Abstract
Abstract 2540
Minimal Residual Disease (MRD) assessment is increasingly used for treatment stratification since it is a strong predictor of outcome in Acute lymphoblastic leukemia (ALL). The most widely used MRD assays include flow cytometric detection of aberrant immunophenotypes and PCR amplification of patient-specific antigen-receptor sequences. The latter approach has proven to provide reliable clinical information but requires the development of patient-specific reagents which is laborious, time-consuming, and generates assays with variable sensitivities. In addition, this methodology may miss clonal changes that can occur during the course of the disease, such as the emergence of subclones as well as genetic evolution. To overcome these limitations, we developed a universal amplification assay with a sequencing readout that eliminates the need for patient-specific reagents, allows the assay to detect leukemic cells that have genetically evolved, and has a higher sensitivity than conventional tests.
To amplify all the IgH sequences, we developed a PCR assay to amplify all alleles of all the V and J segments with very low amplfication bias. Amplified molecules were then subjected to clonal sequencing to obtain >1 million reads to measure the frequency of the different IgH clonotypes in the sample. It should be noted that current next generation sequencing costs of this deep sequencing are similar to those of an MRD test conducted by flow cytometry.
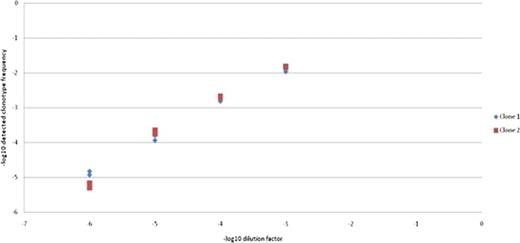
We tested the sensitivity of the method by in serial dilutions of genomic DNA from a leukemia sample known to carry two IgH clonotypes with into genomic DNA obtained from peripheral blood sample from a healthy donor. The material from the dilution series was then sequenced and analyzed to measure the level of these clones. The leukemic clonotypes could be readily detected even when diluted 1 million fold.
Detection of diluted leukemic clonotypes.
The X axis shows the –log10 of the leukemia sample dilution factor and the Y axis depicts the –log10 frequency of the 2 detected clonotypes. All the dilution through sequencing are done in duplicate and plotted. The leukemia clonotypes are reproducibly detected even when diluted 1 million fold.
Detection of diluted leukemic clonotypes.
The X axis shows the –log10 of the leukemia sample dilution factor and the Y axis depicts the –log10 frequency of the 2 detected clonotypes. All the dilution through sequencing are done in duplicate and plotted. The leukemia clonotypes are reproducibly detected even when diluted 1 million fold.
To directly compare the our method to established MRD assays in ALL, we studied diagnostic and follow-up samples from 10 ALL patients whose MRD levels have been previously assessed by both real-time PCR amplification of IgH genes and flow cytometry. The results of these tests were not disclosed until completion of the deep sequencing analysis. The follow up samples were collected during (n = 3) or at the end of remission induction therapy (n = 4), or during continuation therapy (n = 3). Samples were processed similarly to identify the leukemia-specific sequence in the diagnostic samples and determine the level of these sequences in the follow up samples.
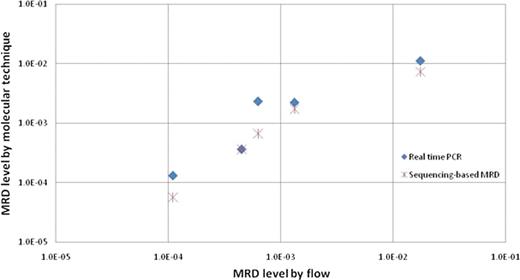
The sequencing-based method identified all 5 samples that were MRD-positive according to flow cytometry and PCR (Figure 2), with highly concordant estimates of MRD levels. Notably, among the remaining 5 samples, scored as MRD-negative by both flow cytometry and conventional PCR, the sequencing method detected residual leukemic sequences at a very low level (∼10−6) in one of the samples. The other 4 samples were MRD-negative by all three methods. Studies with a larger cohort of ALL samples are ongoing.
MRD detection by three different methods
MRD detection by three different methodologies in 5 MRD positive patients is shown in log scale. The X axis depicts the MRD level using flow cytometry. The Y axis shows the MRD level as determined by real time PCR and sequencing.
MRD detection by three different methods
MRD detection by three different methodologies in 5 MRD positive patients is shown in log scale. The X axis depicts the MRD level using flow cytometry. The Y axis shows the MRD level as determined by real time PCR and sequencing.
Contrary to conventional PCR-based MRD testing, the sequencing technology allows for the detection of leukemic clones that evolve by V replacement or other mechanisms. In this study, we identified clonotypes in several of the diagnostic samples that appeared to be the result of V replacement. These and other newly appearing related clones can be monitored in subsequent samples using the generic amplification and sequencing assay.
We developed a highly sensitive and specific MRD detection method based on next-generation sequencing of IgH genes. This method has substantial advantages over conventional PCR MRD in that it eliminates the need for patient-specific reagents, can follow genetic evolution, and has potential for higher sensitivity.
Faham:Sequenta Inc: Employment, Equity Ownership. Willis:Sequenta Inc: Employment, Equity Ownership. Moorhead:Sequenta Inc: Employment, Equity Ownership. Carlton:Sequenta Inc: Employment, Equity Ownership. Zheng:Sequenta Inc: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal