Abstract
The mechanism by which proteins are targeted to neutrophil granules is largely unknown. The intracellular proteoglycan serglycin has been shown to have important functions related to storage of proteins in several types of granules. The possible role of serglycin in the localization of the α-defensin, human neutrophil peptide 1 (HNP-1), a major azurophil granule protein in human neutrophils, was investigated. Murine myeloid cells, stably transfected to express HNP-1, were capable of processing HNP-1, and HNP-1 was found to associate with serglycin in murine and human myeloid cell lines as well as in human bone marow cells. A transgenic mouse expressing HNP-1 in the myeloid compartment was crossed with mice deficient in serglycin or neutrophil elastase to investigate HNP-1 sorting and processing. Neither deficiency affected processing of HNP-1, but the ability to retain fully processed HNP-1 intracellularly was reduced in mice that lack serglycin. Human granulocyte precursors transfected with siRNA against serglycin displayed similar reduced capability to retain fully processed HNP-1, demonstrating a role of serglycin in retaining mature HNP-1 intracellularly, thus preventing potential toxic effects of extracellular HNP-1.
Introduction
Neutrophils are indispensable for our body's defense against microorganisms. Granules of neutrophils contain a variety of proteins that directly or indirectly assist the neutrophil in its microbicidal activities although some of these proteins, such as the proteases neutrophil elastase (NE) and proteinase 3 (PR3), may be harmful to the host. Little is known about the mechanisms that direct proteins from the constitutive secretory pathway into storage organelles such as granules. It has been a long-standing hypothesis that granule proteins are bound to intracellular proteoglycans, which serve not only to sort the proteins into granules, but also to keep the granule proteins, such as proteases, inactive through tight electrostatic binding to the highly negatively charged residues of sulfated proteoglycans.1-4 Some neutrophil proteins have been named for their affinity toward intracellular proteoglycans, for example, heparin-binding protein (also known as azurocidin or cationic antimicrobial protein of 37 kDa [CAP37]). Yet, evidence that proteoglycans bind directly to granule proteins in vivo and play a role in sorting granule proteins to granules is scarce.
We have demonstrated previously that serglycin, the main proteoglycan of hematopoietic cells,2 is absent from mature human neutrophils, but present in the Golgi network and in immature granules of promyelocytes.5 We, among others, have further obtained evidence that serglycin is involved in the sorting of NE to azurophil granules,6,7 and we now examine whether serglycin is involved in targeting the most predominant of the azurophil granule proteins, α-defensins, to granules. α-defensins, also known in neutrophils as human neutrophil peptides (HNPs), are 29- to 34-aa peptides with broad microbial and cytotoxic activities.8 They are generated as propeptides with a 75-aa prosegment having an overall negative charge that is believed to neutralize the highly positively charged C-terminal peptide,9 which is the form stored in azurophil granules of human neutrophils.10 The α-defensins present in Paneth cells of the crypts of Lieberkühn in mice and humans are processed by the serine proteases matrilysin and trypsin, respectively.11,12 The enzyme responsible for processing of neutrophil α-defensins in vivo is currently unknown, but in vitro studies have shown neutrophil α-defensin to be fully processed by NE—and partially processed by cathepsin G and PR3.13 We have previously shown that α-defensins are expressed in both promyelocytes and myelocytes, but only processed to mature α-defensins in promyelocytes.14 With the exception of small amounts of unprocessed α-defensins found in specific granules, only fully processed α-defensins are retained in granules of neutrophils.15,16 Therefore, we investigated whether serglycin might be actively involved in sorting fully mature defensins to granules. Because mouse myeloid cells do not express α-defensins,17 we transfected a mouse myeloid cell line with human prodefensin to ascertain that mouse cells, as previously reported,18 are capable of processing α-defensins. We then investigated the role of serglycin in processing and retention of α-defensin by crossing serglycin knockout (ko) mice with a transgenic mouse strain that expresses human α-defensins in the myeloid compartment. Finally, a human promyelocytic cell line and human primary bone marow (BM) cells were transfected with small interfering RNA (siRNA) against serglycin.
Methods
Cell culture
32D Cl3 (CRL-11346; ATCC), PLB-985 (a kind gift from Dr Peter Newberger, University of Massachusetts Medical School, Worcester, MA), and HL-60 cells (a kind gift from Dr Francois Boulay, Laboratoire de Biochimie et Biophysique des Systèmes Intégrés CEA, Grenoble, France) were cultured in RPMI 1640 medium with glutamax, 10% FCS or 20% FCS (PLB-985), 100 U/mL penicillin, and 100 μg/mL streptomycin (all from Invitrogen) in a humidified incubator with 5% CO2 at 37°C. 32D Cl3 culture was added 1 ng/mL murine IL-3 (Sigma-Aldrich).
Real-time quantitative PCR
RNA isolation and cDNA synthesis were performed as previously described.19 cDNA was subjected to quantitative real-time PCR analysis using TaqMan gene expression assays (Applied Biosystems) on a 7500 Real-Time PCR system, according to the manufacturer's instructions. Assays included: DEFA1 (forward primer: 5′-AGTATGGAGTTAGCAGTGGGTAAGA-3′; reverse primer 5′-CGTGCTAACGGCTGCAATG-3′; FAM reporter primer: 5′-CAGCCAGCATCACCTG-3′) for measurement of genomic HNP-1 in transgenic mice, HNP-1 (Hs00234383_m1), serglycin (Hs00160444_m1), myeloperoxidase (MPO; Mm00447875_g1), neutrophil gelatinase-associated lipocalin (Mm00809552_s1), and matrix metalloproteinase-9 (Mm00600164_g1). Expression levels were normalized to the constitutively expressed housekeeping mouse gene Gapdh (4352339E) or human gene GAPDH (4326317E).
Transfection
Human cDNA encoding HNP-1 (pre-proHNP-1; primers: 5′-GCGGGATCCGCCACCATGAGGACCCTCGCCATCC-3′ and 5′-GCGTCTAGATCAGCAGCAGAATGCCCAG-3′), NE (primers: 5′-ATGGTACCGCCACCATGACCCTCGGCCGCCG-3′ and 5′-ATTCTAGATCAGTGGGTCCTGCTGGC-3′), and PR3 (primers: 5′-ATGGTACCGCCACCATGGCTCACCGGCCCCC-3′ and 5′-ATTCTAGAATCAGGGGCGGCCCTTGG-3′) was amplified by PCR using cDNA from myeloblasts/promyelocytes5 as template. The amplification product for HNP-1 was digested with XbaI and BamHI and cloned into the mammalian expression vector pEF6/myc-His A (Invitrogen) digested with the same enzymes. The amplification products for NE and PR3 were cloned into pCR 2.1-TOPO (Invitrogen), digested with KpnI and XbaI, and cloned into the mammalian expression vector pEF1/V5-His A (Invitrogen) digested with the same enzymes. siRNA specific against serglycin mRNA (s11041; Ambion) was cloned into pSilencer 2.1-U6 puro (Ambion) according to manufacturer's instructions. Plasmids were transformed into Escherichia coli (XL1-blue; Stratagene) and purified using the QIAGEN Plasmid Midi Kit. Sequences were verified by sequencing in both directions (Eurofins MWG Operon). The expression vectors were linearized with ScaI and cleaned for endotoxins using the Endofree Plasmid Maxi Kit (QIAGEN). 32D Cl3 cells and HL-60 cells were washed and resuspended in a 7mM Na2HPO4, 272mM sucrose, pH 7.4 buffer. The cells (0.8 mL at 1 × 107 cells/mL) were then mixed with 20 μg of plasmid DNA and subjected to a single electric pulse of 250 V at a capacitance setting of 960 μF (32D Cl3)/125 μF (HL-60) with a time constant of 22 ms in a 0.4-cm electroporation cuvette (Gene Pulser with capacitance extender; Bio-Rad) at room temperature. Cells were transferred to 25 mL of cell-culture medium and allowed to recover at 37°C in an incubator. After 2 days, 80 mL of culture medium and selection antibiotic were added to a final concentration of 20 μg/mL blasticidin (Invitrogen) or 150 μg/mL G418 (Invitrogen). Transfection of PLB-985 and human primary BM cells was done by resuspending 2 × 106 cells in 100 μL of Ingeneo Electroporation Solution (Mirus Bio). Cells were mixed with 2 μg of plasmid DNA or 100 pmol siRNA and electroporated using Program C-023 on an Amaxa Nucleofector (Lonza). Cells were transferred to 5 mL of cell-culture medium and allowed to recover at 37°C in an incubator. After 2 days, 50 mL of culture medium and selection antibiotic were added to a final concentration of 1 μg/mL puromycin (Invivogen). Cells were plated in wells containing 1 mL each and grown under selection pressure for 3 weeks. Wells positive for living cells after 3 weeks were grown in medium supplemented with selection antibiotics.
Antibodies
The following antibodies (Abs) were used: rabbit anti-proHNP,16 rabbit anti-HNP,15 rabbit anti–human serglycin,5 rabbit anti-murine serglycin,6 rabbit anti-NE (in-house Ab prepared by DAKO), rabbit anti-PR3,20 rabbit anti-GAPDH (14C10; Cell Signaling Technology), rabbit control IgG (DAKO), goat anti-murine MPO (L-20; Santa Cruz Biotechnology), goat anti-murine NE (M-18; Santa Cruz Biotechnology), goat anti-PR3 (P-20; Santa Cruz Biotechnology).
Western blotting
SDS-PAGE21 and immunoblotting22 were performed according to standard procedures. A 14% gel with 3% stacking gel was used, and samples were reduced with mercaptoethanol in Laemmli buffer.21 Protein transfer to polyvinylidene difluoride (PVDF) membranes was performed in 10mM 3-(cyclohexylamino)-1-propanesulfonic acid, pH 11.0, 10% methanol. Blots were blocked in 5% skim milk powder in PBS for 1 hour and incubated overnight with primary Ab in PBS/0.5% BSA. Detection was by HRP-conjugated swine anti–rabbit Ab (DAKO) in PBS/0.5% BSA and 1,4-diaminobutane/metal concentrate in stable peroxide substrate buffer (Pierce).
Biosynthesis of sulfated proteoglycans
Cells were washed and incubated at 2 × 107/mL in RPMI 1640 medium with 200 μCi/mL 35S-sulfate (Perkin Elmer) for 3 hours. Cells were pelleted, resuspended in Laemmli buffer,21 subjected to SDS-PAGE on a 4%/12% gradient gel, stained with Coomassie, soaked in Amplify (GE Healthcare), placed in a Fuji BAS cassette (Fuji Film), and developed by a Fuji BAS2500 PhosphorImager.
Pulse-chase biosynthesis
Cells were pelleted and resuspended at 2 × 107 cells/mL in DMEM without L-methionine/L-cysteine (Invitrogen) containing 100 U/mL penicillin, 100 μg/mL streptomycin, and 10% dialyzed FCS, and incubated for 30 minutes at 37°C. The cells were subsequently pelleted by centrifugation, resuspended at 3 × 107 cells/mL in DMEM to which 35S-methionine/cysteine (without L-methionine/L-cysteine, PerkinElmer) had been added to a final concentration of 200 μCi/mL, and pulsed for 1 hour at 37°C. The pulse was stopped by centrifugation, washing the cells twice in RPMI 1640, and resuspending the cells in RPMI 1640 containing 10% dialyzed FCS at a concentration of 2 × 107 cells/mL. After a chase period of 3 hours at 37°C, the cells were pelleted by centrifugation and resuspended to 107 cells/mL in lysis buffer (150mM NaCl, 30mM HEPES, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS). Complete Mini tablets (Roche) and 1mM PMSF (Sigma-Aldrich) were added to cell lysate and medium. Lysate and medium were incubated overnight at 4°C, and undissolved material was pelleted by centrifugation for 30 minutes at 20 000g. Supernatants were immunoprecipitated as previously described23 with Abs coupled to Sepharose beads in the order: rabbit anti-proHNP followed by rabbit anti-HNP. Beads were pelleted, resuspended in Laemmli buffer, subjected to SDS-Tricine-PAGE on a 16% gel,24 stained with Coomassie, soaked in Amplify, placed in a Fuji BAS cassette, and developed by a Fuji BAS2500 PhosphorImager.
Immunocytochemistry
Cytospins treated as previously described25 were incubated with primary Ab. Detection was done with Envison+ System-HRP (K4010; DAKO) using an Olympus BX51 microscope (60×/1.40 PlanApo oil objective) with an Olympus DP70 camera and the analySIS B 5.0 software package. For fluorescence microscopy, cells were treated as previously described.26 DAPI (4,6 diamidino-2-phenylindole; Invitrogen) was used as nuclear counterstain. Images were acquired on a LSM 700 (63×/1.40 Plan-Apochromat oil objective) microscope and analyzed with the Zen 2009 LE software package (Zeiss).
ELISA
Immunoplates (96-well flat-bottom; Nunc) were incubated overnight at room temperature with affinity-purified Ab in 50mM Na2CO3/NaHCO3, pH 9.6. The plates were subsequently washed in buffer A (500mM NaCl, 3mM KCl, 8mM Na2HPO4/KH2PO4, 1% Triton X-100, pH 7.2) and blocked with buffer B (500mM NaCl, 3mM KCl, 8mM Na2HPO4/KH2PO4, 1% BSA, 1% Triton X-100, pH 7.2). Buffer B was discarded, and the plates were incubated for 1 hour with cells washed twice in PBS and solubilized in buffer B, and incubated with goat anti–rabbit Ab or biotinylated rabbit Ab for 1 hour. After an additional wash in buffer A, the wells were incubated for 1 hour with HRP-conjugated rabbit anti–goat Ab (P0449; DAKO) or HRP-conjugated avidin (P0347; DAKO) in buffer B. After washing, the color reaction was developed with Ortho-Phenylene-Diamine tablets (OPD; KEM-EN-TEC), solubilized in buffer C (100mM Na2HPO4, 100mM citric acid, pH 5.0) with the addition of 0.006% H2O2 immediately before use. The color reaction was stopped with 1M H2SO4, and the absorbance at 492 nm was measured with a Multiscan Ascent ELISA reader (Labsystems). Chondroitinase ABC (cABC) treatment was performed in wells of immunoplates as an extra step after incubation with cell lysates. After washing, bound serglycin was digested with 0.4 U/mL cABC (US Biologic) in 40mM Tris-HCl, 40mM Na-acetate, pH 8.0, 0.01% BSA at 37°C for 1.5 hours.
Mice
C57BL/6 was used as background strain. Generation of serglycin ko mice is previously described.27 Frozen embryos from the transgenic HNP-1 mouse28 were obtained from ATCC with the kind permission of Dr Rose Linzmeier (David Geffen School of Medicine, University of California, Los Angeles, CA). The NE-deficient mouse29 was kindly provided by Dr Jürgen Roes (University College London, London, United Kingdom). The HNP-1 mouse was crossed into NE-deficient and serglycin ko mice. Genotyping was done on DNA from tail tips using primers as described.27,29 Levels of HNP-1 in mice were examined by real-time PCR on tail DNA using a TaqMan Custom gene expression assays specific to the genomic sequence for HNP-1 (DEFA1). Breeding and experiments were performed according to permissions and guidelines from the Danish Animal Experiments Inspectorate.
Isolation of murine BM cells
Mice were euthanized and femora, tibiae, and os ilia were removed by dissection and crushed in 14 mL of PBS/1% BSA in a mortar. BM cells were suspended and filtered through a cell strainer with 70-μm nylon mesh (BD Pharmingen). Erythroid cells were lysed in 300 μL of Pharm Lyse buffer (BD Pharmingen) except for biosynthesis studies. Separation of mature and immature myeloid cells was done by laying cells on a Percoll/PBS solution with a density of 1.072 and centrifugation at 1000g for 20 minutes without break. For fluorescence microscopy, cells were depleted of nongranulocytic cells by immunomagnetic sorting using biotinylated Abs against surface epitopes of T cells (CD3e), B cells (CD45r), and erythroid cells (TER-119; all BD Pharmingen) and the magnetic cell sorting (MACS) system according to instructions of the manufacturer (Miltenyi Biotec).
Isolation of human BM cells
Human BM aspirates were obtained according to the permission and guidelines from the local ethics committee of the National University Hospital (KF 01-060/01). For isolation of granulocytic precursors from BM, erythroid cells were sedimented with dextran, supernatant laid on Lymphoprep (Nycomed Pharma), and centrifuged 400g for 30 minutes. Interphase cells were depleted of nongranulocytic cells by immunomagnetic sorting using biotinylated Abs against CD3, CD7, CD14, CD19, CD235a (all eBioscience), CD56 (BioLegend), and CD61 (AbD Serotec) and the MACS system. Myeloblasts/promyelocytes were isolated by 3-layer density centrifugation as previously described.5
Differential count
Blood samples from mice were collected in Microvette potassium-EDTA tubes (Sarstedt) and analyzed on an ADVIA 120 cell counter (Siemens Healthcare Diagnostics). Statistical calculations were performed with PASW Statistics 18.0 (SPSS).
Mass spectrometry
The identity of mature, processed human HNP-1 and proHNP were verified by peptide mass fingerprinting of a tryptic or Glu-C digest of protein from the transfected 32D Cl3 cells immunoprecipitated with rabbit anti-HNP or anti-proHNP and analyzed by SDS-PAGE. The protein band from the Coomassie-stained gel was excised and subjected to in-gel trypsin or Glu-C digestion followed by matrix-assisted laser desorption-ionization (MALDI) mass spectrometry after micropurification30,31 using a Bruker Ultraflex II instrument. Monoisotopic peptide masses were entered into the database allowing S-carboxyamidomethylation and a mass tolerance of 0.2 Da.
Affinity chromatography
proHNP was purified by affinity chromatography as previously described16 from medium of 32D Cl3 cells transfected with proHNP. Eluted protein was concentrated using Amicon Ultra Centrifugal Filter Units (Millipore) according to the manufacturer's instructions. The purity of the eluted material was ascertained by Western blotting and by SDS-PAGE followed by staining with Coomassie Blue, which showed a single band in the expected molecular weight zone.
Results
Processing and sorting of proHNP by mouse myeloid cells
The mouse myeloid cell line 32D Cl3 was transfected with a plasmid construct containing an expression cassette into which the coding sequence of the HNP-1 gene had been inserted. The transfected cells clearly showed reactivity with Abs against HNPs purified from human neutrophil azurophil granules15 and against the recombinant prosegment of HNP-116 (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Pulse-chase biosynthesis demonstrated that 32D Cl3 cells process the proHNP into a small peptide of similar size as HNPs from human neutrophils (supplemental Figure 1B). Mass spectrometry of affinity purified low-molecular-weight immunoreactive peptide isolated from the transfected 32D Cl3 cells showed this to be fully processed human HNP-1 (supplemental Figure 1C). Pulse-chase biosynthesis also demonstrated that significant amounts of unprocessed HNP-1 was found both in cells and in medium—whereas mature (ie, low molecular weight) HNP-1 was predominantly retained intracellularly, demonstrating the ability of the mouse cells to efficiently retain most processed HNP-1 (supplemental Figure 1B) similar to human myeloid cells (see Figure 5C).
Association of serglycin with mature HNP-1
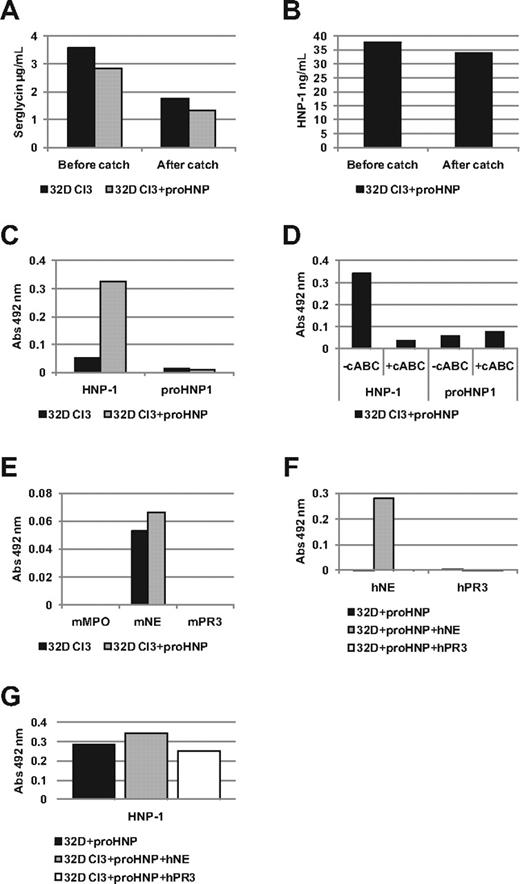
We tested whether serglycin associates with HNP-1 in the 32D Cl3 cells transfected with HNP-1 in a mixed ELISA where serglycin was captured by Ab adsorbed to microtiter plates. Lysates of 32D Cl3 cells transfected with HNP-1 were then added to allow the Ab to bind serglycin present in the wells. Approximately half the serglycin was captured, while only 10% of HNP-1 was removed (Figure 1A-B). The presence of HNP-1 associated with serglycin was probed both with Abs against HNP propeptide, which does not recognize fully processed HNP-1 and with Ab against mature HNPs, which recognizes fully processed HNP-1 and to some extent also unprocessed HNP-1.16 As demonstrated, reactivity was detected against fully mature HNP-1, but not against unprocessed HNP-1 in the material captured by the anti-serglycin Ab (Figure 1C). This indicates that serglycin binds mature HNP-1, but not proHNP. Treatment of cell lysates with cABC eliminated this reactivity (Figure 1D) suggesting that this association relies on serglycin's chondroitin sulfate sidechains. We also probed with Abs against MPO and NE, 2 azurophil granule proteins previously suggested to associate with serglycin,1,6,7 as well as with an Ab against PR3 that has not been found to associate with serglycin (Figure 1E). Of these 3, we were only able to detect relatively weak reactivity with an Ab against murine NE, which was poorly expressed in our 32D Cl3 cells (data not shown). Cells (32D Cl3) already transfected with HNP-1 were transfected with a plasmid expressing either human NE or PR3 to test whether high expression of these proteins would yield a stronger reactivity. Under these conditions, we found a strong signal against human NE in 32D Cl3 cells expressing human NE, but not in control cells (Figure 1F). Probing for PR3 did not yield any signal in 32D Cl3 cells transfected with PR3. Transfection of 32D Cl3 cells with human NE did not displace reactivity when probing for HNP-1 (Figure 1G).
Association of serglycin with fully processed HNP. Immunoplates were incubated with affinity purified Ab. Plates were washed, blocked, and incubated with wild-type or transfected 32D Cl3 cell lysates. (A-B) To assess the amount of serglycin and HNP-1 bound by the anti-serglycin Ab in the mixed ELISA, serglycin and HNP-1 was measured by sandwich ELISA in lysates of 32D Cl3 cells and 32D Cl3 cells stably transfected with proHNP (32D Cl3+proHNP) before and after catching with anti-serglycin Ab. (C-G) Lysates of 32D Cl3 cells were subjected to a mixed ELISA. Serglycin was captured with affinity purified anti-serglycin Ab. Association of a protein with serglycin was probed with a goat or biotinylated rabbit Ab. After incubation with HRP-conjugated rabbit anti–goat Ab or HRP-conjugated avidin, a color reaction was developed using OPD tablets. Color reaction was measured at an absorbance (Abs) of 492 nm and background reactivity subtracted (absorbance in wells with lysis buffer without cells). Absorbance points to an association of a protein with serglycin. (C) Mixed ELISA on lysates of 32D Cl3 cells and 32D Cl3+proHNP using a biotinylated Ab against proHNP, which does not recognize fully processed HNP-1, and a biotinylated Ab against mature HNPs which recognizes fully processed HNP-1. (D) Previous ELISA was repeated, but with cABC treatment as an extra step after incubation with cell lysates. Note that cABC treatment abolished reactivity for the cationic HNP-1 indicating that reactivity is dependent on serglycin's anionic chondroitin sulfate sidechains. (E) Other potential binding candidates of serglycin were tested on a mixed ELISA on lysates of 32D Cl3 cells and 32D Cl3+proHNP using Abs against murine myeloperoxidase (MPO), neutrophil elastase (mNE), and proteinase 3 (mPR3). (F) 32D Cl3+proHNP cells were further transfected with human neutrophil elastase (hNE) or proteinase 3 (hPR3) to test whether signal could be enhanced by greater expression. Probing was done with Abs raised against hNE or hPR3. (G) Serglycin HNP-1 association was probed by mixed ELISA in 32D Cl3 cells transfected with HNP-1 and hNE or hPR3 to test whether high expression of hNE could displace HNP-1.
Association of serglycin with fully processed HNP. Immunoplates were incubated with affinity purified Ab. Plates were washed, blocked, and incubated with wild-type or transfected 32D Cl3 cell lysates. (A-B) To assess the amount of serglycin and HNP-1 bound by the anti-serglycin Ab in the mixed ELISA, serglycin and HNP-1 was measured by sandwich ELISA in lysates of 32D Cl3 cells and 32D Cl3 cells stably transfected with proHNP (32D Cl3+proHNP) before and after catching with anti-serglycin Ab. (C-G) Lysates of 32D Cl3 cells were subjected to a mixed ELISA. Serglycin was captured with affinity purified anti-serglycin Ab. Association of a protein with serglycin was probed with a goat or biotinylated rabbit Ab. After incubation with HRP-conjugated rabbit anti–goat Ab or HRP-conjugated avidin, a color reaction was developed using OPD tablets. Color reaction was measured at an absorbance (Abs) of 492 nm and background reactivity subtracted (absorbance in wells with lysis buffer without cells). Absorbance points to an association of a protein with serglycin. (C) Mixed ELISA on lysates of 32D Cl3 cells and 32D Cl3+proHNP using a biotinylated Ab against proHNP, which does not recognize fully processed HNP-1, and a biotinylated Ab against mature HNPs which recognizes fully processed HNP-1. (D) Previous ELISA was repeated, but with cABC treatment as an extra step after incubation with cell lysates. Note that cABC treatment abolished reactivity for the cationic HNP-1 indicating that reactivity is dependent on serglycin's anionic chondroitin sulfate sidechains. (E) Other potential binding candidates of serglycin were tested on a mixed ELISA on lysates of 32D Cl3 cells and 32D Cl3+proHNP using Abs against murine myeloperoxidase (MPO), neutrophil elastase (mNE), and proteinase 3 (mPR3). (F) 32D Cl3+proHNP cells were further transfected with human neutrophil elastase (hNE) or proteinase 3 (hPR3) to test whether signal could be enhanced by greater expression. Probing was done with Abs raised against hNE or hPR3. (G) Serglycin HNP-1 association was probed by mixed ELISA in 32D Cl3 cells transfected with HNP-1 and hNE or hPR3 to test whether high expression of hNE could displace HNP-1.
Processing and sorting of HNP-1 in vivo
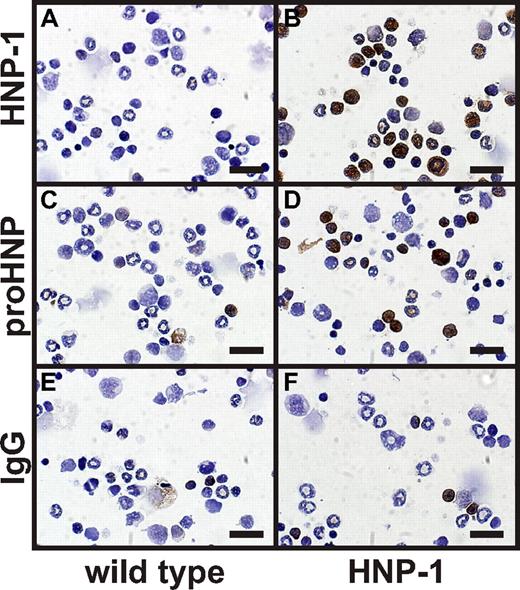
To directly address whether serglycin is involved in retention of HNP-1 in vivo, we crossed a transgenic mouse expressing HNP-1 in the myeloid compartment with the serglycin ko mouse and examined the ability to process HNP-1 and to sort fully processed HNP-1 into the regulated secretory compartment of neutrophils, that is, to granules. The HNP-1 mouse clearly expressed HNP-1 in the myeloid compartment as visualized by immunocytochemistry (Figure 2) and by Western blotting (Figure 3). HNP-1 was processed to mature defensin as judged by Western blotting of myeloid cells from the HNP-1 mouse (Figure 3).
Immunocytochemical staining of murine BM cells from wild type and HNP-1 mice. BM cells were extracted from wild-type and HNP-1 mice. Cells were spun onto slides, fixed, permeabilized, and immunocytochemically stained for (A-B) HNP-1, (C-D) proHNP, and (E-F) IgG control. Bars represent 20 μm.
Immunocytochemical staining of murine BM cells from wild type and HNP-1 mice. BM cells were extracted from wild-type and HNP-1 mice. Cells were spun onto slides, fixed, permeabilized, and immunocytochemically stained for (A-B) HNP-1, (C-D) proHNP, and (E-F) IgG control. Bars represent 20 μm.
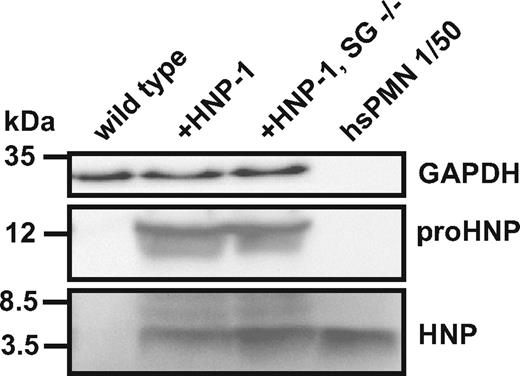
Quantification of HNP and proHNP in HNP-1 mice with or without serglycin. Western blotting of neutrophils isolated by density separation of murine BM cells from wild type mice, HNP-1 mice (+HNP-1), and HNP-1 mice deficient in serglycin (+HNP-1, SG−/−) compared with human polymorphonuclear neutrophils (hsPMN) diluted to a concentration 1/50 of the murine BM cells. Detection was done with Abs against HNP, proHNP, and GAPDH.
Quantification of HNP and proHNP in HNP-1 mice with or without serglycin. Western blotting of neutrophils isolated by density separation of murine BM cells from wild type mice, HNP-1 mice (+HNP-1), and HNP-1 mice deficient in serglycin (+HNP-1, SG−/−) compared with human polymorphonuclear neutrophils (hsPMN) diluted to a concentration 1/50 of the murine BM cells. Detection was done with Abs against HNP, proHNP, and GAPDH.
Murine BM cells from HNP-1 mice were separated on a 1-layer Percoll density gradient. The cell pellet at the bottom of the gradient contained primarily mature neutrophils as shown by May-Grünwald-Giemsa staining (supplemental Figure 2A) and expression profiling of neutrophil differentiation markers as assessed by quantitative real-time PCR (supplemental Figure 2B). HNP-1 expression was observed in both mature and immature BM cells obtained from the Percoll gradient (supplemental Figure 2B). The absolute amount of HNP-1 in the neutrophils of the transgenic mouse was significantly lower than in human neutrophils (Figure 3).
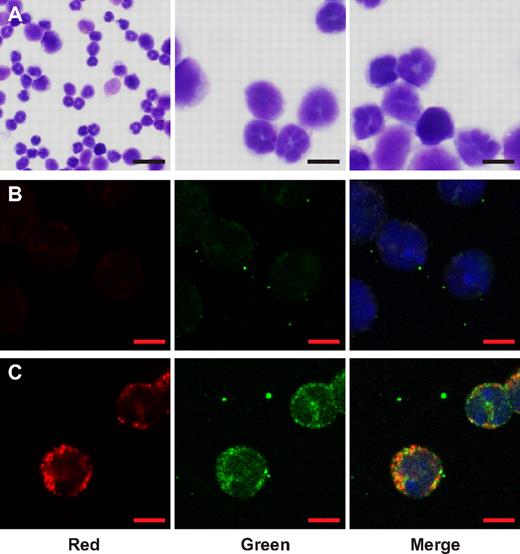
Serglycin has previously been localized to the Golgi apparatus and immature granules of promyelocytes/myelocytes.5 We examined the localization of serglycin in relation to HNP-1 in transgenic HNP-1 mice by fluorescence microscopy on BM cells depleted of nongranulocytic cells by immunomagnetic sorting. The purity of cells was verified by May-Grünwald-Giemsa staining (Figure 4A). Staining for serglycin was strong in a minority of granulocytic cells, but weak/absent in the rest, which correlates well with serglycin mainly being present in promyelocytes/myelocytes. As in human BM, HNP-1 displayed a granular staining pattern consistent with storage of HNP-1 in granules. Interestingly, HNP-1 colocalized with serglycin in cells with strong expression of serglycin (Figure 4C).
Localization of serglycin in relation to HNP-1 in granulocytic BM cells of transgenic HNP-1 mice. Murine BM from transgenic HNP-1 mice was extracted and nongranulocytic cells diminished by ammonium chloride-based lysis and immunomagnetic sorting. Cells were fixed with paraformaldehyde and spun onto slides. (A) May-Grünwald staining of sorted cells: Overview (left) and 2 close-ups. Bars represent 20 μm (left) or 5 μm. (B-C) Immunofluoresence microscopy: Primary Abs were detected with Alexa Fluor 594 or 488 Abs and DNA was stained with DAPI. Red bars represent 5 μm. (B) Cells IgG control (red and green). (C) Cells stained for serglycin (red) and HNP-1 (green). Images were acquired with a Zeiss LSM 700 microscope and merged with Zen 2009 LE software.
Localization of serglycin in relation to HNP-1 in granulocytic BM cells of transgenic HNP-1 mice. Murine BM from transgenic HNP-1 mice was extracted and nongranulocytic cells diminished by ammonium chloride-based lysis and immunomagnetic sorting. Cells were fixed with paraformaldehyde and spun onto slides. (A) May-Grünwald staining of sorted cells: Overview (left) and 2 close-ups. Bars represent 20 μm (left) or 5 μm. (B-C) Immunofluoresence microscopy: Primary Abs were detected with Alexa Fluor 594 or 488 Abs and DNA was stained with DAPI. Red bars represent 5 μm. (B) Cells IgG control (red and green). (C) Cells stained for serglycin (red) and HNP-1 (green). Images were acquired with a Zeiss LSM 700 microscope and merged with Zen 2009 LE software.
We next tested whether serglycin is the major intracellular proteoglycan in mouse myloid cells by comparing the level of sulfated macromolecules in wild-type and serglycin-deficient mice using biosynthesis with 35S-sulfate. This clearly confirmed that serglycin is the dominating cell-associated sulfated proteoglycan in the myeloid compartment in mice (supplemental Figure 3).
The role of serglycin in targeting and processing of HNP-1 was examined in the HNP-1 expression mouse on a serglycin ko background. Comparing the presence of HNP-1 assessed both by the Ab specific for the propeptide and the Ab raised against mature HNPs, no major difference was observed between serglycin ko expressing HNP-1 and serglycin wild-type mice expressing HNP-1 (Figure 3), indicating that serglycin is not involved in the processing of proHNP to mature HNP-1 and that the ability to retain mature HNP-1 is not dependent on serglycin.
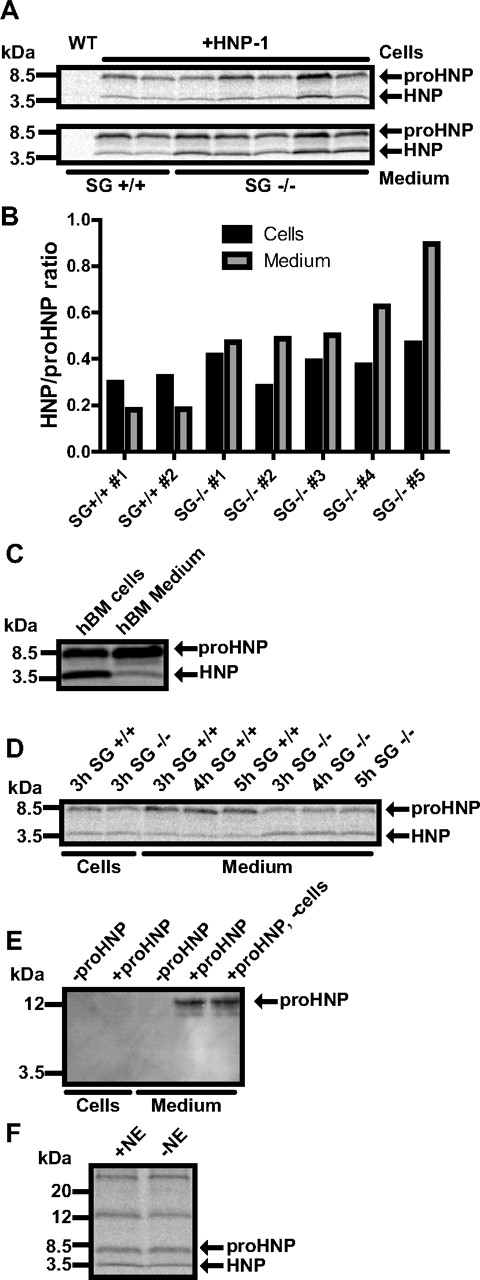
Biosynthesis studies were then performed as a more sensitive approach to test whether serglycin is active in withholding newly processed defensins in cells (Figure 5A). As in human BM (Figure 5C), only minor amounts of mature defensins were released into the medium in HNP-1 mice, while mature defensins were clearly released into the medium in HNP-1 mice deficient in serglycin (Figure 5B), indicating that serglycin diverts mature defensins from constitutive secretion and retains mature defensins in the cells. To exclude that extracellular processing was responsible for the higher amount of extracellular mature defensin in serglycin ko mice, the extracellular medium was separated from cells from the HNP-1 transgenic, serglycin ko mouse after a chase period of 3 hours. The medium was then split in 3 equal parts. Condition 1 was immediately given proteinase inhibitors and put on ice, whereas condition 2 and 3 were chased for 4 and 5 hours, respectively, before addition of protease inhibitors. The extracellular level of defensins did not differ between these conditions (Figure 5D) indicating that processing does not occur extracellularly. To further test that proHNP was not processed extracellularly, we added purified proHNP to the medium of murine BM cells of serglycin-deficient mice and incubated for 3 hours (Figure 5E). No processed HNP-1 or decline in proHNP was detected.
Biosynthesis of proHNP and HNP in HNP-1 mice deficient in serglycin or elastase. (A,C,D,F) BM cells were pulsed with 35S-methionine/cysteine for 1 hour and chased for 3 hours. Cell lysates and medium were immunoprecipitated with Abs against proHNP and HNP. Immunoprecipitates were pooled and analyzed by 16% SDS-Tricine-PAGE and fluorography. (A) proHNP and HNP in BM cells (top lane) and chase medium (bottom lane) of wild-type, HNP-1 mice, and HNP-1 mice deficient in serglycin (SG). (B) Ratio of fully processed HNP compared with proHNP in cells and chase medium. (C) proHNP and HNP in normal human BM. (D) proHNP and HNP-1 in HNP-1 mice and HNP-1 mice deficient in SG. After a 3-hour chase, cells were pelleted and medium split in 3 conditions. Two conditions were incubated at 37°C for another 1 or 2 hours, respectively. (E) BM cells were extracted from 2 SG−/− mice and pooled in chase medium. Cells were split in 2 conditions and purified proHNP was added to one condition and to medium without cells. After 3-hour incubation, cells were pelleted and lysed. Cells and medium were immunoprecipitated with Abs against proHNP and HNP. Precipitates were subjected to SDS-PAGE and immunoblotted for proHNP and HNP. (F) HNP-1 mice and HNP-1 mice deficient in neutrophil elastase (NE).
Biosynthesis of proHNP and HNP in HNP-1 mice deficient in serglycin or elastase. (A,C,D,F) BM cells were pulsed with 35S-methionine/cysteine for 1 hour and chased for 3 hours. Cell lysates and medium were immunoprecipitated with Abs against proHNP and HNP. Immunoprecipitates were pooled and analyzed by 16% SDS-Tricine-PAGE and fluorography. (A) proHNP and HNP in BM cells (top lane) and chase medium (bottom lane) of wild-type, HNP-1 mice, and HNP-1 mice deficient in serglycin (SG). (B) Ratio of fully processed HNP compared with proHNP in cells and chase medium. (C) proHNP and HNP in normal human BM. (D) proHNP and HNP-1 in HNP-1 mice and HNP-1 mice deficient in SG. After a 3-hour chase, cells were pelleted and medium split in 3 conditions. Two conditions were incubated at 37°C for another 1 or 2 hours, respectively. (E) BM cells were extracted from 2 SG−/− mice and pooled in chase medium. Cells were split in 2 conditions and purified proHNP was added to one condition and to medium without cells. After 3-hour incubation, cells were pelleted and lysed. Cells and medium were immunoprecipitated with Abs against proHNP and HNP. Precipitates were subjected to SDS-PAGE and immunoblotted for proHNP and HNP. (F) HNP-1 mice and HNP-1 mice deficient in neutrophil elastase (NE).
Processing and sorting of HNP-1 in human BM
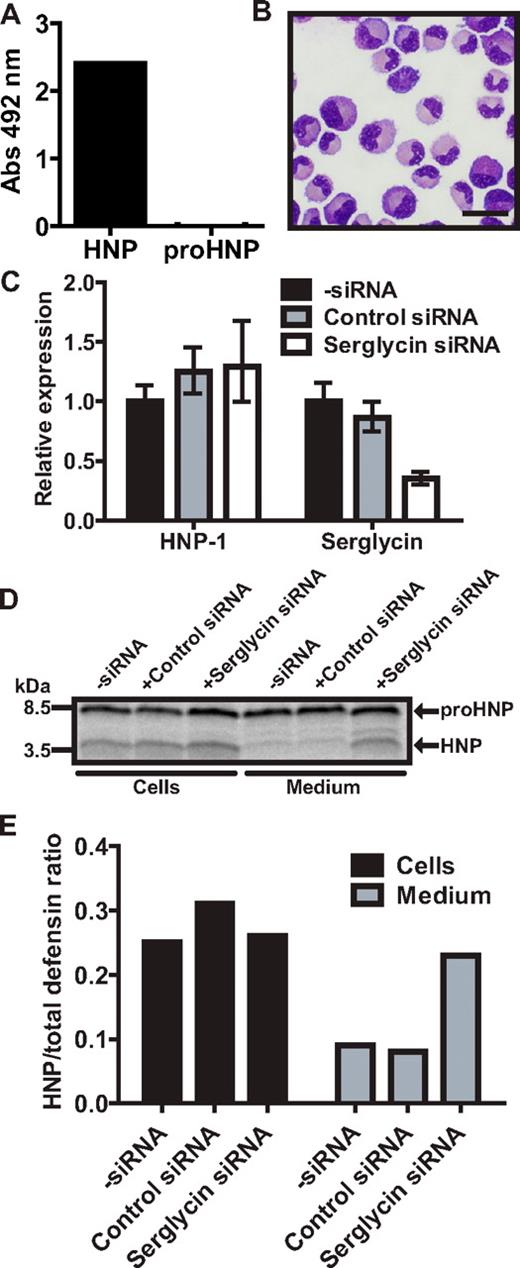
HNP-1 has previously been shown to be retained in myeloblast/promyelocytes, while being secreted in later stages of myelopoiesis.14 Serglycin is mainly expressed in these maturational stages5 and we speculated that serglycin is necessary for retaining mature HNP-1 in the promyelocyte. To find a suitable model for shRNA knockdown of serglycin, we tested the expression of HNP by pulse-chase biosynthesis in the promyelocytic cell line HL-60, its subclone PLB-985,32 and in HL-60 cells stably transfected with the HNP-1–expressing plasmid also used with 32D Cl3 cells (supplemental Figure 4A). As seen, PLB-985 cells produced the largest amount of mature HNP. We therefore stably transfected PLB-985 with a plasmid expressing shRNA against serglycin and achieved a > 90% knockdown of serglycin (supplemental Figure 4B). Because of the clonal nature of the selected cells, HNP-1 mRNA varied greatly between our shRNA clones with most clones producing < 5% compared with wild type and a few producing 4-11 times more. Lysates of PLB-985 cells with high and low HNP-1 expression were subjected to a mixed ELISA and, as in 32D Cl3 cells, serglycin was found to associate with HNP-1, but not proHNP (supplemental Figure 4C). Pulse-chase biosynthesis was performed on wild-type PLB-985 and clones with either shRNA against serglycin or control shRNA (supplemental Figure 4D). Almost no HNP-1 was found in medium of wild-type PLB-985 or shRNA clones with lower expression of HNP-1 than wild type. PLB-985 clones with shRNA against serglycin and a high expression of HNP-1 did have a notable amount of mature HNP in chase medium. Because none of our shRNA controls produced more HNP-1 than wild-type PLB-985, we were not able to discriminate, if the secretion of HNP-1 could be contributed to serglycin knockdown or merely high HNP-1 expression. We therefore tested myeloblasts/promyelocytes from human BM in a mixed ELISA. Again, serglycin was found to associate with HNP-1, but not proHNP (Figure 6A). Next, granulocytic precursors were isolated from human BM (Figure 6B) and transiently transfected with siRNA against serglycin, achieving a > 60% knockdown without affecting HNP-1 expression (Figure 6C). Twenty hours after transfection, cells were subjected to pulse-chase biosynthesis (Figure 6D). Similar to our observations on transgenic mice, the level of mature HNP was increased in medium from human BM myeloid cells with low serglycin expression (Figure 6E).
Biosynthesis of proHNP and HNP in human BM. (A) Myeloblasts/promyelocytes were isolated from human BM by density centrifugation and subjected to a mixed ELISA: immunoplates were incubated with anti-serglycin Ab. Plates were washed, blocked, and incubated with cell lysate. Association of a protein with serglycin was probed with biotinylated Abs against proHNP, which does not recognize fully processed HNP-1, and against HNP-1, which does recognize fully processed HNP-1. After incubation with HRP-conjugated avidin, a color reaction was developed using OPD tablets. Color reaction was measured at an absorbance (Abs) of 492 nm and background reactivity subtracted (absorbance in wells with lysis buffer without cells). Absorbance points to an association of a protein with serglycin. (B) Human BM cells were sedimented with dextran. Supernatant was laid on Lymphoprep and centrifuged at 400g for 30 minutes. Interphase cells were depleted of nongranulocytic cells by immunomagnetic sorting, spun onto slides, and May-Grünwald-Giemsa stained. Bar represents 20 μm. (C) Purified granulocytic precursors were electroporated with siRNA against serglycin, control siRNA, or without siRNA and incubated for 20 hours in a humidified incubator with 5% CO2 at 37°C. Comparative quantification of mRNA for HNP-1 and serglycin. Figure depicts expression levels relative to cells electroporated without siRNA. (D) Transfected granulocyte precursors were pulsed with 35S-methionine/cysteine for 1.5 hours and chased for 3 hours. Cell lysates and medium were immunoprecipitated with Abs against proHNP followed by HNP. Immunoprecipitates were pooled and analyzed by 16% SDS-Tricine-PAGE and fluorography. (E) Ratio of fully processed HNP compared with proHNP in cells and chase medium.
Biosynthesis of proHNP and HNP in human BM. (A) Myeloblasts/promyelocytes were isolated from human BM by density centrifugation and subjected to a mixed ELISA: immunoplates were incubated with anti-serglycin Ab. Plates were washed, blocked, and incubated with cell lysate. Association of a protein with serglycin was probed with biotinylated Abs against proHNP, which does not recognize fully processed HNP-1, and against HNP-1, which does recognize fully processed HNP-1. After incubation with HRP-conjugated avidin, a color reaction was developed using OPD tablets. Color reaction was measured at an absorbance (Abs) of 492 nm and background reactivity subtracted (absorbance in wells with lysis buffer without cells). Absorbance points to an association of a protein with serglycin. (B) Human BM cells were sedimented with dextran. Supernatant was laid on Lymphoprep and centrifuged at 400g for 30 minutes. Interphase cells were depleted of nongranulocytic cells by immunomagnetic sorting, spun onto slides, and May-Grünwald-Giemsa stained. Bar represents 20 μm. (C) Purified granulocytic precursors were electroporated with siRNA against serglycin, control siRNA, or without siRNA and incubated for 20 hours in a humidified incubator with 5% CO2 at 37°C. Comparative quantification of mRNA for HNP-1 and serglycin. Figure depicts expression levels relative to cells electroporated without siRNA. (D) Transfected granulocyte precursors were pulsed with 35S-methionine/cysteine for 1.5 hours and chased for 3 hours. Cell lysates and medium were immunoprecipitated with Abs against proHNP followed by HNP. Immunoprecipitates were pooled and analyzed by 16% SDS-Tricine-PAGE and fluorography. (E) Ratio of fully processed HNP compared with proHNP in cells and chase medium.
NE is not essential for processing of HNP-1
Serglycin has been shown to be essential for expression of NE in myeloid cells in mice.6 The fact that HNP-1–expressing, serglycin-deficient mice seem capable of efficiently processing proHNP, therefore indicates that elastase is not a major protease involved in processing of proHNP in vivo. To address this directly, we crossed the elastase ko mouse into the HNP-1–expressing mouse. We were not able to detect any difference in the capacity for processing of HNP-1 using pulse-chase biosynthesis (Figure 5F), and thus conclude that elastase is not essential for processing of HNP-1 in myeloid cells in this mouse model.
The myelopoietic activity in the BM was not affected by the introduction of HNP-1 (Table 1), nor was there any indication that the presence of HNP-1–induced impairment in the levels of circulating blood cells (Table 1).
Blood picture of mice
| Genotype . | n . | Mean (SD) . | |||
|---|---|---|---|---|---|
| WBC . | RBC . | PMN . | LYMPH . | ||
| Wild type | 4 | 6.73 (2.31) | 9.56 (0.21) | 0.52 (0.23) | 5.93 (2.05) |
| Serglycin ko | 5 | 5.96 (1.29) | 9.01 (0.39) | 0.64 (0.14) | 5.07 (1.14) |
| HNP-1 | 5 | 4.95 (1.00) | 9.19 (0.47) | 0.69 (0.12) | 3.94 (1.03) |
| HNP-1, SG ko | 3 | 6.72 (0.15) | 9.43 (0.19) | 0.75 (0.17) | 5.64 (0.03) |
| Genotype . | n . | Mean (SD) . | |||
|---|---|---|---|---|---|
| WBC . | RBC . | PMN . | LYMPH . | ||
| Wild type | 4 | 6.73 (2.31) | 9.56 (0.21) | 0.52 (0.23) | 5.93 (2.05) |
| Serglycin ko | 5 | 5.96 (1.29) | 9.01 (0.39) | 0.64 (0.14) | 5.07 (1.14) |
| HNP-1 | 5 | 4.95 (1.00) | 9.19 (0.47) | 0.69 (0.12) | 3.94 (1.03) |
| HNP-1, SG ko | 3 | 6.72 (0.15) | 9.43 (0.19) | 0.75 (0.17) | 5.64 (0.03) |
Cell count values are ×109/L.
WBC indicates white blood cells; PMN, neutrophils; LYMPH, lymphocytes; HNP-1, human neutrophil peptide 1; SG, serglycin; and ko, knockout.
Discussion
The significance of serglycin in retaining mature HNP-1 intracellularly in myeloid cells was examined. Our data corroborate that serglycin is the major proteoglycan in hematopoietic cells. Serglycin has been identified in mast cells, cytotoxic lymphocytes, neutrophils, platelets, and macrophages.33 Its function is not fully understood, but its role in packaging of granule proteins has been widely studied. Serglycin is a constituent of the granzyme B and perforin-containing granules of T cells34 as well as granules of endothelial cells,35 mast cells,36 and platelets.37 While the presence in mast cells is critical for retention of several proteases in mast cell granules38 as well as some granule proteins in platelets,37 in T cells, serglycin seems necessary only for retention of granzyme B39 . Serglycin-deficient mice do not have impairment of their ability to combat viral infections known to be dependent on fully functional cytotoxic T cells.40 A discrete phenotype was revealed in later stages of inflammation as contraction of virus-specific CD8+ T cells was slower in serglycin-deficient mice. The hyperproliferation and defective contraction of Ag-specific CD8+ cells was shown to be because of mechanisms extrinsic to the T cells, indicating that serglycin was responsible for intracellular retention of factors regulating normal contraction of the cytotoxic T-cell response.40 In neutrophils, serglycin is only present in early stages of granulopoiesis and has a discrete effect on the content of granules.5,6 So far, elastase is the only neutrophil granule protein which has shown dependency on serglycin for correct localization.6
We demonstrate that serglycin colocalizes with and participates in retaining the most abundant neutrophil granule protein, HNP, intracellularly. Murine myeloid cells possess the machinery for processing of proHNP to mature HNP and have the ability to store mature HNP in granules in accordance with previous findings.18 Using 32D Cl3 myeloid cells, we were able to demonstrate binding of serglycin to mature HNP-1, a small basic peptide, but not to its neutral proform. Chondroitinase treatment abrogated this binding in agreement with serglycin's proposed ability to bind basic proteins electrostatically via its negatively charged glycoaminoglycan sidechains.33 High expression of human NE also provided a notable signal in our binding assay, but did not seem to displace binding of serglycin to mature HNP-1 pointing to a large binding capacity of serglycin. These observations are also in agreement with the ability of defensins to target LDL particles to heparan sulfate containing proteoglycans on endothelial cells.41 Although we could not demonstrate major differences in the amount of mature defensin in myeloid cells of serglycin ko mice transgenic with HNP-1, serglycin participated in retention of mature HNP-1 in cells. Given the relatively small level of HNP-1 expression in the transgenic model used here, it is possible that a major part of the HNP-1 may be retained by binding to partners other than serglycin, in particular when this is absent as in the serglycin ko mouse.
Serglycin was found to associate with serglycin in the human promyelocytic cell lines and in promyelocytic cells isolated from healthy donors. PLB-985 clones with high HNP-1 and low serglycin expression did show significant amount of mature HNP-1 in chase medium, but because of lack of relevant control cells, we could not discriminate whether this effect was because of low serglycin or high HNP-1 expression. To settle this, we transiently transfected purified granulocytic cells from human BM with siRNA against serglycin and showed a substantial increase in mature, extracellular HNP despite the incomplete serglycin knockdown. This indicates an important role for serglycin in retaining mature HNP in human BM.
Mature HNP-1 is a membrane active peptide that may cause cell damage.9,42-44 In addition, neutrophil defensins are known to participate in the binding of lipoproteins to the vasculature and may participate in development of atherosclerosis.41,45,46 Recently, using the same transgenic mouse as we have, neutrophil defensins were shown to induce lung injury by disrupting the capillary-epithelial barrier.28 Effective retention of mature HNP-1 may thus be essential to prevent cell damage and to prevent development of atherosclerosis when the expression of HNP is at a higher level than the expression afforded by this transgenic mouse model.
Serglycin was not found to be essential for processing of proHNP in the transgenic mouse. This indicates that serglycin does not bind and present the proteases necessary for processing of proHNP to mature HNP. More specifically, our results show that elastase is not required for processing of proHNPs in vivo in this mouse model and indicate that the same is true in human myeloid cells. However, we cannot exclude that this may be a matter of the level of HNP expression and pertain only to levels of expression that are significantly lower than in human myeloid cells where mature HNPs constitute 50% of the total protein in azurophil granules.8
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The expert technical assistance of Charlotte Horn is greatly acknowledged. The authors thank the Transgenic Core Facility, Biotech and Research Innovation Center, University of Copenhagen for performing embryo transfer of HNP-1 embryos.
This work was supported by grants from The Danish Medical Research Council (N.B.) and grants from the National University Hospital (A.G.).
Authorship
Contribution: A.G. performed experiments and made the figures; N.H.H. performed and analyzed mass spectrometry; M.T.L. performed experiments; and A.G., J.B.C., and N.B. designed the research, analyzed results, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Niels Borregaard, The Granulocyte Research Laboratory, Department of Hematology–9322, National University Hospital, Blegdamsvej 9, DK-2100 Copenhagen, Denmark; e-mail: borregaard@rh.dk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal