Abstract
Microchimerism is defined by the presence of low levels of nonhost cells in a person. We developed a reliable method for separating viable microchimeric cells from the host environment. For flow cytometric cell sorting, HLA antigens were targeted with human monoclonal HLA antibodies (mAbs). Optimal separation of microchimeric cells (present at a proportion as low as 0.01% in artificial mixtures) was obtained with 2 different HLA mAbs, one targeting the chimeric cells and the other the background cells. To verify purity of separated cell populations, flow-sorted fractions of 1000 cells were processed for DNA analysis by HLA-allele–specific and Y-chromosome–directed real-time quantitative PCR assays. After sorting, PCR signals of chimeric DNA markers in the positive fractions were significantly enhanced compared with those in the presort samples, and they were similar to those in 100% chimeric control samples. Next, we demonstrate applicability of HLA-targeted FACS sorting after pregnancy by separating chimeric maternal cells from child umbilical cord mononuclear cells. Targeting allelic differences with anti-HLA mAbs with FACS sorting allows maximal enrichment of viable microchimeric cells from a background cell population. The current methodology enables reliable microchimeric cell detection and separation in clinical specimens.
Introduction
Microchimerism refers to the state of a person harboring 2 genetically distinct cell populations, whereby the chimeric population constitutes < 1% of the total number of cells. Several situations can lead to the occurrence of microchimerism. Chimerism between mother and child is observed after cell transfer during pregnancy and after delivery.1-5 Microchimerism in the mother is associated with autoimmune disease later in life,6-9 but, in contrast, fetal10 and maternal microchimerism11,12 may favor the health of the mother and offspring postpartum.5 Microchimerism also can be found after solid-organ transplantation as a result of the migration of passenger hematopoietic cells from the donor graft into the peripheral circulation and tissues of the recipient. Some reports show that microchimerism is associated with graft acceptance13 and that it correlates with lower incidence of rejection.14,15 Other investigators, however, do not confirm a beneficial effect of microchimerism on graft acceptance.16-18
Endothelial cell chimerism after renal transplantation, resulting from the replacement of donor endothelial cells with cells from the recipient, is mainly observed in grafts of patients who have experienced rejection.19 After (intrauterine) blood transfusion, donor cell–derived DNA can be detected in the recipient's periphery.20-23 Microchimerism may be one of the mechanisms contributing to the beneficial effect of pretransplantation blood transfusions on transplantation outcome.24 In summary, the detection of peripheral blood microchimerism may aid in understanding the immunologic effects that occur during and after pregnancy, transplantation, and blood transfusion.
The low frequency of chimeric cells in peripheral blood samples necessitates a technical approach of detection that is reproducible and sensitive. Both classic and (real-time) quantitative PCR (qPCR) techniques have been used to detect (micro)chimerism.25-28 The detection of Y-chromosome–localized genes by qPCR offers a reproducible and sensitive technique,2,7,9,13,26 but it is restricted to sex-mismatched combinations only. HLAs represent a broader applicable target for the detection of microchimerism8,25,26,29 because they are highly polymorphic30 and thus frequently mismatched between 2 persons. The oligonucleotide primers that are suitable for PCR-based HLA typing of patients who undergo transplantation can be applied to detect microchimerism.
Increased sensitivity is achieved when the preliminary PCR is followed by second-round PCR (nested PCR) with sequence-specific primers that target the first amplicon.31-34 However, the detection of microchimerism by DNA amplification has drawbacks. Direct, single PCR, but especially nested PCR, may be prone to decreased specificity and to contamination by aspecific PCR products, resulting in false-positive results.35,36 Furthermore, once processed for qPCR purposes, functional studies of chimeric cells are precluded. The technique presented in the current study makes use of flow cytometric cell sorting on the basis of phenotypic HLA differences and yields potentially viable, highly enriched microchimeric cell populations. Their purity was verified by qPCR analysis, which targeted HLA class I and II alleles. The methodology presented enables molecular and functional studies of microchimeric cells, even in sex-matched samples.
Methods
Monoclonal anti-HLA antibodies
Human hybridomas were established from B lymphocytes of HLA antibody–seropositive, multiparous women by EBV transformation. This was followed by electrofusion, HAT-ouabain selection of antibody-secreting EBV lines, and rigorous subcloning. HLA specificities of mAbs were determined by complement-dependent cytotoxicity against large (n > 240) panels of serologically HLA typed PBMCs. Some mAbs react with epitopes shared between several allelic products.37 HLA mAbs of IgG isotype were purified from hybridoma supernatants by protein A chromatography (Pharmacia). Ammonium sulfate cuts (30%-50% saturation) of supernatants containing IgM isotype HLA mAbs were dialyzed against PBS and loaded on a high-load 16/26 Sephadex G-75 column (preparation grade; Pharmacia). Fractions with a molecular weight exceeding 75 kDa were collected, pooled, and dialyzed against PBS. Protein concentrations of purified mAb preparations were determined by the bicinchoninic acid method (Pierce). The purity of IgM-mAb preparations was assessed by the use of PAGE under reducing conditions.
Purified HLA mAbs were labeled with biotin (Pierce biotinylation kit no. 21430)38 according to the manufacturer's instructions, and their reactivity was validated by flow cytometry. Selected (for HLA phenotype) PBMCs (5 × 105) were incubated with 1 μg of biotine-labeled HLA-mAbs, followed by phycoerythrin-conjugated streptavidin and FITC-labeled anti-CD3 mAb (BD Biosciences) and analysis on a FACS Calibur (BD Biosciences Immunocytometry Systems) equipped with CellQuest. All biotine mAbs demonstrated homogeneous, HLA allelic product–specific staining on CD3-positive cells (not shown). Alexa 647–conjugated HLA-A2–specific mouse mAb was purchased from AbD Serotec (MCA2090).
Preparation of chimeric cell mixtures
Artificially spiked samples, simulating microchimerism, were established by preparing cell dilution series of PBMCs from one adult donor (chimeric cells) in PBMCs from another donor (background cells), ranging from 0.1% (1:1000) to 0.01% (1:10 000). Donor cells were molecularly typed for HLA, and serologic equivalents were used for the selection of mAbs. Because there was a sex difference between the 2 donors in each of the combinations, male-specific DYZ1, in addition to HLA alleles, could be targeted with specific primers. In each experiment, a 10% mixture was included, which was used to set gates in the FACS sort. Donor selection was such that at least one antigen from each of 2 donors was uniquely addressable with a mAb and by qPCR analysis for verification of cell sorting. Cells from suitable donors were thawed in the presence of Benzonase nuclease (E1014; Sigma-Aldrich) to limit cell clumping.
Labeling of cells with mAbs
Cell suspensions were incubated for 10 minutes at 4°C with 1 μL of FcR blocking reagent (130-059-901; Miltenyi Biotec) per 106 cells. Cell mixtures were stained for 30 minutes at 4°C with anti-CD45-FITC (340664) and anti-CD14-Percp (345786) from BD Biosciences Pharmingen. Biotine-labeled mAbs were used to target background cells, and mAbs directly labeled with PE were used to target chimeric cells. Streptavidin Alexa Fluor R647 conjugate (S32357; Invitrogen) was used to visualize the biotine-labeled mAbs. mAbs were conjugated with R-phycoerythrin (Z25455) via use of the Zenon HuIgG labeling kit (Invitrogen). Labeled mAbs were used within 30 minutes.
Single versus double antibody labeling
Efficiency of separation of cells from 2 donors by FACS was compared between single labeling (only donor 1) and double labeling (both donors). Cells from donor 1 (HLA-A2, -A33, -B62, -B17) were diluted in cells from donor 2 (A3, A11, B7, B35) in the following percentages: 50, 25, 12.5, 6.25, 3.13, 1.56, 0.78, 0.39, 0.20, 0.098, 0.049, 0.024, 0.012, and 0.006. For single labeling, only anti-HLA-A2 (MCA2090) was used, which targets cells from donor 1 (chimeric). For double labeling, mAb MCA2090 was used in combination with mAb BRO11F6-biotine, which targets HLA-A3 and A11 on cells from donor 2 (background).
Flow cytometry-based cell sorting
Cell sorting was performed with the BD FACSAriatm cell sorter (BD Biosciences), equipped with a 576/26-nm filter for PE-labeled cells, a 530/30-nm filter for FITC-labeled cells, and a 660/20-nm filter for Alexa 647–labeled cells. Cell suspensions were passed through a 70-μm filter before sorting. In the 0.01%-0.05% cell mixtures, 20-40 × 106 cells and in ≥ 0.1% cell mixtures at least 1 × 106 cells were analyzed by the use of FACS sorting.
For cell lysis and qPCR, 1000 cells from the cell mixture (presort sample), the positive fraction (chimeric cells), and the negative fraction (background cells) were collected in a PCR plate. Yield sort settings were used in the cell sorter, which means that a minimal number of 1000 positive cells were sorted, whereas some contamination by negative cells could not be excluded. As positive and negative controls, 100% cell fractions of each of the 2 separate donors were sorted.
Sample preparation for cell lysis
One thousand cells from the FACS were collected in PCR plates with 5 μL of 2 × lysis mixture containing 0.8 μg/μL of Proteinase K (Gibco BRL) and 0.01% SDS (Sigma-Aldrich). The PCR plates with collected cells were centrifuged for 1 minute at 100g. The total volume was adjusted to 10 μL with water, and plates were closed with caps. For complete cell lysis and DNA release, the following protocol was run in a thermo cycler: 1 hour at 50°C, 30 minutes at 99°C, and 10 minutes at 10°C.
Real-time qPCR primers
To determine sensitivity of the PCR assays, DNA containing the HLA genotype, which corresponds with the specificity of the particular primer set, was diluted in background DNA. To determine specificity of the primers, amplification signals (Ct values) from DNA of 2 different donors with the HLA genotype corresponding with the specificity of the particular primer set were compared with signals from DNA of at least 4 different donors not containing that particular HLA genotype. This was tested with 3 different concentrations of primers (10, 20, and 30 pmol/μL). The difference in Ct value (ΔCt) between positive and negative DNA controls served as a measure of specificity. The condition with the greatest ΔCt indicated the optimal primer concentration for subsequent experiments.
Real-time qPCR assays
For real-time qPCR 1 μL of the cell lysate was used (corresponding to ∼ 0.7 ng DNA) together with IQ SybrGreen supermix (Bio-Rad) and HLA allele-specific or Y-chromosome–targeting primers (10-30 pmol/μL) in a reaction volume of 20 μL. Primers for the human hematopoietic cell kinase (HCK) gene were used for normalization of the DNA content (internal standard) in each sample. Amplifications were carried out with an iCycler from Bio-Rad.
To obtain optimal specificity of the primers, a “touchdown” PCR profile was used: denaturation at 94°C for 8.5 minutes, followed by 40 cycles of 15 seconds at 94°C, 45 seconds at 68°C (decreasing with 0.2°C each cycle), and 30 seconds at 72°C. This was followed by a melting profile: 1 minute at 95°C and then 80 cycles, each for 10 seconds, starting at 55°C and increasing 0.5°C each cycle.
Results
Human mAbs
A limited set of 10 human HLA mAbs (8 shown in Table 1) was selected for this study on microchimerism in pregnancy from a larger list (n = 120) of in-house developed mAbs.37 The current selection of mAbs enables mother/child distinction on the basis of HLA class I in > 90% of pregnancies of white women. On biotin-labeling of purified mAb and flow cytometric analysis of single PBMC suspensions carrying the relevant antigens, the geometric means of PE fluorescence intensity reached levels in the third decade in fluorescence histograms (data not shown), ensuring that microchimeric cells would be identifiable unambiguously. The amount of biotine mAb used for 500 000 cells ranged from 0.05 to 0.4 μg (Table 1).
Characteristics of human monoclonal anti-HLA antibodies
| Name . | HLA specificity . | Type . | Amount, μg* . |
|---|---|---|---|
| BRO11F6 | A11/A3/A24 | IgG1,L | 0.2 |
| BVK1F9 | B8 | IgG1,K | 0.2 |
| DK7C11 | B12 | IgG1,K | 0.1 |
| GV5D1 | A1/A9 (except A*2403) | IgG1,L | 0.4 |
| HDG8D9 | B51/B35 | IgG1,L | 0.1 |
| SN607D8 | A2/A28 | IgG1,K | 0.05 |
| VTM1F11 | B27/B7/B60 | IgG1,K | 0.2 |
| OK2F3 | A3 | IgM,K | 0.6 |
| Name . | HLA specificity . | Type . | Amount, μg* . |
|---|---|---|---|
| BRO11F6 | A11/A3/A24 | IgG1,L | 0.2 |
| BVK1F9 | B8 | IgG1,K | 0.2 |
| DK7C11 | B12 | IgG1,K | 0.1 |
| GV5D1 | A1/A9 (except A*2403) | IgG1,L | 0.4 |
| HDG8D9 | B51/B35 | IgG1,L | 0.1 |
| SN607D8 | A2/A28 | IgG1,K | 0.05 |
| VTM1F11 | B27/B7/B60 | IgG1,K | 0.2 |
| OK2F3 | A3 | IgM,K | 0.6 |
Per 500 000 cells.
Single versus double antibody labeling
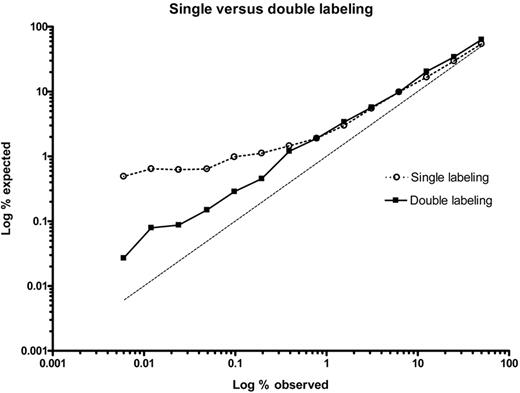
Efficiency of separation by FACS of cells from 2 donors was investigated for single labeling (only donor 1) and double labeling (both donors). In both cases the relationship between expected percentage chimeric cells and observed percentage chimeric cells in the positive fraction was plotted. Because single labeling targeted only HLA-A2 on the microchimeric cell fraction, and with chimeric cell frequencies of 0.4% or lower, the observed percentage of separated chimeric cells diverged from the expected percentage (Figure 1 open circles). In contrast, with double labeling, where one mAb targets the chimeric cell population and the second mAb targets the background cell population, the line representing relation between expected and observed cell frequency ran parallel to that for the expected situation. This was seen for microchimeric cell frequencies down to 0.006% (Figure 1 black squares). These observations show that double labeling is beneficial with respect to optimal separation of 2 cell populations, one of which that is present at a low frequency.
Single labeling versus double labeling of cell populations by HLA mAbs. Cells from donor 1 (HLA-A2, A33, B62, B17) were diluted in cells from donor 2 (HLA-A3, A11, B7, B35) in the following percentages: 50, 25, 12.5, 6.25, 3.13, 1.56, 0.78, 0.39, 0.20, 0.098, 0.049, 0.024, 0.012, and 0.006. For single labeling, MCA2090-A647 (anti–HLA-A2) mAb was used. For double labeling, MCA2090-A647 was combined with the mAb BRO11F6-biotine (anti–HLA-A3/A11).
Single labeling versus double labeling of cell populations by HLA mAbs. Cells from donor 1 (HLA-A2, A33, B62, B17) were diluted in cells from donor 2 (HLA-A3, A11, B7, B35) in the following percentages: 50, 25, 12.5, 6.25, 3.13, 1.56, 0.78, 0.39, 0.20, 0.098, 0.049, 0.024, 0.012, and 0.006. For single labeling, MCA2090-A647 (anti–HLA-A2) mAb was used. For double labeling, MCA2090-A647 was combined with the mAb BRO11F6-biotine (anti–HLA-A3/A11).
Sensitivity and specificity of qPCR primers
Before the application of cell separation by HLA mAbs, qPCR assays first needed to be developed as a tool for verifying the suitability of the cell sorting approach. Characteristics of HLA-specific and DYZ1 primer combinations used in the study are summarized in Table 2. These include concentration used in the PCR, sensitivity, and specificity.
Characteristics of HLA- and Y-specific primer sets and PCR assays
| PCR target . | Amplicon, base pairs . | Concentration, pmol/μL . | Sensitivity,*†‡ copies/% . | Specificity,‡§ Ct value pos/neg . | PCR efficiency, % . |
|---|---|---|---|---|---|
| HLA-A*01 | 205 | 30 | 1.7 ± 1.0 (0.006% ± 0.004%) | 20.7 / 39.1 (Δ = 18.5) | 117.2 ± 15.3 |
| HLA-A*02 | 205 | 10 | 1.7 ± 1.0 (0.006% ± 0.004%) | 21.7 / 40.0 (Δ = 18.3) | 107.4 ± 12.0 |
| HLA-A*24 | 125 | 30 | 2.0 ± 2.2 (0.007% ± 0.008%) | 22.9 / 37.4 (Δ = 14.5) | 127.1 ± 9.0 |
| HLA-B*51/52 | 120 | 10 | 2.2 ± 0.9 (0.008% ± 0.003%) | 22.6 / 36.4 (Δ = 13.8) | 124.9 ± 15.0 |
| HLA-B*08 | 105 | 20 | 1.7 ± 1.0 (0.006% ± 0.004%) | 21.6 / 38.2 (Δ = 16.6) | 120.3 ± 8.8 |
| HLA-DRB1*04 | 100 | 50 | 4.0 ± 1.6 (0.014% ± 0.006%) | 23.8 / 40.0 (Δ = 16.2) | 131.2 ± 12.0 |
| HLA-DRB1*10 | 90 | 20 | 1.3 ± 0.9 (0.005% ± 0.003%) | 20.0 / 37.4 (Δ = 17.3) | 131.0 ± 6.2 |
| HLA-DRB1*13 | 130 | 20 | 2.0 ± 2.2 (0.007% ± 0.008%) | 22.8 / 38.5 (Δ = 15.7) | 145.4 ± 3.9 |
| HLA-DRB1*16 | 145 | 20 | 1.7 ± 1.0 (0.006% ± 0.004%) | 20.3 / 34.2 (Δ = 13.9) | 116.4 ± 5.4 |
| DYZ1 | 86 | 20 | ≤ 1.0 ± 0.0 (≤ 0.004% ± 0.0%) | 19.4 / 33.4 (Δ = 14.0) | 117.9 ± 6.0 |
| HCK | 81 | 20 | 20.0 / – (–) |
| PCR target . | Amplicon, base pairs . | Concentration, pmol/μL . | Sensitivity,*†‡ copies/% . | Specificity,‡§ Ct value pos/neg . | PCR efficiency, % . |
|---|---|---|---|---|---|
| HLA-A*01 | 205 | 30 | 1.7 ± 1.0 (0.006% ± 0.004%) | 20.7 / 39.1 (Δ = 18.5) | 117.2 ± 15.3 |
| HLA-A*02 | 205 | 10 | 1.7 ± 1.0 (0.006% ± 0.004%) | 21.7 / 40.0 (Δ = 18.3) | 107.4 ± 12.0 |
| HLA-A*24 | 125 | 30 | 2.0 ± 2.2 (0.007% ± 0.008%) | 22.9 / 37.4 (Δ = 14.5) | 127.1 ± 9.0 |
| HLA-B*51/52 | 120 | 10 | 2.2 ± 0.9 (0.008% ± 0.003%) | 22.6 / 36.4 (Δ = 13.8) | 124.9 ± 15.0 |
| HLA-B*08 | 105 | 20 | 1.7 ± 1.0 (0.006% ± 0.004%) | 21.6 / 38.2 (Δ = 16.6) | 120.3 ± 8.8 |
| HLA-DRB1*04 | 100 | 50 | 4.0 ± 1.6 (0.014% ± 0.006%) | 23.8 / 40.0 (Δ = 16.2) | 131.2 ± 12.0 |
| HLA-DRB1*10 | 90 | 20 | 1.3 ± 0.9 (0.005% ± 0.003%) | 20.0 / 37.4 (Δ = 17.3) | 131.0 ± 6.2 |
| HLA-DRB1*13 | 130 | 20 | 2.0 ± 2.2 (0.007% ± 0.008%) | 22.8 / 38.5 (Δ = 15.7) | 145.4 ± 3.9 |
| HLA-DRB1*16 | 145 | 20 | 1.7 ± 1.0 (0.006% ± 0.004%) | 20.3 / 34.2 (Δ = 13.9) | 116.4 ± 5.4 |
| DYZ1 | 86 | 20 | ≤ 1.0 ± 0.0 (≤ 0.004% ± 0.0%) | 19.4 / 33.4 (Δ = 14.0) | 117.9 ± 6.0 |
| HCK | 81 | 20 | 20.0 / – (–) |
neg indicates negative; and pos, positive.
Sensitivity was determined by diluting positive DNA in 2 different negative, background DNA samples, each in duplicate. Column represents geometric means ± SD.
One copy represents 7.1 pg of DNA; 0.01% means detection of 1 copy in a background of 10 000; 0.005% means 1 in 20 000.
A threshold of 75.0 was used in the qPCR software.
For each primer set 0.2 μg of 2 different positive DNA samples and 4 different negative DNA samples was amplified. Column represents geometric means ± SD.
Separation of cell mixtures by HLA monoclonal antibodies and verification by qPCR
We established artificially spiked samples that simulated microchimerism during pregnancy by preparing cell dilutions of PBMCs from one donor in PBMCs from another donor. The first combination of 2 adult donors was investigated by the use of FACS sorting and qPCR in 3 separate experiments. HLA-A2 in the chimeric cell population and HLA-B35 in the background cell population were targeted by mAbs MCA2090 and HDG8D9, respectively. Cells from positive and negative fractions after sorting (Figure 2A top left and bottom right quadrants, respectively) were collected for qPCR analysis.
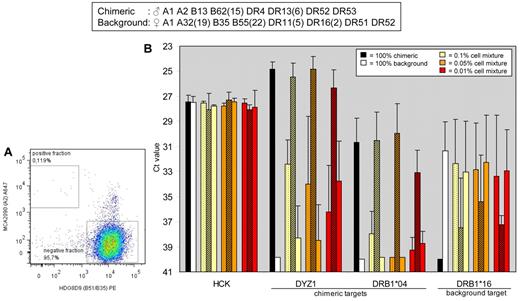
FACS-based separation of cells of 2 donors by HLA mAbs and verification by qPCR analysis. (A) Flow cytometric analysis. Monoclonal Abs MCA2090 (anti-HLA-A2) and HDG8D9 (anti-HLA-B51/B35) were used for cell sorting. Graph denotes 100 000 cells analyzed by the FACS in the 0.1% cell mixture. (B) Presort samples and sorted samples were processed for qPCR analysis of the reference gene HCK, the chimeric DNA markers DYZ1 and HLA-DRB1*04, and the background DNA marker HLA-DRB1*16. Bars show means ± SD of 3 separate experiments. Dotted bars, presort samples; shaded bars, positive fractions after sorting; open bars, negative fractions after sorting.
FACS-based separation of cells of 2 donors by HLA mAbs and verification by qPCR analysis. (A) Flow cytometric analysis. Monoclonal Abs MCA2090 (anti-HLA-A2) and HDG8D9 (anti-HLA-B51/B35) were used for cell sorting. Graph denotes 100 000 cells analyzed by the FACS in the 0.1% cell mixture. (B) Presort samples and sorted samples were processed for qPCR analysis of the reference gene HCK, the chimeric DNA markers DYZ1 and HLA-DRB1*04, and the background DNA marker HLA-DRB1*16. Bars show means ± SD of 3 separate experiments. Dotted bars, presort samples; shaded bars, positive fractions after sorting; open bars, negative fractions after sorting.
In 0.1%, 0.05%, and 0.01% cell mixtures, the PCR signal of the chimeric target DYZ1 in the positive fractions after sort (Figure 2B, shaded bars) was significantly enhanced compared with that in the presorted samples (dotted bars; Ct values 25.5 ± 1.1 vs 32.4 ± 1.9, P = .006; 24.8 ± 1.0 vs 34.0 ± 5.4, P = .04; and 26.3 ± 1.4 vs 36.2 ± 3.7, P = .002, respectively). Moreover, the PCR signals in the positive fractions after sorting were similar to those in the 100% chimeric, positive control samples (black bars; P = NS). Signals for reference gene HCK were not significantly different between conditions. Results for a second chimeric DNA marker (HLA-DRB1*04) corresponded with the results for DYZ1: in the positively sorted fractions PCR signals were significantly enhanced compared with those in the presort samples (P < .015).
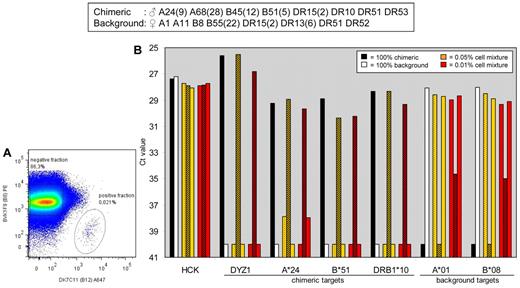
To validate results obtained from the first combination of donors, experiments were performed on a different combination of 2 donors (Figure 3). This time HLA-B45(12) on the chimeric cells and HLA-B8 on the background cells were targeted by mAbs DK7C11 and BVK1F9, respectively. Positive and negative cell fractions (Figure 2B bottom right and top left quadrants, respectively) were analyzed for 4 chimeric DNA markers (DYZ1, HLA-A*24, HLA-B*05, HLA-DRB1*10) and 2 background DNA markers (HLA-A*01, HLA-B*08) by qPCR. For all targets the PCR signal in the positive fractions (shaded bars) was similar to the signal seen for the 100%-chimeric cell populations (black bars). The aforementioned results demonstrate that microchimeric cell populations of 0.01% or greater can be enriched considerably to maximal extent by FACS by the use of HLA mAbs.
FACS-based separation of cells derived from a second combination of 2 donors by the use of HLA mAbs and verification by qPCR analysis. (A) Flow cytometric analysis. mAbs DK7C11 (anti-HLA-B12) and BVK1F9 (anti-HLA-A8) were used for cell sorting. Graph denotes 10% of the cells analyzed by the FACS in the 0.05% cell mixture. (B) Presort samples and sorted samples were processed for qPCR analysis of the reference gene HCK, the chimeric DNA markers DYZ1, HLA-A*24, HLA-B*51 and HLA-DRB1*10, and the background DNA markers HLA-A*01 and HLA-B*08. Dotted bars, presort samples; shaded bars, positive fractions after sorting; open bars, negative fractions after sorting.
FACS-based separation of cells derived from a second combination of 2 donors by the use of HLA mAbs and verification by qPCR analysis. (A) Flow cytometric analysis. mAbs DK7C11 (anti-HLA-B12) and BVK1F9 (anti-HLA-A8) were used for cell sorting. Graph denotes 10% of the cells analyzed by the FACS in the 0.05% cell mixture. (B) Presort samples and sorted samples were processed for qPCR analysis of the reference gene HCK, the chimeric DNA markers DYZ1, HLA-A*24, HLA-B*51 and HLA-DRB1*10, and the background DNA markers HLA-A*01 and HLA-B*08. Dotted bars, presort samples; shaded bars, positive fractions after sorting; open bars, negative fractions after sorting.
Application of HLA-directed cell sorting on clinical material
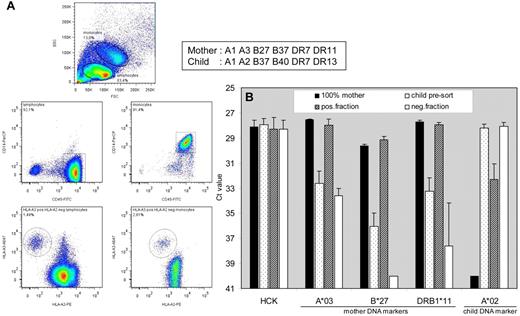
We next investigated applicability of HLA-targeted cell sorting on blood cells obtained after pregnancy (Figure 4). In unsorted cells from umbilical cord blood (dotted bars) maternal DNA markers HLA-A*03 (Ct: 32.6 ± 1.0), HLA-B*27 (Ct: 36.1 ± 1.1), and HLA-DRB1*11 (Ct: 33.2 ± 1.1) could be detected just above background by qPCR. After FACS sorting with 2 HLA mAbs (mAb SN607D8-PE against child's HLA-A2 and mAb OK2F3-biotine against mother's HLA-A3), PCR signals in the positive fractions (shaded bars) were significantly enhanced for HLA-A*03 (28.0 ± 0.5, P < .0005), HLA-B*27 (29.2 ± 0.3, P < .0001), and HLA-DRB1*11 (27.9 ± 0.2, P < .0001), and they were comparable with signals seen for 100% mother (positive control) samples (dark bars). In contrast, child DNA marker HLA-A*02 was not detected in the mother (Ct 40.0 ± 0.0), was maximally detectable in unsorted umbilical cord (Ct 28.2 ± 0.3), but showed a significantly lower signal in the positively sorted fraction (Ct 32.3 ± 1.3, P < .001). The aforementioned results demonstrate that HLA-directed FACS sorting allows enrichment of microchimeric cells from clinical material.
FACS-based separation of maternal cells from umbilical cord blood. FACS-sorted cells, acquired with 2 HLA mAbs (mAb SN607D8-PE against child's HLA-A2 and mAb OK2F3-biotine against mother's HLA-A3), were processed for qPCR analysis. (A) In both the lymphocyte (CD14−CD45+) and monocyte (CD14+CD45+) population (2 middle plots), gated from the forward-sideward scatter (top plot), microchimeric cells could be detected, which are HLA-A3pos and HLA-A2neg (2 bottom plots). (B) Reference gene HCK, maternal DNA markers HLA-A*03, HLA-B*27 and HLA-DRB1*11, and child DNA marker HLA-A*02 were analyzed in quadruplicate with qPCR on an equivalent of 100 unsorted and sorted PBMCs.
FACS-based separation of maternal cells from umbilical cord blood. FACS-sorted cells, acquired with 2 HLA mAbs (mAb SN607D8-PE against child's HLA-A2 and mAb OK2F3-biotine against mother's HLA-A3), were processed for qPCR analysis. (A) In both the lymphocyte (CD14−CD45+) and monocyte (CD14+CD45+) population (2 middle plots), gated from the forward-sideward scatter (top plot), microchimeric cells could be detected, which are HLA-A3pos and HLA-A2neg (2 bottom plots). (B) Reference gene HCK, maternal DNA markers HLA-A*03, HLA-B*27 and HLA-DRB1*11, and child DNA marker HLA-A*02 were analyzed in quadruplicate with qPCR on an equivalent of 100 unsorted and sorted PBMCs.
Discussion
For the current study we used a set of human monoclonal anti-HLA antibodies for the isolation of microchimeric cells. By FACS-based cell sorting and a combination of 2 mAbs, microchimeric cell populations in a frequency of 0.01% or greater could be separated from background cells in artificial spiked samples. This enrichment of microchimeric cell fractions is reached to maximal extent, as shown by qPCR for HLA class I, HLA class II, and Y-chromosome–localized genes. Furthermore, we demonstrated applicability of HLA-targeted FACS sorting after pregnancy by separating microchimeric maternal cells from child umbilical cord. Our unique set of in-house developed mAbs directed against human HLA antigens potentially allows FACS-based cell enrichment in > 90% of fetal-maternal combinations in pregnancies of white women. For those cases, where the current set is not discriminative, we can select additional mAbs from our large file of 120 HLA-specific mAbs.
Many years ago Herzenberg et al39 described a FACS enrichment procedure of chimeric cells whereby an anti–HLA-A2 mAb was used to target fetal cells among maternal peripheral blood lymphocytes. As shown in the present study, single HLA mAb labeling of cell populations in a frequency of < 0.4% leads to less sufficient separation, and the use of double HLA mAb labeling is crucial for optimal separation of low-frequency microchimeric cells. Single labeling of only the chimeric cells, when present in a frequency < 0.4%, resulted in more cells observed by FACS in the positive fraction than expected. Judged by the corresponding scatter plots (data not shown), this was the result of a high proportion of background cells that surreptitiously had ended up in the chimeric fraction. Simultaneous labeling of the chimeric and background cell population before FACS solved this problem. Our observations with regard to double labeling correspond with those from a previous study.40 More recently, flow cytometry with anti-HLA mAbs was presented as a suitable monitoring tool for chimerism after HLA-mismatched stem cell transplantation.41 The commercial mAbs used in that study allowed a threshold for detection that was not lower than 0.1%.
We observed that PCR signals of chimeric DNA markers in the positive fractions after cell sorting were significantly enhanced compared with those in the presort samples and that they were similar to those in the 100% chimeric, positive control samples. The latter observation indicates that chimeric cells could be obtained to full extent from genetically different background cells. We did observe slightly elevated PCR signals of background DNA markers in the positive fractions, especially with the 0.01% cell mixture. This finding indicates the presence of background cells within the chimeric cell fraction after sorting, which can probably be explained by the fact that yield sort settings rather than pure sort settings were used during FACS analysis. Pure sort settings would result into a minimal number of background cells in the positive fractions at the expense of a lower total number of chimeric cells being available for PCR analysis.
With respect to the sensitivity of HLA-targeted FACS sorting, we found that 0.01% cell dilutions are the lower limit for optimal cell separation. The eventual number of positively sorted cells was generally 2-fold lower than expected from the number of starting cells for the FACS. It is unclear what the cause is of this discrepancy. Adequate separation of microchimeric cells from a 0.001% mixture (ie, 1 in 100 000 cells) would require an excessive number of cells (at least 200 million) to start with for FACS to acquire 1000 positively sorted cells for PCR analysis (allowing 10 PCRs). Analysis of cell volumes of such size by FACS would take too long, increasing the risk of cell clumping during the procedure, and causing suboptimal cell separation. These aforementioned observations imply that application of current methodology may be cumbersome in pregnancy situations with fetal microchimerism in a frequency < 0.01%.
However, in pregnancy samples we (B.M.K., C.v.d.K., J.J.M.D., M.E., unpublished observations, February-April 2011) and others42 have frequently encountered relatively high percentages of maternal microchimerism. This indicates that acquisition of viable microchimeric cells by HLA-targeted FACS sorting is feasible in a considerable proportion of pregnancy samples, which enables further phenotypical characterization and investigation of functional relationship between chimeric and host-cell populations at the immunologic level. Indeed, in the current study we demonstrated applicability of HLA-targeted FACS sorting in the pregnancy setting by separating chimeric maternal cells from umbilical cord. Furthermore, stem cell transplantation and liver transplantation represent situations in which chimeric cell frequencies generally are relatively high, and monitoring of chimerism on these therapies would benefit from current methodology.
In summary, we have developed a technique that makes use of flow cytometry-based cell sorting and HLA-allele–specific human mAbs. This technique enables enrichment of potentially viable, low-frequency microchimeric cells from a background of cells that are genetically different. The current approach opens the way to investigation of the functional interplay between microchimeric cells and host cells, and it allows reliable microchimerism detection in clinical specimens.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: J.J.M.D. performed research and wrote the manuscript; C.v.d.K. performed the FACS sorting experiments; B.M.K. optimized and performed the qPCR assays; A.M. is head of the team that developed the HLA monoclonal antibodies, and he interpreted the data; S.A.S. contributed clinical material after pregnancy and interpreted the data; F.H.J.C. interpreted the data; M.E. analyzed and interpreted the data and wrote the manuscript; and all authors read the manuscript and commented on it.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael Eikmans, Department of Immunohematology and Blood Transfusion, Leiden University Medical Center, Building 1, E3-Q Albinusdreef 2, 2333 ZA Leiden, The Netherlands; e-mail: m.eikmans@lumc.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal