In the hope of identifying key events that drive malignant transformation, researchers have been aiming to compare tumor cells with the corresponding pretransformed cell population of origin. This approach is attractive because, once established, such key events can then become the focus of new therapies that interfere with the transformation process as it happens, thus preventing it.
Different Cre transgenes initiate Sleeping-Beauty (SB) transposon-mutagenesis at distinct stages during T-cell development, causing T-ALL in mice. Distinct combinations of common insertion sites (CISs), equivalent to driver mutations, are selected for in the tumors, depending on when mutagenesis is initiated. Surprisingly, if mutagenesis is initiated late during development, at the DP stage, the resulting tumors show features of human ETP-ALL, raising the possibility that this high-risk leukemia originates from more mature cells that revert to an immature immune phenotype on transformation.
Different Cre transgenes initiate Sleeping-Beauty (SB) transposon-mutagenesis at distinct stages during T-cell development, causing T-ALL in mice. Distinct combinations of common insertion sites (CISs), equivalent to driver mutations, are selected for in the tumors, depending on when mutagenesis is initiated. Surprisingly, if mutagenesis is initiated late during development, at the DP stage, the resulting tumors show features of human ETP-ALL, raising the possibility that this high-risk leukemia originates from more mature cells that revert to an immature immune phenotype on transformation.
In this issue of Blood, Berquam-Vrieze et al directly show that the biologic state at which a population of cells first experiences oncogenic mutations strongly influences the type of driver mutations selected for in the resulting tumor in a mouse model of T-cell acute lymphoblastic leukemia (T-ALL).1 T-ALL is a malignant clonal expansion of immature T cells that accounts for 10% to 15% of pediatric and 25% of adult ALL cases. While 80% of patients with T-ALL can currently be cured with intensive chemotherapy, a subset of cases has a poor prognosis. A recent study found early T-cell precursor (ETP) leukemia to be a high-risk subset of T-ALL characterized by an immature surface immune-phenotype, increased genomic instability, a high frequency of remission failure or hematologic relapse, and a distinct gene expression profile.2 Because of its ETP-like immune-phenotype and gene expression profile, ETP-ALL was proposed to originate from very early thymic immigrants. However, the current work of Berquam-Vrieze and colleagues introduces an alternative scenario.
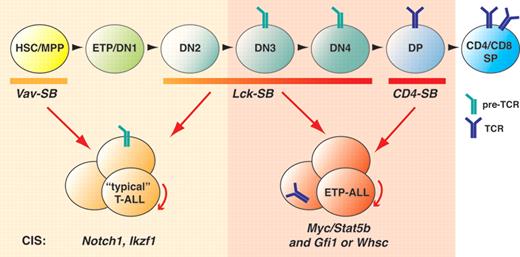
The authors designed a forward-genetic screen, using a Sleeping-Beauty (SB)–based transposon system that allows Cre-dependent activation of the SB transposase. This allows transposon-induced mutagenesis to be initiated under the control of stage-specific Cre transgenes.3 Mutagenesis was induced either early in hematopoietic stem cells (Vav-SB) or at 2 subsequent developmental stages of thymocyte development: the CD4−CD8− double-negative (DN) compartment (Lck-SB), and the β-selected, CD4+CD8+ double-positive (DP) compartment (CD4-SB). Tumors develop as a result of transposon insertions into or near tumor suppressors or oncogenes, either activating or disrupting their expression. By mapping common insertion sites (CISs), that is, transposon insertion sites frequently selected for, one can thus identify bona fide driver mutations for T-ALL originating from different developmental states (see figure).
Berquam-Vrieze et al sequenced the insertion sites of 101 lymphomas and deduced a total of 94 driver mutations. As a proof of principle, 75% of these mutations have previously been described to cause cancer. On the other hand, 25% of these candidate driver mutations have no previous entry in the literature linking them to cancer development and thus invite follow-up studies.
Do differences or commonalities exist between the models as well as between mouse and human T-ALL? Interestingly, yes. When transposon-induced mutation was initiated in hematopoietic progenitors, under the control of Vav1-Cre, a large fraction (72%) of the resulting T-cell tumors had mutations in Notch1, a hallmark of human T-ALL.4,5 The second most common mutation affected the Ikzf1 locus encoding for the transcription factors Ikaros, which is a rare target for mutation in human T-ALL. However, Ikzf1 mutations have frequently been observed to cooperate with Notch1 in murine T-ALL models. Interestingly, loss of Ikaros function was recently shown to result in increased activated Notch1 levels through alternative promoter usage in the Notch1 locus.6,7
When transposon-induced mutations were initiated much later during T-cell development, however, Berquam-Vrieze and colleagues note very different mutation profiles in the CD4-Cre–dependent mouse strain: Notch1 mutations were no longer significantly overrepresented. Instead, the tumors now predominantly harbored mutations of Myc and Stat5b, often in a mutually exclusive combination with additional CISs affecting either Gfi1 or Whsc1, Akt2, Jak1, and/or Sos1. Inappropriate overexpression of Myc drives many cancers and has also been implicated in human T-ALL as a downstream target of Notch1.8 Myc is also required for mouse T-ALL induced by up-regulation of the Wnt/β-catenin signaling pathway.9
The Lck-SB tumors, finally, segregated into 2 categories. In the one group, mutually exclusive Gfi1 versus Whsc1 mutations in combination with Myc and Stat5b were observed similar to CD4-SB tumors. In the other group, tumors carried Notch1 and Ikzf1 mutations similar to the Vav-SB model. This dichotomy might reflect a major change in cell state on completion of β-selection at the DN3 stage. Extensive chromatin reorganization associated with pre-TCR expression and signaling might result in very different genetic selection events becoming predominant. And a narrow window of time between Lck-Cre expression and β-selection might explain the emergence of these distinct subsets. It is also interesting in this regard that, to the best of our knowledge, the only 2 mouse models of T-ALL described to date that are Notch1-independent, conditional activation of β-catenin9 and loss of Pten,10 depend on TCR rearrangement, activate Myc, and can be induced after β-selection under the control of CD4Cre.
Interestingly, gene expression profiling of Vav-SB and CD4-SB tumors revealed an unexpected correlation between the CD4-SB model and human ETP-ALL and between the Vav-SB model and non-ETP (ie, more typical) T-ALL. In a second approach, unsupervised clustering of the 54 most differentially expressed genes between Vav-SB and CD4-SB also identified a gene set that was up-regulated in CD4-SB and ETP-ALL and a second that was up-regulated in Vav-SB and typical T-ALL. This important observation raises the possibility that ETP-ALL or at least a subset of ETP-ALL might originate from more differentiated cells than the ETP-like surface phenotype suggests. In fact, approximately 50% of the human ETP-ALL cases studied previously had rearrangements of TCR genes, likewise suggesting a more mature origin.2
In conclusion, this study by Berquam-Vrieze and colleagues confirms the notion that the biologic state of origin could be reflected in the genetic selection events observed in the resulting tumor. The study further demonstrates the power of the Sleeping Beauty transposon system to derive valuable new animal models that provide insights into the etiology of cancers less likely to be gained from gain or loss of function of single genes in mice and impossible to be gained in humans.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■
REFERENCES
National Institutes of Health

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal