Among the 140 patients, 66 were in the Favorable genetic group and 74 in the Intermediate-I genetic group. Lower MN1 expression levels were found more frequently in the Favorable than Intermediate-I group patients (57% vs 43%, P = .03; Table 1).
Within the ELN Favorable group, we observed no significant differences in CR rates (P = .24), DFS (P = .84), OS (P = .81), or EFS (P = .69) between low and high MN1 expressers (Table 4). In contrast, within the Intermediate-I genetic group, CN-AML patients with low MN1 expression had better CR rates (77% vs 43%, P = .008), a trend toward longer DFS (P = .15; 3-year DFS rates, 13% vs 0%), and significantly longer OS (P = .05; 3-year OS rates, 10% vs 2%) and EFS (P = .003; 3-year EFS rates, 10% vs 0%; Table 4). The CR rate of 77% in patients with low MN1 expression in the Intermediate-I genetic group was comparable to CR rates of patients with both low and high MN1 expression in the ELN Favorable group (83% and 69%, respectively). Likewise, the EFS of low MN1 expressers in the Intermediate-I genetic group was not significantly different from the EFS of patients in the ELN Favorable genetic group (Table 4; Figure 3B).
Outcomes according to MN1, expression in older CN-AML within the ELN genetic groups
| End point . | All . | Low MN1 . | High MN1 . | P* . | OR/HR (95% CI) . |
|---|---|---|---|---|---|
| ELN Favorable group | n = 66 | n = 40 | n = 26 | ||
| CR, no. (%) | 51 (77) | 33 (83) | 18 (69) | .24 | 2.10 (0.65, 6.72) |
| DFS | .84 | ||||
| Median, y | 0.9 | 1.0 | 0.7 | 1.07 (0.56, 2.03) | |
| Disease-free at 3 y, % (95% CI) | 22 (12-34) | 21 (9-36) | 22 (7-43) | ||
| OS | .81 | 0.94 (0.54, 1.61) | |||
| Median, y | 1.5 | 1.5 | 1.4 | ||
| Alive at 3 y, % (95% CI) | 29 (18-40) | 32 (19-47) | 23 (9-40) | ||
| EFS | .69 | 0.90 (0.53, 1.53) | |||
| Median, y | 0.8 | 0.9 | 0.6 | ||
| Event-free at 3 y, % (95% CI) | 17 (9-27) | 18 (8-31) | 15 (5-31) | ||
| ELN Intermediate-I group | n = 74 | n = 30 | n = 44 | ||
| CR, no. (%) | 42 (57) | 23 (77) | 19 (43) | .008 | 4.32 (1.54, 12.17) |
| DFS | .15 | 0.63 (0.33, 1.20) | |||
| Median, y | 0.5 | 0.6 | 0.5 | ||
| Disease-free at 3 y, % (95% CI) | 7(2-17) | 13 (3-30) | 0 (NA) | ||
| OS | .05 | 0.61 (0.38, 1.00) | |||
| Median, y | 0.7 | 0.9 | 0.7 | ||
| Alive at 3 y, % (95% CI) | 5 (2-12) | 10 (3-24) | 2 (.1-10) | ||
| EFS | .003 | 0.49 (0.30, 0.80) | |||
| Median, y | 0.3 | 0.6 | 0.2 | ||
| Event-free at 3 y, % (95% CI) | 4 (1-10) | 10 (3-24) | 0 (NA) |
| End point . | All . | Low MN1 . | High MN1 . | P* . | OR/HR (95% CI) . |
|---|---|---|---|---|---|
| ELN Favorable group | n = 66 | n = 40 | n = 26 | ||
| CR, no. (%) | 51 (77) | 33 (83) | 18 (69) | .24 | 2.10 (0.65, 6.72) |
| DFS | .84 | ||||
| Median, y | 0.9 | 1.0 | 0.7 | 1.07 (0.56, 2.03) | |
| Disease-free at 3 y, % (95% CI) | 22 (12-34) | 21 (9-36) | 22 (7-43) | ||
| OS | .81 | 0.94 (0.54, 1.61) | |||
| Median, y | 1.5 | 1.5 | 1.4 | ||
| Alive at 3 y, % (95% CI) | 29 (18-40) | 32 (19-47) | 23 (9-40) | ||
| EFS | .69 | 0.90 (0.53, 1.53) | |||
| Median, y | 0.8 | 0.9 | 0.6 | ||
| Event-free at 3 y, % (95% CI) | 17 (9-27) | 18 (8-31) | 15 (5-31) | ||
| ELN Intermediate-I group | n = 74 | n = 30 | n = 44 | ||
| CR, no. (%) | 42 (57) | 23 (77) | 19 (43) | .008 | 4.32 (1.54, 12.17) |
| DFS | .15 | 0.63 (0.33, 1.20) | |||
| Median, y | 0.5 | 0.6 | 0.5 | ||
| Disease-free at 3 y, % (95% CI) | 7(2-17) | 13 (3-30) | 0 (NA) | ||
| OS | .05 | 0.61 (0.38, 1.00) | |||
| Median, y | 0.7 | 0.9 | 0.7 | ||
| Alive at 3 y, % (95% CI) | 5 (2-12) | 10 (3-24) | 2 (.1-10) | ||
| EFS | .003 | 0.49 (0.30, 0.80) | |||
| Median, y | 0.3 | 0.6 | 0.2 | ||
| Event-free at 3 y, % (95% CI) | 4 (1-10) | 10 (3-24) | 0 (NA) |
OR indicates the odds of achieving a CR for low MN1 vs high MN1 expressers; HR, the hazard of having an event for low MN1 vs high MN1 expressers; CI, confidence interval; and NA, not applicable (CI could not be attained).
P values for categorical variables are from Fisher exact test. P values for time-to-event variables are from the log-rank test (OS, DFS, and EFS).
Outcome of CN-AML patients 60 years of age or older with respect to MN1 expression within the ELN Favorable and ELN Intermediate-I genetic groups. (A) OS. (B) EFS.
Outcome of CN-AML patients 60 years of age or older with respect to MN1 expression within the ELN Favorable and ELN Intermediate-I genetic groups. (A) OS. (B) EFS.
Because each of the ELN genetic groups is composed of specific molecular subsets, the ELN guidelines recommend reporting outcome measures also by these specific subsets. There was no impact of MN1 expression on either of the 2 CN-AML molecular subsets within the ELN Favorable genetic group (data not shown). The situation was different when we analyzed the impact of MN1 expression in the 3 molecular subsets composing the Intermediate-I genetic group. As seen in supplemental Table 1, all patients in the subset characterized by wild-type NPM1 genes and the presence of FLT3-ITD had high MN1 expression, thus precluding assessment of the prognostic significance of MN1 expression in this subset. Of the remaining 2 subsets, significant differences in CR rates, OS, and EFS between the low and high MN1 expressers were observed only among patients with mutated NPM1 who harbored FLT3-ITD, whereas these outcome measures did not differ significantly in the subset encompassing patients with wild-type NPM1 and no LT3-ITD (supplemental Table 1). However, because the numbers of patients in each subset composing the ELN Intermediate-I genetic group were relatively small (29, 11, and 34 patients, respectively), our analyses should be considered preliminary and of a descriptive nature, and the results have to be confirmed by larger studies.
Genome-wide gene-expression profiling
To gain insights into the biology of older CN-AML patients differentially expressing MN1, we derived a genome-wide gene expression signature. The MN1-associated gene expression signature consisted of 507 probe sets, representing 323 annotated genes (Figure 4).
Heat map of the derived gene expression signature associated with MN1 expression in the group of CN-AML patients 60 years of age or older. The patients are ordered from left to right by increasing expression of MN1. Expression values of the probe sets are represented by color: blue represents expression less than the median value for the given probe set; and red, expression greater than the median value for the given probe set. Up- and down-regulated genes that are mentioned in the text are indicated along the side.
Heat map of the derived gene expression signature associated with MN1 expression in the group of CN-AML patients 60 years of age or older. The patients are ordered from left to right by increasing expression of MN1. Expression values of the probe sets are represented by color: blue represents expression less than the median value for the given probe set; and red, expression greater than the median value for the given probe set. Up- and down-regulated genes that are mentioned in the text are indicated along the side.
In low MN1 expressers, 258 probe sets, representing 158 genes, were found underexpressed, and 249 probe sets, representing 164 genes, overexpressed compared with high MN1 expressers. The probe set representing MN1 was among the most underexpressed probe sets in the low MN1-expressing patients, corroborating the quantification of MN1 expression obtained by real-time RT-PCR (Figure 4). All microarray gene expression data are available on ArrayExpress under accession number E-TABM-1189.
Consistent with our observation in younger patients,18 patients with low MN1 expression had lower expression of genes previously associated with worse outcome in AML, such as BAALC,19,29,30 the surface marker CD200,35 the growth factor HGF,36 and CD34, as well as the adhesion molecule CD44, a key regulator of AML leukemic stem cells necessary for the stem cells to interact with their microenvironment (Figure 4).37,38 We also observed lower expression of ABCB1 (MDR1), a gene encoding the multidrug resistance protein, whose high expression also has been associated with worse outcome in older AML patients.39 Furthermore, patients with low MN1 expression had lower expression of AKT3, a member of the AKT kinase family, which has a central role in cell proliferation, survival, and drug resistance in AML,40 and of the transcription factor STAT5B. Indeed, Heuser et al41 previously showed that STAT5 signaling is critical for leukemia stem cell self-renewal in an MN1 and HOXA9-expressing leukemia model.
Highly expressed in low MN1-expressing patients were the HOXA and HOXB cluster genes, as well as the HOX cofactor MEIS1, which are important for developmental processes and hematopoietic stem cell function (Figure 4).42 We also observed higher expression of the tumor suppressor TP53BP2, which is known to interact with and inhibit the antiapoptotic protein BCL2.43
Genome-wide microRNA expression profiling
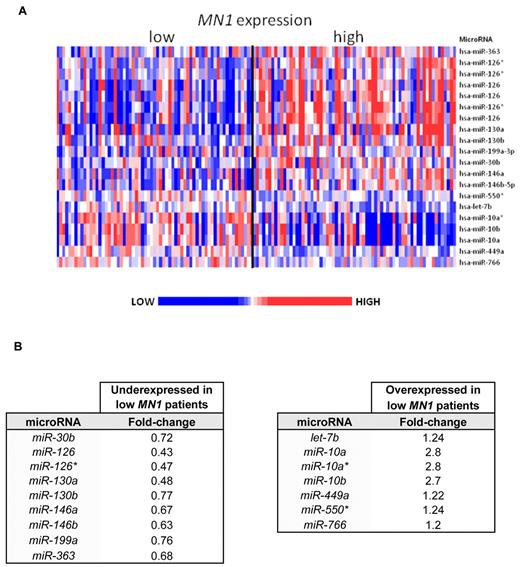
To further elucidate the biologic features associated with low MN1 expression, we derived a microRNA expression signature. The MN1-associated microRNA expression signature was composed of 20 probes (Figure 5), 13 of which, representing 9 microRNAs, were underexpressed, and 7, representing 7 microRNAs, overexpressed in low MN1 expressers compared with high MN1 expressers. All microRNA data are available on ArrayExpress under accession number E-TABM-1190.
Heat map and table of the derived microRNA expression signature associated with MN1 expression in the group of CN-AML patients 60 years of age or older. (A) Heat map of the derived microRNA expression signature associated with MN1 expression in the group of CN-AML patients 60 years of age or older. The patients are ordered from left to right by increasing expression of MN1. Expression values of the probe sets are represented by color: blue represents expression less than the median value for the given probe; and red, expression greater than the median value for the given probe. Up- and down-regulated genes and microRNAs are indicated along the side. (B) Tables showing fold changes of the microRNAs that form the signature with corresponding fold changes.
Heat map and table of the derived microRNA expression signature associated with MN1 expression in the group of CN-AML patients 60 years of age or older. (A) Heat map of the derived microRNA expression signature associated with MN1 expression in the group of CN-AML patients 60 years of age or older. The patients are ordered from left to right by increasing expression of MN1. Expression values of the probe sets are represented by color: blue represents expression less than the median value for the given probe; and red, expression greater than the median value for the given probe. Up- and down-regulated genes and microRNAs are indicated along the side. (B) Tables showing fold changes of the microRNAs that form the signature with corresponding fold changes.
In the low MN1-expressing patients, we found miR-126 and its passenger strand miR-126* among the most underexpressed microRNAs, which is consistent with our previous findings in younger patients.18 In addition, underexpressed were miR-146a, low expression of which has been associated with the 5q− syndrome,44 and miR-146b. Furthermore, we observed a lower expression of miR-30b, whose amplification and overexpression have been linked to medulloblastoma.45
Consistent with the higher expression of HOX genes in low MN1 expressers, we observed higher expression of the HOX-gene embedded microRNAs miR-10a and miR-10b. We also observed higher expression of let-7b, a member of a known tumor-suppressor microRNA family, which has been found down-regulated in AML with favorable cytogenetics [ie, t(8;21), inv(16) and t(15;17)].46 We observed higher expression of miR-449a, shown to target HDAC1 and induce growth arrest in prostate cancer.47
Discussion
The majority of patients with AML are older than 60 years at diagnosis. Although our knowledge of molecular prognostic markers is most extensive in younger CN-AML patients,7 recently there has been progress in our understanding of the role molecular alterations play in prognostication of older patients.9-12,27,28 The main objective of this study was to elucidate the prognostic impact of MN1 expression in older CN-AML patients, and to determine whether this knowledge can be integrated into the landscape of other established molecular markers.
We demonstrate here that CN-AML patients 60 years of age or older with low MN1 expression have higher CR rates and that their OS and EFS are longer than those of patients with high MN1 expression. However, we did not observe a significant difference in DFS, which is somewhat different from our findings in younger patients, where low MN1 expression associated with higher CR rates and longer DFS, OS, and EFS.18 This discrepancy might be related to the differences in the intensity of treatment regimens administered to the younger and older patients. As in younger CN-AML patients, we observed an association of low MN1 expression with mutated NPM1 and lower BAALC expression. In multivariable analyses, MN1 expresser status remained a significant prognosticator for CR attainment, even in the context of other molecular markers, including NPM1 mutation and BAALC expresser status. Indeed, the expresser status of MN1 and BAALC were the only molecular markers associated with CR achievement in the patients investigated in the current study. We did not find an independent impact of MN1 expression status on OS in the entire cohort of older patients that we analyzed, in contrast to younger patients.18 This might be accounted for by differences in disease biology but might also be related to the lower intensity of postremission treatment administered to older AML patients compared with younger patients.
To our knowledge, only one recent study of the prognostic significance of MN1 expression in CN-AML included patients 60 years of age or older among those analyzed.48 This study showed that high MN1 expression was associated with lower probability of CR achievement and shorter relapse-free survival, OS, and EFS.48 However, in contrast to our findings, MN1 expression was not an independent prognostic factor in the entire cohort of 210 patients analyzed by Metzeler et al,48 and the outcome data were not reported separately for a subgroup of 101 patients 60 years of age or older.48
Our group recently reported age-related differences with respect to the impact on outcome of 2 molecular markers in older CN-AML patients. We found a stronger impact of NPM1 mutations in patients 70 years of age or older as opposed to those 60-69 years of age,9 and a stronger impact of FLT3-ITD in patients 60-69 years of age as opposed to those 70 years of age and older.11 The current study provides evidence that the prognostic impact of MN1 expression is also influenced by the patients' age. Although low MN1 expressers in both age subgroups had higher probability of achieving a CR, MN1 expression was prognostic with respect to OS and EFS only in the subgroup 70 years of age or older. Moreover, our data suggest that our finding of better outcome associated with NPM1 mutations in patients 70 years of age or older does not pertain to all such patients but mostly to those who, in addition to NPM1 mutation, have low MN1 expression. Consequently, if our findings are confirmed and a standardized method of MN1 expression quantification is established, testing for both NPM1 mutations and MN1 expression could be recommended to achieve the best prognostic stratification of CN-AML patients 60 years of age or older. The reasons for the age-related differences in the impact of MN1 expression, NPM1 mutations, or FLT3-ITD on outcome of older patients remain unknown.
Recently, the ELN expert panel proposed a novel risk classification for AML based on cytogenetics and molecular markers.8 Within this classification, CN-AML patients are assigned to Favorable or Intermediate-I genetic groups based on the mutational status of the CEBPA, NPM1, and FLT3 genes.8 To evaluate whether determination of MN1 expression levels can improve this classification, we analyzed the prognostic significance of MN1 expression status separately within the ELN Favorable and the Intermediate-I genetic groups of CN-AML. In our patient cohort, MN1 expression did not impact on outcome of the Favorable group. However, we observed a strong impact of MN1 expression status on patients belonging to the Intermediate-I genetic group. Within this group, patients with low expression of MN1 had better outcome than those with high MN1 expression, and their CR rates and EFS were not significantly different from those in the ELN Favorable group. However, DFS and OS of low MN1 expressers in the Intermediate-I genetic group were better than high MN1 expressers, but not comparable with those in the ELN Favorable group. If our findings are confirmed, MN1 expression status might become a molecular marker that will help refine the ELN classification. Furthermore, our analysis of molecular subsets within the Intermediate-I genetic group suggests that patients who benefit most from having low MN1 expression are those who harbor both NPM1 mutation and FLT3-ITD. This finding requires corroboration in a larger set of patients.
The molecular mechanisms by which MN1 contributes to leukemia remain elusive. To gain deeper insights into the biology of the disease, we derived gene- and microRNA-expression signatures associated with MN1 expression. The genome-wide microarray profiling supports the prognostic significance of low MN1 expression levels by demonstrating concurrent underexpression of genes and microRNAs associated with biologic features of aggressive phenotypes. Not surprisingly, we found the gene expression signature associated with MN1 expression derived in older CN-AML patients to be similar to the one we reported in patients younger than 60 years.18 It is known that MN1 expression not only negatively impacts on cell differentiation but also affects chemotherapy response,49 which is in line with our finding of the importance of low MN1 expression for CR achievement. Heuser et al50 previously showed that genes that are associated with undifferentiated hematologic precursor cells also associate with a poor response to induction therapy. Consistently, in both younger and older patients, low MN1 expression was associated with higher CR rates and lower expression of known adverse outcome predictors and of genes involved in chemotherapy resistance, such as ABCB1. These findings may, at least in part, explain the observed, independent association with better treatment response of low MN1 expressers in older CN-AML. We also found overlapping microRNA expression features in younger and older patients with low MN1 expression, including down-regulation of miR-126 and miR-130b, and this may indicate that these microRNAs play an important role in modifying patients' response to therapy.
In conclusion, we show that MN1 expression is an important predictor of treatment response in older de novo CN-AML patients. Prognostic impact of MN1 expression is especially strong in patients 70 years of age or older, and a combination of low MN1 expression and mutated NPM1 identifies a subset of these patients with a particularly good outcome. Furthermore, the gene- and microRNA-expression profiles we derived may help to shed light on the complex biology of MN1-associated disease. Once a standardized method of expression quantification is established (eg, by digital mRNA quantification technologies) and absolute cutpoints are defined, measurements of pretreatment MN1 expression may be included in diagnostic panels and used to improve risk stratification of older CN-AML patients and to guide treatment decisions in clinical trials testing new agents targeting genes, such as ABCB1 or even MN1 itself.
The online version of this article contains a data supplement.
Presented in part at the 101st annual meeting of the American Association for Cancer Research, Washington, DC, April 19, 2010, and published in abstract form.51
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Kelsi B. Holland for statistical support; Donna Bucci of the CALGB Tissue Bank at The Ohio State University Comprehensive Cancer Center for sample processing and storage services; Lisa J. Sterling and Colin G. Edwards for data management; and The Ohio State University Comprehensive Cancer Center's Nucleic Acid and Microarray Shared Resources for technical support.
This work was supported in part by the National Cancer Institute (grants CA101140, CA114725, CA140158, CA31946, CA33601, CA16058, CA77658, and CA129657), the Coleman Leukemia Research Foundation, and the Deutsche Krebshilfe–Dr Mildred Scheel Cancer Foundation (H.B.).
Authorship
Contribution: S.S., G.M., J.K., M.D.R., K. Mrózek, K. Maharry, M.A.C., and C.D.B. contributed to the design and analysis of this study and the writing of this manuscript; S.S., H.B., K.H.M., S.P.W., and Y.-Z.W. carried out laboratory-based research; J.E.K., M.D.R., and K. Maharry performed statistical analyses; G.M., B.L.P., M.R.B., J.E.K., A.J.C., R.A.L., M.A.C., and C.D.B. were involved directly or indirectly in care of patients and/or sample procurement; and all authors agreed on the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Clara D. Bloomfield, The Ohio State University, James Cancer Hospital and Solove Research Institute, 300 West 10th Ave, Columbus, OH 43210; e-mail: clara.bloomfield@osumc.edu; and Michael A. Caligiuri, The Ohio State University, Comprehensive Cancer Center, Cancer Hospital and Solove Research Institute, 300 W 10th Ave, Columbus, OH 43210; e-mail: michael.caligiuri@osumc.edu.
References
Author notes
S.S. and G.M. contributed equally to this study.
M.A.C. and C.D.B. are co-senior authors and contributed equally to this study.