Abstract
Aberrations in protein ubiquitination have recently been identified in the pathogenesis of acute myeloid leukemia (AML). We studied whether expression changes of more than 1600 ubiquitination related genes correlated with clinical outcome in 525 adult AML patients. High expression of one of these genes, BRE, was observed in 3% of the cases and predicted favorable prognosis independently of known prognostic factors (5-year overall survival: 57%). Remarkably, unsupervised expression profiling showed that 86% of high BRE-expressing patients were confined to a previously unrecognized cluster. High BRE expression was mutually exclusive with FLT3 ITD, CEBPA, IDH1, and IDH2 mutations, EVI1 overexpression, and favorable karyotypes. In contrast, high BRE expression co-occurred strongly with FAB M5 morphology and MLL-AF9 fusions. Within the group of MLL-AF9–positive patients, high BRE expression predicted superior survival, while normal BRE expression predicted extremely poor survival (5-year overall survival of 80% vs 0%, respectively, P = .0002). Both the co-occurrence of high BRE expression with MLL-AF9 and its prognostic impact were confirmed in an independent cohort of 436 AML patients. Thus, high BRE expression defines a novel subtype of adult AML characterized by a favorable prognosis. This work contributes to improved risk stratification in AML, especially among MLL-AF9–positive patients.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease characterized by recurrent genetic lesions that have different prognostic impact. These lesions affect various biologic processes like gene transcription, nuclear transport, epigenetics, and signal transduction.1-3 Recently, mutations in genes that are involved in ubiquitination have been identified in AML pathogenesis as well.4 Ubiquitination is a posttranslational modification of cellular proteins. In this process, the small ubiquitin protein is conjugated to substrate proteins as a single moiety or as ubiquitin chains. This may mark substrates for degradation by the 26S proteasome or may alter the substrate's functional activity, depending on the type of ubiquitin chain that is attached.4 Substrate ubiquitination is catalyzed via a 3-step cascade involving E1-activating enzymes, E2-conjugating enzymes, and E3 ligases. Like other posttranslational modifications, ubiquitination is a reversible process, as deubiquitinases (DUBs) remove ubiquitin chains from substrates. Because of its versatile role in many biologic pathways, it is not surprising that changes in protein ubiquitination contribute to cancer development including malignant hematopoiesis.4,5 For example, mutations in the E3 ligase C-CBL have been described in AML and other myeloid neoplasms.6,7 Mutations in this important negative regulator of receptor tyrosine kinases lead to prolonged receptor signaling and hence enhanced proliferation. Apart from gene mutations, altered expression of proteins that regulate ubiquitination can also contribute to leukemogenesis. An example is TRIB2 overexpression which triggers proteasomal degradation of CEBPα. This results in impaired myeloid differentiation and the onset of leukemia in mouse models.8 Independent research lines have shown the potential of interfering with the ubiquitination process as anticancer therapy. Several of these therapies are currently tested in clinical trials for hematologic malignancies.9-12
Recurrent mutations in AML have an impact on disease outcome. Treatment decisions are therefore dependent on the presence or absence of these mutations. Because expression levels of certain genes, like EVI113 , are significantly associated with disease outcome, the type of treatment is also guided by expression levels of individual genes. In the last decade, gene expression profiles of large AML cohorts have been generated. By combining these expression data with clinical data, new prognostic AML subgroups have been identified.14-16 Despite improved genetic-based risk stratification, the survival within prognostic subgroups still shows large variation. The identification of novel recurrent mutations or expression markers with prognostic value may therefore improve individualized treatment strategies.
Here we studied whether changes in expression of genes involved in ubiquitination (hereafter referred to as Ub genes) correlated with disease outcome using outlier gene expression analysis. With this type of analysis, genes that are differentially expressed in a subset of patients are nominated. By comparing the expression data of these genes with clinical parameters of the patients, correlations with survival can be studied. Because gene expression data of large AML cohorts are available nowadays, outlier analysis has great potential in the identification of novel prognostic factors. Indeed, by applying this approach for Ub genes, we identified a novel prognostic factor in AML. This work may add to the risk stratification of AML patients.
Methods
Selection of Ub genes
The interpro database (www.ebi.ac.uk/interpro/) was used to generate a list of structural protein domains involved in ubiquitination, using several enzyme classes as search terms (eg, E2-conjugating enzyme, E3 ligase, deubiquitinating enzyme, proteasome), as well as the overall term “ubiquitin.” The list of interpro codes was subsequently filtered manually for domains with a clear link to ubiquitination, including UBA (ubiquitin-associated) and UBL (ubiquitin-like) domains, resulting in a total of 121 individual codes (supplemental Table 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Using the gene ontology (GO) database, GO terms involving ubiquitination were collected as well (75 GO terms, supplemental Table 1B).
By use of the biomart database (www.biomart.org/biomart/martview/), genes containing the relevant interpro domains and GO terms were identified. To ensure maximal completeness, this list of unique refseq entries was merged with a list of Ub genes previously published by Harper and colleagues17 and an overview of human DUBs generated by S. Urbé and S. Hayes (Physiological Laboratory, University of Liverpool; e-mail communication, September 2009).
Gene expression profiling of AML samples
For the present study, our previously published cohort of 525 AML patient samples, 11 CD34+ donor samples, and 5 normal BM (NBM) control samples was used.18 Details on clinical data and experimental procedures of the microarray profiling on the Affymetrix HG-U133 plus 2.0 arrays can be found elsewhere.18 The array data are accessible online at the Gene Expression Omnibus (GSE14468).
To validate observed findings in an independent cohort, 40k cDNA array data (arrays manufactured by the Stanford Functional Genomics Facility) of a second previously published cohort of 436 AML patients were analyzed, including 11 MLL-AF9–positive patients (GSE16432).19 In addition, Affymetrix HG-U133 2.0 plus array data of 14 adult MLL-AF9–positive AML patients were used for data validation. These samples were provided by the German-Austrian AML Study Group (AMLSG) with patient informed consent obtained in accordance with the Declaration of Helsinki and institutional review board approval from all participating centers. Patients were entered into AML HD93, AML HD98A, AMLSG 06-04, and AMLSG 07-04 treatment protocols.
For the validation MLL-AF9 dataset (n = 25), BRE expression data of the HG-U133 plus 2.0 arrays (11 MLL-AF9 samples) and the 40k cDNA arrays (14 MLL-AF9 samples) were combined. Data normalization and filtering for well expressed genes was performed separately for the cDNA and Affymetrix datasets as previously described.20,21 Subsequently, datasets were combined following (1) the averaging of the expression of multiple clones/probe sets measuring the same gene (based on the gene symbols), and (2) the mean-centering of the cDNA and Affymetrix data to reduce platform specific effects. Normalized BRE expression data of these 25 patients are available in supplemental Table 2.
Cell isolation
For the isolation of primary donor blood cells, mononuclear cells were first isolated from buffy coats using ficoll 1077 gradients (GE Healthcare). Subsequently, T cells were isolated by magnetic cell sorting (MACS) using anti-CD3 microbeads (Miltenyi Biotec). From the remaining fraction, monocytes, NK cells, and B cells were isolated by FACS using CD14 (BD Biosciences), CD19 (DAKO), and CD56 (BD Biosciences) Abs, respectively. Granulocytes were isolated from total blood samples by MACS using anti-CD14 microbeads (Miltenyi Biotec). Cell purity of the hematopoietic fractions was confirmed to be 90% or higher by flow cytometric analyses. In addition to primary cells, the t(9;11)–positive cell lines Molm13, Monomac-6, and THP1 were used. Isolation of AML cells, RNA purification, and cDNA preparation have been described previously.18,20
BRE QPCR and MLL-AF9 fusion gene detection
BRE expression was measured by QPCR on cDNA samples using a commercially available primer/probe set (Applied Biosystems; Hs01046283_m1), and data were normalized by PBGD or β-ACTIN quantitative PCR (QPCR).22 MLL-AF9 breakpoint PCR on the cohort of 525 patients was performed as described previously.23,24 For the validation cohort, conventional chromosome banding and FISH for MLL-AF9 translocations were performed as previously described.25
Statistical analyses
Genes with outlier expression were defined as probe sets that showed expression values of more than 4× the SD above or below the mean expression of all AML samples for at least one sample. As this analysis does not involve a discernable null hypothesis, a P value is not obtained. It is therefore unattainable to perform corrections for multiple testing. For survival analyses, we selected probe sets detecting outlier expression in at least 10 patients because this accounts for at least 2% of the patients, a percentage that we consider biologically and clinically relevant. In addition, the likelihood that correlations are found by chance would increase in smaller groups.
The Cox proportional hazard model was used for multivariate analyses to identify whether genes were independent prognostic factors for overall survival (OS) and event-free survival (EFS; with events defined as: relapse, death by any cause, and no complete remission), including known prognostic factors in the model (ie, age [continuous variable], white blood cell count [WBC; continuous variable], FAB M5 morphology [positive vs negative], stem cell transplantation status, favorable karyotypes [t(8;21), t(15;17), inv(16)], CEBPA double mutations, FLT3 ITD, and NPM1 mutations). Before performing multivariate analyses, the proportional hazard assumption was tested for high BRE expression and no indication of nonproportionality was found.
Differences between variables of patient subgroups were statistically tested by performing χ2 tests, Fisher exact tests, or Mann-Whitney U tests. Survival differences were visualized using Kaplan-Meier plots. Significance of survival differences between patient groups was calculated using the log-rank method. All analyses were performed 2-tailed. Because we anticipated that only small numbers of patients were identified with outlier expression per probe set, we considered P < .05 as statistically significant.
Results
Identification of genes with prognostic impact in AML using outlier analysis
To study the expression of genes encoding proteins involved in ubiquitination, we selected these proteins based on the presence of catalytic domains used in the ubiquitin pathway and relevant GO terms. In total, 1618 proteins (based on HUGO Gene Nomenclature Committee [HGNC] nomenclature) were selected, which are encoded by 2608 refseq entries (Table 1, supplemental Table 3). The Ub genes are represented by 2890 probe sets on the used Affymetrix HG-U133 plus 2.0 microarray.
Representation of important ubiquitination core enzyme families in the human genome
| Protein family . | Structural domains . | No. of Refseq IDs . | No. of HGNC proteins* . |
|---|---|---|---|
| E1-activating enzymes | Ubiquitin-activating domain40 | 9 | 4 |
| E2-conjugating enzymes | UBC core domain41 | 78 | 43 |
| E3 ligases | RING42 | 562 | 334 |
| TRIAD43 | 33 | 22 | |
| HECT44 | 45 | 28 | |
| PHD45 | 290 | 173 | |
| U box46 | 42 | 31 | |
| E3 complex formation | SOCS box47 | 54 | 39 |
| Substrate recruitment | F-box48 | 115 | 72 |
| BTB49 | 273 | 188 | |
| Proteasome complex | 85 | 68 | |
| DUB | UCH, USP, OUT, Josephin, JAMM50 | 98 | 86 |
| Protein family . | Structural domains . | No. of Refseq IDs . | No. of HGNC proteins* . |
|---|---|---|---|
| E1-activating enzymes | Ubiquitin-activating domain40 | 9 | 4 |
| E2-conjugating enzymes | UBC core domain41 | 78 | 43 |
| E3 ligases | RING42 | 562 | 334 |
| TRIAD43 | 33 | 22 | |
| HECT44 | 45 | 28 | |
| PHD45 | 290 | 173 | |
| U box46 | 42 | 31 | |
| E3 complex formation | SOCS box47 | 54 | 39 |
| Substrate recruitment | F-box48 | 115 | 72 |
| BTB49 | 273 | 188 | |
| Proteasome complex | 85 | 68 | |
| DUB | UCH, USP, OUT, Josephin, JAMM50 | 98 | 86 |
DUB indicates deubiquitinating enzyme.
Number of proteins based on protein nomenclature by the HGNC:HUGO Gene Nomenclature Committee.
Because the introduction of microarrays to study gene expression in AML, novel prognostic subclasses based on similarities in expression profiles have been identified. Here, we conducted outlier expression analysis on a cohort of 525 AML patients as an alternative approach to study whether distinct expression of individual Ub genes in subsets of patients correlated with survival. Outlier expression was defined as an expression level of more than 4× the SD above or below the mean expression of all AML samples (see “Statistical analysis”). In total, 15 probe sets were identified that showed outlier expression in at least 10 patients (supplemental Table 4). Subsequently, overall survival of patient groups exhibiting outlier expression was compared with the remaining AML patients. Of these 15 probe sets, 3 showed a significant correlation with OS (supplemental Figure 1). One of these probe sets represented EVI1, which is a known prognostic factor in AML.13 The other 2 represented BRE (brain and reproductive organ-expressed), a member of the BRCA1 E3 ubiquitin ligase complex.
High BRE expression correlates with FAB M5 morphology and the MLL-AF9 fusion
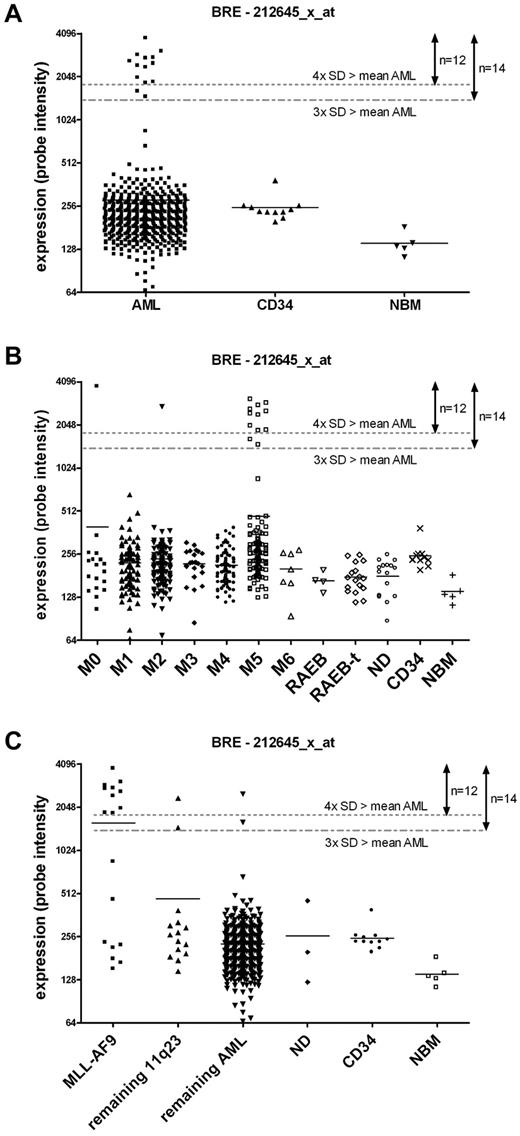
By outlier analysis, 12 high BRE expressing samples were identified based on the cutoff used for outlier expression. Figure 1 shows that these 12 samples actually belong to a distinct group of 14 patients with high BRE expression (which all showed BRE expression of 3× the SD above the mean), representing 3% of the total AML cohort of 525 samples (Figure 1A). Two samples showed intermediate expression and the remaining patients showed limited variation in BRE expression. To validate the BRE array expression data, a BRE-specific QPCR was performed. The QPCR confirmed the array data as a good correlation between the 2 types of data was observed (linear regression r2 > 0.8, P < .001, supplemental Figure 2). We also studied whether BRE was differentially expressed during normal hematopoiesis. Its expression was measured in CD34+ cells, total BM, and several mature blood cell fractions from healthy donors (T, B, NK cells, granulocytes, and monocytes). All fractions showed BRE expression comparable with levels observed in the majority of the AML samples, and no high BRE expression was found (supplemental Figure 3).
High BRE expression correlated strongly with FAB M5 morphology and MLL-AF9 rearrangements. (A) Gene expression profiling showed that high BRE expression, as represented by the probe set intensity of 212645_x_at, occurred in 14 of 525 AML patients, accounting for 3% of the cohort. Donor CD34+ and normal BM cells do not show high BRE expression. (B) High BRE expression correlated with FAB M5. BRE expression is visualized per FAB morphology class. (C) High BRE expression co-occurred with the presence of the MLL-AF9 fusion. BRE expression is represented by the fluorescent intensity of probe set 212645_x_at. Other probe sets representing BRE (205550_s_at and 211566_x_at) showed comparable results (data not shown). The mean per subclass is represented by a horizontal line. Dotted lines represent the mean of all AML samples plus 3 or 4× the SD of BRE expression, used as values for outlier analysis and cutoff for high BRE expression, assigning 14 or 12 samples with high expression, respectively.
High BRE expression correlated strongly with FAB M5 morphology and MLL-AF9 rearrangements. (A) Gene expression profiling showed that high BRE expression, as represented by the probe set intensity of 212645_x_at, occurred in 14 of 525 AML patients, accounting for 3% of the cohort. Donor CD34+ and normal BM cells do not show high BRE expression. (B) High BRE expression correlated with FAB M5. BRE expression is visualized per FAB morphology class. (C) High BRE expression co-occurred with the presence of the MLL-AF9 fusion. BRE expression is represented by the fluorescent intensity of probe set 212645_x_at. Other probe sets representing BRE (205550_s_at and 211566_x_at) showed comparable results (data not shown). The mean per subclass is represented by a horizontal line. Dotted lines represent the mean of all AML samples plus 3 or 4× the SD of BRE expression, used as values for outlier analysis and cutoff for high BRE expression, assigning 14 or 12 samples with high expression, respectively.
Evaluating co-occurrence of high BRE expression with known parameters in AML showed high co-occurrence with FAB M5 morphology (P < .001, Figure 1B, Table 2). Furthermore, co-occurrence was found with the t(9;11) translocation (P < .001), and a relatively young age (P = .006). In contrast to these positive correlations, high BRE expression was mutually exclusive with FLT3 ITD mutations (P = .027). High BRE expression was also mutually exclusive with favorable karyotypes, CEBPA mutations, EVI1 overexpression, and IDH1 and IDH2 mutations. The corresponding P values were not significant in this cohort, likely because of low sample numbers (Table 2).
Characteristics of patients with high BRE expression (14 patients)
| . | Normal BRE-expressing patients . | High BRE-expressing patients* . | P . | ||
|---|---|---|---|---|---|
| Sex (N = 518), no. (%) | .788† | ||||
| Male | 252 | (50) | 6 | (42.9) | |
| Female | 252 | (50) | 8 | (57.1) | |
| Age (N = 518), y | .006‡ | ||||
| Median (range) | 47 | (14-77) | 37.5 | (17-49) | |
| WBC (N = 518), ×109/L | .500‡ | ||||
| Median (range) | 28.6 | (0.3-510) | 68 | (1-240) | |
| Transplantation status (N = 525), no. (%) | .130† | ||||
| No transplantation | 302 | (59.1) | 5 | (35.7) | |
| Autologous transplantation | 68 | (13.3) | 2 | (14.3) | |
| Allogeneic transplantation | 141 | (27.6) | 7 | (50) | |
| FAB classification (N = 509), no. (%) | < .001§ | ||||
| M0 | 17 | (3.4) | 1 | (7.1) | .400† |
| M1 | 100 | (20.2) | 0 | (0) | .083† |
| M2 | 130 | (26.3) | 1 | (7.1) | .130† |
| M3 | 23 | (4.6) | 0 | (0) | 1.000† |
| M4 | 93 | (18.8) | 0 | (0) | .084† |
| M5 | 104 | (21.0) | 12 | (85.7) | < .001† |
| M6 | 7 | (1.4) | 0 | (0) | 1.000† |
| Other/unknown | 21 | (4.2) | 0 | (0) | 1.000† |
| Cytogenetics, no. (%) | |||||
| t(15;17) (N = 522) | 25 | (4.9) | 0 | (0) | 1.000† |
| t(8;21) (N = 522) | 38 | (7.5) | 0 | (0) | .613† |
| inv(16) (N = 522) | 42 | (8.3) | 0 | (0) | .617† |
| 11q23 (N = 522) | 21 | (4.1) | 12 | (85.7) | < .001† |
| MLL-AF9 (N = 522) | 8 | (1.6) | 10 | (71.4) | < .001† |
| MLL-AF10 (N = 522) | 2 | (0.4) | 1 | (7.1) | .080† |
| MLL-ENL (N = 522) | 0 | (0) | 1 | (7.1) | .027† |
| Other genetic aberrations, no. (%) | |||||
| FLT3 ITD mutations (N = 525) | 143 | (28.0) | 0 | (0) | .015† |
| FLT3 TKD mutations (N = 523) | 51 | (10.0) | 2 | (14.3) | .643† |
| NPM1 mutations (N = 525) | 157 | (30.7) | 1 | (7.1) | .075† |
| CEBPA single mutations (N = 525) | 12 | (2.3) | 0 | (0) | 1.000† |
| CEBPA double mutations (N = 525) | 26 | (5.1) | 0 | (0) | 1.000† |
| EVI1 overexpression (N = 518) | 52 | (10.3) | 0 | (0) | .380† |
| IDH1 mutations (N = 522) | 38 | (7.5) | 0 | (0) | .613† |
| IDH2 mutations (N = 522) | 49 | (9.6) | 0 | (0) | .382† |
| . | Normal BRE-expressing patients . | High BRE-expressing patients* . | P . | ||
|---|---|---|---|---|---|
| Sex (N = 518), no. (%) | .788† | ||||
| Male | 252 | (50) | 6 | (42.9) | |
| Female | 252 | (50) | 8 | (57.1) | |
| Age (N = 518), y | .006‡ | ||||
| Median (range) | 47 | (14-77) | 37.5 | (17-49) | |
| WBC (N = 518), ×109/L | .500‡ | ||||
| Median (range) | 28.6 | (0.3-510) | 68 | (1-240) | |
| Transplantation status (N = 525), no. (%) | .130† | ||||
| No transplantation | 302 | (59.1) | 5 | (35.7) | |
| Autologous transplantation | 68 | (13.3) | 2 | (14.3) | |
| Allogeneic transplantation | 141 | (27.6) | 7 | (50) | |
| FAB classification (N = 509), no. (%) | < .001§ | ||||
| M0 | 17 | (3.4) | 1 | (7.1) | .400† |
| M1 | 100 | (20.2) | 0 | (0) | .083† |
| M2 | 130 | (26.3) | 1 | (7.1) | .130† |
| M3 | 23 | (4.6) | 0 | (0) | 1.000† |
| M4 | 93 | (18.8) | 0 | (0) | .084† |
| M5 | 104 | (21.0) | 12 | (85.7) | < .001† |
| M6 | 7 | (1.4) | 0 | (0) | 1.000† |
| Other/unknown | 21 | (4.2) | 0 | (0) | 1.000† |
| Cytogenetics, no. (%) | |||||
| t(15;17) (N = 522) | 25 | (4.9) | 0 | (0) | 1.000† |
| t(8;21) (N = 522) | 38 | (7.5) | 0 | (0) | .613† |
| inv(16) (N = 522) | 42 | (8.3) | 0 | (0) | .617† |
| 11q23 (N = 522) | 21 | (4.1) | 12 | (85.7) | < .001† |
| MLL-AF9 (N = 522) | 8 | (1.6) | 10 | (71.4) | < .001† |
| MLL-AF10 (N = 522) | 2 | (0.4) | 1 | (7.1) | .080† |
| MLL-ENL (N = 522) | 0 | (0) | 1 | (7.1) | .027† |
| Other genetic aberrations, no. (%) | |||||
| FLT3 ITD mutations (N = 525) | 143 | (28.0) | 0 | (0) | .015† |
| FLT3 TKD mutations (N = 523) | 51 | (10.0) | 2 | (14.3) | .643† |
| NPM1 mutations (N = 525) | 157 | (30.7) | 1 | (7.1) | .075† |
| CEBPA single mutations (N = 525) | 12 | (2.3) | 0 | (0) | 1.000† |
| CEBPA double mutations (N = 525) | 26 | (5.1) | 0 | (0) | 1.000† |
| EVI1 overexpression (N = 518) | 52 | (10.3) | 0 | (0) | .380† |
| IDH1 mutations (N = 522) | 38 | (7.5) | 0 | (0) | .613† |
| IDH2 mutations (N = 522) | 49 | (9.6) | 0 | (0) | .382† |
WBC indicates white blood cell count; and FAB, French-American-British.
Cutoff used for high BRE expression was mean of all AML + 3× SD.
P values are based on Fisher exact tests.
P values are based on Mann-Whitney U tests.
P values are based on χ2 tests.
High BRE expression was strongly associated with t(9;11) translocations as explained in the previous paragraph. This translocation, leading to the MLL-AF9 fusion gene, can be missed by routine cytogenetic analysis. Therefore, we reanalyzed the complete cohort for the presence of the MLL-AF9 fusion by RT-PCR. All samples that were t(9;11)–positive based on cytogenetic findings were also positive for the MLL-AF9 fusion in the PCR. However, 8 additional MLL-AF9–positive samples were identified by PCR (supplemental Figure 4). This yielded a total incidence of 3.5% of AML cases with MLL-AF9 fusions (Table 2). Five of the newly identified MLL-AF9–positive samples showed high BRE expression. In total, 10 of 14 samples with high BRE expression contained the MLL-AF9 fusion, one had an MLL-ENL translocation, one had an MLL-AF10 translocation and the other 2 were 11q23 negative (Table 2, Figure 1C).
High BRE expression but not MLL-AF9 is an independent factor for favorable OS and EFS
To determine whether high BRE expression is an independent predictor for favorable OS, a multivariate analysis with the Cox proportional hazard model was performed, including known prognostic factors (see “Statistical analyses”). First, analyses were performed comparing the survival of the 12 patients with outlier-high BRE expression to the survival of the remaining cohort. Using this cutoff, high BRE expression was an independent factor for both OS and EFS (hazard ratios: 0.15 and 0.13, P values = .003 and .002 for OS and EFS, respectively; Table 3, High BRE expression based on mean of all AML + 4× SD [12 patient samples]). Afterward, the analysis was repeated, but now by comparing the 14 patients with the highest BRE expression (which showed distinct BRE expression) to the rest of the cohort. Importantly, in this analysis, high BRE expression was an independent factor for OS and EFS as well (hazard ratios: 0.3 and 0.26, P values = .047 and .023 for OS and EFS, respectively). In contrast, the presence of the MLL-AF9 fusion did not correlate significantly with OS or EFS in the complete AML cohort (Table 3).
High BRE expression is an independent prognostic factor for OS and EFS, based on the multivariate Cox regression hazard model
| . | OS . | EFS . | ||||
|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| High BRE expression based on mean of all AML + 4× SD (12 patient samples) | ||||||
| High BRE | 0.15 | 0.042-0.52 | .003 | 0.13 | 0.038-0.47 | .002 |
| MLL-AF9 | 1.82 | 0.81-4.1 | .15§ | 1.52 | 0.68-3.4 | .31§ |
| Favorable karyotype* | 0.30 | 0.21-0.43 | < .001 | 0.32 | 0.23-0.44 | < .001 |
| CEBPA double mut | 0.26 | 0.14-0.50 | < .001 | 0.28 | 0.16-0.50 | < .001 |
| NPM1 mut | 0.45 | 0.34-0.59 | < .001 | 0.41 | 0.31-0.53 | < .001 |
| FLT3 ITD | 1.60 | 1.2-2.1 | < .001 | 1.55 | 1.2-2.0 | .001 |
| Transplantation status† | 0.68 | 0.60-0.78 | < .001 | 0.75 | 0.66-0.85 | < .001 |
| FAB M5 | 1.05 | 0.80-1.4 | .73 | 1.00 | 0.77-1.3 | .98 |
| Age, y‡ | 1.01 | 1.0-1.0 | .080 | 1.00 | 1.0-1.0 | .52 |
| WBC‡ | 1.00 | 1.0-1.0 | .097 | 1.00 | 1.0-1.0 | .027 |
| High BRE expression based on mean of all AML + 3× SD (14 patient samples) | ||||||
| High BRE | 0.30 | 0.094-0.98 | .047 | 0.26 | 0.08-0.83 | .023 |
| MLL-AF9 | 1.29 | 0.52-3.22 | .58§ | 1.11 | 0.46-2.7 | .82§ |
| Favorable karyotype* | 0.31 | 0.22-0.45 | < .001 | 0.33 | 0.24-0.46 | < .001 |
| CEBPA double mut | 0.28 | 0.15-0.52 | < .001 | 0.30 | 0.17-0.52 | < .001 |
| NPM1 mut | 0.47 | 0.36-0.62 | < .001 | 0.43 | 0.33-0.56 | < .001 |
| FLT3 ITD | 1.54 | 1.2-2.0 | .001 | 1.48 | 1.1-1.9 | .003 |
| Transplantation status† | 0.70 | 0.61-0.80 | < .001 | 0.76 | 0.67-0.87 | < .001 |
| FAB M5 | 1.09 | 0.83-1.4 | .54 | 1.05 | 0.81-1.4 | .72 |
| Age, y‡ | 1.01 | 1.0-1.0 | .059 | 1.00 | 1.0-1.0 | .43 |
| WBC‡ | 1.00 | 1.0-1.0 | .32 | 1.00 | 1.0-1.0 | .13 |
| . | OS . | EFS . | ||||
|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| High BRE expression based on mean of all AML + 4× SD (12 patient samples) | ||||||
| High BRE | 0.15 | 0.042-0.52 | .003 | 0.13 | 0.038-0.47 | .002 |
| MLL-AF9 | 1.82 | 0.81-4.1 | .15§ | 1.52 | 0.68-3.4 | .31§ |
| Favorable karyotype* | 0.30 | 0.21-0.43 | < .001 | 0.32 | 0.23-0.44 | < .001 |
| CEBPA double mut | 0.26 | 0.14-0.50 | < .001 | 0.28 | 0.16-0.50 | < .001 |
| NPM1 mut | 0.45 | 0.34-0.59 | < .001 | 0.41 | 0.31-0.53 | < .001 |
| FLT3 ITD | 1.60 | 1.2-2.1 | < .001 | 1.55 | 1.2-2.0 | .001 |
| Transplantation status† | 0.68 | 0.60-0.78 | < .001 | 0.75 | 0.66-0.85 | < .001 |
| FAB M5 | 1.05 | 0.80-1.4 | .73 | 1.00 | 0.77-1.3 | .98 |
| Age, y‡ | 1.01 | 1.0-1.0 | .080 | 1.00 | 1.0-1.0 | .52 |
| WBC‡ | 1.00 | 1.0-1.0 | .097 | 1.00 | 1.0-1.0 | .027 |
| High BRE expression based on mean of all AML + 3× SD (14 patient samples) | ||||||
| High BRE | 0.30 | 0.094-0.98 | .047 | 0.26 | 0.08-0.83 | .023 |
| MLL-AF9 | 1.29 | 0.52-3.22 | .58§ | 1.11 | 0.46-2.7 | .82§ |
| Favorable karyotype* | 0.31 | 0.22-0.45 | < .001 | 0.33 | 0.24-0.46 | < .001 |
| CEBPA double mut | 0.28 | 0.15-0.52 | < .001 | 0.30 | 0.17-0.52 | < .001 |
| NPM1 mut | 0.47 | 0.36-0.62 | < .001 | 0.43 | 0.33-0.56 | < .001 |
| FLT3 ITD | 1.54 | 1.2-2.0 | .001 | 1.48 | 1.1-1.9 | .003 |
| Transplantation status† | 0.70 | 0.61-0.80 | < .001 | 0.76 | 0.67-0.87 | < .001 |
| FAB M5 | 1.09 | 0.83-1.4 | .54 | 1.05 | 0.81-1.4 | .72 |
| Age, y‡ | 1.01 | 1.0-1.0 | .059 | 1.00 | 1.0-1.0 | .43 |
| WBC‡ | 1.00 | 1.0-1.0 | .32 | 1.00 | 1.0-1.0 | .13 |
Mut indicates mutation; FAB, French-American-British; WBC, white blood cell count; OS, overall survival; EFS, event-free survival; HR, hazard ratio; and CI, confidence interval.
Favorable karyotype: t(8;21), t(15;17), and inv(16).
Transplantation status: either no transplantation, autologous transplantation, or allogeneic transplantation.
Continuous variables.
The P value of MLL-AF9 is mainly affected by co-occurrence with high BRE expression (based on correlation of regression coefficients, data not shown).
Among intermediate and high-risk AML groups, patients with MLL-AF9 translocations have been reported to show a relatively good prognosis,26,27 although others have not been able to confirm this.28 To address this issue in the cohort studied here, we determined the effect of the presence of an MLL-AF9 fusion on OS and EFS in intermediate- and high-risk patients (excluding patients with favorable karyotypes and CEBPA double mutations). This showed that MLL-AF9 positivity did not correlate significantly with favorable OS or EFS (Figure 2A). As all 14 patients with high BRE expression also belonged to the intermediate- and high-risk prognostic groups, we tested the predictive value of high BRE expression on survival among these patients as well. In contrast to MLL-AF9 positivity, both OS and EFS of the 14 patients with high BRE expression were significantly better compared with the remaining patients (5-year OS of 27.5 ± 2.3 vs 57.1 ± 13.2%, P value = .04 and 5-year EFS of 20.0 ± 2.1 vs 57.1 ± 13.2%, P value = .01 for patients with normal and high BRE expression, respectively; Figure 2B). In conclusion, high BRE expression correlated with favorable outcome in poor- and intermediate-risk prognostic groups and was an independent factor for clinical outcome in AML.
High BRE expression correlated with superior OS in intermediate- and poor-risk patients. (A) Kaplan-Meier plots for OS (left panels) and EFS (right panels) showed that MLL-AF9 positivity did not correlate significantly with a good OS or EFS among the intermediate- and poor-risk AML patients [ie, excluding patients with t(15;17), t(8;21), inv(16), and CEBPA double mutations; 5-year OS of 27.2 ± 2.4 vs 44.4 ± 11.7%, P = .17 and 5-year EFS of 20.2 ± 2.1 vs 44.4 ± 11.7%, P = .067 for MLL-AF9–negative and –positive patients, respectively]. (B) In the group of intermediate- and poor-risk AML patients, patients with high BRE expression exhibited a significantly better OS and EFS (5-year OS of 27.5 ± 2.3 vs 57.1 ± 13.2%, P = .04 and 5-year EFS of 20.0 ± 2.1 vs 57.1 ± 13.2%, P = .01 for patients with normal and high BRE expression, respectively). (C) Within the group of MLL-AF9–positive patients, high BRE expression correlated with a significantly better OS and EFS (5-year OS of 0% vs 80%, P = .0002 and 5-year EFS of 0% vs 80%, P = .0001 for patients with normal and high BRE expression, respectively). P values were determined with the log-rank test. The number of patients included in the analyses is shown in brackets.
High BRE expression correlated with superior OS in intermediate- and poor-risk patients. (A) Kaplan-Meier plots for OS (left panels) and EFS (right panels) showed that MLL-AF9 positivity did not correlate significantly with a good OS or EFS among the intermediate- and poor-risk AML patients [ie, excluding patients with t(15;17), t(8;21), inv(16), and CEBPA double mutations; 5-year OS of 27.2 ± 2.4 vs 44.4 ± 11.7%, P = .17 and 5-year EFS of 20.2 ± 2.1 vs 44.4 ± 11.7%, P = .067 for MLL-AF9–negative and –positive patients, respectively]. (B) In the group of intermediate- and poor-risk AML patients, patients with high BRE expression exhibited a significantly better OS and EFS (5-year OS of 27.5 ± 2.3 vs 57.1 ± 13.2%, P = .04 and 5-year EFS of 20.0 ± 2.1 vs 57.1 ± 13.2%, P = .01 for patients with normal and high BRE expression, respectively). (C) Within the group of MLL-AF9–positive patients, high BRE expression correlated with a significantly better OS and EFS (5-year OS of 0% vs 80%, P = .0002 and 5-year EFS of 0% vs 80%, P = .0001 for patients with normal and high BRE expression, respectively). P values were determined with the log-rank test. The number of patients included in the analyses is shown in brackets.
High BRE expression correlates with superior survival within MLL-AF9–positive AML patients
Of the 18 MLL-AF9–positive AML samples, 10 cases showed high BRE expression (Table 2, Figure 1C). We therefore questioned whether BRE expression predicted disease outcome within the group of MLL-AF9–positive patients. This analysis showed that MLL-AF9–positive patients with high BRE expression exhibited a superior survival while the remaining MLL-AF9–positive patients did very poor (Figure 2C), both with respect to OS and EFS (5-year OS and EFS of 0% vs 80% for patients with normal and high BRE expression, respectively, P = .0002 [OS] and P = .0001 [EFS]).
Validation of the prognostic value of BRE expression in an independent AML cohort
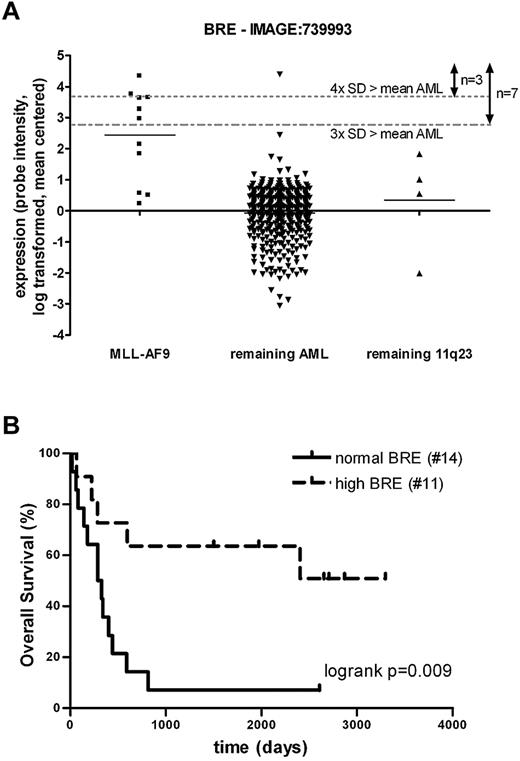
When studying the statistical impact of large numbers of genes, correlations can be found by chance. Therefore, we studied an independent cohort of 436 AML patients.19 All nonpredictive outliers identified in the first cohort did not predict outcome in the second cohort either (data not shown). With regard to BRE, outlier expression was also observed in a subset of this second cohort (Figure 3A). Patients with high BRE expression (above the mean plus 3× the SD of the complete cohort) had a favorable outcome compared with the remaining intermediate- and high-risk patients in this cohort as well (5-year OS of 25.6 ± 2.6 vs 71.4 ± 17.1%, P = .039 for patients with normal and high BRE expression, respectively; supplemental Figure 5). Despite the clear prognostic effect, we were unable to confirm the independent prognostic value of high BRE expression in this cohort. This could be explained by the low number of high BRE expressing patients and lack of sufficient molecular data of > 50% of the patients. Furthermore, the co-occurrence with MLL-AF9 in this cohort was even stronger than in the primary cohort (6 of 7 high BRE patients were MLL-AF9–positive, P < .001, Figure 3A). To validate the effect of BRE expression on survival within MLL-AF9 patients, we combined the BRE expression data of the 11 MLL-AF9–positive patients in this cohort with an additional group of 14 MLL-AF9–positive patients. These 25 patients were divided according to normal and high BRE expression based on the cutoff used in the second AML cohort of 436 samples. Also in this MLL-AF9 cohort, the OS of the patients with high BRE expression was significantly better than the OS of the remaining MLL-AF9–positive patients (5-year OS of 7.1 ± 6.9 vs 63.6 ± 14.5%, P = .0089 for patients with normal and high BRE expression, respectively; Figure 3B). We conclude that high BRE expression is a good prognostic factor, especially among MLL-AF9–positive cases, based on data from 2 large independent AML cohorts.
An independent AML cohort confirmed the positive effect of high BRE expression on prognosis. (A) BRE expression was high in a subset of a second cohort of 436 AML patients and co-occurred highly with MLL-AF9 translocations (P < .001 based on Fisher exact tests). cDNA (40k) expression array data of BRE (clone: IMAGE:739993) were log transformed and mean centered. Patients were subdivided into 3 groups based on MLL-AF9 and 11q23 positivity. Dotted lines represent the mean of all AML samples plus 3 or 4× the SD of BRE expression, used as values for outlier analysis and cutoff for high BRE expression, assigning 7 or 3 samples with high expression, respectively. (B) High BRE expression accounts for good OS within an independent MLL-AF9 cohort (5-year OS of 7.1 ± 6.9 vs 63.6 ± 14.5%, P = .0089 for patients with normal and high BRE expression, respectively). BRE array expression data of 2 cohorts of MLL-AF9–positive patients were combined by log transformation and mean centering of the data. Based on the cutoff used for high BRE expression in panel A (mean of 436 AML samples + 3 × SD), MLL-AF9 patients were subdivided into 2 groups: high and normal BRE expression (see supplemental Table 2). P values were determined with the log-rank test. The number of patients included in the analyses is shown in brackets.
An independent AML cohort confirmed the positive effect of high BRE expression on prognosis. (A) BRE expression was high in a subset of a second cohort of 436 AML patients and co-occurred highly with MLL-AF9 translocations (P < .001 based on Fisher exact tests). cDNA (40k) expression array data of BRE (clone: IMAGE:739993) were log transformed and mean centered. Patients were subdivided into 3 groups based on MLL-AF9 and 11q23 positivity. Dotted lines represent the mean of all AML samples plus 3 or 4× the SD of BRE expression, used as values for outlier analysis and cutoff for high BRE expression, assigning 7 or 3 samples with high expression, respectively. (B) High BRE expression accounts for good OS within an independent MLL-AF9 cohort (5-year OS of 7.1 ± 6.9 vs 63.6 ± 14.5%, P = .0089 for patients with normal and high BRE expression, respectively). BRE array expression data of 2 cohorts of MLL-AF9–positive patients were combined by log transformation and mean centering of the data. Based on the cutoff used for high BRE expression in panel A (mean of 436 AML samples + 3 × SD), MLL-AF9 patients were subdivided into 2 groups: high and normal BRE expression (see supplemental Table 2). P values were determined with the log-rank test. The number of patients included in the analyses is shown in brackets.
Patients with high BRE expression are confined to a new subclass in AML
Unsupervised cluster analysis of gene expression profiles in the first AML cohort revealed that patients with high BRE expression were confined almost completely to a previously uncharacterized cluster (12 of the 14 patients with high BRE expression; Figure 4). Two additional samples without high BRE expression belonged to this cluster as well. MLL-AF9 positivity was overrepresented in this cluster (10 of the 18 MLL-AF9–positive patients) as was expected because of its co-occurrence with high BRE expression. However, the correlation of the cluster with MLL-AF9 positivity was less than the correlation with high BRE expression. The OS of the patients within this cluster was significantly better than the remaining cohort, as could be anticipated based on the overrepresentation of patients with high BRE expression (data not shown).
Patients with high BRE expression were confined to a novel subclass in AML. Pairwise correlations of 525 AML patients based on optimal clustering as described before16 identified several clusters of patients with similar expression profiles which can be recognized by the red blocks showing high correlation along the diagonal. One of these clusters was represented by a high incidence (86%) of patients with high BRE expression; the left 3 black bars show the relative intensity of the 3 probe sets representing BRE expression on the array. The other 3 black bars show the relative expression intensity of the probe set for MRPL33 and 2 probe sets for RBKS as indicated in the figure, revealing coexpression of these chromosomal neighbors with BRE. The MLL-AF9 status of the samples is represented in the green bar (green: negative; red: positive; blue: ND).
Patients with high BRE expression were confined to a novel subclass in AML. Pairwise correlations of 525 AML patients based on optimal clustering as described before16 identified several clusters of patients with similar expression profiles which can be recognized by the red blocks showing high correlation along the diagonal. One of these clusters was represented by a high incidence (86%) of patients with high BRE expression; the left 3 black bars show the relative intensity of the 3 probe sets representing BRE expression on the array. The other 3 black bars show the relative expression intensity of the probe set for MRPL33 and 2 probe sets for RBKS as indicated in the figure, revealing coexpression of these chromosomal neighbors with BRE. The MLL-AF9 status of the samples is represented in the green bar (green: negative; red: positive; blue: ND).
Discussion
Several mutations and changes in expression of Ub genes are implicated in the pathogenesis of AML.4,6-8 By correlating outlier expression of Ub genes with clinical outcome in 525 AML patients, we identified differentially high expression of EVI1 and BRE as prognostic factors. Previous research has already shown that high EVI1 expression is an independent poor prognostic factor,13 providing proof-of-principle that outlier analysis is a powerful approach to identify prognostic factors.
BRE is a subunit of the BRCA1-BARD1 DNA damage repair ubiquitin E3 ligase complex and enhances its activity.29,30 BRE also down regulates TNFα signaling by binding death receptors31 and overexpression of BRE inhibits apoptosis.31-33 Differentially high BRE expression occurred in 3% of the AML patients, while the remaining patients showed only small differences in BRE expression. No high BRE expression was found in donor CD34+ cells, total BM or several mature blood cells, nor in several t(9;11) cell lines tested by us (supplemental Figure 3) or others.23 Importantly, the differential expression found in AML was an independent factor for both favorable OS and EFS (Table 3). High BRE expression co-occurred with FAB M5 morphology, a relatively young age and MLL-AF9 fusions. In contrast, high expression was mutually exclusive with favorable karyotypes, CEBPA mutations, FLT3 ITD, EVI1 overexpression, and IDH1 and IDH2 mutations in the studied cohort (Table 2). The co-occurrence of high BRE expression with MLL-AF9 fusions and it is favorable prognostic value in AML was validated in an independent cohort of 436 patients.
t(9,11) translocations can be missed by routine cytogenetic analysis. Indeed, reanalysis of the 525 AML samples by MLL-AF9 RT-PCR identified 8 additional MLL-AF9–positive samples (total = 18 samples), indicating that 44% of the cases were missed by cytogenetic analysis. Previous research in various cohorts showed that the prognostic value of MLL-AF9 positivity in AML is controversial.26-28 In the cohort studied here, the total group of MLL-AF9–positive patients showed an intermediate survival, which was not significantly different compared with the remaining intermediate and high risk patients. Importantly, our data suggest that the MLL-AF9–positive group actually consists of 2 prognostic groups that are characterized by the level of BRE expression: MLL-AF9–positive patients with high BRE expression showed a superior survival while patients without high BRE expression showed a very poor outcome (Figure 2C). The prognostic impact of high BRE expression was reinforced by showing the favorable effect of high BRE expression on overall survival in an independent cohort of MLL-AF9–positive patients. Thus, patients with high BRE expression constitute a novel favorable prognostic group in AML. During the preparation of this manuscript, Balgobind et al showed correlations of high BRE expression with favorable relapse free survival (RFS) in pediatric AML.23 They did not observe correlations with OS and EFS, as described here for adult AML. In their study, high BRE expression was associated with FAB M5 morphology and 11q23 translocations (primarily t(9;11)) as well. They showed a favorable RFS of MLL rearranged pediatric patients with high BRE expression, compared with the remaining MLL rearranged patients without high BRE expression. However, they did not address survival among MLL-AF9 patients, as was done in this study. Nevertheless, high BRE expression is a prognostic factor both in pediatric and adult AML, although the effect of high BRE expression on clinical outcome is different for adults and children. Additional studies are required to determine whether this is caused by differences in treatment regimens.
Genome-wide expression profiling supported the fact that patients with high BRE expression constitute a new subtype in AML, because unsupervised cluster analysis showed that most patients with high BRE expression were confined to a distinct cluster (Figure 4). This indicates that these patients shared global expression profiles. In this cluster no recurrent genetic aberrations were identified other than the expected partial co-occurrence with the MLL-AF9 fusion. Within this cluster, high BRE expression was more frequently observed than MLL-AF9 positivity, proposing a better correlation of high BRE expression than MLL-AF9 positivity with this cluster. In addition, BRE appeared to be among the most discriminatively expressed genes comparing this cluster to the other AML samples (data not shown).
The frequent co-occurrence of high BRE expression and MLL-AF9 fusions could suggest that BRE is a direct target of the oncogenic fusion protein. However, independent mouse studies did not identify BRE as MLL-AF9 target in untransformed or transformed cells.34,35 We observed that approximately half of the MLL-AF9–positive patients did not show high BRE expression, strongly suggesting that it is not a direct target in all cases. Furthermore, the unique expression profile characteristic for patients with high BRE expression co-occurred only partially with MLL-AF9 positivity (Figure 4). This could suggest that other factors than MLL-AF9 determine this profile. Although we observed no correlations between BRE expression and known MLL-AF9 target genes as HoxA7, HoxA9, and MLL (data not shown), we did observe an inverse correlation with the MLL-AF9 target MEIS1 (supplemental Figure 6). This could suggest that MLL-AF9 uses separate tumor-dependent cofactors to activate different gene programs, one including BRE, the other including MEIS1. Whether MLL-AF9 binds different regulatory DNA sequences depending on the availability of cofactors needs to be addressed in future research.
By studying genes that showed coexpression with high BRE expression, we found that 2 coexpressed genes, RBKS and MRPL33, are located directly upstream of BRE on chromosome 2 (2p23.2; Figure 4). High BRE expression was inextricably linked with high RBKS and MRPL33 expression. Reversely, high RBKS or MRPL33 expression did not necessarily co-occur with high BRE expression. Genes more upstream or downstream of BRE did not show this co-expression (data not shown). These data might indicate that a larger chromosomal locus is deregulated in patients with high BRE expression. To our knowledge, no small aberrations in the chromosomal region comprising the BRE locus have been identified in AML patients so far.36-39 Therefore, it will be important to determine in more detail whether the high expression of the 3 consecutive genes on this locus is because of local changes in transcriptional regulation or whether this is caused by chromosomal aberrations.
We conclude that unbiased statistical outlier analyses of Ub gene expression distributions in AML allowed us to identify high BRE expression as a novel prognostic factor. Furthermore, we identified a previously unrecognized subtype in AML with favorable outcome, represented by the differentially high BRE expression. These findings provide new insights into the risk stratification of AML, especially within the group of MLL-AF9–positive patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank S. Urbé and S. Hayes for sharing their overview of human DUBs with them.
This work was supported by The Vanderes Foundation.
Authorship
Contribution: S.M.N. and B.A.v.d.R. designed the present study; K.D. and L.B. provided BRE expression data of the second AML cohort; experiments were performed by S.M.N., E.T., and A.v.d.H.; data were analyzed by S.M.N., M.A.S., and C.G. and data were interpreted by S.M.N., M.A.S., K.D., L.B., J.H.J., P.J.M.V., and B.A.v.d.R.; and S.M.N. and B.A.v.d.R. wrote the manuscript which was critically revised by M.A.S., K.D., L.B., J.H.J., and P.J.M.V.
Conflict-of-interest disclosure: P.J.M.V. has declared ownership interests in Skyline, a spin-off company of Erasmus University Medical Center (Erasmus, MC), held in a Special Purpose Foundation of Erasmus MC. The remaining authors declare no competing financial interests.
Correspondence: Bert A. van der Reijden, Geert Grooteplein 8, 6525 GA Nijmegen, The Netherlands; e-mail: b.vanderreijden@labgk.umcn.nl.

![Figure 2. High BRE expression correlated with superior OS in intermediate- and poor-risk patients. (A) Kaplan-Meier plots for OS (left panels) and EFS (right panels) showed that MLL-AF9 positivity did not correlate significantly with a good OS or EFS among the intermediate- and poor-risk AML patients [ie, excluding patients with t(15;17), t(8;21), inv(16), and CEBPA double mutations; 5-year OS of 27.2 ± 2.4 vs 44.4 ± 11.7%, P = .17 and 5-year EFS of 20.2 ± 2.1 vs 44.4 ± 11.7%, P = .067 for MLL-AF9–negative and –positive patients, respectively]. (B) In the group of intermediate- and poor-risk AML patients, patients with high BRE expression exhibited a significantly better OS and EFS (5-year OS of 27.5 ± 2.3 vs 57.1 ± 13.2%, P = .04 and 5-year EFS of 20.0 ± 2.1 vs 57.1 ± 13.2%, P = .01 for patients with normal and high BRE expression, respectively). (C) Within the group of MLL-AF9–positive patients, high BRE expression correlated with a significantly better OS and EFS (5-year OS of 0% vs 80%, P = .0002 and 5-year EFS of 0% vs 80%, P = .0001 for patients with normal and high BRE expression, respectively). P values were determined with the log-rank test. The number of patients included in the analyses is shown in brackets.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/20/10.1182_blood-2011-06-359182/4/m_zh89991181470002.jpeg?Expires=1765893156&Signature=YBZitc5UPp1ap5LWtXjL47yIVhmGa6W1yEhpm6W3NasUKqEz5c6vmATVl36gEPc5LH-4VPbOaZHbTuOT5ioD4drwVGPhYob82n75l8~L-PgnPYnbhibXrYbCxMe-R31NTmz6t~K4iUylHqK1mcq8r~FotQm1H069jM-jqkNMGXldR9mDsGGNHEcxVjiWtk58lrgHqZlVuPyUiYDTBVJmKv6nJl368nrXfBHnJE6H~V1tDDxbihC4ZMdifApdL9gs-l45rTaZY7v53xoMdkuhvogOP48ruw-AaWYipthCJ3jKWZJzAXtRT8-db2ykPeMg25EYhPuN0Vci7ONs-HYYCQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)