Abstract
Iron regulatory proteins (IRPs) 1 and 2 are RNA-binding proteins that control cellular iron metabolism by binding to conserved RNA motifs called iron-responsive elements (IREs). The currently known IRP-binding mRNAs encode proteins involved in iron uptake, storage, and release as well as heme synthesis. To systematically define the IRE/IRP regulatory network on a transcriptome-wide scale, IRP1/IRE and IRP2/IRE messenger ribonucleoprotein complexes were immunoselected, and the mRNA composition was determined using microarrays. We identify 35 novel mRNAs that bind both IRP1 and IRP2, and we also report for the first time cellular mRNAs with exclusive specificity for IRP1 or IRP2. To further explore cellular iron metabolism at a system-wide level, we undertook proteomic analysis by pulsed stable isotope labeling by amino acids in cell culture in an iron-modulated mouse hepatic cell line and in bone marrow-derived macrophages from IRP1- and IRP2-deficient mice. This work investigates cellular iron metabolism in unprecedented depth and defines a wide network of mRNAs and proteins with iron-dependent regulation, IRP-dependent regulation, or both.
Introduction
Iron homeostasis in mammalian cells is maintained through posttranscriptional regulation by the IRP/IRE regulatory system.1,2 In iron-deficient cells, active iron regulatory protein (IRP) 1 and IRP2 recognize iron-responsive elements (IREs), conserved RNA structures located in the untranslated regions (UTRs) of mRNAs that encode proteins involved in iron metabolism.3 IRP/IRE-regulated mRNAs include those encoding proteins for iron acquisition (transferrin receptor 1 [Tfrc], divalent metal transporter 1 [Slc11a2]), storage (ferritin H [Fth1] and ferritin L [Ftl]), use (erythroid 5-aminolevulinic acid synthase [ALAS2], mitochondrial aconitase [Aco2], Drosophila succinate dehydrogenase [Sdh]), and export (ferroportin [Slc40a1]). A typical IRE is composed of a 6-nucleotide apical loop (5′-CAGWGH-3′, where W stands for A or U and H for A, C, or U) on a stem of 5 paired nucleotides, an unpaired asymmetrical cytosine bulge on the 5′strand of the stem, and an additional lower stem of variable length (depicted in Figure 3A); the nucleotide composition forming the 2 stem segments may vary considerably.4,5 The mRNAs of Fth1, Ftl, Alas2, Aco2, dSdh, and Slc40a1 contain one single IRE in their 5′UTRs, whereas the Slc11a2 mRNA harbors a single IRE in its 3′UTR; Tfrc mRNA is the only currently known mRNA with multiple (5) IREs, and all of them are located in its 3′UTR. Depending on the location of the IRE, IRP binding regulates gene expression by different mechanisms. Both IRPs inhibit translation initiation when bound to 5′UTR IREs (eg, Fth1 and Ftl mRNAs), whereas their association with the 3′UTR IREs of the Tfrc mRNA mediates mRNA stabilization by preventing endonucleolytic cleavage.6-8
IRP binding activity to IREs is differentially regulated by intracellular iron levels and other stimuli, including nitric oxide, oxidative stress, and hypoxia. IRP1 and IRP2 share 60% to 70% overall amino acid identity, depending on the studied species. Notably, not all amino acids of IRP1 that directly contact the IRE in the IRP1/H-ferritin IRE crystal structure9 are conserved in IRP2, which may be relevant in the context of IRP-specific mRNAs. In response to the cellular labile iron pool, distinct mechanisms control the activities of IRP1 and IRP2, which is high in iron-deficient cells and low in iron-replete cells. Under iron-replete conditions, an iron-sulfur cluster (4Fe-4S) assembles in IRP1, preventing IRE binding and converting it into the cytosolic aconitase. In iron deficiency, IRP1 undergoes conformational changes that allow it to bind to IREs as an apoprotein.9-11 In contrast, IRP2 does not contain an Fe-S cluster and is regulated by ubiquitination and degradation mediated by the iron-regulated ubiquitin ligase FBXL5.12,13 The IRP/IRE regulatory system is essential, as demonstrated by the embryonic lethality of mice lacking both IRPs.14,15 From a cell biology perspective, the IRPs are critical for securing sufficient iron supplies to mitochondria.16 Overall, the regulation of the IRE-binding activities of IRP1 and IRP2 ensures the appropriate expression of IRP target genes and cellular iron balance.
Over the past 25 years, IREs have been identified successively in less than a dozen mRNAs by coincidence or after diverse directed approaches (eg, study of iron-regulated genes, specific bioinformatics searches). To systematically define the IRP/IRE regulatory network on a genome-wide and proteomic scale, we followed 2 approaches. First, we isolated complexes formed between IRP1 or IRP2 and mRNAs isolated from 5 different murine tissues, and then we identified their mRNA constituents using genome-wide microarrays. As a proof of concept for this method, we recently identified 2 novel IRE-containing mRNAs in the oxygen-sensing transcription factor Epas1 (Hif2α) and in the human cell cycle phosphatase Cdc14A using a small, custom-made cDNA microarray, the IronChip.17,18 Second, we used bone marrow-derived macrophages from control mice or animals lacking both macrophage IRPs as well as an iron-perturbed hepatocytic cell line, and then we analyzed the impact of IRP expression, activity, and iron-induced changes on cellular protein synthesis by pulsed stable isotope labeling by amino acids in cell culture (pSILAC).19
Methods
Mice and RNA extraction
Brain and bone marrow (BM) samples were obtained from 8- to 10-week-old C57BL6 mice fed with a standard chow. BM cells were flushed out from the femur with ice-cold Hanks balanced salt solution (HBSS) and pelleted at 300g for 10 minutes at 4°C for RNA extraction. Duodenum, liver, and spleen tissues were obtained from C57BL6 mice fed with a low iron (< 10-mg/kg) diet (C1000; Altromin) for 3 weeks, starting at weaning age; iron deficiency was functionally validated by decreased hematocrit and hemoglobin values compared with animals that received a control diet.
Mice with selective ablation of both IRP1 and IRP2 in macrophages were generated using Cre/Lox technology. Mice homozygous for floxed Aco1 (Irp1) and Ireb2 (Irp2) alleles (Aco1flox/flox, Ireb2flox/flox)15 were bred to a knockin strain (LysM+/Cre) with an insertion of the Cre recombinase cDNA into the LysozymeM locus.20 Aco1flox/flox, Ireb2flox/flox, LysM+/Cre animals [designated IrpLysM::Cre(+)] were born at mendelian ratios and were physically indistinguishable from Aco1flox/flox,Ireb2flox/flox, LysM+/+ control littermates [IrpLysM::Cre(−); Ferring-Appel et al, manuscript in preparation]. Animals were housed under a constant light/dark cycle in the European Molecular Biology Laboratory specific-pathogen-free mouse barrier unit and had access to food and water ad libitum. They were killed by CO2 inhalation. Animal handling was in accordance with institutional guidelines.
Total RNA used for immunoprecipitations (IPs) was extracted from mouse tissues using TRIzol reagent (Invitrogen) following the manufacturer's protocol.
Immunoprecipitations
The IP experiments were performed as described previously.18 In brief, 50 μg of total RNA was combined with purified, His6-tagged recombinant IRP1 or His-tagged IRP2 produced in Escherichia coli and a rabbit polyclonal anti-IRP1 antibody (for IRP1 IPs) or a mouse monoclonal anti-His tag antibody (for IRP2 IPs). A control reaction in which the recombinant IRP was omitted was performed in parallel (mock IP). The IPs were done in duplicate using 2 independent pools of total RNA, each one composed of a pool of total RNA extracted from 4 to 6 mice. Coimmunoprecipitated RNAs from the messenger ribonucleoproteins (mRNPs) were isolated by proteinase K digestion and ethanol precipitation.
Quantitative real-time PCR
Quantitative (q)PCR was performed in an ABI PRISM 7500 Real Time PCR system (Applied Biosystems) with SYBR Green and ROX as a passive reference dye. Seventy nanograms of RNA recovered from the immunoprecipitation reactions was used for qPCR analysis. Genes analyzed were as follows: Fth1, Tfrc, Slc11a2-IRE, Slc11a2-nonIRE, Slc40a1, Epas1, Gapdh, and ACtb. Levels of Actb, Gapdh, and Slc11a2-nonIRE were used as negative controls, for normalization, or both. PCR product quality was monitored by post-PCR melt curve analysis. Fold enrichments were calculated using the relative expression software tool.21 Primer sequences are available on request.
Microarray experiments
In total, 40 Affymetrix GeneChip Mouse Genome 430 2.0 arrays were used to determine the mRNA composition of mRNPs obtained by immunoprecipitation with the recombinant IRPs. Immunopreciptated RNA (120 ng) was used as input for a 2-step amplification procedure to generate biotin-labeled RNA fragments for hybridization to the Affymetrix microarray according to the Standard Affymetrix 2 Cycle protocol (Eukaryotic Sample and Array Processing manual 701024 Rev.3). The amplified material was verified for specificity by qPCR before labeling and hybridization (data not shown). Intensity values for the hybridizations were obtained either using robust multichip average (RMA), with calculations done in bioconductor (www.bioconductor.org) or Affymetrix Microarray Suite 5 (MAS5), with calculations done using the Affymetrix GCOS package. MAS5-calculated intensities were further quantile normalized using bioconductor. Both methods are complementary and commonly used normalization procedures in the context of established algorithms for microarray evaluation. Ratios between the intensities from immunoprecipitate and mock reactions were calculated to obtain the fold enrichment level for each probe. Genes were considered “positive” if both independent biologic replicas yielded significant enrichment values above the cut-off threshold, based in the lowest log2 ratio obtained from probes of known IRE-containing mRNAs or a 0.6 log2 ratio value as threshold (1.5-fold enrichment). Microarray data reported here have been deposited within Gene Expression Omnibus (National Center for Biotechnology Information; GSE17096, GSE17097).
Plasmids
Mouse full-length cDNA clones for Fth1, Slc40a1, Ppp1r1b, Gyg, Gstm6, Cxcl16, Pfn2, and Pdcl3 were obtained from Origene Technologies or Riken FATOM 3 clone collections. Clones containing the H-ferritin wild-type and mutant IRE followed by the chloramphenicol-acetyltransferase (CAT) mRNA (wt and mut in this paper, originally pI-12.CAT and pI-19.CAT clones)10 and the Renilla luciferase control plasmid were described previously.22 All plasmids were verified by DNA sequencing.
Competitive EMSAs
Competitive EMSAs were done using from 15 000 to 30 000 cpm of 32P-radiolabeled H-ferritin IRE probe mixed with appropriate molar excess of trace-labeled competitor (1×, 2×, 5×, 10×, and 40× fold molar excess) and 10 to 60 ng of recombinant IRP1 in cell lysis buffer as described previously.17 The H-ferritin mutant with a deletion (δ-C, N14) in the IRE loop was used as a negative control. An extensive description of competitive EMSAs with H-ferritin IRE mutants and with full-length transcripts is provided in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Tissue culture, pSILAC, and protein detection
Cells were grown at 37°C in a 5% CO2 atmosphere.
The murine hepatocellular carcinoma cell line Hepa 1-6 was purchased from DSMZ and grown in Dulbecco modified Eagle medium with 4.5 g/L glucose supplemented with 10% of heat-inactivated FCS (HyClone Laboratories), and 1% penicillin + streptomycin (Invitrogen). Hepa 1-6 cells were adapted to SILAC light medium that contains dialyzed FCS for 1 week. On the day of the experiment, cells were incubated with 100μM hemin (Leiras Oy) or 200μM desferrioxamine (DFO; Sigma-Aldrich) or left untreated for 2 hours, followed by 1 hour of amino acid starvation to enhance the incorporation of isotopes, and 6 hours of labeling with M SILAC medium for control cells and H SILAC medium for hemin- or DFO-treated cells; total treatment time was therefore 9 hours. Cells were washed twice, harvested on ice with cold PBS, and then centrifuged; the pellets were frozen in liquid nitrogen.
Time and concentration of the hemin or DFO treatments for pSILAC experiments were optimized in [35S]methionine/cysteine metabolic labeling studies monitoring ferritin L and H protein levels and transferrin receptor 1 mRNA levels (data not shown). Cell toxicity of the treatments was assessed by the colorimetric MTT assay, and no adverse effects were observed under the chosen conditions (data not shown).
Bone marrow–derived macrophages (BMDMs) were recovered from the femur of IrpLysM::Cre(+) and IrpLysM::Cre(−) animals using ice-cold HBSS. The cell suspension was filtered through an 80-μm cell strainer (Falcon, BD Biosciences Discovery Labware), and cells were seeded at a density of ∼ 5 × 104 cells/cm2 in RPMI 1640 + GlutaMAX (Invitrogen) supplemented with 20% of heat-inactivated FCS (HyClone Laboratories), 1% penicillin + streptomycin (Invitrogen), and 100 ng/mL of macrophage colony-stimulating factor (M-CSF; Sigma-Aldrich). After 4 days, nonadherent cells were removed by washing with HBSS, and the medium was subsequently replaced daily with light SILAC medium containing M-CSF for 2 days. Cells were further incubated in the presence of medium (control BMDM) versus heavy (IRP-deficient cells) SILAC medium containing M-CSF for 24 hours. After 3 washes with ice-cold HBSS, cells were harvested using a rubber policeman and pelleted by centrifugation at 300g for 10 minutes at 4°C, and pellets were frozen in liquid nitrogen. The proportion of macrophages typically exceeds 90% as assessed by labeling with an Alexa Fluor 488–coupled rat monoclonal antibody against the F4/80 macrophage-specific marker (Serotec); consistent with its reported efficiency,20 the LysM+/Cre deletor strain yields 70% to 80% recombination of the floxed Aco1 and Ireb2 alleles as assessed by Southern blotting (data not shown).
Cell pellets were lysed in SDS-sample buffer, and then appropriate samples were combined, separated by SDS-PAGE, processed, and analyzed by liquid chromatography-tandem mass spectrometry.19 A detailed description is fully provided in supplemental Methods. Data analysis for mass spectrometry was performed as described previously19 ; a cut-off for protein regulation of 1.4-fold was chosen based on results with the control protein Tfrc.
Bioinformatic analysis
The searching for iron-responsive elements (SIREs) web-server tool for prediction of iron-responsive elements has been described previously.23
Statistics
The data are reported as mean ± SEM. All statistical analyses were performed using Prism version 5.0 (GraphPad Software). Student t tests were used to compare results between groups of 2. A value of P < .05 is considered statistically significant.
Results
Isolation and identification of mRNAs associated with IRP1 and IRP2
To systematically elucidate the IRP regulatory network, we developed a strategy to specifically immunoprecipitate IRP-containing mRNP particles and to identify the copurified mRNAs by microarray analysis. Total RNA from 5 different mouse tissues (duodenum, liver, brain, spleen, and bone marrow) was incubated with recombinant IRP1 or IRP2 and suitable antibodies (see “Methods”). In parallel, a reaction omitting the recombinant protein was analyzed (mock IP) to assess the background level of the system.
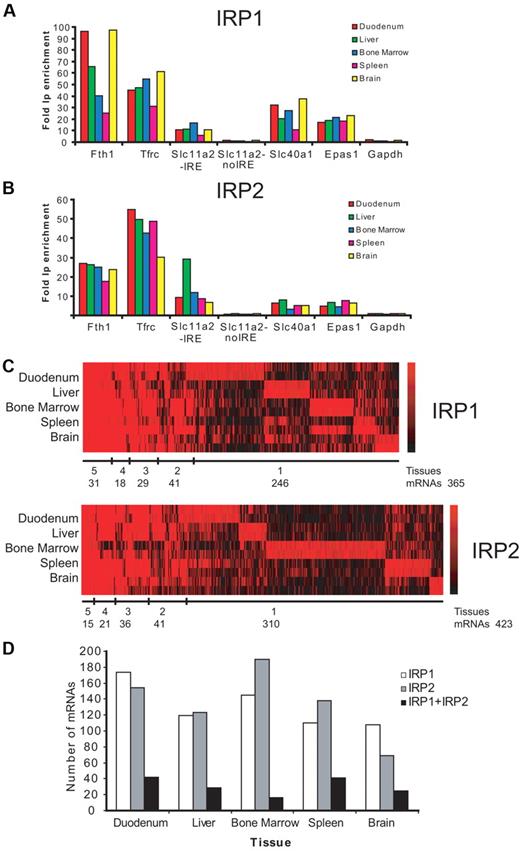
The IPs were tested for specificity by analyzing the enrichment of known IRE-containing mRNAs in the IP versus the mock IP reactions by qPCR. The mRNAs of Fth1, Tfrc, Slc11a2, Slc40a1, and Epas1 are all strongly enriched (from 3- to 97-fold) in the IP fractions with recombinant IRP1 or IRP2 from RNA samples of all 5 analyzed tissues (Figure 1A-B). No enrichment over the background was detected for Gapdh mRNA and for the non-IRE form of Slc11a2 mRNA (Figure 1A-B). Thus, IRP1- and IRP2-binding mRNAs are consistently and specifically detected by this procedure.
Transcriptomic identification of IRP1- and IRP2-binding mRNAs. Fold enrichment of known IRE-containing mRNAs (Fth1, Tfrc, Slc11a2, Slc40a1, and Epas1) in IRP1 (A) or IRP2 (B) IPs versus mock controls was determined by qPCR. Gapdh and the non-IRE form of Slc11a2 were used as negative controls. Data show the values obtained with the first biologic replica used in microarray analysis; similar data were obtained with the second biologic replica (data not shown). (C) Heatmap visualization of microarray data (RMA analysis) for mRNAs copurified with IRP1 or IRP2. Fold-change values from 2 independent replicates for each tissue tested are given for probe sets that were detectable above background showing at least a 1.5-fold enrichment (log2 ratio > 0.6). Red and black lines indicate positive and negative IP enrichment, respectively, relative to mock IPs. The color scales to the right indicate the magnitude of the fold change (base 2 logarithm) for a particular transcript. Number of mRNAs clustered by tissues is shown below each heatmap. (D) Number of mRNAs detected in each particular tissue bound by IRP1, IRP2, or IRP1 + IRP2.
Transcriptomic identification of IRP1- and IRP2-binding mRNAs. Fold enrichment of known IRE-containing mRNAs (Fth1, Tfrc, Slc11a2, Slc40a1, and Epas1) in IRP1 (A) or IRP2 (B) IPs versus mock controls was determined by qPCR. Gapdh and the non-IRE form of Slc11a2 were used as negative controls. Data show the values obtained with the first biologic replica used in microarray analysis; similar data were obtained with the second biologic replica (data not shown). (C) Heatmap visualization of microarray data (RMA analysis) for mRNAs copurified with IRP1 or IRP2. Fold-change values from 2 independent replicates for each tissue tested are given for probe sets that were detectable above background showing at least a 1.5-fold enrichment (log2 ratio > 0.6). Red and black lines indicate positive and negative IP enrichment, respectively, relative to mock IPs. The color scales to the right indicate the magnitude of the fold change (base 2 logarithm) for a particular transcript. Number of mRNAs clustered by tissues is shown below each heatmap. (D) Number of mRNAs detected in each particular tissue bound by IRP1, IRP2, or IRP1 + IRP2.
Genome-wide identification of the immunoprecipitated mRNAs within the IRP mRNP complexes was achieved using Affymetrix microarrays (GeneChip Mouse Genome 430 2.0) that cover >39 000 mRNAs and RNA variants. Affymetrix array data were normalized and analyzed using 2 independent and commonly used statistical algorithms, MAS5 and RMA, to perform stringent data analysis. The total number of IRP1- or IRP2-binding mRNAs clustered by tissues is shown below the heatmap in Figure 1C (RMA analysis), and the total number of mRNAs detected in each particular tissue is shown in Figure 1D (RMA analysis). A similar analysis was done with the MAS5 data (supplemental Figure 1A-B). The majority of IRP-associated mRNAs is detected in a tissue-specific way (for IRP1, 67.4% or 70.3% and for IRP2, 73.3% or 80.3%, respectively, depending on the microarray algorithm used).
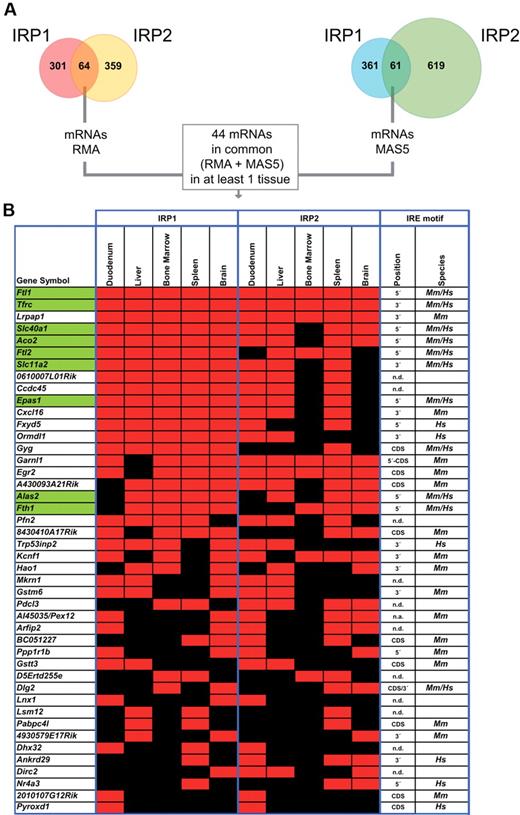
The RMA microarray data analysis revealed that 64 mRNAs bind to both IRPs in at least one tissue; 61 mRNAs were detected using the MAS5 analysis method (Figure 2A). Combining both analyses, we identified 44 mRNAs that are significantly enriched in both IRP1 and IRP2 mRNPs (Figure 2A-B). Importantly, all 9 previously known murine IRE-containing mRNAs (Ftl1, Ftl2, Fth1, Tfrc, Slc40a1, Slc11a2, Alas2, Aco2, and Epas1; marked in green in Figure 2B) were reidentified by our experiments. Therefore, the genome-wide search for IRP-associated mRNAs reveals 35 novel mRNAs able to bind both IRP1 and IRP2 in at least 1 of the 5 tested tissues (Figure 2B).
Novel mRNA targets for IRP1 and IRP2. (A) Number of mRNAs positively detected as enriched in IRP1, IRP2, or both IPs by microarrays with MAS5, RMA, or a combined (MAS5 + RMA) mathematical data analysis. (B) Virtual heatmap representing the 44 identified mRNAs that are bound by both IRPs in at least one of the 5 studied tissues. Affymetrix-positive probes were grouped by gene and reported in each of the rows. Known IRE-containing mRNAs are shown in green. Red and black squares indicate positive or lacking IP enrichment, respectively, relative to mock IPs in 2 independent biologic replicas. Predicted IREs by SIREs program in mouse (Mus musculus, Mm.) or human (Homo sapiens, Hs.) databases/species and their positions also are indicated. N.d. denotes that an IRE motif was not detected using the SIREs bioinformatic program; n.a. denotes nonavailable information; and 3′ or 5′ denotes the 3′ or 5′UTR.
Novel mRNA targets for IRP1 and IRP2. (A) Number of mRNAs positively detected as enriched in IRP1, IRP2, or both IPs by microarrays with MAS5, RMA, or a combined (MAS5 + RMA) mathematical data analysis. (B) Virtual heatmap representing the 44 identified mRNAs that are bound by both IRPs in at least one of the 5 studied tissues. Affymetrix-positive probes were grouped by gene and reported in each of the rows. Known IRE-containing mRNAs are shown in green. Red and black squares indicate positive or lacking IP enrichment, respectively, relative to mock IPs in 2 independent biologic replicas. Predicted IREs by SIREs program in mouse (Mus musculus, Mm.) or human (Homo sapiens, Hs.) databases/species and their positions also are indicated. N.d. denotes that an IRE motif was not detected using the SIREs bioinformatic program; n.a. denotes nonavailable information; and 3′ or 5′ denotes the 3′ or 5′UTR.
In addition to these mRNAs that are bound by both IRPs, the microarray data (combining MAS5 and RMA analysis methods) detect 101 mRNAs as exclusive interactors of IRP1 and 113 IRP2-binding mRNAs in at least one tissue (supplemental Tables 1-2). To the best of our knowledge, this is the first time that exclusive IRP1- or IRP2-interacting cellular mRNAs are identified.
Experimental refinement of and bioinformatic analysis for IRE motifs in novel IRP target mRNAs
Earlier bioinformatic searches of mammalian transcriptomes for IRE motifs24,25 had not identified any of the novel mRNAs that we found to bind IRP1 and IRP2 in the experiments described so far. In part, such failure probably results from limitations of the definition of an “IRE motif” that was used for these searches. To address some of these limitations experimentally, we first considered previous systematic evolution of ligands by exponential enrichment experiments that reported that the 6-nucleotide apical loop of an IRE can differ from the canonical CAG(U/A)GN sequence26-28 or that the C-bulge can be replaced by a G-bulge28,29 (supplemental Table 3). In addition, we also accommodated atypical IRE structures that are present in the validated Slc11a2, Epas1, and Hao1 mRNAs.30-32
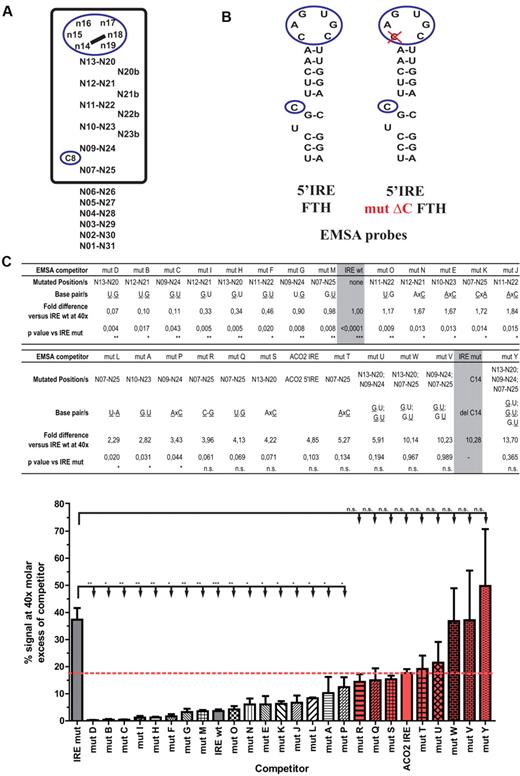
On the basis of these published data, we designed a series of additional experiments to further define IRP1-binding RNA motifs by competitive EMSA (Figure 3A; supplemental Table 3). Specifically, we subjected the upper stem and the position below the bulged cytosine (C8) of the H-ferritin IRE to further analysis, testing 24 new variants (Figure 3C) together with the H-ferritin IRE controls (Figure 3B) in competitive EMSAs with recombinant IRP1.
Experimental definition of IRP1-binding sites using EMSA. (A) Schematic representation of an IRE motif. Squared region indicates the IRE core region predicted by SIREs software. (B) Schematic representation of the H-ferritin wild-type and mutant IRE used for electrophoretic mobility shift assays. Deletion of C14 in the IRE is indicated by a red cross. (C) Competitive EMSA analyses with 24 different H-ferritin IRE variants (mutants A to Y). Values and standard errors for competition with the H-ferritin wild-type and the δ-C mutant IRE are highlighted in gray. In the table, mutated positions are indicated according to the nomenclature shown in panel A, and mutated nucleotides are underlined. Values for fold difference between the different variants versus the H-ferritin IRE wild-type (IRE wt) at 40× fold molar excess are reported. P values comparing each variant with the H-ferritin IRE mutant, (IRE mut) are reported (***P < .001, **P < .01, *P < .05; unpaired 2-tailed Student t test). Mutants that do not reach statistical significance are filled in red. The red dashed line indicates the value for Aco2 5′IRE. Graph shows percentage of raw signal for competitive EMSAs at 40× molar excess of the indicated competitor. Data are presented as mean ± SEM from a minimum of 3 experiments.
Experimental definition of IRP1-binding sites using EMSA. (A) Schematic representation of an IRE motif. Squared region indicates the IRE core region predicted by SIREs software. (B) Schematic representation of the H-ferritin wild-type and mutant IRE used for electrophoretic mobility shift assays. Deletion of C14 in the IRE is indicated by a red cross. (C) Competitive EMSA analyses with 24 different H-ferritin IRE variants (mutants A to Y). Values and standard errors for competition with the H-ferritin wild-type and the δ-C mutant IRE are highlighted in gray. In the table, mutated positions are indicated according to the nomenclature shown in panel A, and mutated nucleotides are underlined. Values for fold difference between the different variants versus the H-ferritin IRE wild-type (IRE wt) at 40× fold molar excess are reported. P values comparing each variant with the H-ferritin IRE mutant, (IRE mut) are reported (***P < .001, **P < .01, *P < .05; unpaired 2-tailed Student t test). Mutants that do not reach statistical significance are filled in red. The red dashed line indicates the value for Aco2 5′IRE. Graph shows percentage of raw signal for competitive EMSAs at 40× molar excess of the indicated competitor. Data are presented as mean ± SEM from a minimum of 3 experiments.
Fifteen of the 24 H-ferritin IRE mutants were used to test whether the number and position of G.U or U.G wobble base pairs in the upper stem are relevant for IRP binding. The results show that the presence of more than 2 G.U or U.G base pairs in the IRE motif dramatically impairs its ability to compete with the wild-type probe (Figure 3C mutants U, V, W, and Y), whereas variants bearing 1 (which is naturally present in the H-ferritin IRE) or 2 G.U or U.G base pairs compete well with the wild-type probe (Figure 3C mutants A, B, C, D, F, G, H, I, M, O). Mutants R and Q are unable to significantly compete with the wild-type IRE probe, probably because both of these variants contain a G at position 25 that may interact with the critical and unpaired C8 nucleotide.
Seven additional mutants were used to study the effect of a base mismatch in the IRE structure. In the context of an H-ferritin IRE-backbone, one single AxC mismatch at positions other than n13-n20 (mutant S) or n07-n25 (mutant T) is well tolerated, as demonstrated by mutants E, J, N, and P. The mismatch at position n13-n20 (mutant S) was not tolerated, because it will probably disturb the physiologic definition of the apical loop. Mutant K (CxA mismatch at position n07-n25), but not mutant T (AxC position n07-n25), is able to efficiently compete with the wild-type IRE for binding to IRP1. We suspect that A25 in mutant K may base pair with the bulged uridine present in the H-ferritin IRE (Figure 3B U6), which would be not possible for mutant T. This explanation was not evaluated further. These experimental results were interpreted into our newly developed SIREs algorithm23 (Figure 3A).
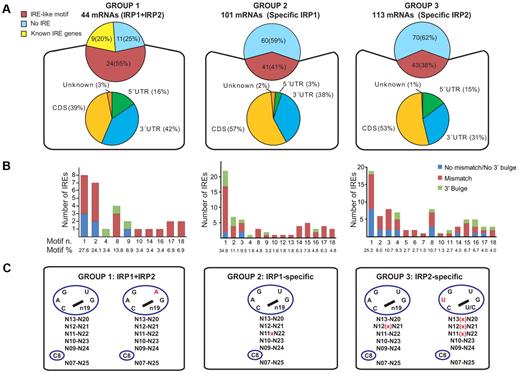
Applying SIREs to identify IREs within the mRNAs that coimmunoprecipitate with IRPs, we not only identify all known IREs but also at least 1 IRE-like motif in 24 of the 35 (68.6%) novel IRP1 and IRP2 murine target mRNAs or its human orthologs (Table 1; Figure 4A). Moreover, the prediction of IRE-like motifs is significantly enriched in the pool of 35 novel IRP target genes (P 6 × 10^-13, Fisher exact test) compared with 150 mouse mRNA sequences that were arbitrarily selected and randomly shuffled to avoid the selection of true positive hits by chance. In total, 29 IRE motifs were detected by SIREs in these 24 mRNAs (Table 1). The motif distribution of these predicted IREs shows that motif 1 and 2 together with motif 8 (supplemental Table 3) constitute more than 65% of newly detected motifs (27.6%, 24.1%, and 13.8%, respectively; Figure 4B). These new IRE-like motifs are located in all parts of the mRNAs, the 5′UTR (16%), 3′UTR (42%), and coding sequence region (CDS, 39%; Figure 4A; Table 1).
Bioinfomatic prediction of IREs by SIREs in novel IRP1 and IRP2 target mRNAs
| Gene name . | Description . | Reference ID . | IRE sequence . | Position . | Motif . | Mismatch . | Bulge . |
|---|---|---|---|---|---|---|---|
| 1 2010107G12Rik | RIKEN cDNA 2010107G12 gene | NM_001025573 | GTTCCGCTTC.C.C. TTACC.CAGGGC.AGTGGG.GCCTCCCCAC | CDS | 8 | N13-N20:C_A | |
| 2 4930579E17Rik | RIKEN cDNA 4930579E17 gene | NM_178629 | GTGGGCAAAC.T.C.TTGCC.TAGTAT.GGATAGA.GCGACATCCC | 3′UTR | 9 | N21b:A | |
| 3 8430410A17Rik | RIKEN cDNA 8430410A17 gene | NM_173737 | GCCGGCGGCG.G.C.TCCCG.CAGTGG.AGGGAC.CCCGACAAGT | CDS | 1 | N13-N20:G_A | |
| 4 A430093A21Rik | RIKEN cDNA A430093A21 gene | NM_001081436 | CTTGTATAAG.C.C.GCCTG.CAGAGA.CTGGTG.AAACTGTGCA | CDS | 2 | N12-N21:T_T | |
| 5 AI450353 | Expressed sequence AI450353 | AK134743 | TGTGAGAGTA.G.C.TTTTT.GAGTGT.GTAAGC.CTACATTTGA | N/A | 17 | N12-N21:T_T | |
| 6 ANKRD29 | Ankyrin repeat domain 29 | NM_173505 | ATCTTGAGTT.T.C.CAACA.CCGTGC.TGCTTGA.TAGAATGACT | 3′UTR | 4 | N21b:C | |
| 7 BC051227 | cDNA sequence BC051227 | NM_183170 | TGCGCGCGCG.G.C.CTGGA.CAGAGA.TTCAGC.GCGCGGCGCT | CDS | 2 | ||
| 8 Cxcl16 | Chemokine (C-X-C motif) ligand 16 | NM_023158 | AGCAGGCTCG.T.C.TCCAT.CAGTGA.ATGGAA.CCTGAGCTCA | 3′UTR | 1 | ||
| 9 Dlg2 | Discs, large homolog 2 (Drosophila) | NM_011807 | GACTACGAAG.T.C.GACGG.CAGAGA.CTATCA.CTTTGTCATT | CDS | 2 | N11-N22:C_A | |
| DLG2 | Discs, large homolog 2 (Drosophila) | NM_001364 | AAGAGATAGA.G.C.CATCA.GAGTGA.TGGGGC.TTCTTCACAG | 3′UTR | 17 | N10-N23:A_G | |
| 10 Egr2 | Early growth response 2 | NM_010118 | TGGCAGTGGG.T.C.TGCAG.CAGTGA.CTGCCA.CCCCTTATAA | CDS | 1 | N09-N24:T_C | |
| 11 FXYD5 | FXYD domain containing ion transport regulator 5 | NM_144779 | CCTCCTTGCG.T.C.AGTCC.CAGTGA.GGGATA.AGCGCCTGGC | 5′UTR | 1 | N10-N23:G_A | |
| 12 Garnl1 | GTPase activating RANGAP domain-like 1 | NM_019994 | TTTACACTCT.C.C.AGATT.CAGAGC.GGTCTT.CTAAACTGCA | CDS | 2 | N07-N25:C_T | |
| Garnl1 | GTPase activating RANGAP domain-like 1 | NM_001003719 | GTCGTCGGGA.C.C.CGGCC.GGGAGG.GGCCGC.GGCGGCCGCA | 5′UTR | 16 | N07-N25:C_C | |
| 13 Gstm6 | Glutathione S-transferase, mu 6 | NM_008184 | TATGCAGGCT.C.C.ATCTC.CAGAGT.GGGAAG.GCCCAGTCTT | 3′UTR | 2 | N09-N24:A_A | |
| 14 Gstt3 | Glutathione S-transferase, theta 3 | NM_133994 | TGACTTGGTG.G.C.CATCA.CAGAGC.TGATGC.ATCCTGTCAG | CDS | 2 | ||
| 15 Gyg | Glycogenin | NM_013755 | ATCAGCAGCA.C.C.AGACC.CAGGGT.GGCCTG.ACTGTTTCAA | CDS | 8 | N11-N22:A_C | |
| GYG1 | Glycogenin-1 | NM_004130 | GTCAGCAGCA.C.C.AGACC.CAGGGT.GGCCTG.ACTGCTTCAA | CDS | 8 | N11-N22:A_C | |
| GYG1 | Glycogenin-1 | NM_004130 | CAGATGAGAG.G.C.TTTTT.TAGGAT.AAGAGG.TGAGAACTGG | 3′UTR | 10 | N07-N25:G_G | |
| 16 Hao1 | Hydroxyacid oxidase 1, liver | NM_010403 | TCATTTATAG.T.C.ACATT.CAGTGT.AAAGTA.CATATTTTGT | 3′UTR | 1 | N11-N22:A_A | |
| 17 Kcnf1 | Potassium voltage-gated channel, subfamily F, member 1 | NM_201531 | ATTTTGTGAA.T.C.GTGAA.TAGTAC.TTTACA.TTCAAAATTT | 3′UTR | 9 | ||
| Kcnf1 | Potassium voltage-gated channel, subfamily F, member 1 | NM_201531 | CAAAGGCTAA.G.C.TGGGC.CAGTGG.GCCTAA.CCCTTTATGG | 3′UTR | 1 | N07-N25:G_A | |
| 18 Lrpap1 | Low-density lipoprotein receptor-related protein associated protein 1 | NM_013587 | CTTCCCAGAA.C.C.CTCAG.CAGTGT.CTGAGG.CTCAGAGAAA | 3′UTR | 1 | ||
| 19 NR4A3 | Nuclear receptor subfamily 4, group A, member 3 | NM_173199 | CCGCTCACCG.C.C.TCCGG.GAGCCG.CTGGGC.TTGTACACCG | 5′UTR | 14 | N07-N25:C_C | |
| 20 ORMDL1 | ORM1-like 1 (Saccharomyces. cerevisiae) | NM_016467 | TAAGTTAAAG.G.G.CATCA.CAGTGA.GGGTGT.AGTAGATAAA | 3′UTR | 18 | N13-N20:A_G | |
| 21 Pabpc4l | Poly(A) binding protein, cytoplasmic 4-like | NM_001101479 | GTCTTTGGGT.C.C.ATTTG.CAGGGT.TAAAGTG.ATGCAGGAAG | CDS | 8 | N22b:A | |
| 22 Ppp1r1b | Protein phosphatase 1, regulatory subunit 1B | NM_144828 | TTTCCTGGGT.G.C.GGGGA.CAGTGC.TCCTCC.TCCTCCTCCG | 5′UTR | 1 | ||
| 23 PYROXD1 | Pyridine nucleotide-disulfide oxidoreductase domain 1 | NM_024854 | GTCAGTTACA.G.C.TGATA.CAGAGA.TGTGGC.CTGTCTATGT | CDS | 2 | N10-N23:G_G | |
| 24 TP53INP2 | Tumor protein p53 inducible nuclear protein 2 | NM_021202 | TAGGTCAGCG.T.G.CAGAG.CAGTGA.TGCTGG.AGGACACACC | 3′UTR | 18 | N12-N21:A_G |
| Gene name . | Description . | Reference ID . | IRE sequence . | Position . | Motif . | Mismatch . | Bulge . |
|---|---|---|---|---|---|---|---|
| 1 2010107G12Rik | RIKEN cDNA 2010107G12 gene | NM_001025573 | GTTCCGCTTC.C.C. TTACC.CAGGGC.AGTGGG.GCCTCCCCAC | CDS | 8 | N13-N20:C_A | |
| 2 4930579E17Rik | RIKEN cDNA 4930579E17 gene | NM_178629 | GTGGGCAAAC.T.C.TTGCC.TAGTAT.GGATAGA.GCGACATCCC | 3′UTR | 9 | N21b:A | |
| 3 8430410A17Rik | RIKEN cDNA 8430410A17 gene | NM_173737 | GCCGGCGGCG.G.C.TCCCG.CAGTGG.AGGGAC.CCCGACAAGT | CDS | 1 | N13-N20:G_A | |
| 4 A430093A21Rik | RIKEN cDNA A430093A21 gene | NM_001081436 | CTTGTATAAG.C.C.GCCTG.CAGAGA.CTGGTG.AAACTGTGCA | CDS | 2 | N12-N21:T_T | |
| 5 AI450353 | Expressed sequence AI450353 | AK134743 | TGTGAGAGTA.G.C.TTTTT.GAGTGT.GTAAGC.CTACATTTGA | N/A | 17 | N12-N21:T_T | |
| 6 ANKRD29 | Ankyrin repeat domain 29 | NM_173505 | ATCTTGAGTT.T.C.CAACA.CCGTGC.TGCTTGA.TAGAATGACT | 3′UTR | 4 | N21b:C | |
| 7 BC051227 | cDNA sequence BC051227 | NM_183170 | TGCGCGCGCG.G.C.CTGGA.CAGAGA.TTCAGC.GCGCGGCGCT | CDS | 2 | ||
| 8 Cxcl16 | Chemokine (C-X-C motif) ligand 16 | NM_023158 | AGCAGGCTCG.T.C.TCCAT.CAGTGA.ATGGAA.CCTGAGCTCA | 3′UTR | 1 | ||
| 9 Dlg2 | Discs, large homolog 2 (Drosophila) | NM_011807 | GACTACGAAG.T.C.GACGG.CAGAGA.CTATCA.CTTTGTCATT | CDS | 2 | N11-N22:C_A | |
| DLG2 | Discs, large homolog 2 (Drosophila) | NM_001364 | AAGAGATAGA.G.C.CATCA.GAGTGA.TGGGGC.TTCTTCACAG | 3′UTR | 17 | N10-N23:A_G | |
| 10 Egr2 | Early growth response 2 | NM_010118 | TGGCAGTGGG.T.C.TGCAG.CAGTGA.CTGCCA.CCCCTTATAA | CDS | 1 | N09-N24:T_C | |
| 11 FXYD5 | FXYD domain containing ion transport regulator 5 | NM_144779 | CCTCCTTGCG.T.C.AGTCC.CAGTGA.GGGATA.AGCGCCTGGC | 5′UTR | 1 | N10-N23:G_A | |
| 12 Garnl1 | GTPase activating RANGAP domain-like 1 | NM_019994 | TTTACACTCT.C.C.AGATT.CAGAGC.GGTCTT.CTAAACTGCA | CDS | 2 | N07-N25:C_T | |
| Garnl1 | GTPase activating RANGAP domain-like 1 | NM_001003719 | GTCGTCGGGA.C.C.CGGCC.GGGAGG.GGCCGC.GGCGGCCGCA | 5′UTR | 16 | N07-N25:C_C | |
| 13 Gstm6 | Glutathione S-transferase, mu 6 | NM_008184 | TATGCAGGCT.C.C.ATCTC.CAGAGT.GGGAAG.GCCCAGTCTT | 3′UTR | 2 | N09-N24:A_A | |
| 14 Gstt3 | Glutathione S-transferase, theta 3 | NM_133994 | TGACTTGGTG.G.C.CATCA.CAGAGC.TGATGC.ATCCTGTCAG | CDS | 2 | ||
| 15 Gyg | Glycogenin | NM_013755 | ATCAGCAGCA.C.C.AGACC.CAGGGT.GGCCTG.ACTGTTTCAA | CDS | 8 | N11-N22:A_C | |
| GYG1 | Glycogenin-1 | NM_004130 | GTCAGCAGCA.C.C.AGACC.CAGGGT.GGCCTG.ACTGCTTCAA | CDS | 8 | N11-N22:A_C | |
| GYG1 | Glycogenin-1 | NM_004130 | CAGATGAGAG.G.C.TTTTT.TAGGAT.AAGAGG.TGAGAACTGG | 3′UTR | 10 | N07-N25:G_G | |
| 16 Hao1 | Hydroxyacid oxidase 1, liver | NM_010403 | TCATTTATAG.T.C.ACATT.CAGTGT.AAAGTA.CATATTTTGT | 3′UTR | 1 | N11-N22:A_A | |
| 17 Kcnf1 | Potassium voltage-gated channel, subfamily F, member 1 | NM_201531 | ATTTTGTGAA.T.C.GTGAA.TAGTAC.TTTACA.TTCAAAATTT | 3′UTR | 9 | ||
| Kcnf1 | Potassium voltage-gated channel, subfamily F, member 1 | NM_201531 | CAAAGGCTAA.G.C.TGGGC.CAGTGG.GCCTAA.CCCTTTATGG | 3′UTR | 1 | N07-N25:G_A | |
| 18 Lrpap1 | Low-density lipoprotein receptor-related protein associated protein 1 | NM_013587 | CTTCCCAGAA.C.C.CTCAG.CAGTGT.CTGAGG.CTCAGAGAAA | 3′UTR | 1 | ||
| 19 NR4A3 | Nuclear receptor subfamily 4, group A, member 3 | NM_173199 | CCGCTCACCG.C.C.TCCGG.GAGCCG.CTGGGC.TTGTACACCG | 5′UTR | 14 | N07-N25:C_C | |
| 20 ORMDL1 | ORM1-like 1 (Saccharomyces. cerevisiae) | NM_016467 | TAAGTTAAAG.G.G.CATCA.CAGTGA.GGGTGT.AGTAGATAAA | 3′UTR | 18 | N13-N20:A_G | |
| 21 Pabpc4l | Poly(A) binding protein, cytoplasmic 4-like | NM_001101479 | GTCTTTGGGT.C.C.ATTTG.CAGGGT.TAAAGTG.ATGCAGGAAG | CDS | 8 | N22b:A | |
| 22 Ppp1r1b | Protein phosphatase 1, regulatory subunit 1B | NM_144828 | TTTCCTGGGT.G.C.GGGGA.CAGTGC.TCCTCC.TCCTCCTCCG | 5′UTR | 1 | ||
| 23 PYROXD1 | Pyridine nucleotide-disulfide oxidoreductase domain 1 | NM_024854 | GTCAGTTACA.G.C.TGATA.CAGAGA.TGTGGC.CTGTCTATGT | CDS | 2 | N10-N23:G_G | |
| 24 TP53INP2 | Tumor protein p53 inducible nuclear protein 2 | NM_021202 | TAGGTCAGCG.T.G.CAGAG.CAGTGA.TGCTGG.AGGACACACC | 3′UTR | 18 | N12-N21:A_G |
Motif refers to type of apical loop as defined in supplemental Table 3.
Analysis of IRE motifs bound by IRP1, IRP2, or both IRPs. (A) Bioinformatic prediction of IRE motifs in IRP1 and IRP2 target mRNAs, IRP1-specific target mRNAs and IRP2-specific target mRNAs. (B) Motif distribution of novel IREs (also see supplemental Table 3). Motif number and percentage (motif n. and motif %) are shown in the 3 IRP-target mRNAs lists. IREs without a mismatch or a 3′ bulge are depicted in blue, IREs with a mismatch are in red, and IREs with a 3′ bulge are shown in green. (C) Enrichment of IRE motifs in IRP-target mRNAs. After normalization for n19 multiple IRE options in each IRE type, observed and percentage expected frequencies of IRE motifs were calculated (χ2 test, 1 df) and statistically significantly overrepresented motifs are shown. Red font, cross sign “x” or bracket cross sign “(x)” indicate differences compared with the canonical motif 1 IRE sequence. The cross sign or bracket cross sign indicate mismatches.
Analysis of IRE motifs bound by IRP1, IRP2, or both IRPs. (A) Bioinformatic prediction of IRE motifs in IRP1 and IRP2 target mRNAs, IRP1-specific target mRNAs and IRP2-specific target mRNAs. (B) Motif distribution of novel IREs (also see supplemental Table 3). Motif number and percentage (motif n. and motif %) are shown in the 3 IRP-target mRNAs lists. IREs without a mismatch or a 3′ bulge are depicted in blue, IREs with a mismatch are in red, and IREs with a 3′ bulge are shown in green. (C) Enrichment of IRE motifs in IRP-target mRNAs. After normalization for n19 multiple IRE options in each IRE type, observed and percentage expected frequencies of IRE motifs were calculated (χ2 test, 1 df) and statistically significantly overrepresented motifs are shown. Red font, cross sign “x” or bracket cross sign “(x)” indicate differences compared with the canonical motif 1 IRE sequence. The cross sign or bracket cross sign indicate mismatches.
SIREs also detects IRE motifs in 41% (41 mRNAs) and 38% (43 mRNAs) of the IRP1- or IRP2-specific targets, respectively, when searching mouse and human databases (Figure 4A; supplemental Tables 1-2). The motif distribution and localization of these predicted IREs also are indicated (Figure 4A-B).
We observed that the 3 different groups of mRNAs identified as IRP targets—group 1, IRP1 + IRP2-binding mRNAs; group 2, IRP1-specific mRNAs; and group 3, IRP2-specific mRNAs—display slightly different IRE motif preference (Figure 4C). Group 1 shows a predominance of the canonical IRE sequence without mismatches or bulges in the upper stem and with the classic apical loop nucleotide sequence CAGUGN (motif 1; P < .0001) or CAGAGN (motif 2; P = .0359). By contrast, group 2 (IRP1-specific binders) is enriched in IRE motifs with a mismatch in the upper stem at a central position n11-n22 (P = .0261). Group 3 (IRP2 binders) is enriched in IREs with a canonical loop presenting or not a mismatch at position n12-n21 of the upper stem, or CUGUGN19 loop with a mismatch at either of the 3 upper base pairs of the stem (P < .0001 and P = .0359, respectively; Figure 4C).
To complete the characterization of IRP binding to the transcriptome, we finally chose 16 of the 35 newly identified mRNAs that bind both IRP1 and IRP2, and we generated in vitro RNA transcripts for competitive IRP1 binding assays. These 16 mRNAs include tissue-specific and multitissue IRP binders (Figures 1C and 2B), as well as transcripts within which IRE motifs could or could not be identified (Figures 2B and 4A; Table 1). Six full-length IRP target mRNAs (Ppp1r1b, Gyg, Gstm6, Cxcl16, Pfn2, and Pdcl3) significantly compete for IRP1 binding at 40-fold molar excess over an H-ferritin IRE probe (P < .001 or P < .01; Figure 5). Ten of the novel IRP-associated mRNAs (0610007L01Rik, 2010107G12Rik, 8430410A17Rik, BC051227, Dhx32, Dirc2, Gstt3, Lsm12, Ormdl1, and Pyrodx1) did not display significant competition but neither did 2 positive controls with weak IREs (Slc11a2 and Aco2; data not shown). Several reasons may explain these observations: (1) like the Slc11a2 and Aco2 mRNAs, these mRNAs also may interact weakly with IRP1; (2) the IRP1 binding sites are part of the cellular mRNAs but excluded from the in vitro transcript; or (3) IRP1 binding to the candidate mRNA and the labeled ferritin IRE probe occurs to different sites of the protein and is noncompetitive. In spite of all the specificity controls included in the experiments, we also cannot formally exclude the possibility that non–IRP-binding mRNAs specifically copurified by (eg, hydrogen bounding) interactions with IRE-containing mRNAs.
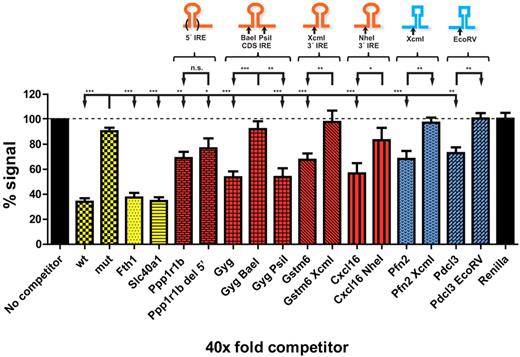
Validation of select novel IRP target mRNAs with in vitro transcripts. In vitro transcribed full-length mRNAs were incubated at a 40× molar excess against an H-ferritin IRE-radiolabeled probe for the binding of recombinant IRP1. Wt corresponds to an ∼ 1-kb reporter mRNA bearing the 5′ IRE of H ferritin mRNA; mut is the same reporter with a deltaC14 mutation of the IRE. Black bars represent negative control mRNAs (Renilla) or no competitor signal. Yellow bars represent positive controls (H-ferritin reporters and full-length Fth1 and Slc40a1 mRNAs). Red and blue bars correspond to novel IRP target mRNAs with or without a bioinformatically predicted IRE-like motif, respectively. Tested full-length and indicated 5′ restriction enzyme truncation mRNAs are grouped together. Above each group a schematic representation is shown indicating the restriction enzyme used to assess truncated forms and the location of the predicted IRE motif (round hairpin) or the putative IRP-binding RNA region (squared hairpin). P values are reported (***P < .001, **P < .01, *P < .05, unpaired 2-tailed Student t test compared with the mutant H-ferritin IRE construct, mut, or with each corresponding non-IRE construct). Data are presented as mean ± SEM.
Validation of select novel IRP target mRNAs with in vitro transcripts. In vitro transcribed full-length mRNAs were incubated at a 40× molar excess against an H-ferritin IRE-radiolabeled probe for the binding of recombinant IRP1. Wt corresponds to an ∼ 1-kb reporter mRNA bearing the 5′ IRE of H ferritin mRNA; mut is the same reporter with a deltaC14 mutation of the IRE. Black bars represent negative control mRNAs (Renilla) or no competitor signal. Yellow bars represent positive controls (H-ferritin reporters and full-length Fth1 and Slc40a1 mRNAs). Red and blue bars correspond to novel IRP target mRNAs with or without a bioinformatically predicted IRE-like motif, respectively. Tested full-length and indicated 5′ restriction enzyme truncation mRNAs are grouped together. Above each group a schematic representation is shown indicating the restriction enzyme used to assess truncated forms and the location of the predicted IRE motif (round hairpin) or the putative IRP-binding RNA region (squared hairpin). P values are reported (***P < .001, **P < .01, *P < .05, unpaired 2-tailed Student t test compared with the mutant H-ferritin IRE construct, mut, or with each corresponding non-IRE construct). Data are presented as mean ± SEM.
IRE-like motifs were found in 4 of the 6 mRNAs that positively compete in the EMSA experiments (Ppp1r1b, Gyg, Gstm6, and Cxcl16; Table 1). No IRE-like motif was recognized by SIREs in Pfn2 and Pdcl3 mRNAs. The mRNAs of Gstm6 and Cxcl16 contain one IRE-like motif in their 3′UTR, Ppp1r1b contains one IRE in its 5′UTR, and Gyg contains one IRE in its coding region. Exclusion of the region predicted to bear the IRE-like motif prevents the observed competition in all but the Ppp1r1b mRNA (Figure 5), supporting the notion that IRP binding occurs mainly via the newly recognized motifs. Pfn2 and Pdcl3 clones lacking 3′-terminal sequences beyond the XcmI and EcoRV sites, respectively, fail to compete, suggesting that the RNA element responsible for IRP interaction is located within this region of the transcripts (Figure 5).
Proteomics of cellular iron regulation
In addition to defining the target mRNAs of IRP1 and IRP2 in the global transcriptome, we also wanted to explore the role of the IRE/IRP network in shaping the cellular proteome. To this end, we used the recently described method of pSILAC,19 a method that quantifies relative differences in de novo protein biosynthesis, on IRP-deficient BMDM cells from macrophage-specific IRP1 and IRP2 knockout mice. Furthermore, we investigated iron regulation of the proteome of mouse hepatoma Hepa1-6 cells by the same method; the IRE/IRP regulatory system is intact in these cells and iron regulation integrates IRP-dependent and IRP-independent proteomic changes.
In Hepa1-6 cells, we performed 2 independent pSILAC experiments to compare untreated cells with iron supplemented (100μM hemin) or iron-deficient cells (200μM DFO, an iron chelator), respectively. In total, we confidently detected 2364 proteins in the iron-replete cells and 3594 proteins in their iron-starved counterparts. The results show that the biosynthetic labeling of 73 (3.1%) proteins is increased and of 480 (20.3%) proteins is decreased by hemin treatment; DFO treatment up-regulates the labeling of 67 (1.86%) proteins and down-regulates 920 (25.6%) proteins. Opposite regulation by iron is seen for 22 proteins, with 12 proteins more highly expressed after hemin treatment and repressed by DFO, and 10 proteins with reciprocal regulation (supplemental Table 4). Ferritin H and L are among the 12 proteins that are increased by iron and decreased by iron deficiency, displaying a 14- and a 8.6-fold increase by hemin and a 1.5- and a 1.6-fold decrease under iron-chelating conditions, respectively. The Tfrc labeling is strongly decreased by hemin (−5.4 fold) and induced by DFO (1.4-fold) treatment. Of these 22 proteins, only the mRNAs encoding Ftl1, Fth1, and Tfrc bear IREs that are recognized by SIREs. Moreover, none of the 19 remaining bidirectionally iron-regulated proteins correspond to IRP-associated mRNAs, suggesting that iron regulation of these proteins occurs by IRP-independent or only indirectly IRP-dependent mechanisms.
In pSILAC experiments with IRP-deficient BMDM cells, the absence of both IRPs causes the up-regulation of 63 (2.03%) proteins and the down-regulation of 188 (6.06%) proteins (supplemental Table 5). These changes may be a direct consequence of IRP ablation or a secondary consequence of complete IRP deficiency, which causes marked cellular and mitochondrial iron deficiency in hepatocytes.16 Detectably IRP-regulated proteins include the positive control IRP-target mRNAs ferritin L and H (6.4- and 5.3-fold up-regulated, respectively) and Tfrc (−2.5 fold down-regulated). Of the 248 remaining IRP-regulated proteins, 3 proteins are encoded by mRNAs that were found to be directly bound by IRP1 or IRP2 (Lpl, Psmd10, and Tomm40), and IREs were predicted in 54 mRNAs by SIREs. This result raises the possibility that cellular responses to the IRE/IRP network reach beyond the currently known targeted mRNAs.
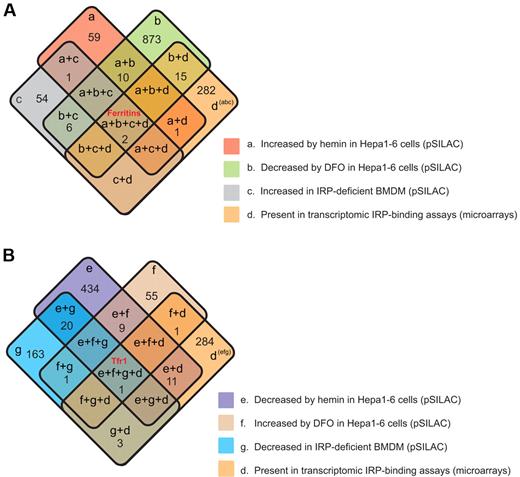
Finally, we integrated all global datasets (IRP-binding transcriptome and iron/IRP-regulated proteome) in a Venn diagram analysis, generating 2 4-set Venn diagrams (Figure 6A-B). Apparently, the majority of iron-regulated proteins are not likely to be direct IRP targets. Ferritin L and H proteins/mRNAs were detected in all assays, showing an opposite iron regulation in Hepa 1-6 cells (group a and b), an increase in their expression in BMDMs lacking both IRPs (group c), and positively detected in IRP immunopurification experiments (group d; Figure 6A). Thirty-three additional proteins/mRNAs meet 2 of these criteria (supplemental Table 6). The Tfrc protein/mRNA was oppositely iron-regulated in Hepa 1-6 cells (groups e and f), down-regulated in BMDM cells lacking both IRPs (group g), and positively IRP immunopurified (group d; Figure 6B). As such, it is the only protein/mRNA that displays this complete regulatory response. Forty-five additional proteins meet 2 of the 4 criteria (supplemental Table 6).
Integrative data analysis combining iron proteomics and microarray data. (A) Combination of categories a, b, c, and d. (B) Combination of categories e, f, g, and d. Category definitions are as follows: a, proteins up-regulated by hemin in Hepa1-6 cells in pSILAC experiments; b, proteins down-regulated by DFO in Hepa1-6 cells in pSILAC experiments; c, proteins up-regulated in IRP-deficient BMDMs (pSILAC experiments); d, IRP target mRNAs detected by IPs and microarrays; e, proteins down-regulated by hemin in Hepa1-6 cells in pSILAC experiments; f, proteins up-regulated by DFO in Hepa1–6 cells in pSILAC experiments; and g, proteins down-regulated in IRP-deficient BMDMs (pSILAC experiments).
Integrative data analysis combining iron proteomics and microarray data. (A) Combination of categories a, b, c, and d. (B) Combination of categories e, f, g, and d. Category definitions are as follows: a, proteins up-regulated by hemin in Hepa1-6 cells in pSILAC experiments; b, proteins down-regulated by DFO in Hepa1-6 cells in pSILAC experiments; c, proteins up-regulated in IRP-deficient BMDMs (pSILAC experiments); d, IRP target mRNAs detected by IPs and microarrays; e, proteins down-regulated by hemin in Hepa1-6 cells in pSILAC experiments; f, proteins up-regulated by DFO in Hepa1–6 cells in pSILAC experiments; and g, proteins down-regulated in IRP-deficient BMDMs (pSILAC experiments).
These data place the ferritins and Tfrc at the center of cellular iron- and IRP-dependent regulation. More importantly, they uncover and specifically identify numerous proteins and mRNAs through which iron regulation, IRP regulation, or both connect with other aspects of cell biology and physiology.
Discussion
Posttranscriptional regulation of gene expression by the IRP/IRE regulatory system plays a central role in the control of cellular iron metabolism. In the course of the past 25 years, research on this system has laid foundations for understanding of cellular iron homeostasis, even if the list of IRP-target mRNAs is still limited to few that encode core iron metabolism proteins. With this work, we begin to connect this core with other cellular functions that need to respond to changes in iron metabolism. To this end, we have deciphered the whole-genome repertoire of mRNAs associated with IRP1 and IRP2 from 5 tissues, and we defined the proteomic changes in IRP-deficient cells and in an iron-perturbed hepatic cell line. In isolation and in combination, these experiments have yielded a wealth of new information that will help instruct future experiments.
We have identified new mRNAs that can interact either with both IRPs or specifically only with one of them. Each of the 3 classes considerably enlarges the IRP regulatory repertoire, whereas the latter 2 classes offer first examples of IRP-specific cellular target mRNAs. These mRNAs encode proteins involved in different cellular functions, including metal ion binding proteins, transferases, ligases, helicases, and transcription or DNA binding factors, according to functional annotation clustering analyses using Database for annotation, visualization and integrated discovery (supplemental Table 7). Interestingly, 9 of the novel IRP1 + 2 target mRNAs (Lnx1, Mkrn1, Egr2, Nr4a3, Pex12, Garnl1, Cxcl16, Kcnf1, and Dhx32) fall into the category “metal ion binding” that also includes the already known IRP-regulated mRNAs Fth1, Ftl1, Aco2, Slc11a2, and Slc40a1. For 7 of these genes (Lnx1, Mkrn1, Egr2, Nr4a3, Pex12, Garnl1, and Cxcl16), the encoded proteins are reported to interact selectively and noncovalently with zinc ions. Iron and zinc are known to compete for the absorptive pathway through binding to the IRE-containing Slc11a2 metal transporter.33 Our results raise the possibility of an extensive fine tuning coordination of iron and zinc metabolism via the IRE/IRP system.
Concerning exclusive IRP1 or IRP2 targets, both lists are enriched in functional term categories related to RNA splicing, regulation of cell migration, and zinc finger (RING type) motifs. IRP2-associated mRNAs are exclusively enriched in functional terms related to ubiquitin-mediated proteolysis/ligase activity (Pml, Ube2j2, Ube2q2, and Herc4) and the cytokine receptor/Jak-STAT signaling pathway (Il6ra, Il6st, Csf2rb, Tnfrsf1B, and Cx3cr1; supplemental Table 7). The discovery of common and specific subsets of IRP target mRNAs implicated in a wide spectrum of functions may, at least in part, explain phenotypic differences between IRP1 and IRP2 knockout mice34,35 and connects the regulation of intracellular iron homeostasis with these functions, defining starting points for future explorations.
Previous in vitro studies reported IRP1- or IRP2-specific RNA sequences generated by systematic evolution of ligands by exponential enrichment 26-28 (in this study, motifs 9, 10, 11, 12, and 17 for IRP1 and motifs 4, 5, and 15 for IRP2). Our data confirm and extend these results: mRNAs containing IREs with motif 4 CN(5)CCGUG(A/U/C) are specifically enriched in immunoprecipitation for IRP2, representing 9.3% of the total of predicted IREs for this group (Figure 4B). Although many of the newly identified IRP-associated mRNAs bear IRE-like motifs, some lack such elements that we could recognize and entirely different IRP-binding sites or other not considered IRE structures29 may exist. Indeed, this applies to 2 of the mRNAs for which we validated IRP1 binding with in vitro transcripts (Pfn2 and Pdcl3).
None of the previously known IRE motifs were identified entirely within the coding region. Notably, one of the specifically validated mRNAs identified here (encoding glycogenin, the scaffold protein for glycogen synthesis) contains a conserved IRE motif within its CDS. Other functional cis-acting regulatory elements have been found previously within the CDS of mRNAs.36,37 Additional IRE-like motifs have been reported in different species (eg, humans, primates, rats) that seem not to be conserved in the mouse (CDC14A, APP, AHSP, and CDC42BPA).17,38-40 None of these mRNAs were identified as IRP-target mRNAs in this study using mouse total RNA as starting material. Although we believe that this work identifies IRP-binding mRNAs in a way that satisfies stringent biochemical criteria, it is important to point out that further in vivo studies are needed to evaluate the functionality and physiologic relevance of these candidate IREs and IRP-binding mRNAs and to elucidate the underlying regulatory mechanisms.
We also complemented the exploration of transcriptomic IRP-binding targets by a proteomic approach by pSILAC, a method that assesses overall changes in proteins synthesis (including changes in de novo protein synthesis or profound changes in protein stability). This work, for example, shows that the synthesis of the heme b-binding protein succinate dehydrogenase (Sdh) subunit C is strongly increased by hemin (> 84 fold). Interestingly, a functional IRE is present in the 5′UTR of the Drosophila melanogaster Sdh subunit B mRNA,41 but no IRE-like motif is found in the murine or human Sdh subunit C mRNA, and neither of these RNAs was found to be associated with IRPs in our experiments. These observations suggest that contrasting with flies, the induction of these proteins in mammals is not driven by an IRP-dependent mechanism, even if the regulatory outcomes are similar.
Although other membrane-associated transporters are identified in our pSILAC experiments, Slc11a2 and Slc40a1 were not detected. Possible explanations include low expression levels in the tested cells, insufficient amount of de novo synthesis, or proteomic methodologic limitations. The Aco2 protein was detected but not found to be regulated by iron manipulation in Hepa1-6 cells or by IRP deficiency in BMDMs. This observation is probably explained by the well recognized low binding affinity of the 5′ IRE of Aco2 mRNA for IRP1, and this mRNA is known to be less stringently regulated at the translational level compared with ferritin mRNAs.42
Analysis of the regulated proteins using the STRING algorithm43 reveals the down-regulation of the mitochondrial electron transport chain, including 24 NADH dehydrogenases (complex I) and all 4 proteins of complex II (Sdha, Sdhb, Sdhc, and Sdhd) under conditions of IRP deficiency (supplemental Methods; data not shown). These findings are very interesting in the context of recent data showing that a general biologic function of IRPs lies in securing mitochondrial iron sufficiency and function.16
The IRP-regulated proteins and mRNAs identified in this study could physiologically respond to iron changes as well as to iron-independent signals that alter the activity of the IRPs. Collectively, our results begin to interconnect the well-characterized core IRP regulon with key aspects of cell biology and physiology and provide opportunities to further deepen the molecular and cellular understanding of iron homeostasis; iron-related diseases; and other IRP-dependent, iron-independent pathways.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dunja Ferring-Appel (EMBL) for excellent technical assistance with mouse work. They are grateful to Monica Campillos (EMBL) and Ildefonso Cases (IMPPC) for support in the development of the bioinformatic web-server SIREs.
This work was supported by the Young Investigator Award of the Medical Faculty, University of Heidelberg (Germany) and the postdoctoral fellowship Beatriu de Pinós, Generalitat de Catalunya (Spain) and Ramón y Cajal Program, Spanish Ministry of Science and Innovation (RYC-2008-02352) to M.S. and by grants from the Forschungsschwerpunktprogramm des Landes Baden-Württemberg (RNA and disease) to M.W.H. and M.U.M.
Authorship
Contribution: M. Sanchez designed and performed experiments and analyzed data; B.G. performed animal experiments, BMDM work, pSILAC labeling of BMDMs, and assisted in manuscript preparation; B.S. performed proteomics experiments and data analysis; T.B.-I. performed microarray experiments; J.B. performed microarray data analysis; M. Selbach, V.B., M.U.M., and M.W.H. oversaw the study and designed experiments; M.U.M. assisted in manuscript preparation; and M. Sanchez and M.W.H. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Matthias W. Hentze, EMBL, Meyerhofstrasse 1, 69117 Heidelberg, Germany; e-mail: hentze@embl.de; or Martina U. Muckenthaler, Department of Pediatric Oncology, Haematology and Immunology, University Hospital of Heidelberg, and Molecular Medicine Partnership Unit, Im Neuenheimer Feld 156, 69120 Heidelberg, Germany; e-mail: martina.muckenthaler@med.uni-heidelberg.de.
References
Author notes
M.U.M. and M.W.H. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal