Abstract
Interactions between histone deacetylase inhibitors (HDACIs) and decitabine were investigated in models of diffuse large B-cell lymphoma (DLBCL). A number of cell lines representing both germinal center B-like and activated B-cell like DLBCL, patient-derived tumor cells and a murine xenograft model were used to study the effects of HDACIs and decitabine in this system. All explored HDACIs in combination with decitabine produced a synergistic effect in growth inhibition and induction of apoptosis in DLBCL cells. This effect was time dependent, mediated via caspase-3 activation, and resulted in increased levels of acetylated histones. Synergy in inducing apoptosis was confirmed in patient-derived primary tumor cells treated with panobinostat and decitabine. Xenografting experiments confirmed the in vitro activity and tolerability of the combination. We analyzed the molecular basis for this synergistic effect by evaluating gene-expression and methylation patterns using microarrays, with validation by bisulfite sequencing. These analyses revealed differentially expressed genes and networks identified by each of the single treatment conditions and by the combination therapy to be unique with few overlapping genes. Among the genes uniquely altered by the combination of panobinostat and decitabine were VHL, TCEB1, WT1, and DIRAS3.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common type of lymphoid neoplasm in the world, representing 30% to 40% of all lymphomas.1 Standard immunochemotherapy (cyclophosphamide, doxorubicin, vincristine, prednisone, and rituximab) ensures cure in 60% to 65% of patients, whereas the remainder progress or relapse.2,3 Although advances have been made in the treatment of DLBCL, especially with the addition of rituximab, it is now apparent that there are significant differences in prognosis related to the cell of origin and that this disease is heterogeneous. The prospect of pharmacologically modulating the oncogene-tumor suppressor gene balance in DLBCL, including critical targets such as BCL-6, p53, HSP, and nuclear factor-κB, using epigenetic strategies offers the possibility of targeting these diseases at their genomic roots.4 Histone deacetylase inhibitors (HDACIs) have primarily emerged as promising new agents for the treatment of T-cell lymphomas.5 In contrast, HDACIs have not been shown to be very active in B-cell lymphomas, for reasons that are not entirely clear.6 Although these agents are known to induce cell cycle arrest, apoptosis, and cell differentiation, recent evidence suggests that HDACIs may require rational partners to produce activity in B-cell lymphoma. Data for important epigenetic modifications in DLBCL7,8 suggest that the identification of synergistic second agents for use with HDACIs will be necessary to move epigenetic therapy forward in this disease.
One of the potential therapeutic partners are drugs that affect cytosine methylation in genomic DNA.9-11 Both hypermethylation and hypomethylation12 are well documented in cancer cells compared with their normal cells of origin. In a self-perpetuating loop, hypermethylated CpG islands attract methyl-CpG binding domain proteins (MBDP: MBD1, MBD2, MBD3, MeCP2 i Kaiso)13,14 and recruit HDACs, all of which inhibits transcription,15,16 and MBDPs and HDACs can bind histone methyltransferases and the polycomb group of proteins leading to metastable gene silencing.12 5-Azacytidine and decitabine have produced an increased rate of clinical response and improved quality of life in clinical trials in acute myeloid leukemia17 and myelodysplastic syndrome (MDS),18 which led to their approval for the treatment of MDS.19
It has been hypothesized that the combination of HDACI and hypomethylating agents might be synergistic by disrupting the previously mentioned transcription repressor complex consisting of MBDPs and HDACs. The marked synergistic in vitro effect of HDACIs and hypomethylating agents in inducing apoptosis has been shown in preclinical models of acute myeloid leukemia,20,21 mantle cell lymphoma,22 and solid tumors.23 Although the precise mechanism of these effects remains to be elucidated, the emerging experience from the clinical trials in support of the combination has largely been confined to the treatment of myeloid malignancies with little to no preclinical or clinical experiences in lymphoid diseases. Studies by Soriano et al,24 Garcia-Manero et al25 and Voso et al,26 to name only a few, reported encouraging effects in patients with acute myeloid leukemia and MDS treated with combinations of various HDACI and hypomethylating agents. Data from a study by Leshchenko et al22 showed that the combination of vorinostat and decitabine was synergistic for killing mantle cell lymphoma cells, with reexpression of MLF1 and PCDH8. These data strongly suggest that this combination might represent a novel treatment option for patients with relapsed or refractory mantle cell lymphoma.22
Here we evaluate the efficacy of several HDAC inhibitors (panobinostat, belinostat, depsipeptide, and vorinostat) and decitabine in combination in well-characterized lymphoma cell lines and a xenograft model of DLBCL. In addition, we analyze the molecular basis for this synergistic effect by evaluating gene expression and CpG methylation using microarrays for the cells treated with the single and combination agents in DLBCL: (1) to identify genes and pathways affected by panobinostat and decitabine alone and their combination; (2) to assess the relative contribution of individual drugs to methylation and gene-expression phenotype; and (3) to identify biomarkers that could potentially be used in clinical studies evaluating the efficacy of these drugs in patients with relapsed or refractory DLBCL.
Methods
Materials
Panobinostat (LBH589) was obtained from Novartis, Vorinostat (SAHA) from Merck, Belinostat (PXD101) from CuraGen, Romidepsin (depsipeptide) from Gloucester, and decitabine, 5-azacytidine, MS-275, and Scriptaid from Sigma-Aldrich. Reagents for Western blotting were purchased from Bio-Rad and Invitrogen. Reagents for immunofluorescence were purchased from Fisher Scientific. Dimethyl sulfoxide was obtained from Sigma-Aldrich.
Cell lines
OCI-Ly1, OCI-Ly7, and Su-DHL6 are germinal center DLBCL cell lines; OCI-Ly10, RIVA, and Su-DHL2 are activated B-cell DLBCL lines. Ly1, Ly7, Ly10, RIVA, and Su-DHL2 were grown in Iscove modified Dulbecco medium with 10% FCS; Su-DHL6 was grown in RPMI medium with 10% FCS. Fresh medium was added every 2 to 3 days, and the cells were kept at a cell concentration of 0.5 to 1 × 106/mL.
Cytotoxicity assays
Cytotoxicity was evaluated using the Cell-Titer-Glo Reagent (Promega) as reported previously.27
Flow cytometry
To study apoptosis, Yo-Pro-1 and propidium iodide were used (Vybrant apoptosis assay kit 4, Invitrogen) as previously described.27 The activation of caspase 3 was assayed using the CaspGLOW Green Caspase Staining Kit (BioVision) according to the manufacturer's instructions. Cell cycle analysis was assessed after 24, 48, and 72 hours of incubation with each agent. Cells were harvested in 0.3 mL of cold PBS and were then fixed with cold 70% ethanol, left incubating on ice for 1 hour, and washed twice with cold PBS. RNase A was added to a final concentration of 10 mg/mL to each sample and incubated at 37°C for 1 hour. Cells were analyzed by flow cytometry using the FL2-A channel.
Western blotting
Western blotting was performed as previously described.28 The following primary antibodies were used: anti-acetylated H3, anti-skp2, anti-p27, and anti–β-actin (all from Cell Signaling Technology). Goat anti–rabbit (Santa Cruz Biotechnology) secondary antibody was used.
Immunofluorescence
After incubation with panobinostat and decitabine for 48 hours, the samples were placed on the slides using the cytospin. After fixation in 10% formalin and 100% methanol, the slides were transferred to the blocking buffer (10% nonfat dry milk) and incubated with primary antibody in the humidity chamber overnight. The slides were then incubated with fluorochrome-conjugated secondary antibody for 45 minutes and mounted with 4,6-diamidino-2-phenylindole. The images were collected using Nikon Eclipse TE 2000-E inverted epifluorescent microscope, a 40×/0.60 oil objective, and a Nikon Photometrics Coolsnap HQ2 camera. The images were analyzed using NIS-Elements AR 3.2 software and Volocity 5.5.1 software. Antiacetylated H3 primary antibody (Cell Signaling Technology) and Cy3-conjugated donkey anti–rabbit IgG (Jackson ImmunoResearch Laboratories) secondary antibody were used.
Isolation of primary DLBCL samples
Sample collection and laboratory studies were in compliance with institutional review board and Helsinki protocols. Based on the immunostaining algorithm of CD10, BCL6, and MUM1/IRF4, the samples were classified as germinal center DLBCL, and all 3 samples were from de novo diagnosed patients and were used immediately after harvest. Mononuclear cells were isolated by Ficoll-Hypaque density gradient and negative cell selection using B-cell isolation kit II (MACS). After negative cell selection, primary cell lines were treated with drugs following the same procedure as in permanent cell lines, and the results were analyzed by flow cytometry.
In vivo tumor model
Five- to 7-week-old SCID beige mice (Harlan Sprague-Dawley) were injected with Ly1 line (107 cells) in the posterior flank subcutaneously. When the tumors approached 50 mm3, the mice were divided into 5 groups of 8 mice: (1) control group, which received saline with 10% dimethyl sulfoxide every day for 5 consecutive days; (2) decitabine alone group, which received a dose of 4.5 mg/kg divided into 3 equal doses on days 1, 3, and 5; (3) panobinostat alone, which received a total dose of 75 mg/kg divided into 5 equal doses on days 1 to 5; (4) combination group (treatment schedule 1), which received decitabine at a dose of 3 mg/kg (2 equal daily doses on days 1 and 5) and panobinostat at a dose of 75 mg/kg (5 equal daily doses on days 1-5); and (5) combination group (treatment schedule 2), which received decitabine at a dose of 4.5 mg/kg (3 daily doses on days 1, 3, and 5) and panobinostat at a dose of 75 mg/kg (5 daily doses on days 1-5). The data were expressed as average tumor volume (mm3) per group as a function of time. Tumors were assessed as previously described.27 Animals were maintained as previously described28 in accordance with the principles of laboratory animal care under an Institutional Animal Care and Use Committee–approved protocol.
Statistical analysis
IC50 (half the maximal inhibitory concentration) for each cell line was calculated using the Calcusyn Version 2.0 software (Biosoft).29
Relative risk ratio (RRR) was used as a model for establishing synergy between 2 drugs. RRR is based on calculating the ratio between the actual value and expected value (EV). In the case of 2 cytotoxic compounds, EV is calculated by formula: EV = NA × NB/100, where NA represents the percentage of viable cells in the sample treated with drug A and NB represents the percentage of viable cells in the sample treated with drug B. RRR values < 1 represent the synergistic effect of the 2 drugs: values equal to 1 indicate the mean additive effect of the drugs; and values > 1 represent an antagonistic effect.
For each of the experiments, synergy was confirmed by Calcusyn Version 2.0 software by determining combinational index (CI). Values < 1 represent synergistic effect of the 2 drugs, values equal to 1 indicate the mean additive effect of the drugs, and values > 1 represent an antagonistic effect.30 Wilcoxon rank-sum test was used to calculate the statistical significance in vivo for both actual and relative tumor volume. Relative tumor volume was calculated by comparing the actual volumes divided by the volumes on day 1 between the groups. A global test for significance was conducted by calculating the area under the actual (or relative) tumor volume curves using the trapezoidal rule.
Semiquantitative multiplex reverse-transcribed PCR
Reverse transcription was performed in a 20-μL reaction system with a total of 2 mg of RNase free DNase-treated RNA using Omniscript RT kit (QIAGEN) and oligo-d(T). Multiplex polymerase chain reactions (PCRs) were run in 20, 25, and 30 thermal cycles, and then the PCR products were observed by electrophoresis on 2% agarose gel and visualized after staining with ethidium bromide. The primers of von Hippel Lindau (VHL) were as follows: forward, 5′-TGAGCCTTCAGTCAGGGTTTGTCA-3′; and reverse, 5′-AGCACTTCAACAAGAGTGGACCGA-3′. The PCR product is 216 bp. β-Actin primers are as follows: forward, 5′-TCACCCACACTGTGCCCATCTACGA-3′; and reverse, 5′-CAGCGGAACCGCTCATTGCCAATGG-3′. The PCR product is 295 bp. Images were analyzed using ImageQuant TL Version 7.0 software (GE Healthcare), and band densities were normalized to β-actin.
Bisulfite sequencing
Genomic DNA, 0.5 to 1 μg, was bisulfite converted using the EpiTect Bisulfite Kit (QIAGEN) following the manufacturer's protocol. Converted DNA was amplified for the region chromosome 11:32 408 696–32 409 069 (National Center for Biotechnology Information36/hg18) by PCR using primers designed in MethPrimer: forward, TTGTATAATTTTTAGAGTTTTGTTT; and reverse, ACTTTACCTCCCTAACCCCTA. DNA was amplified using a 60°C touchdown for 9 cycles and 51°C for 29 cycles of amplification. For each sample, the PCR reactions were performed in duplicate and then pooled and purified using the Wizard SV Gel and PCR Clean-Up System (Promega) following the manufacturer's protocol. For bisulfite sequencing, the purified products were cloned using TOPO TA Cloning Kit for Sequencing (Invitrogen) based on the manufacturer's protocol. Plasmid DNA was isolated using the Wizard SV 96 Plasmid DNA Purification Kit (Promega), and multiple plasmids for each treatment condition were sequenced and analyzed for CpG methylation patterns.
Gene-expression microarrays
RNA was extracted using TRIZOL reagent (Invitrogen) from the samples collected after 48 hours of incubation with panobinostat and decitabine. Concentration was determined by NanoDrop spectrophotometer (Thermo Scientific), and the quality was determined by electrophoresis on agarose gel with ethidium bromide dye. After fragmentation and cDNA synthesis, gene-expression profiling analysis was performed using Illumina HumanHT-12 Version 3 Expression platform (Illumina). Arrays represent more then 25 000 annotated genes with > 48 000 probes. Expression intensity measures went through background correction, log2 transformation, and quantile normalization methods executed by BeadStudio Version 3.2 software. Data are available in National Center for Biotechnology Information Gene Expression Omnibus database (accession number GSE27226)
Methylation microarrays
DNA was extracted using the phenol chloroform method from the samples collected after 48 hours of incubation with panobinostat and decitabine. Concentration was determined by NanoDrop spectrophotometer (Thermo Scientific), and the quality was determined by electrophoresis on agarose gel with ethidium bromide dye. Bisulfite conversion of the DNA was performed using a conversion kit (Zymo Research). Genome-wide methylation analysis was performed using Illumina Human methylation27 platform (Illumina), which enables the direct investigation of 27 578 individual cytosines at CpG loci throughout the genome, which are focused on the promoter regions of 14 495 genes. Methylation scores represented as β values are generated for each site using BeadStudio Version 3.2 software (Illumina) and are computed based on the ratio of methylated to methylated plus unmethylated signal outputs. Thus, the β values range from 0 (unmethylated) to 1 (fully methylated) on a continuous scale. Intra-array normalization was performed by the Illumina BeadStudio Version 3.2 software. The resulting β-valued data matrix was normalized further using a quantile-normalization strategy, designed to reduce unwanted interarray variation using CLC Bio Genomics Workbench 3 Version 3.7.1 software package. Data are available in National Center for Biotechnology Information Gene Expression Omnibus database (accession number GSE27226)
Microarray data analysis
Unsupervised clustering of methylation and gene-expression data by principal component analysis were performed with the use of GenePattern Version 8.6 software (http://www.broad.mit.edu/cancer/software/genepattern/). Supervised analysis of the gene-expression and methylation data was carried out with a moderated t test with a significance level (P) < .05.
Gene network and gene ontology analysis
Analysis of the canonical pathways was performed using Ingenuity Pathway Analysis 8.6 software (Ingenuity Systems) using a P value cut-off of .05 to define the network eligible genes.
Results
HDACIs synergize with hypomethylating agents in DLBCL cells
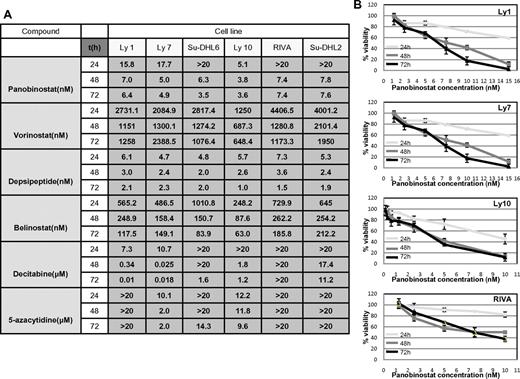
RRR and CI calculations were used to explore the synergy between the 2 classes of drugs as described in “Methods.” Before exploring cell viability with the combination of drugs, the IC50 values were determined for each of the 2 hypomethylating agents and 4 HDACIs at 3 time points across the spectrum of 6 DLBCL lines as shown in Figure 1A. All drugs demonstrated a concentration- and time-dependent effect (example of panobinostat in 4 DLBCL lines is shown in Figure 1B), which was more evident with hypomethylating agents, especially in the case of decitabine (data not shown). IC50 values for the HDACIs revealed that depsipeptide and panobinostat were the most potent HDACIs, followed by belinostat and vorinostat. Panobinostat exhibited a broad range of concentration-dependent effects and was therefore chosen for all subsequent experiments. Decitabine was slightly more potent than 5-azacytidine, with select cell lines being resistant to concentrations of hypomethylating agents as high as 20μM (Figure 1A).
IC50 values: luminometric assays. (A) Growth inhibition IC50 mean values in 6 DLBCL cell lines at 3 time points explored for 4 HDACI and 2 hypomethylating agents. (B) Panobinostat induces growth inhibition in a spectrum of DLBCL lines. In 4 shown DLBCL lines, panobinostat induced a concentration and time-dependent growth inhibition. Values represent means expressed as percentages compared with the untreated control; error bars represent SD.
IC50 values: luminometric assays. (A) Growth inhibition IC50 mean values in 6 DLBCL cell lines at 3 time points explored for 4 HDACI and 2 hypomethylating agents. (B) Panobinostat induces growth inhibition in a spectrum of DLBCL lines. In 4 shown DLBCL lines, panobinostat induced a concentration and time-dependent growth inhibition. Values represent means expressed as percentages compared with the untreated control; error bars represent SD.
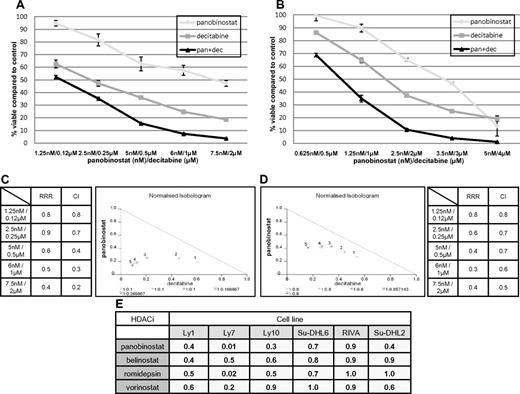
Figure 2A-B demonstrates the synergistic interaction for panobinostat and decitabine in the Ly1 and Ly10 lines. In both cell lines at all explored concentrations, the RRR and CI values were significantly < 1 and isobolograms clearly reveal synergy (Figure 2C-D). RRR values across the spectrum of explored lines show strong synergy or, in the case of romidepsin in RIVA and Su-DHL2 and vorinostat in Su-DHL6, an additive effect (Figure 2E). This synergy was observed in experiments with 2 additional HDACIs: MS-275 and Scriptaid in Ly1 and Ly10 DLBCL lines. Calculated RRR and CI values for these 2 HDACIs in combination with decitabine were < 1 (data not shown).
Synergy between panobinostat and decitabine in luminometric assays. (A) Combination of panobinostat and decitabine in Ly1 DLBCL line after 72 hours of incubation. Values represent means expressed as percentages compared with the untreated control; error bars represent SD. (B) Combination of panobinostat and decitabine in the Ly10 DLBCL line after 72 hours of incubation. Values represent means expressed as percentages compared with the untreated control; error bars represent SD. (C) RRR and CI values for combination of decitabine and panobinostat in Ly1 DLBCL line after 72 hours of incubation. Also shown is a normalized isobologram. (D) RRR and CI values for combination of decitabine and panobinostat in the Ly10 DLBCL line after 72 hours of incubation. Also shown is a normalized isobologram. (E) RRR values across the spectrum of DLBCL lines for 4 explored HDACIs in combination with decitabine.
Synergy between panobinostat and decitabine in luminometric assays. (A) Combination of panobinostat and decitabine in Ly1 DLBCL line after 72 hours of incubation. Values represent means expressed as percentages compared with the untreated control; error bars represent SD. (B) Combination of panobinostat and decitabine in the Ly10 DLBCL line after 72 hours of incubation. Values represent means expressed as percentages compared with the untreated control; error bars represent SD. (C) RRR and CI values for combination of decitabine and panobinostat in Ly1 DLBCL line after 72 hours of incubation. Also shown is a normalized isobologram. (D) RRR and CI values for combination of decitabine and panobinostat in the Ly10 DLBCL line after 72 hours of incubation. Also shown is a normalized isobologram. (E) RRR values across the spectrum of DLBCL lines for 4 explored HDACIs in combination with decitabine.
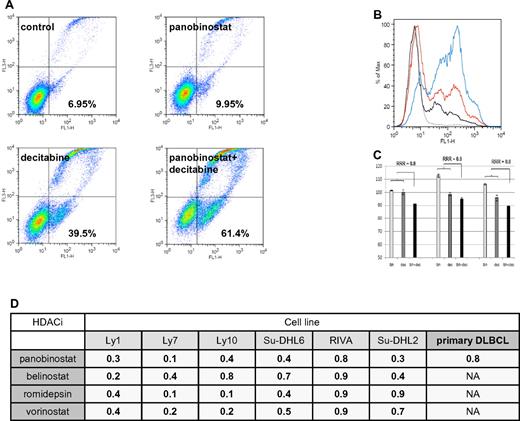
Flow cytometry revealed that the HDACIs and decitabine synergize in inducing apoptosis in DLBCL lines as well. As shown in Figure 3A-B, the combination of panobinostat and decitabine induced apoptosis in 61.4% of Ly1 cells compared with 9.95% for panobinostat alone and 39.5% for decitabine alone, resulting in synergistic RRR values of 0.6. Similarly, synergy was observed across the spectrum of DLBCL lines (Figure 3D). To validate these observations in primary cells, CD19+ tumor cells from patients with DLBCL were treated with panobinostat (2.5nM) and decitabine (2.5μM), and the extent of apoptosis was determined by flow cytometry. These data revealed that neither panobinostat nor decitabine alone induced apoptosis in DLBCL cells, whereas in combination the calculated RRR values were 0.8 (Figure 3C-D).
Assessment of apoptosis by Yo-Pro-1 and propidium iodide in DLBCL lines. (A) Ly1 DLBCL line was incubated with decitabine alone (5μM), panobinostat alone (5nM), or their combination for 48 hours. Compared with the untreated control, panobinostat alone induced apoptosis in a minimal amount of cells. Decitabine alone induced apoptosis in 39.5% of cells and the combination of drugs at the same concentrations induced apoptosis in 61.4% of lymphoma cells. After normalization to the untreated control, RRR value is 0.6. (B) Histogram depiction of the experiment shown under panel A. (C) Panobinostat (2.5nM) and decitabine (2.5μM) in combination induce apoptosis in primary DLBCL lines. (D) RRR values across the spectrum of DLBCL lines for 4 explored HDACIs in combination with decitabine in flow cytometric measurement of apoptosis. Also shown are the RRR values for the combination of panobinostat and decitabine in primary DLBCL lines.
Assessment of apoptosis by Yo-Pro-1 and propidium iodide in DLBCL lines. (A) Ly1 DLBCL line was incubated with decitabine alone (5μM), panobinostat alone (5nM), or their combination for 48 hours. Compared with the untreated control, panobinostat alone induced apoptosis in a minimal amount of cells. Decitabine alone induced apoptosis in 39.5% of cells and the combination of drugs at the same concentrations induced apoptosis in 61.4% of lymphoma cells. After normalization to the untreated control, RRR value is 0.6. (B) Histogram depiction of the experiment shown under panel A. (C) Panobinostat (2.5nM) and decitabine (2.5μM) in combination induce apoptosis in primary DLBCL lines. (D) RRR values across the spectrum of DLBCL lines for 4 explored HDACIs in combination with decitabine in flow cytometric measurement of apoptosis. Also shown are the RRR values for the combination of panobinostat and decitabine in primary DLBCL lines.
To determine the impact of schedule on the activity of the combination, cell viability of Ly1 and Ly10 cells was measured by flow cytometry after treatment with 5 or 10nM of panobinostat and 5 or 10μM of decitabine as follows: (1) simultaneous exposure; (2) 24 hours of panobinostat pretreatment followed by 24- and 48-hour exposure to decitabine; and (3) 24 hours of decitabine pretreatment followed by 24- and 48-hour exposure to panobinostat. In both Ly1 and Ly10 lines, the most synergy was obtained when panobinostat and decitabine were added simultaneously (RRR = 0.6 for Ly1; RRR = 0.4 for Ly10) as shown in supplemental Figure 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Pretreatment with panobinostat or decitabine produced synergy coefficients of 0.8 for both panobinostat → decitabine and decitabine → panobinostat combinations.
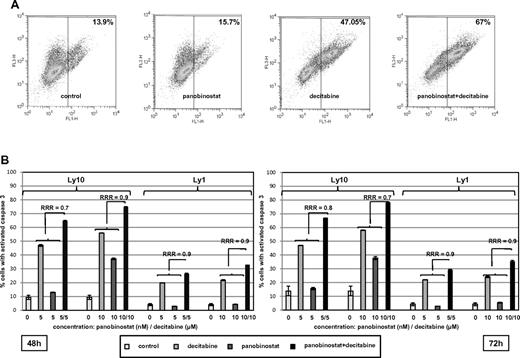
In addition, activation of caspase by the single agents and the combination are presented in Figure 4. Figure 4A shows a representative experiment where the combination of decitabine and panobinostat activated caspase 3 in 67% of Ly1 cells compared with 15.7% for panobinostat and 47.05% for decitabine. Figure 4B summarizes RRR values for caspase 3 activation in Ly1 and Ly10 cells at 2 time points and confirms observations from other assays supporting synergy of the combination. The combination of panobinostat and decitabine did not exhibit a specific growth arrest. Supplemental Figure 2 shows that panobinostat alone increased the percentage of cells in the G1 population, whereas decitabine alone and the combination of drugs had no effect on cell cycle, besides a slight increase in G2/M phase. The decrease in G1 and S phases in these 2 samples was consistent with an increase in apoptotic sub-G1 population.
Assessment of caspase 3 activation. (A) Percentage of Ly10 cells with activated caspase 3 at the 72-hour time point. Flow cytometric analysis reveals higher percentage of cells with activated caspase 3 in a sample treated with the combination of decitabine and panobinostat compared with samples treated with single agents (RRR = 0.8; P < .05). (B) Summary of caspase 3 activation in Ly1 and Ly10 cell lines with RRR at 2 time points (48 and 72 hours) and 2 concentrations of each drug (decitabine 5 and 10μM, panobinostat 5 and 10nM). Values represent means expressed as percentages compared with the untreated control; error bars represent SD.
Assessment of caspase 3 activation. (A) Percentage of Ly10 cells with activated caspase 3 at the 72-hour time point. Flow cytometric analysis reveals higher percentage of cells with activated caspase 3 in a sample treated with the combination of decitabine and panobinostat compared with samples treated with single agents (RRR = 0.8; P < .05). (B) Summary of caspase 3 activation in Ly1 and Ly10 cell lines with RRR at 2 time points (48 and 72 hours) and 2 concentrations of each drug (decitabine 5 and 10μM, panobinostat 5 and 10nM). Values represent means expressed as percentages compared with the untreated control; error bars represent SD.
The combination of panobinostat and decitabine enhances histone acetylation
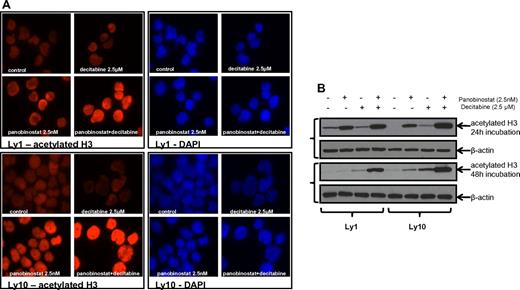
Immunofluorescent microscopy revealed an increase in histone H3 acetylation in the samples that were treated with the combination of panobinostat and decitabine compared with either single drug exposure. Immunofluorescent images were collected at 24 hours (Figure 5A) and 48 hours. All the results were confirmed by Western blot analysis of H3 acetylation (Figure 5B) at the same time points. Both experiments were performed for Ly1 and Ly10 DLBCL lines at concentrations previously shown to have a synergistic effect (2.5nM of panobinostat and 2.5μM of decitabine). The same effect on the histone acetylation was confirmed in the Su-DHL6 line (data not shown).
Histone acetylation in DLBCL lines after treatment with panobinostat and decitabine. (A) Immunofluorescence analysis of H3 acetylation in Ly1 and Ly10 lines after treatment with panobinostat and decitabine alone or their combination for 24 hours. The same exposure times reveal increased H3 acetylation in the combination group. (B) Western blot analysis of the H3 acetylation after treatment with panobinostat and decitabine alone or their combination in Ly1 and Ly10 lines at 2 time points (24 and 48 hours).
Histone acetylation in DLBCL lines after treatment with panobinostat and decitabine. (A) Immunofluorescence analysis of H3 acetylation in Ly1 and Ly10 lines after treatment with panobinostat and decitabine alone or their combination for 24 hours. The same exposure times reveal increased H3 acetylation in the combination group. (B) Western blot analysis of the H3 acetylation after treatment with panobinostat and decitabine alone or their combination in Ly1 and Ly10 lines at 2 time points (24 and 48 hours).
The combination of panobinostat and decitabine is synergistic in a xenograft model of DLBCL
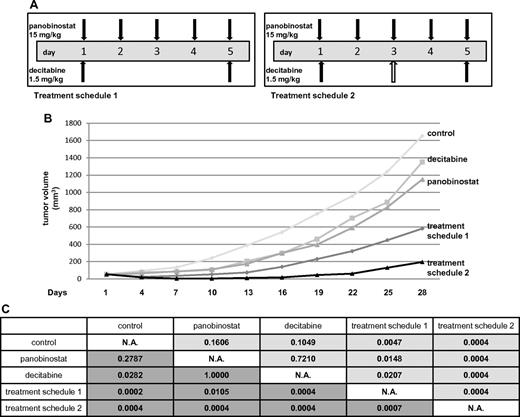
The maximum tolerated dose of each of the drugs and the combination was determined in SCID beige mice. These experiments revealed the following single-drug schedules to be tolerable in murine SCID beige animals: (1) panobinostat 15 mg/kg on days 1 to 5 and (2) decitabine 1.5 mg/kg on days 1, 3, and 5.
In the Ly1 xenograft experiment, 5 treatment cohorts were studied. Besides vehicle control, decitabine alone, and panobinostat alone, the combination of drugs was evaluated in 2 different treatment cohorts to compare the toxicity of different drug schedules. One combination group received panobinostat and decitabine in the same doses as in the single-drug groups (treatment schedule 2 in Figure 6A). The other combination group received panobinostat in the same dose as in the single-drug group while the total dose of decitabine was reduced (treatment schedule 1 in Figure 6A) because decitabine exhibited more toxicity as measured by weight loss in the initial maximum tolerated dose experiment. Figure 6B depicts mean tumor volumes of the treatment cohorts. Weight loss > 10% was observed in some of the animals in all the groups except for the control (Table 1). More than 10% of weight loss was observed in treatment schedule 2 (5 mice) and decitabine alone (3 mice) groups. Only one animal in the panobinostat alone and treatment schedule 1 groups experienced > 10% loss of body weight. Three animals in the control group and one in decitabine alone cohort had to be killed because of progressive disease (tumor volume exceeding 2000 mm3) on day 28. In the combination treatment cohort receiving schedule 2, 3 animals died because of combined drug toxicity: 2 on day 10 and 1 on day 13. The only group in which a complete remission of tumors was observed was actually the treatment schedule 2 cohort where 5 of 8 animals achieved complete remission: 2 on day 4 and 3 on day 7. The statistical significance between the groups is shown in Figure 6C. Both of the combination groups had a statistically significant tumor growth inhibition compared both with control and cohorts treated with single drugs. The same statistical significance was observed comparing the area under the tumor volume for each of the cohorts (P < .05). We also applied RRR calculation to confirm the synergy between the drugs. The EV was calculated by dividing each of the tumor volumes from the treated groups with the control on the day of measurement and multiplying the 2 values. Actual value (tumor volume in the treated group divided by the tumor volume of the control) was divided by the EV to obtain the RRR values, which were 0.4 and 0.06 for the treatment schedule 1 and schedule 2, respectively (data not shown).
Panobinostat and decitabine synergize in the SCID beige DLBCL xenograft model. (A) Treatment schedules for groups of animals receiving both panobinostat and decitabine. Total decitabine dose in treatment schedule 1 was 3 mg/kg, and in treatment schedule 2 was 4.5 mg/kg. (B) Tumor volumes for treatment cohorts expressed as means. (C) Statistical significance between the groups. The upper right triangle of the table (pale gray) represents P values for actual tumor volumes between the groups; and lower left triangle (dark gray), P values for relative tumor volumes (calculated by dividing the actual volume of the tumor with its volume on the treatment day 1).
Panobinostat and decitabine synergize in the SCID beige DLBCL xenograft model. (A) Treatment schedules for groups of animals receiving both panobinostat and decitabine. Total decitabine dose in treatment schedule 1 was 3 mg/kg, and in treatment schedule 2 was 4.5 mg/kg. (B) Tumor volumes for treatment cohorts expressed as means. (C) Statistical significance between the groups. The upper right triangle of the table (pale gray) represents P values for actual tumor volumes between the groups; and lower left triangle (dark gray), P values for relative tumor volumes (calculated by dividing the actual volume of the tumor with its volume on the treatment day 1).
Summary of toxicity and response to therapy at day 28 from the start of the treatment
| . | Weight loss > 10% . | Death-drug toxicity . | Death-tumor volume . | Complete remission . |
|---|---|---|---|---|
| Control | 0/8 | 0/8 | 3/8 | 0/8 |
| Panobinostat | 1/8 | 0/8 | 0/8 | 0/8 |
| Decitabine | 3/8 | 0/8 | 1/8 | 0/8 |
| Schedule 1 | 1/8 | 0/8 | 0/8 | 0/8 |
| Schedule 2 | 5/8 | 3/8 | 0/8 | 5/8 |
| . | Weight loss > 10% . | Death-drug toxicity . | Death-tumor volume . | Complete remission . |
|---|---|---|---|---|
| Control | 0/8 | 0/8 | 3/8 | 0/8 |
| Panobinostat | 1/8 | 0/8 | 0/8 | 0/8 |
| Decitabine | 3/8 | 0/8 | 1/8 | 0/8 |
| Schedule 1 | 1/8 | 0/8 | 0/8 | 0/8 |
| Schedule 2 | 5/8 | 3/8 | 0/8 | 5/8 |
The combination of panobinostat and decitabine has unique effects on gene-expression and gene-specific CpG methylation
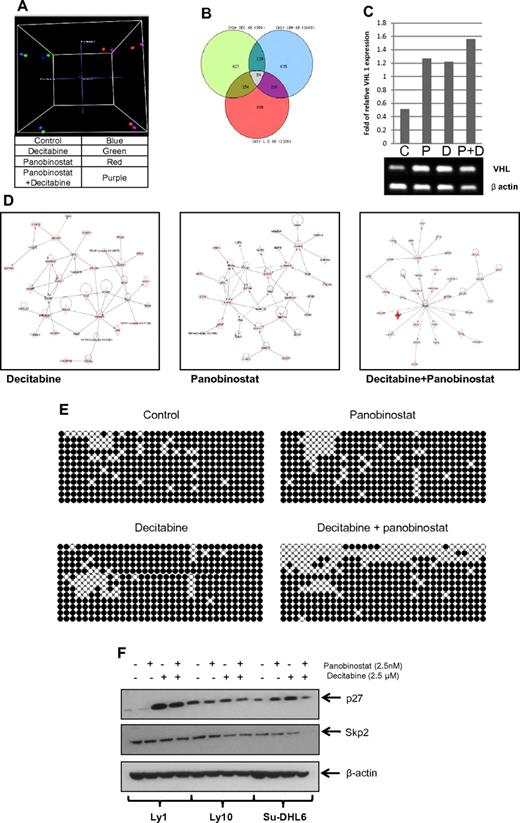
To clarify the molecular basis for synergy between these 2 types of drugs, 3 DLBCL lines were treated with panobinostat and decitabine alone and in combination. Gene-expression profiling (Figure 7) of all 3 DLBCL cell lines revealed a specific pattern of genes whose expression was altered in the samples treated by the combination and not altered by the single drugs. A principal component analysis clearly clusters the samples treated with panobinostat and the combination together and at greater distances from untreated samples and samples treated by decitabine alone (Figure 7A). A Venn diagram analysis (Figure 7B) illustrates the number of significantly altered genes (P < .05) that overlap between each of the 3 treatment conditions. There is a separate subset of genes altered uniquely by the combination and not altered by single-drug treatments. The list of specific genes altered by each treatment modality and their overlap is shown in supplemental Table 1. To identify pathways affected by different treatments, the Ingenuity Pathway Analysis was used. The top pathways influenced were significantly different between the different samples (Figure 7D). The top network of genes differentially expressed (P < .05) by panobinostat involved critical transcription factors, such as GATA1, GATA4, SMAD, and DNMT3A. In addition, a network-functional analysis of genes perturbed by the combination treatments was enriched for critical pathways involved in cell death, cell development, and cellular proliferation.
Panobinostat and decitabine affect the expression of distinct sets of genes in DLBCL cell lines. Gene-expression profiling of DLBCL cell lines 48 hours after treatment with decitabine, panobinostat, and the combination of both drugs was performed using Illumina HT-12 arrays. (A) Three-dimensional principal component analysis clusters panobinostat-treated cells distinctly compared with decitabine-treated cells. (B) Venn diagram of the overlap in differentially expressed genes (P < .05) between the 3 treatment groups. (C) Semiquantitative PCR of Su-DHL6 samples treated with decitabine (7.5μM), panobinostat (7.5nM), and their combination after 48 hours showing increase in VHL levels. (D) Top networks enriched by Ingenuity Pathway Analysis by decitabine, panobinostat, and the combination of both drugs. (E) WT1 intron 1 CpG island bisulfite sequencing in Ly10 cell line after treatment with either drug alone or in combination. (F) The effect of epigenetic therapy in 3 DLBCL cell lines on p27 and skp2 levels.
Panobinostat and decitabine affect the expression of distinct sets of genes in DLBCL cell lines. Gene-expression profiling of DLBCL cell lines 48 hours after treatment with decitabine, panobinostat, and the combination of both drugs was performed using Illumina HT-12 arrays. (A) Three-dimensional principal component analysis clusters panobinostat-treated cells distinctly compared with decitabine-treated cells. (B) Venn diagram of the overlap in differentially expressed genes (P < .05) between the 3 treatment groups. (C) Semiquantitative PCR of Su-DHL6 samples treated with decitabine (7.5μM), panobinostat (7.5nM), and their combination after 48 hours showing increase in VHL levels. (D) Top networks enriched by Ingenuity Pathway Analysis by decitabine, panobinostat, and the combination of both drugs. (E) WT1 intron 1 CpG island bisulfite sequencing in Ly10 cell line after treatment with either drug alone or in combination. (F) The effect of epigenetic therapy in 3 DLBCL cell lines on p27 and skp2 levels.
Genome-wide methylation analysis showed similar results with a specific subset of genes hypomethylated in the combination sample compared with the single-drug treated samples (supplemental Figure 3; supplemental Table 2). Correlation between genome-wide methylation analysis and gene-expression profiling identified 16 overlapping genes in the samples treated by combination of panobinostat and decitabine. These genes included: ABCD3, C20orf75, CD19, CEACAM5, CHKA, DIRAS3, FUBP1, GALT, HSPC138, HSPC268, METAP1, PGM2L1, TCEB1, VHL, WT1, and ZFP95. Increase in VHL mRNA in Su-DHL6 samples after treatment with panobinostat and decitabine is shown in Figure 7C. Subsequently, we analyzed the effect on VHL downstream molecules, skp2 and p27.30 In Su-DHL6, there was a decrease in skp2 levels at 24 hours and a subsequent increase in p27 (Figure 7F). The levels of Skp2 did not change in Ly1 and decreased in Ly 10 at 48 hours (data not shown). The levels on p27 increased significantly in Ly1 cell line treated with decitabine alone and combination of decitabine and panobinostat. Knockdown of VHL with shRNA in Su-DHL6 cell line did not change the effect panobinostat and decitabine had on cell viability. RRR values in Su-DHL6 cell with and without VHL shRNA were the same (data not shown). Furthermore, bisulfite sequencing of a CpG island in WT1 intron 1 revealed a significant increase in the number of hypomethylated CpG dinucleotides in the combination sample, thus confirming the synergistic effect of this combination compared with decitabine alone and panobinostat alone (Figure 7E).
Discussion
Epigenetic therapy represents a novel approach for treating hematologic malignancies that is changing the current treatment paradigms for many of these diseases. Recent examples include the Food and Drug Administration approval of vorinostat and romidepsin for the treatment of cutaneous T-cell lymphoma,31,32 and decitabine and 5-azacitidine for MDS.33 Before the Food and Drug Administration approval of hypomethylating agents for MDS, the disease was managed with supportive measures and the only curative option was allogeneic stem cell transplantation.34 Recent reports have shown that 5-azacitidine improved overall survival in patients with MDS.35 Even more interesting is the potential value of using these agents in combination. For example, several publications have demonstrated that combinations of HDACIs and hypomethylating agents in myeloproliferative diseases are associated with even more significant response rates and duration of response.36 In the treatment of cutaneous T-cell lymphoma, both single-agent romidepsin and vorinostat have demonstrated overall response rates in the range of 30% to 40% in heavily treated patients.37 What remains unclear is why these agents have not emerged in similar fashion for the treatment of B-cell malignancies.
Although the single-agent activity of various epigenetic therapies in B-cell malignancies is limited at best,6 there is a paucity of data exploring epigenetic combinations in B-cell lymphomas, certainly little in DLBCL. The synergy of vorinostat and decitabine in killing mantle cell lymphoma cells is one example of an effective epigenetic combination in a B-cell malignancy.22 We have explored the effect of different HDACIs and decitabine combinations in in vitro and in vivo models of DLBCL. These data suggest a class effect, with all 4 HDACIs (panobinostat, belinostat, vorinostat, and depsipeptide) synergizing with decitabine in cytotoxicity assay across the spectrum of DLBCL cells. This effect was observed even in the cell lines considered relatively resistant to decitabine. RRR calculations from these experiments reveal that the synergistic effect of HDACIs and decitabine is most prominently present in Ly1 and Ly7 cell lines (germinal center B-like [GCB] DLBCL subtype), whereas the values closest to 1 (the least synergistic) are present in RIVA cell line (activated B-cell like [ABC] DLBCL subtype). This might point to a trend that this combination might be more potent in GCB subtype of the disease, further corroborated by the synergistic trend shown in primary GCB-like samples. However, there were also ABC lines in which the combination yielded synergistic effect (highly synergistic in Ly10 and intermediately synergistic in Su-DHL2) and GCB lines that were less sensitive to the combination (Su-DHL6). This might point to the fact that in vitro conditions might mute the differences between 2 disease subtypes resulting in nuclear factor-κB activation not only in ABC but also in GCB subtype, as well as shown in significant proportion of cell lines analyzed by Compagno et al.38 For these reasons, a wider analysis of the outcome of patients with both subtypes of the disease treated with panobinostat and decitabine within a setting of a clinical trial is warranted. Especially interesting would be to analyze primary samples taken from these patients before and after epigenetic therapy with genome-wide methylation and gene-expression arrays as shown by Shaknovich et al.39
Synergy was validated in a number of other assays, including a caspase 3 activation and apoptosis. The combination of panobinostat and decitabine induced markedly increased histone acetylation. This augmented effect on protein acetylation might partially account for the synergistic effects seen in cytotoxicity assays. Disruption of a repressor complex formed by a family of methyl binding proteins bound to DNA with DNA methyltransferases, histone methyltrasferases, and the polycomb group of proteins could yield increased histone acetylation and reexpression of epigenetically silenced genes.15 The in vitro observations were confirmed in an in vivo murine xenograft experiment with the Ly1 DLBCL line. These data demonstrated that the combination of panobinostat and decitabine was synergistically effective and tolerable in inhibiting tumor growth in a biologic model of the disease.
Genome-wide methylation analysis and gene-expression profiling demonstrated that the combination of these 2 epigenetic therapies produced a unique gene-expression profile compared with the samples treated with single drugs. Among the genes specifically altered by the combination were VHL, TCEB1, WT1, and DIRAS3. In a study by Amara et al,40 the authors reported VHL methylation to be a common event in DLBCL. Methylation of VHL produced a negative impact on disease-free survival and overall survival, in a similar fashion to that seen with methylation of p16 and death-associated protein kinase.40 Patients with unmethylated VHL had a median survival of 58 months compared with those with methylated VHL, whose median survival was only 16 months.40 VHL acts by down-regulating HIF41 and skp2.30 In one of DLBCL cell lines, VHL down-regulated skp2 subsequently increasing p27 levels. These data suggest that the combination of these 2 classes of epigenetic drugs might be able to overcome the adverse prognostic impact of VHL methylation. Besides DLBCL, other hematologic diseases are known to have the VHL gene hypermethylated, including multiple myeloma42 and chronic lymphocytic leukemia.43 However, in our model, knockdown of VHL with shRNA did not change the effect panobinostat and decitabine had on cell viability. This and the fact that other target genes identified from the genome-wide analyses in this study included TCEB1, WT1, and DIRAS 3 mean that these drugs in DLBCL cells influence various targets and yield effect on multiple genes, which are important in cell survival. This might also explain why decitabine and panobinostat are effective in both GCB and ABC models.
These data strongly support the potential therapeutic role of a combinatorial epigenetic platform for the treatment of B-cell lymphomas, in particular in patients with DLBCL. The established safety profile of several combinations,44 coupled with the strong preclinical evidence, should make these early-phase trials a priority. Clearly, future studies will need to focus on integrating the appropriate correlative studies, with an effort to identify and/or validate biomarkers of activity with these combinations, which would be optimal for each disease context.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Riccardo Dalla Favera for kindly providing DLBCL cell lines and Dr Jeffrey Conroy (Roswell Park Cancer Center Genomics Shared Resource) for assistance with the microarray analysis.
This work was supported by Leukemia & Lymphoma Society grant LLS 7017-09 (O.A.O.), and National Institutes of Health grant R21CA125461 (B.T.).
National Institutes of Health
Authorship
Contribution: M.K. performed the majority of the experiments; M.K. and O.A.O. designed the study, analyzed the data, and wrote the paper; E.M. contributed with the cytotoxicity experiments and mouse xenograft experiments; L.S. conducted the RT-PCR experiment; J.A. contributed with in vitro experiments; V.E.S. conducted the statistical analysis, G.B. provided primary tissue samples and contributed in the writing of the paper; N.U., V.V.L., and S.P. performed microarray data and gene network analysis; S.P. contributed in the writing of the paper; B.T. contributed in designing the microarray and bisulfite sequencing experiments and editing the manuscript; and A.M.T. contributed with bisulfite sequencing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Owen A. O'Connor, New York University Cancer Institute, New York University Langone Medical Center, 522 First Avenue, Smilow 1201, New York, NY 10016; e-mail: owen.o'connor@nyumc.org.