Abstract
ETV6-RUNX1 gene fusion is usually an early, prenatal event in childhood acute lymphoblastic leukemia (ALL). Transformation results in the generation of a persistent (> 14 years) preleukemic clone, which postnatally converts to ALL after the acquisition of necessary secondary genetic alterations. Many cancer cells show some expression of the erythropoietin receptor (EPOR) gene, although the “functionality” of any EPOR complexes and their relevant signaling pathways in nonerythroid cells has not been validated. EPOR mRNA is selectively and ectopically expressed in ETV6-RUNX1+ ALL, but the presence of a functional EPOR on the cell surface and its role in leukemogenesis driven by ETV6-RUNX1 remains to be identified. Here, we show that ETV6-RUNX1 directly binds the EPOR promoter and that expression of ETV6-RUNX1 alone in normal pre-B cells is sufficient to activate EPOR transcription. We further reveal that murine and human ETV6-RUNX1+ cells expressing EPOR mRNA have EPO ligand binding activity that correlates with an increased cell survival through activation of the JAK2-STAT5 pathway and up-regulation of antiapoptotic BCL-XL. These data support the contention that ETV6-RUNX1 directly activates ectopic expression of a functional EPOR and provides cell survival signals that may contribute critically to persistence of covert premalignant clones in children.
Introduction
The t(12;21) is the most frequent chromosomal lesion in childhood B cell precursor acute lymphoblastic leukemia (ALL), occurring with an incidence of 25% overall and generating the ETV6-RUNX1 fusion gene.1,2 The fusion gene develops predominantly in utero as a prenatal and likely initiating event in childhood ALL and results in the generation of a persistent preleukemic clone, which converts, at low frequency, to ALL after the acquisition of necessary secondary genetic alterations.3-5 Observations on clinical samples (normal cord blood6 and monozygotic twins,7 plus animal modeling8-12 ) indicate that ETV6-RUNX1 can induce a preleukemic phenotype that can remain covert for up to 15 years but is insufficient for overt or clinical leukemia. The additional genetic abnormalities, including multiple gene deletions,13 are secondary to ETV6-RUNX1 fusion and probably postnatal in origin.14 This pattern of natural history raises important functional questions as to the nature of the signaling pathways corrupted by ETV6-RUNX1 and their contribution to a persistent preleukemic state.
The t(12;21) translocation fuses protein dimerization domains of ETV6 with essentially all of the DNA binding and activating regions of RUNX1.1 The latter is a key component of the core binding factor complex (ie, core binding factor-α/RUNX1) that is critical for normal hematopoiesis; genes encoding proteins in this complex are highly selected as targets for mutational change initiating acute leukemias.15,16 The chimeric transcription factor ETV6-RUNX1 can bind to RUNX1 target sequences and potentially deregulate gene expression in cells with the translocation. ETV6-RUNX1 has been shown to inhibit RUNX1-dependent transcription converting RUNX1 to a predominantly negative transcriptional regulator impeding differentiation and involving the recruitment of corepressor molecules, such as NCOR and Sin3A.17-19 Alternative mechanisms of ETV6-RUNX1 activity have also been suggested because ETV6-RUNX1 can disrupt the activity of wild-type ETV6 by dimerization via the helix-loop-helix domains.20,21 Recently, ETV6-RUNX1 has also been shown to interfere with apoptosis, affecting antiapoptotic genes such as survivin,9,22 and is proposed to act as a dominant negative transcription factor that may reduce expression of tumor suppressor genes while increasing expression of antiapoptotic genes.23
Taken together, such transcriptional deregulation might lead to a sustained preleukemic clone that is additionally vulnerable to further mutation, possibly under proliferative stress.24 There is evidence that ETV6-RUNX1 can generate a population of self-renewing human cord blood cells with a unique phenotype (CD34+/CD19+/CD38−), compatible with a very early stage of B-cell lineage commitment,25 and recently we demonstrated that the expression of ETV6-RUNX1 in preleukemic models deregulates the TGF-β pathway, suggesting a plausible mechanism by which deregulated immune response to infection might promote the malignant evolution of preleukemic clones.10 ETV6-RUNX1 expression may also allow quiescent, preleukemic stem cells to persist in the bone marrow.26
We hypothesized that survival of ETV6-RUNX1–expressing leukemia cells might be mediated in part by erythropoietin (EPO). The use of PCR and microarray technology has revealed the EPO receptor (EPOR) to be consistently selectively expressed, ectopically, in ETV6-RUNX1+ ALL, although the presence of a functional receptor on the cell surface and its role, if any, in leukemogenesis driven by ETV6-RUNX1 remains uncertain.27-29 Inthal et al have shown that EPO enhances proliferation of ETV6-RUNX1+ leukemia and attenuates the sensitivity to induced apoptosis.28 However, these studies used the REH cell line and primary leukemia samples, where the expression of ETV6-RUNX1 is already coupled with secondary mutations. We therefore sought to demonstrate the presence of a functional, ligand binding receptor on ETV6-RUNX1+ ALL cells and to directly assess the impact of ETV6-RUNX1 protein on EPOR activity. We developed 3 experimental systems where the impact of ETV6-RUNX1 protein itself can be assessed in a “preleukemic” scenario. We have used an in vitro murine pro-B progenitor cell line with hormone inducible ETV6-RUNX1 and a transgenic E μ-ETV6-RUNX1 murine model10 ; and to provide a system that more closely approximates to preleukemogenesis in children, we have also exploited a system for expressing ETV6-RUNX1 in human cord blood progenitor cells.25
Methods
Detection of cell surface EPOR
Quantitative determination of cells expressing EPOR was performed using Fluorokine Biotinylated Human Erythropoietin (R&D Systems). Briefly, cells were washed with PBS and incubated with biotinylated EPO for 60 minutes. Cells were directly either avidin-FITC or avidin-PE (Santa Cruz Biotechnology) labeled for 30 minutes and then washed and resuspended in 0.2 mL for final flow cytometric analysis using FACS.
Survival and apoptosis assays
BaF3 clone 1 control cells and BaF3 1/27 cells have been described before10 ; the latter cells can express ETV6-RUNX1 after induction with mifepristone. Control cells (clone 1) and cells inducible for ETV6-RUNX1 (1/27) were treated with 12.5pM mifepristone (Invitrogen) for 2, 4, and 8 days. A total of 5 × 105 cells were seeded in T-6 well plates in the absence of IL-3, presence of EPO 20 U/mL (R&D Systems) or JAK2 inhibitor AG490 10 μM where indicated. A total of 30 μg of siRNA-EPOR mix was added in some cases (supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Cells were harvested and counted at each of time points for growth curve analysis.
Apoptosis and cell cycle changes were evaluated with propidium iodide (Invitrogen) staining and FACS analysis. For the analysis of quiescence, exponentially growing clone 1 and 1/27 cells were induced with mifepristone for 3 days; and where indicated, IL-3 was removed from the media and substituted by 20 U/mL EPO. After 4 days in culture, cells were incubated with 10 μg/mL Hoechst 33342 (Sigma-Aldrich) in Hoechst staining buffer (HBSS + 3% FBS + 10mM HEPES) at 37°C. After 45 minutes, 1 μg/mL of pyronin Y (Sigma-Aldrich) was added to the cell culture and incubated for an additional 45 minutes. Subsequently, cells were trypsinized, washed 3 times with PBS, and analyzed by FACS.
Isolation of human hematopoietic progenitor cells, lentiviral transduction, and B-lineage priming
Human cord blood (CB) CD34+ hematopoietic stem/progenitor cells were isolated from human mononuclear CB cells purchased from Stem Cell Technologies. CD34+ hematopoietic stem/progenitor cells were positively selected using a MACS CD34 MicroBead kit (Miltenyi Biotec). Isolated CD34+ cells were maintained for < 24 hours before infection in IMDM containing 10% FCS and cytokines.30 The cytokines included human recombinant IL-3 (10 ng/mL), Flt3 ligand (10 ng/mL), thrombopoietin (10 ng/mL), SCF (5 ng/mL), and G-CSF (5 ng/mL). All cytokines were obtained from R&D Systems. All experiments were approved by the Institute of Cancer Research and the United Kingdom Home Office.
Lentivirus was generated by transfection of HEK 293 cells through calcium phosphate precipitation as described.31 Lentiviral vectors used were: pWPI (empty vector) and pWPI-ETV6-RUNX1-myc in which the expression of the ETV6-RUNX1 is coupled with GFP expression. Two sequential infections of 0.4 × 106 CD34+ cells/well by concentrated lentiviral particles were carried out in 48-well plates precoated with retronectin (Takara) at 50 μg/mL. Cells were collected 3 days after infection, and GFP (ETV6-RUNX1) expression was analyzed by flow cytometry.
Luciferase reporter assays
A 200-bp fragment of the upstream region of the EPOR gene, including the putative ETV6-RUNX1 binding site, was amplified from human genomic DNA and inserted into pGL4.10 luciferase vector (Promega). Primers used for fragment amplification were: reverse 5′-AGTCACCTGTCCAGGGCCC-3′; forward 5′-GCGCCTCTAAGTGGCAGAT-3′. The first putative RUNX1 binding site was subsequently mutated using QuikChange XL Site-Directed Mutagenesis Kit (Stratagene): 5′-CACACGTTTTTTTTTTCTTTGTGGCCCTGGACAGGTGACTTACC-3′.
The putative RUNX1 binding site is underlined and the 3 bases mutated to ACC in the mutant vector are in bold italics.
HEK293 cells were transiently cotransfected using calcium phosphate transfection methods with 3 μg pEF-MCIneoGATA1 and pWPI-ETV6-RUNX1, 2 μg promoter firefly luciferase vectors (pGL4.10 EPOR 200 and pGL4.10 constructs), and 0.5 μg Renilla luciferase vector for normalization. The cells were lysed 48 hours after transfection, and luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega) and fluorescence reader Berthold Junior LB 9509.
Western blotting and immunoblotting
Cells were pelleted and lysed for 30 minutes at 4°C in lysis buffer (50mM Tris pH 8.0, 1mM EDTA, 1% NP-40, 250mM NaCl, and 1× protease and phosphatase inhibitor cocktail, Sigma-Aldrich). Lysates were cleared by centrifugation at 4°C for 15 minutes at 14 000g, and total protein was separated through an SDS-polyacrylamide gel (NuPAGE 4%-12% gels, Invitrogen) and transferred to a polyvinylidene difluoride membrane (Millipore). The membranes were blocked and incubated overnight with primary antibodies. After a final incubation with secondary antibodies conjugated with peroxidase (1/10 000 dilution; GE Healthcare), immune complexes were detected by chemiluminescence using the ECL Plus Kit (GE Healthcare). Antibodies used were: phospho-JAK2 (Tyr1007-1008), phospho-Akt (ser463), phospho STAT5 (Tyr694), STAT5, and Bcl-xL, all from Cell Signaling Technology, and α-tubulin (Sigma-Aldrich). For Western blot detection of human EPOR, A82 antibody was used according to Amgen protocols.32
Results
ETV6-RUNX1+ ALL is associated with increased levels of ligand binding EPOR
It has previously been described that leukemic blasts in ETV6-RUNX1+ ALL patients, as well as the ETV6-RUNX1+ leukemic cell line REH, show increased levels of EPOR gene expression,27-29 but the presence of a functional ligand-binding receptor on the surface has not been validated.
We confirmed the expression of EPOR protein in the pre-B ALL ETV6-RUNX1+ cell line REH by Western blot analysis using the well-established EPOR antibody A82.33 K562 and HEL cells were used as positive controls for EPOR protein expression, and ETV6-RUNX1–negative pre-B ALL cell lines SEM and Nalm6 were used as negative controls along with HeLa cells (Figure 1A-B). The expression level of EPOR seen in REH cells was comparable with that observed in the erythroleukemia cell line K562. Given that some cell types, such as K562,34 do not respond to EPO despite gene expression of the receptor, we wished to confirm that the expression of EPOR detected in REH cells would correlate with functionality of the receptor. To this end, we first used biotinylated EPO and flow cytometric analysis, which allowed us to study the ligand binding of EPO to its receptor. We found that REH cells have higher cell surface levels of ligand-binding EPOR than other non-ETV6-RUNX1 pre-B ALL cell lines or indeed K562 cells (Figure 1C; supplemental Figure 1), although similar levels of EPOR protein were detected in these 2 cell lines by Western blot (Figure 1A). Next, when we “blind screened” CD19+ cells isolated from 12 patients diagnosed with pre-B ALL, we identified 8 cases that showed expression of EPOR by Western blot, 4 of which were subsequently shown to be ETV6-RUNX1+ by FISH (Figure 1B; supplemental Figure 1; and data not shown). Furthermore, when these patient samples were screened using biotinylated EPO, only 5 from the 8 EPOR-protein positive samples showed expression of ligand-binding EPOR on the cell surface, suggesting that protein detection does not directly correlate with functional EPOR on the cell surface. Four of these 5 samples were the cases identified as ETV6-RUNX1+, and the remaining sample was a case of relapsed ALL (Figure 1C; supplemental Figure 1). Our data show, for the first time, the presence of functional ligand binding EPOR in ETV6-RUNX1+ pre-B ALL.
Association of functional EPOR expression with ETV6-RUNX1+ leukemic cell lines and patient samples. (A-B) Western blot analysis of EPOR expression using Amgen antibody A82 in different ALL cell lines and CD19+ cells isolated from patients diagnosed with ALL. K562 and HeLa cell lines were used as controls for low and negative expression of EPOR, respectively, and HEL cells for high expression. As a direct comparison with REH cells, the SEM, Nalm6, and Raji cell lines were used as pre-B cell lymphoid ETV6-RUNX1-negative cell lines. ETV6-RUNX1-positive and -negative patients are designated ER+ and ER−, respectively. (C) Functional EPOR is ectopically expressed on the cell surface of CD19+ cells isolated from ETV6-RUNX1+ pre-B ALL patients (ER+) and leukemic cell lines but not ETV6-RUNX1-negative cases (ER−). Washed cell lines and CD19+ sorted patient cells were incubated with biotinylated EPO that binds to the EPOR. Avidin-fluorescein was added and the amount of EPO measured as the increase in fluorescence analyzed by FACS compared with controls (see also supplemental Figure 1).
Association of functional EPOR expression with ETV6-RUNX1+ leukemic cell lines and patient samples. (A-B) Western blot analysis of EPOR expression using Amgen antibody A82 in different ALL cell lines and CD19+ cells isolated from patients diagnosed with ALL. K562 and HeLa cell lines were used as controls for low and negative expression of EPOR, respectively, and HEL cells for high expression. As a direct comparison with REH cells, the SEM, Nalm6, and Raji cell lines were used as pre-B cell lymphoid ETV6-RUNX1-negative cell lines. ETV6-RUNX1-positive and -negative patients are designated ER+ and ER−, respectively. (C) Functional EPOR is ectopically expressed on the cell surface of CD19+ cells isolated from ETV6-RUNX1+ pre-B ALL patients (ER+) and leukemic cell lines but not ETV6-RUNX1-negative cases (ER−). Washed cell lines and CD19+ sorted patient cells were incubated with biotinylated EPO that binds to the EPOR. Avidin-fluorescein was added and the amount of EPO measured as the increase in fluorescence analyzed by FACS compared with controls (see also supplemental Figure 1).
Expression of ETV6-RUNX1 alone is sufficient to increase expression of EPOR in “preleukemic” mouse models
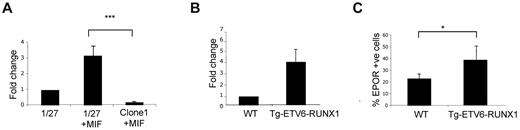
To ask whether ETV6-RUNX1 protein was able to directly influence EPOR expression, we first used an inducible system whereby the expression of ETV6-RUNX1 can be up-regulated in pro-B lymphocyte clone BaF3 1/27 cells by the addition of mifepristone.10 After induction of ETV6-RUNX1, a significant increase in EPOR mRNA expression was observed compared with controls (Figure 2A). We next analyzed changes in EPOR expression, both mRNA and protein, in a murine transgenic E μ-ETV6-RUNX1 model.10 For that purpose, bone marrow B220+ and Sca1+, IL7R+ lymphoid populations were isolated for quantitative PCR and FACS analysis of EPOR expression. In concordance with the BaF3 cell line model, the results showed an increase in EPOR expression in cells from ETV6-RUNX1+ transgenic mice compared with cells from wild-type littermates (Figure 2B-C).
ETV6-RUNX1 expression in mouse preleukemic models is associated with increased levels of EPOR. (A) Quantitative PCR detection of EPOR in a murine BaF3 cell line (1/27) induced to express ETV6-RUNX1 (1/27 + MIF) or a mock-induced control (clone 1 + MIF; n = 4; P = .007). (B) Quantitative PCR detection of EPOR in a Sca1+ Kit+ population isolated from a transgenic E μ-ETV6-RUNX1 mouse model (n = 6). (C) Detection of functional ligand-binding EPOR in the E μ-ETV6-RUNX1 mouse model by FACS (n = 22; P = .033).
ETV6-RUNX1 expression in mouse preleukemic models is associated with increased levels of EPOR. (A) Quantitative PCR detection of EPOR in a murine BaF3 cell line (1/27) induced to express ETV6-RUNX1 (1/27 + MIF) or a mock-induced control (clone 1 + MIF; n = 4; P = .007). (B) Quantitative PCR detection of EPOR in a Sca1+ Kit+ population isolated from a transgenic E μ-ETV6-RUNX1 mouse model (n = 6). (C) Detection of functional ligand-binding EPOR in the E μ-ETV6-RUNX1 mouse model by FACS (n = 22; P = .033).
ETV6-RUNX1 directly regulates EPOR expression
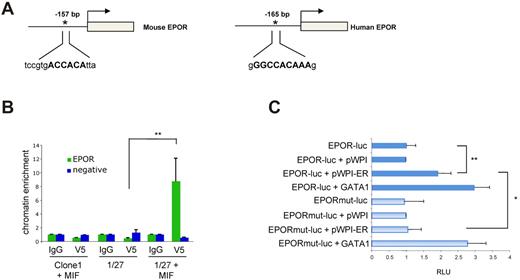
Next, we asked whether the observed up-regulation of EPOR by ETV6-RUNX1 could be a direct effect of ETV6-RUNX1 itself binding to EPOR promoter regulatory regions. We first performed in silico sequence analysis on the EPOR promoter to determine putative ETV6-RUNX1 binding sites. As the ETV6-RUNX1 fusion protein retains the DNA binding domain of RUNX1, we hypothesized that any putative RUNX1 sites observed in the promoter would also be putative binding sites for ETV6-RUNX1. We analyzed the entire upstream region of human and mouse EPOR gene using Chip Mapper, a Web-based tool for multigenome analysis of positions and patterns of regulation elements. For the mouse EPOR upstream region, Chip Mapper found 3 putative binding sites for RUNX1, the one located at 157 bp upstream of the initiation of transcription being the most significant in terms of the highest probability of RUNX1 binding (Figure 3A). When the human EPOR gene was analyzed, 3 analogous RUNX1 binding sites were found; and again, that closest to the initiation of transcription was the most significant (Figure 3A).
ETV6-RUNX1 binds to the EPOR gene promoter region and activates its transcription. (A) In silico analysis of mouse and human upstream genomic regions of the EPOR gene revealed the existence of a highly conserved putative RUNX1 binding site located ∼ 200 bp upstream of transcription initiation. (B) ChIP assays at the putative RUNX1 binding site located in the EPOR promoter in mouse 1/27 BaF3 cells induced to express ETV6-RUNX1 (+ MIF) or mock-induced control cells (clone 1 + MIF, n = 6; P = .0048). α-V5 indicates V5 epitope antibody used for ETV6-RUNX1 immunoprecipitation in BaF3 1/27 cells and IgG is isotype control; and negative, an EPOR genomic region negative for ETV6-RUNX1 binding sites (see also supplemental Figure 2). (C) The first 200 bp of the human EPOR promoter was cloned upstream of a pGL4 luciferase vector and transfected into 293T cells with or without expression vectors for GATA1 (positive control) or ETV6-RUNX1 (ER). Both GATA1 (as expected) and ETV6-RUNX1 were able to enhance basal luciferase activity of the promoter (P = .0076). This effect was dependent on an intact RUNX1 binding site as the mutation (mut) of the putative site abolished the increase in luciferase activity observed with ETV6-RUNX1. RLU indicates relative luciferase units, normalized to EPOR promoter vector (EPOR-luc). Data are from 4 independent experiments.
ETV6-RUNX1 binds to the EPOR gene promoter region and activates its transcription. (A) In silico analysis of mouse and human upstream genomic regions of the EPOR gene revealed the existence of a highly conserved putative RUNX1 binding site located ∼ 200 bp upstream of transcription initiation. (B) ChIP assays at the putative RUNX1 binding site located in the EPOR promoter in mouse 1/27 BaF3 cells induced to express ETV6-RUNX1 (+ MIF) or mock-induced control cells (clone 1 + MIF, n = 6; P = .0048). α-V5 indicates V5 epitope antibody used for ETV6-RUNX1 immunoprecipitation in BaF3 1/27 cells and IgG is isotype control; and negative, an EPOR genomic region negative for ETV6-RUNX1 binding sites (see also supplemental Figure 2). (C) The first 200 bp of the human EPOR promoter was cloned upstream of a pGL4 luciferase vector and transfected into 293T cells with or without expression vectors for GATA1 (positive control) or ETV6-RUNX1 (ER). Both GATA1 (as expected) and ETV6-RUNX1 were able to enhance basal luciferase activity of the promoter (P = .0076). This effect was dependent on an intact RUNX1 binding site as the mutation (mut) of the putative site abolished the increase in luciferase activity observed with ETV6-RUNX1. RLU indicates relative luciferase units, normalized to EPOR promoter vector (EPOR-luc). Data are from 4 independent experiments.
To study the direct regulation of EPOR transcription by ETV6-RUNX1, ChIP assays were performed on uninduced murine BaF3 1/27 cells and 1/27 cells induced to express ETV6-RUNX1. Cells were fixed, sonicated, and incubated with the indicated antibodies (taking advantage of the specific ETV6-RUNX1-V5 epitope tag in our inducible system). After isolation of the immunoprecipitated DNA, quantitative PCR was performed using specific primers that covered the proximal (most significant) RUNX1 binding site within the mouse EPOR promoter. The results shown in Figure 3B confirmed the in vivo occupancy of ETV6-RUNX1 protein on this proximal site with ∼ 9-fold increase in chromatin enrichment compared with control cells. In addition, using human REH cells (ETV6-RUNX1+) and antibodies specific for RUNX1 and ETV6, we confirmed the in vivo occupancy of ETV6-RUNX1 within the human EPOR promoter (supplemental Figure 2). The deregulation of EPOR expression by ETV6-RUNX1 was also confirmed by luciferase reporter assays. The first 200 bp upstream of the human EPOR promoter, including the proximal putative ETV6-RUNX1 binding site, was cloned into the pGL4.10 luciferase vector and transfected into human nonerythroid 293T cells. Luciferase activity was normalized to the minimal luciferase activity from the EPOR promoter. Cotransfection of the EPOR promoter with a pEF-MCIneo-GATA1 expression vector increased this activity to nearly 3-fold, demonstrating the functionality of the EPOR promoter fragment (Figure 3C), whereas overexpression of ETV6-RUNX1 alone enhanced minimal EPOR promoter activity by > 2-fold. Moreover, the effect on EPOR transcription by ETV6-RUNX1 is dependent on an intact RUNX1 binding site, as mutation of only 3 bp within the binding site abrogates ETV6-RUNX1's effect on EPOR promoter activity (Figure 3C). Altogether, these results demonstrate that ETV6-RUNX1 directly binds to and can positively regulate transcription from the EPOR promoter.
EPO sustains cell survival of ETV6-RUNX1–expressing cells through the JAK2-STAT5 pathway
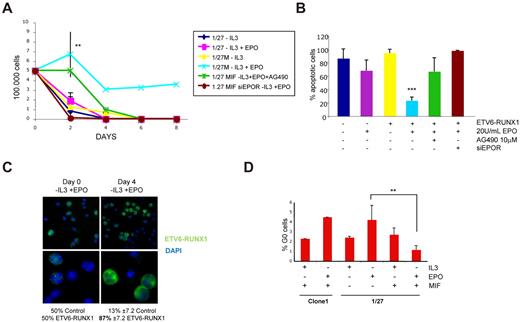
Given the proposed prosurvival properties of EPO on nonerythroid cells,35 we next asked whether the observed increase in EPOR expression via ETV6-RUNX1 could correlate with an increased cell survival in the presence of EPO alone. Cell survival experiments, including growth curves, propidium iodide staining, and cell cycle analysis, revealed that only 1/27 cells expressing ETV6-RUNX1 (normally IL-3 dependent) showed a prolonged survival in the presence of EPO after withdrawal of IL-3 (Figure 4A). Of note, this effect was probably dependent on EPOR expression and an intact JAK2-STAT5 pathway, as silencing of the receptor using siRNA or treatment with the nonspecific JAK2 inhibitor AG490 abolished EPO-induced survival in ETV6-RUNX1–expressing cells (Figure 4A; supplemental Figure 3). In agreement with these results, analysis of cell death by propidium iodide staining revealed a clear reduction of apoptotic cells in the ETV6-RUNX1–expressing population grown in the presence of EPO, especially at longer stages (8 days) of growth (Figure 4B). To confirm that EPO alone can sustain survival of ETV6-RUNX1+ cells, we performed growth competition experiments using the inducible system with a 50:50 mix of ETV6-RUNX1–expressing and nonexpressing cells. Equal amounts of mock-induced control cells and cells induced to express ETV6-RUNX1 were mixed at day 0 and grown for 4 days in the absence of IL-3 and the presence of 20 U/mL EPO. After 4 days, the remaining cells were immunostained with V5-FITC antibody to detect and count ETV6-RUNX1+ cells. As shown in Figure 4C and supplemental Figure 4, ∼ 87% of surviving cells were ETV6-RUNX1+ and only 13% were ETV6-RUNX1–negative control cells, demonstrating that, under these culture conditions, EPO can indeed aid survival of ETV6-RUNX1+ cells.
EPO alone sustains cell survival in ETV6-RUNX1–expressing cells. (A) Growth curves: A total of 500 000 uninduced and ETV6-RUNX1–expressing BaF3 1/27 cells were cultured in the absence of IL-3 and absence or presence of 20 U/mL human EPO for up to 8 days (n = 4 P = .0085). Only in ETV6-RUNX1–expressing cells was EPO able to maintain cell survival in the absence of IL-3 but not in the absence of EPOR (siRNA knockdown) or intact JAK2 activity (AG490). (B) Apoptosis analysis: On day 8, cells were stained with propidium iodide, and the sub G1 population (apoptotic cells) was analyzed. In the long-term assay, only cells expressing ETV6-RUNX1 maintained low levels of apoptosis. (C) Growth competition experiment: A total of 105 cells of mifepristone-induced clone 1 control cells and 1/27 cells expressing ETV6-RUNX1 were mixed and grown for 4 days in the absence of IL-3 and the presence of 20 U/mL EPO. Cells were stained at day 0 and day 4 with α-V5-FITC antibody and 4,6-diamidino-2-phenylindole, and a minimum of 200 cells were counted for ETV6-RUNX1 expression. Images were taken with an Axioscope microscope (Zeiss) with 63×/1.25 oil and 100×/1.30 oil objectives in a Vecta shield mounting medium H-1000 (Vector Laboratories Inc). A Hamamatsu digital camera ORLA-ER (C-4747-80) was used and images were captured with smart Capture X Version 2.6.2 and Adobe Photoshop CS Version 8.0. (D) Analysis of cell cycle: On day 4 of growth in the absence or presence of IL-3 or EPO, cells were stained with Hoechst 33342 and pyronin Y (n = 3; P = .0095). Only in ETV6-RUNX1–expressing cells could EPO induce G1 arrest of cell cycle, but not quiescence, measured as a percentage of cells in G0 (see also supplemental Figure 5).
EPO alone sustains cell survival in ETV6-RUNX1–expressing cells. (A) Growth curves: A total of 500 000 uninduced and ETV6-RUNX1–expressing BaF3 1/27 cells were cultured in the absence of IL-3 and absence or presence of 20 U/mL human EPO for up to 8 days (n = 4 P = .0085). Only in ETV6-RUNX1–expressing cells was EPO able to maintain cell survival in the absence of IL-3 but not in the absence of EPOR (siRNA knockdown) or intact JAK2 activity (AG490). (B) Apoptosis analysis: On day 8, cells were stained with propidium iodide, and the sub G1 population (apoptotic cells) was analyzed. In the long-term assay, only cells expressing ETV6-RUNX1 maintained low levels of apoptosis. (C) Growth competition experiment: A total of 105 cells of mifepristone-induced clone 1 control cells and 1/27 cells expressing ETV6-RUNX1 were mixed and grown for 4 days in the absence of IL-3 and the presence of 20 U/mL EPO. Cells were stained at day 0 and day 4 with α-V5-FITC antibody and 4,6-diamidino-2-phenylindole, and a minimum of 200 cells were counted for ETV6-RUNX1 expression. Images were taken with an Axioscope microscope (Zeiss) with 63×/1.25 oil and 100×/1.30 oil objectives in a Vecta shield mounting medium H-1000 (Vector Laboratories Inc). A Hamamatsu digital camera ORLA-ER (C-4747-80) was used and images were captured with smart Capture X Version 2.6.2 and Adobe Photoshop CS Version 8.0. (D) Analysis of cell cycle: On day 4 of growth in the absence or presence of IL-3 or EPO, cells were stained with Hoechst 33342 and pyronin Y (n = 3; P = .0095). Only in ETV6-RUNX1–expressing cells could EPO induce G1 arrest of cell cycle, but not quiescence, measured as a percentage of cells in G0 (see also supplemental Figure 5).
Cell-cycle analysis determined that live ETV6-RUNX1+ cells cultured in the presence of EPO are held in the G1 phase of the cell cycle (supplemental Figure 5). Classically, cell-cycle exit from G1 into G0 phase is accompanied by low rates of transcription (RNA content) and reduced cell size, as measured by forward scatter and pyronin Y staining of polyribosomal RNA and might protect cells from accumulating genetic changes that could result in malignancy. In cell culture, entry into G0 has been modeled using nutrient limitation, such as cytokine withdrawal.36,37 Because ETV6-RUNX1 expression can maintain hematopoietic stem cells in a stage of cellular quiescence (G0),26 we asked whether, in the presence of EPO, ETV6-RUNX1 was able to deregulate the normal G1 to G0 transition induced by withdrawal of IL-3 in the in vitro BaF3 murine model.
Mock-induced BaF3 control cells and cells induced to express ETV6-RUNX1 were cultured in the absence of IL-3 and absence or presence of EPO. After 4 days of culture, cells were stained with Hoechst 33342 and pyronin Y, and the percentage of cells in G0 and the transcription rate (RNA staining) were analyzed by FACS. On withdrawal of IL-3, there is an increase in the percentage of cells in G0 phase and reduction of RNA staining in both control and ETV6-RUNX1–expressing cells, followed by massive cell death (Figure 4D; supplemental Figures 5 and 6). The addition of EPO to ETV6-RUNX1–expressing cells was able to block this effect, as the percentage of cells in G0 significantly decreased (Figure 4D; supplemental Figure 6), accompanied by an increase in the rate of transcription (supplemental Figure 5). In concordance with growth curve data, the addition of the JAK2 inhibitor AG490 abolished this effect of EPO on ETV6-RUNX1–expressing cells.
To substantiate the block of entry into G0 of ETV6-RUNX1–expressing cells treated with EPO, we also studied the expression of genes implicated in quiescence induction and entry into cell cycle. Quantitative PCR data from EPO-treated ETV6-RUNX1–expressing cells showed an increased expression of proliferation-associated genes Myc and Ki67 but not of the cell cycle inhibitor p27, the increase of which is associated with stage of quiescence, indicating that, in terms of gene expression, these cells are not in a quiescent stage of the cell cycle (supplemental Figure 7).
Taken together, these results show that, under conditions of stress, such as withdrawal of nutrients, EPO signaling through JAK2 kinase sustains cell survival of ETV6-RUNX1–expressing cells, holding them in the G1 stage of the cell cycle. This arrest is translated into high transcription rates and a block of cell cycle exit from G1 into G0, and so, may overcome the barrier that protects cells from possible genetic and metabolic alterations.
Activation of STAT5 as a mechanism of cell survival in the presence of EPO
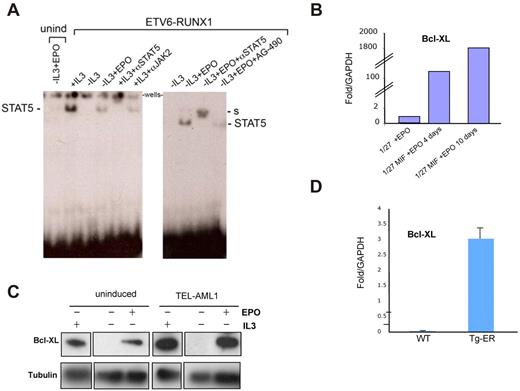
To further elucidate the mechanism by which EPO enhances cell survival in ETV6-RUNX1–expressing cells, we analyzed the EPOR signaling cascade triggered by EPO through the classic JAK2-STAT5 pathway, having already shown (Figure 4) that treatment with a JAK2 inhibitor abolished the survival effect. Uninduced and ETV6-RUNX1–expressing cells were cultured for 8 hours in the absence of FBS and IL-3 and then incubated with 20 U/mL EPO for 20 minutes. Endogenous EPOR signaling was analyzed by Western blot; and in concordance with previous reports,28 an increase in levels of JAK2 and AKT phosphorylation was observed in cells expressing ETV6-RUNX1 in the presence of EPO (supplemental Figure 8). To confirm the functional activation of the JAK2-STAT5 pathway by EPO in these cells, we performed electrophoretic mobility assays (EMSAs) with a probe to a known STAT5 binding element in the β-casein promoter38 using nuclear extracts prepared from cells either not expressing or expressing ETV6-RUNX1 and treated with or without EPO for 2 days. Normally, STAT5 only translocates to the nucleus after tyrosine phosphorylation and dimerization after activation through a variety of ligands and receptors. As a positive control for activation of STAT5, we used the IL-3 dependence of BaF3 cells to confirm binding of STAT5 to the probe via the observed bandshift (Figure 5A). STAT5 antisera could super-shift the complex; and, as expected, withdrawal of IL-3 blocked the translocation of STAT5 to nucleus and inhibited the STAT5 binding complex. As seen in Figure 5A (left panel), only cells expressing ETV6-RUNX1, and consequently EPOR, were able to activate STAT5 after incubation with EPO; uninduced cells showed no binding complex. Preincubation of cells with the selective JAK2 inhibitor AG490 reduced the STAT5 binding complex observed in the presence of ETV6-RUNX1 and EPO, further suggesting that this was a JAK2-dependent mechanism. We next asked whether the ability of EPO to signal activation of STAT5 in this system was directly related to its ability to promote survival, protection from apoptosis, and block of entry into quiescence. We studied changes in expression of BCL-XL, a well-known STAT5 target gene that plays an important role in apoptosis inhibition and quiescence enhancement.39 Western blot and quantitative PCR analysis demonstrated an increase in mouse Bcl-XL protein and mRNA expression in EPO-treated ETV6-RUNX1+ cells (Figure 5B-C). Up-regulation of Bcl-XL mRNA was also detected in B220+ bone marrow populations isolated from ETV6-RUNX1+ mice compared with wild-type (Figure 5D).
EPO promotes STAT5 signaling and increases Bcl-XL expression in ETV6-RUNX1+ mouse cells. (A) EMSA assay. Uninduced and ETV6- RUNX1-expressing BaF3 1/27 cells were incubated with 20 U/mL EPO for 2 days. Equal amounts of nuclear extracts were incubated with a γ-32P labeled β-casein promoter probe to analyze EPOR signaling through STAT5 DNA binding activity. In cells expressing ETV6-RUNX1, EPO triggers activated STAT5 complexes (marked) compared with non–ETV6-RUNX1–expressing cells. S indicates supershift (see also supplemental Figure 4). (B) Quantitative PCR analysis of Bcl-XL expression in uninduced (1/27) and ETV6-RUNX1–expressing BaF3 1/27 cells (1/27M) grown in the presence of 20 U/mL EPO for 4 or 10 days, respectively. (C) Western blot analysis of Bcl-XL expression in uninduced and ETV6-RUNX1–expressing BaF3 1/27 cells grown for 2 days in the presence or absence of IL-3 or EPO. (D) Quantitative PCR analysis of Bcl-XL expression in bone marrow Sca1+ cells isolated from wild-type and ETV6-RUNX1+ transgenic mice (n = 6).
EPO promotes STAT5 signaling and increases Bcl-XL expression in ETV6-RUNX1+ mouse cells. (A) EMSA assay. Uninduced and ETV6- RUNX1-expressing BaF3 1/27 cells were incubated with 20 U/mL EPO for 2 days. Equal amounts of nuclear extracts were incubated with a γ-32P labeled β-casein promoter probe to analyze EPOR signaling through STAT5 DNA binding activity. In cells expressing ETV6-RUNX1, EPO triggers activated STAT5 complexes (marked) compared with non–ETV6-RUNX1–expressing cells. S indicates supershift (see also supplemental Figure 4). (B) Quantitative PCR analysis of Bcl-XL expression in uninduced (1/27) and ETV6-RUNX1–expressing BaF3 1/27 cells (1/27M) grown in the presence of 20 U/mL EPO for 4 or 10 days, respectively. (C) Western blot analysis of Bcl-XL expression in uninduced and ETV6-RUNX1–expressing BaF3 1/27 cells grown for 2 days in the presence or absence of IL-3 or EPO. (D) Quantitative PCR analysis of Bcl-XL expression in bone marrow Sca1+ cells isolated from wild-type and ETV6-RUNX1+ transgenic mice (n = 6).
BCL-XL, together with BCL-2, can prolong survival by inhibition of apoptosis, but can also show antiproliferative effects through an ability to enhance quiescence by increasing p27 levels and decreasing cell size and RNA content.39 As expected, the expression of mouse Bcl-XL (and Bcl-2, data not shown) was increased by EPO in ETV6-RUNX1–expressing cells (Figure 5), but this rise was not accompanied by a decrease in cell size and RNA content, nor increase of p27 expression (supplemental Figure 7). The reduced levels of p27 may also explain why the increased levels of Bcl-XL do not correlate with an induction of quiescence. A similar block in p27 transcription in the presence of ETV6-RUNX1 has been described in response to TGF-β.10 Taken together, these results confirm that EPO can supply a JAK-STAT signal in mouse ETV6-RUNX1–expressing cells, with the concomitant up-regulation of the antiapoptotic protein Bcl-XL.
ETV6-RUNX1 regulates the EPOR-STAT5-BCL-XL pathway in human CD34+ progenitor cells
In our third model of “preleukemia,” human CD34+ cells were isolated from CB and transduced in vitro with a lentivirus capable of expressing both ETV6-RUNX1 and GFP (Figure 6A). CD34+ CB cells were subsequently “primed” for pre-B lineage commitment over 5 days, as described by Luo et al.30 The expression of EPOR was again analyzed using biotinylated EPO and immunoblotting with A82 EPOR antibody. Sorted GFP+ (ETV6-RUNX1+) cells also showed increased levels of functional EPOR compared with empty vector controls (Figure 6B), and increased levels of EPOR protein were also detectable by Western blot (Figure 6C). Interestingly, when the expression of BCL-XL was analyzed, ETV6-RUNX1+ B-cell primed CD34+ cells also showed higher levels of the antiapoptotic protein compared with empty vector-transduced cells (Figure 6D). These data accord with our murine preleukemic models and confirm the deregulation of EPOR and BCL-XL by the ETV6-RUNX1 fusion protein in the relevant cellular context.
ETV6-RUNX1 deregulates EPOR expression, activates STAT5 signaling pathways, and enhances BCL-XL expression in a human preleukemic cell model. (A) Experimental schema: human CD34+ cells were isolated from normal cord blood and infected with an ETV6-RUNX1–expressing lentivirus. Cells were primed for B-cell differentiation for up to 5 days. (B) GFP+ (ETV6-RUNX1+) cells were analyzed for EPOR expression by FACS. EV indicates empty vector. (C) GFP+ (ETV6-RUNX1+) cells were analyzed for EPOR expression by Western blot. (D) GFP+ (ETV6-RUNX1+) cells were analyzed for BCL-XL expression and Western blot.
ETV6-RUNX1 deregulates EPOR expression, activates STAT5 signaling pathways, and enhances BCL-XL expression in a human preleukemic cell model. (A) Experimental schema: human CD34+ cells were isolated from normal cord blood and infected with an ETV6-RUNX1–expressing lentivirus. Cells were primed for B-cell differentiation for up to 5 days. (B) GFP+ (ETV6-RUNX1+) cells were analyzed for EPOR expression by FACS. EV indicates empty vector. (C) GFP+ (ETV6-RUNX1+) cells were analyzed for EPOR expression by Western blot. (D) GFP+ (ETV6-RUNX1+) cells were analyzed for BCL-XL expression and Western blot.
EPO triggers STAT5 signaling in human ETV6-RUNX1+ ALL samples
To further study the functionality of the EPO-EPOR axis in ETV6-RUNX1+ models, we asked whether the findings in our “preleukemic” models could be extrapolated to patients with ALL. We isolated CD19+ cells from different ETV6-RUNX1+ and ETV6-RUNX1–negative ALL cases and incubated them with 20 U/mL EPO for 1 hour. Analyses of the STAT5 signaling pathway revealed a significant increase in STAT5 phosphorylation in ETV6-RUNX1+ ALL after EPO treatment compared with ETV6-RUNX1–negative ALL (Figure 7). In addition and corroborating the preleukemic cell data, higher expression of BCL-XL was observed in ETV6-RUNX1+ ALL samples, although no changes between nontreated and EPO-treated samples were observed (Figure 7). Although incubation with EPO for 1 hour may be enough for phosphorylation of STAT5, it may not be sufficient for induction of a full transcriptional effect.
EPO triggers STAT5 signaling in human ETV6-RUNX1+ ALL patient samples. CD19+ cells were isolated from ALL patients, grown in RPMI medium with or without EPO 20 U/mL for 1 hour, and lysates were analyzed by Western blot for STAT5 phosphorylation and BCL-XL expression. Only in ETV6-RUNX1+ patients with higher expression of EPOR was its ligand EPO able to trigger STAT5 signaling. BCL-XL expression was always higher in ETV6-RUNX1+ samples, although there was no difference in the presence or absence of EPO.
EPO triggers STAT5 signaling in human ETV6-RUNX1+ ALL patient samples. CD19+ cells were isolated from ALL patients, grown in RPMI medium with or without EPO 20 U/mL for 1 hour, and lysates were analyzed by Western blot for STAT5 phosphorylation and BCL-XL expression. Only in ETV6-RUNX1+ patients with higher expression of EPOR was its ligand EPO able to trigger STAT5 signaling. BCL-XL expression was always higher in ETV6-RUNX1+ samples, although there was no difference in the presence or absence of EPO.
Discussion
Data from mouse models,8,10,12,26 monozygotic twin studies,14,25 and a large cohort of newborn cord bloods6 have collectively verified that ETV6-RUNX1 fusion itself is insufficient to induce a full leukemic phenotype and that additional mutations are required for overt ALL. We have developed a minimal “2-hit,” prenatal/postnatal model for the natural history and molecular pathogenesis of ETV6-RUNX1+ ALL.7 This model predicts that, in the absence of the necessary secondary mutations, preleukemic (“one-hit”) clones of fetal origin can persist subclinically in blood at a relatively constant level throughout childhood. A clue to the mechanism that might be responsible for maintenance of this “silent” phase comes from expression array data on cases of ETV6-RUNX1+ ALL where essentially every case ectopically expresses EPOR.27 But to date, there has only been limited evidence to suggest that the receptor is functional and directly regulated by ETV6-RUNX1.28
Expression of the human EPOR is regulated primarily by hypoxia and is controlled by both transcriptional and post-translational modifications.40 Small amounts of receptor appear on the cell surface as a preformed homodimer and, after activation by EPO, can function to prevent apoptosis of erythroid progenitor cells as well as to induce their expansion and proliferation. The EPOR gene is expressed in many nonerythroid cell types, including normal and malignant lymphoid cells,27-29 but cell surface protein expression or activation of signal transduction pathways through EPO interaction has not been confirmed.
In the present study, we have demonstrated, for the first time, the presence of functional, ligand-binding EPOR on the surface of ETV6-RUNX1+ ALL cells and show that ETV6-RUNX1 directly regulates the expression of the receptor. Using 3 different ETV6-RUNX1+ preleukemic models with both murine and human cells, we have shown that ETV6-RUNX1 itself (ie, in the absence of secondary genetic alterations) is sufficient to increase the expression of EPOR, both at the RNA level and functional protein. (These experiments were performed using A82 antibody, the recommended EPOR specific antibody described by Elliott et al for analysis of low levels of EPOR detection in nonerythroid cell types32 ).
We identified a putative, highly conserved, ETV6-RUNX1 binding site within the human and mouse EPOR promoters. Using ChIP, luciferase reporter assays, and EMSA, we confirmed the in vivo occupancy of ETV6-RUNX1 on the EPOR promoter and the in vitro regulation of its transcription. ETV6-RUNX1 can bind to RUNX1 target sequences and potentially deregulate gene expression, as well as functioning in a dominant negative manner.17,41 Under physiologic conditions, as might occur in preleukemia where RUNX1 and ETV6-RUNX1 are coexpressed, the fusion protein could relieve an inhibitory effect of RUNX1 on EPOR expression, thus allowing its expression in a cell type where it would not normally be expressed.
On binding of EPO, the EPOR undergoes a conformational change that results in the phosphorylation of a number of intracellular signaling proteins, followed by rapid EPO/EPOR endocytosis and degradation. The ensuing signal cascade can activate the JAK-STAT, MAP kinase, or PI3/AKT pathways, all of which have critical effects on erythroid progenitor cell survival and differentiation.42 Here, we show that only in the presence of ETV6-RUNX1 is EPO able to activate JAK2 and AKT phosphorylation in B cells, as well as induce STAT5 binding activity. The latter is dependent on JAK2 phosphorylation because incubation with the JAK2 inhibitor AG490 prevented STAT5 activation. In erythroid cells, EPOR-STAT5 signaling in combination with GATA-143 induces transcription of the prosurvival or antiapoptotic protein BCL-XL. We found that the EPO-dependent survival of early B cells occurs through a similar JAK2- and AKT-dependent mechanism. However, the concomitant increase in BCL-XL occurs in the presence of little or no GATA1. Expression of BCL-XL is also coupled with entry into G0 of the cell cycle through a p27-dependent mechanism.39 BCL-XL facilitates G0 quiescence by decreasing RNA content, cell size, and up-regulating p27 protein. However, our data suggest that, in ETV6-RUNX1–expressing B-cell precursors, the EPO-driven increase in BCL-XL is not coupled with entry into G0 quiescence. On the contrary, in the presence of EPO, there is an increase in cell size and RNA content without up-regulation of the cell cycle inhibitor p27. The antiproliferative factor TGF-β maintains hematopoietic cells (including B cells) in quiescence by inhibiting cycling.44 We have previously described that the increase in endogenous p27 transcription in response to TGF-β in B precursors is also blocked in the presence of ETV6-RUNX1.10
Activated AKT is also known to play a role in the cell cycle, and its phosphorylation of p27 usually serves to overcome cell cycle arrest in G1.45 However, in the presence of ETV6-RUNX1 and EPO, p27 is not up-regulated and the cells remain active but in G1 arrest. Taken together, our current data suggest that ETV6-RUNX1–expressing cells may have a deregulated p27 response to cell cycle stimuli through the EPO-EPOR axis. Although we cannot disregard that other signal cascades may be activated by ETV6-RUNX1, including up-regulation of the common β-receptor thought to form a “tissue-protective” heterodimer with EPOR,46 we propose EPOR-STAT5-BCL-XL signaling to be one of the mechanisms responsible for maintenance of preleukemic ETV6-RUNX1+ clones generated in utero. Activation of constitutive JAK-STAT signaling also occurs in other subtypes of ALL via, for example, mutations in JAKs47,48 or upstream CRLF2.49,50 Although rare cases of ALL are described in which the EPOR gene is activated via fusion with the IGH@ locus,49,50 the co-option of an ectopic EPOR for maintenance of preleukemic clones in ETV6-RUNX1+ ALL is novel.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Susan Colman for FISH analysis, Prof Javier Leon for pEF-MCIneo-GATA1 expression vector, Dr Ian Titley and Gowri Vijayaraghavan for flow cytometry assistance, and Dr Steve Elliott for helpful discussions on EPOR protein expression and signaling. The A82 EPOR antibody was a kind gift of Amgen Inc.
This work was supported by Leukaemia & Lymphoma Research United Kingdom (A.F., J.P., and M.G.) and the Kay Kendall Leukaemia Fund United Kingdom (V.T., P.C., and M.G.).
Authorship
Contribution: M.G. and A.F. conceived the studies and wrote the manuscript; V.T., J.P., P.C., and A.F. designed and performed the research and analyzed the data; A.F. supervised the studies; and V.T. cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anthony M. Ford, Haemato-Oncology Research Unit, Division of Molecular Pathology, Institute of Cancer Research, Brookes Lawley Building, 15 Cotswold Road, Sutton, Surrey SM2 5NG, United Kingdom; e-mail: tony.ford@icr.ac.uk.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal