Abstract
This article reviews the regulation of production of RBCs at several levels. We focus on the regulated expansion of burst-forming unit-erythroid erythroid progenitors by glucocorticoids and other factors that occur during chronic anemia, inflammation, and other conditions of stress. We also highlight the rapid production of RBCs by the coordinated regulation of terminal proliferation and differentiation of committed erythroid colony-forming unit-erythroid progenitors by external signals, such as erythropoietin and adhesion to a fibronectin matrix. We discuss the complex intracellular networks of coordinated gene regulation by transcription factors, chromatin modifiers, and miRNAs that regulate the different stages of erythropoiesis.
Introduction
In mammals, definitive erythropoiesis first occurs in the fetal liver with progenitor cells from the yolk sac.1 Within the fetal liver and the adult bone marrow, hematopoietic cells are formed continuously from a small population of pluripotent stem cells that generate progenitors committed to one or a few hematopoietic lineages (Figure 1). In the erythroid lineage, the earliest committed progenitors identified ex vivo are the slowly proliferating burst-forming unit-erythroid (BFU-E). Early BFU-E cells divide and further differentiate through the mature BFU-E stage into rapidly dividing colony-forming unit-erythroid (CFU-E).2 CFU-E progenitors divide 3 to 5 times over 2 to 3 days as they differentiate and undergo many substantial changes, including a decrease in cell size, chromatin condensation, and hemoglobinization, leading up to their enucleation and expulsion of other organelles.3
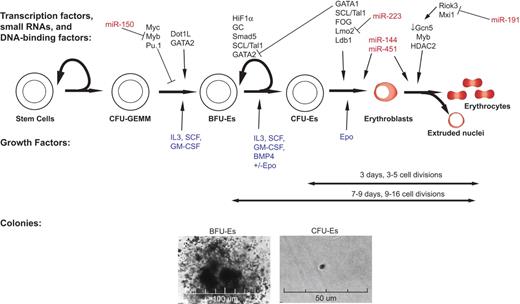
An overview of erythropoiesis: regulation at multiple levels by multiple proteins and miRNAs. Formation of RBCs from HSCs is regulated by signaling through both external factors (blue), such as cytokines and fibronectin, as well as intracellular factors, such as transcription factors (black) and miRNAs (red). Below the differentiation network, a timeline and images are shown for in vitro methylcellulose colony formation of murine BFU-E and CFU-E.
An overview of erythropoiesis: regulation at multiple levels by multiple proteins and miRNAs. Formation of RBCs from HSCs is regulated by signaling through both external factors (blue), such as cytokines and fibronectin, as well as intracellular factors, such as transcription factors (black) and miRNAs (red). Below the differentiation network, a timeline and images are shown for in vitro methylcellulose colony formation of murine BFU-E and CFU-E.
In humans, the life span of RBCs is 120 days. Under normal conditions, approximately 1% of RBCs are synthesized each day but RBC production can increase substantially during times of acute or chronic stress, such as acute trauma or hemolysis. Exquisite short-term control of erythropoiesis is regulated by the kidney-derived cytokine erythropoietin (Epo), which is induced under hypoxic conditions and stimulates the terminal proliferation and differentiation of CFU-E progenitors.4 BFU-E cells respond to many hormones in addition to Epo, including SCF, insulin like growth factor 1 (IGF-1), glucocorticoids (GCs), and IL-3, and IL-6. In cases of chronic erythroid stress, such as hemolysis, the number of CFU-E progenitors is insufficient to produce the needed RBCs, even under high Epo levels, and the body responds by producing more of these progenitors from BFU-E.5 It is not entirely known which cells in the fetal liver or adult bone marrow produce these and other regulatory cytokines, or how they interact to regulate the division of BFU-E cells and control their self-renewal and their ability to differentiate into more mature CFU-E progenitors.
At each stage of RBC production, intracellular signal transduction proteins and transcription factors activated downstream of these hormones interact with a group of DNA-binding and other transcription factors and chromatin modifiers as well as with multiple noncoding regulatory RNAs, such as microRNAs (miRNA); many of these transcription factors and noncoding RNAs are essential for the function and/or identity of these progenitor cells. Here we summarize the ways in which RBC production is regulated at each differentiation stage: through cytokines, transcription factors, and cofactors, post-translational modifications of histones, and miRNAs. We start with the terminal proliferation and differentiation of CFU-E erythroid progenitors, as this step is very well understood, and then work backward toward the less understood processes of formation and function of BFU-E progenitors.
Extracellular signals regulating proliferation and differentiation of CFU-E progenitors
Erythropoietin has long been understood to be the major factor governing erythropoiesis and its role in regulating the expansion, differentiation, apoptosis, and activation of erythroid specific genes is well characterized.4 The first phase of CFU-E erythroid differentiation is highly Epo dependent, whereas later stages are no longer dependent on Epo.6-8 Consistent with this, Epo receptors are lost as erythroid progenitors undergo terminal proliferation and differentiation.9
The extracellular matrix protein fibronectin is also important for erythropoiesis10 ; fibronectin and Epo regulate erythroid proliferation in temporally distinct steps. During the first day in culture, CFU-E erythroid progenitor cells undergo 2 divisions, up-regulate the transferrin receptor, and begin expression of Ter119 and several hundred other erythroid-important genes. This stage requires Epo but is independent of fibronectin. During the second day, there are 2 or 3 rapid cell divisions with short or absent G1 and G2 stages; most cells are in S or M. There is then complete repression of all gene transcription, chromatin condensation, nuclear condensation, and enucleation. Adhesion to fibronectin, but not the presence of Epo, is essential for the last one or 2 terminal cell divisions and promotes enucleation. α4, α5, and β1 are the principal integrins expressed on erythroid progenitors, and fibronectin fragments that engage α4β1 (but not α5β1) integrin support normal terminal proliferation. In the absence of fibronectin, a fraction of cultured erythroblasts enucleate, but these generate larger-than-normal “reticulocytes.” We do not know whether this is related to macrocytic anemias where large poorly hemoglobinized RBCs are produced. Taken together, these data suggest a 2-phase model for growth factor and extracellular matrix regulation of erythropoiesis, with an early Epo-dependent, integrin-independent phase followed by an Epo-independent, α4β1 integrin-dependent phase.
Binding of Epo to Epo receptors (EpoRs) on the surface of erythroid progenitors triggers activation of multiple intracellular signal transduction pathways, including the signal transducer and activator of transcription 5 (Stat5), phosphoinositide-3 kinase/Akt, and Shc/Ras/mitogen-activated kinase (MAPK) pathways. Elimination of either of the first 2 pathways leads to significant apoptosis of early progenitors and reduced output of erythrocytes.11,12 In contrast, blocking the Ras/ MAPK pathway has only subtle effects on terminal erythropoiesis.13
Disruption of the Stat5 pathway, in contrast, revealed not only the cellular role of this factor in erythropoiesis, but also the regulation of RBC production by expansion of earlier progenitor and stem cell populations. Although many (albeit hypomorphic) adult Stat5a−/−5b−/− mice have a normal or near-normal steady-state hematocrit, they are deficient in generating high erythropoietic rates in response to stress and have very high endogenous levels of Epo in the blood.14 Stat5 is essential for the high erythropoietic rate during fetal development; the double-knockout embryos are severely anemic; and erythroid progenitors are present in low numbers, show higher levels of apoptosis, and are less responsive to Epo.15
Jak2 stimulates proliferation of erythroid precursors in part through activation of Id1 by binding of Stat5 to a downstream enhancer of Id1.16 Id1 expression levels also correlate with the levels of the constitutively active mutant Jak2V617F in both transgenic cell lines as well as patients with polycythemia vera. The role of Stat5 in iron metabolism has also been established by evaluation of HSC-targeted knockdown of Stat5; these mice displayed microcytic, hypochromic anemia indicative of iron deficiency, and showed 50% decreased levels of the transferrin receptor (Tfr1) on their erythroblasts and 80% decreased Tfr1 mRNA levels. Tfr1 was shown to be a direct transcriptional target of Stat5A by chromatin immunoprecipitation studies.17
Stat5 is activated by other homodimeric cytokine receptors (the thrombopoietin receptor in megakaryocytes and the prolactin receptor in mammary glands) but activates very different sets of genes in these cells compared with erythroid progenitors. Clearly, Stat5 interacts with different resident transcription factors and chromatin-modifying enzymes in different cell types. In addition, using reintroduction of minimal Epo receptor chimeras that lack many of the intracellular tyrosines required for known downstream signaling via Jak2 docking into EpoR-knockout tissue, it was found that Epo might actually signal through other pathways. Direct Epo-EpoR targets include tyrosine phosphatase Prl1 and Rank (receptor activator of NF-κB) as well as 3 regulators of protein synthesis (EF1α, eIF3-p66, and Nat1).18
Transcriptional regulators of erythroid proliferation and function
In erythroid cells, Stat5 and other Epo-regulated transcriptional regulators interact with a relatively small number of lineage-restricted transcriptional regulators, including GATA-1, SCL/Tal1, LMO2, LDB1, Klf1, and Gfi-1b, to produce mRNAs essential for erythropoiesis (Figures 1 and 2). These crucial regulators are found in numerous combinations of multiprotein complexes; their functions have been established by gene-targeting knockout mouse models as well as by studies of rare diseases of ineffective erythropoiesis.19,20 Other transcriptional regulators, such as Bcl-11a, have been identified that mainly affect the transition between fetal γ- and adult β-globins.21 Dysregulation of signaling pathways downstream of many of these regulators leads to the development of specific leukemias and myeloproliferative disorders.22,23 However, how distinct complexes interact to repress or activate specific gene expression programs is still poorly understood. Recent techniques, such as ChIP coupled with massive parallel sequencing (ChIP-seq), combined with gene expression profiling and bioinformatic analysis, have begun to uncover additional interactions between known regulators as well as some understanding of the interplay between these complexes and the local nucleosome environment.24
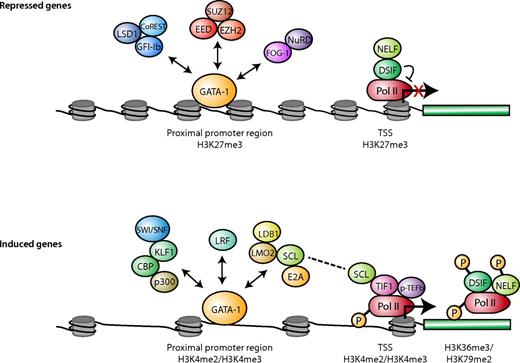
Transcription factors, Pol II status, and histone modifications associated with actively transcribed and repressed genes in erythroid cells. For genes activated during differentiation (bottom panel), the GATA-1 activation-associated proteins exist in different complexes (as indicated by the double arrows) through binding to both distal regulatory regions and promoter regions near the TSS. H3K4me2 and H3K4me3 are enriched in some of these bound regions. TIF-1 and the SCL complex recruit pTEF-b to promote Pol II elongation by phosphorylating DSIF, NELF, and Pol II. SCF could be the link between the GATA-1 complex and Pol II elongation machinery. H3K79me2 and H3K36me3 are often enriched in the transcribed portion of the gene. Among repressed genes (top panel), the GATA-1 repressor complexes are less clear and could exist in different forms as indicated by the double arrows. H3K27me3 is enriched near the TSS. Pol II is either not bound or is paused around the TSS. In the absence of TIF-1, recruitment of p-TEFb is impaired, and DSIF and NELF inhibit the phosphorylation of Pol II and Pol II elongation.
Transcription factors, Pol II status, and histone modifications associated with actively transcribed and repressed genes in erythroid cells. For genes activated during differentiation (bottom panel), the GATA-1 activation-associated proteins exist in different complexes (as indicated by the double arrows) through binding to both distal regulatory regions and promoter regions near the TSS. H3K4me2 and H3K4me3 are enriched in some of these bound regions. TIF-1 and the SCL complex recruit pTEF-b to promote Pol II elongation by phosphorylating DSIF, NELF, and Pol II. SCF could be the link between the GATA-1 complex and Pol II elongation machinery. H3K79me2 and H3K36me3 are often enriched in the transcribed portion of the gene. Among repressed genes (top panel), the GATA-1 repressor complexes are less clear and could exist in different forms as indicated by the double arrows. H3K27me3 is enriched near the TSS. Pol II is either not bound or is paused around the TSS. In the absence of TIF-1, recruitment of p-TEFb is impaired, and DSIF and NELF inhibit the phosphorylation of Pol II and Pol II elongation.
GATA1, discovered as the first member of the family of factors to bind the DNA consensus sequence (A/T)GATA(A/G) through 2 zinc fingers characteristic of the family,25 has been extensively studied. Yet, it turns out that the annotation of GATA consensus sites is a poor predictor of in vivo GATA1 binding to erythroid-specific genes.26 Indeed, the bulk of GATA1 binding occurs at distal regulatory elements, with very few (only ∼ 10%-15%) in the proximal promoter regions.27,28 These GATA1-bound sites were all enriched for monomethylated H3K4 (H3K4me1), a marker found predominately at distal enhancer elements,29,30 providing further evidence that GATA1 acts primarily through distal enhancers.31 Motif analysis of the activated genes revealed highly enriched consensus sequences for SCL/Tal1 at GATA1-bound sites,27,28,31-33 suggesting that GATA1 activation occurs in concert with the SCL/Tal1 coregulator. In erythroid cells, SCL/Tal1 forms a complex with the ubiquitous bHLH protein E2A, and also the LIM domain containing factors LMO2 and Ldb1.19 This activating complex, along with GATA1, binds to composite GATA1/E-box domains spaced approximately 9 to 11 nt apart.20 The components of the GATA1 repressor complexes are much less clear and probably include FOG1, the repressor Gfi-1b, and/or the chromatin modifiers EZH2 and core PRC2 component EED (Figure 2).20
Similar to GATA1, other components of the activation complex are also necessary for RBC development; Lmo2, GATA1, and SCL/Tal1 knockout mice are all anemic and die of lack of primitive erythropoiesis during fetal development.19 Embryos lacking Ldb1 show defective primitive erythropoiesis, and targeted deletion of Ldb1 via Mx-Cre expression in adult mice results in severe anemia and ultimate death, confirming that Ldb1 is also required for adult definitive erythropoiesis. Genome-wide location analysis of SCL/Tal1 binding of components of this complex confirmed the reciprocal findings of GATA1 binding described in the previous paragraph: that the majority of SCL/Tal1 sites are also outside the proximal promoter region.32 After combining Ldb1 ChIP-seq with chromosome conformation capture sequencing (3C), one group discovered that Ldb1 binds directly to the HS2, HS3, and HS4 DNA hypersensitive sites of the globin locus control region, despite the fact that there are no direct Ldb1 binding sites there.34
Klf1 (formerly called EKLF) is a zinc finger transcription factor that recognizes a subset of extended CACC box motifs and is remarkably erythroid-restricted35 ; its essential role in erythropoiesis has been established for quite some time.19 Similar to the GATA1 activation complex, genome-wide location analysis of Klf1 binding has revealed that the majority of Klf1-bound sites are more than 10 kb away from any known gene.36 Comparing the Klf and GATA1 binding site maps, one group36 discovered that almost half (48%) of the Klf1 sites were within 1 kb of GATA1 sites and few contained the SCL/Tal1 consensus sequence or overlapped with SCL/Tal1-GATA1 co-occupied sites. This suggested that GATA1 and Klf1 may activate genes in a complex distinct from that which contains Tal1.20
Even more interesting, Klf1 may actually be responsible for recruiting regions of chromatin with active transcription into transcription factories rather than the machinery moving to the chromatin. Several studies in the past decade have shown that nascent transcription actually occurs within a limited number of nuclear foci containing high concentrations of active RNA polymerase II (Pol II) and transcriptional machinery37 and that even distant actively transcribed chromatin colocalizes to these active areas rather than moving the machinery itself. In erythroid tissue, Klf1 is found to colocalize to these areas of active transcription; and through looping experiments using the 4C technique, it was confirmed that Klf can recruit active chromatin to these transcription factories.38
Other transcription factors have emerged as critical determinants of erythropoiesis. Similar to SCL/TAL1, LYL-1 is a transcription factor containing a basic helix-loop-helix motif. The defect in erythropoiesis in Lyl-1 knockout hematopoietic cells can be partially explained by their higher rate of apoptosis associated with a decreased level of Bcl-xL.39 Loss of the related POZ-Kruppel family transcription factor, LRF (or Zbtb7a/Pokemon), leads to lethal embryonic anemia and causes an Epo-unresponsive macrocytic anemia in adult mice.40 Apoptosis of erythroid cells because of deficiency of LRF can be rescued by loss of the proapoptotic factor, Bim. Although Lrf knockout mice share a similar phenotype with Stat5a/5b knockout mice,15 Epo-STAT5 signaling remains intact in the absence of LRF, given the observation that Bcl-xL is induced on Epo stimulation and also the MAPK and PI3K pathways are activated normally in Lrf-null erythroblasts.40 Lrf is a direct target of GATA1, and also GATA-1 physically interacts with LRF in activating direct target genes.28 A positive feedback loop by which GATA-1 mediates LRF activation could be critical for erythropoiesis.
Regulated pausing of RNA Pol II during erythroid differentiation
Genome-wide studies in several cell types showed that Pol II is frequently stalled at promoters shortly after transcription initiation. In some cells, the percentage of pausing may be as high as 90% of expressed genes, supporting the concept that regulation of Pol II elongation is a critical step in gene expression.41-43 Recruitment of P-TEFb (positive transcription elongation factor b) kinase promotes elongation by phosphorylating Spt5 and Pol II. Studies on the murine globin gene cluster illustrate the regulation of transcriptional elongation through release of paused Pol II; globin gene transcription is dramatically increased only after significant binding of NF-E2, TFIIB, and Pol II to the promoter. Deletion of the locus control region reduces phosphorylation of Pol II and subsequent Pol II elongation, resulting in a 90% decrease of globin transcription.44 Zebrafish bearing a loss-of-function mutation in transcriptional intermediary factor 1 γ (TIF-1γ) are extremely anemic. TIF-1 modulates the outcome of erythroid or myeloid decision from HSCs by controlling the balance of gata-1 and pu.1 level.45 In the absence of TIF-1γ, recruitment of p-TEFb and FACT is impaired, which in turn reduces the phosphorylation level of Pol II and Pol II elongation.46
Epigenetic changes in chromatin during erythroid differentiation
In many developmental systems, post-translational modifications of histones are crucial in regulating gene expression (Figure 2).47,48 Although some modifications tend to be associated with gene activation or repression states, the actual situation is generally more complex.
For example, the level of the epigenetic histone mark H3K79me2, added by the H3K79 methyltransferase Dot1, is correlated with transcriptional activation and elongation in Drosophila cells49 as well as in higher eukaryotes, such as human ES cells.41 However, location analysis on human T cells revealed that the H3K79me2 mark showed no preferential association with either gene activation or repression,50 raising the possibility that there is tissue specificity of the regulation of gene expression by H3K79me2 levels. H3K79 methylation occurs within the globin locus, suggesting a role in erythropoiesis. Indeed, loss of Dot1 greatly impairs both primitive and definitive yolk sac erythropoiesis, and BFU-E colonys number and size are significantly reduced in Dot1 knockout mice.51 Importantly, the relative levels of 2 crucial regulators, GATA2 and Pu.1 (Sfp1), are critical in determining the cell fate of hematopoietic progenitors: high Pu.1 and low GATA2 result in myeloid differentiation, low Pu.1 and high GATA2 lead to erythroid differentiation. The ratio of Pu.1 to GATA2 was reversed by Dot1L knockdown, resulting in high Pu.1 and low GATA2 levels in Dot1L-deficient yolk-sac erythroid progenitors. The effect of H3K79me2 on the level of Pu.1 is probably indirect because loss of H3K79me2 leads to high levels of Pu.1.
As a second example, the H3K4me3 modification at transcriptional start sites (TSSs) is commonly associated with gene activation and transcription initiation by RNA Pol II. However, the H3K4 modification often colocalizes with H3K27me3 modifications, which are associated with repressed genes. Such chromatin regions marked both by H3K27me3 and H3K4me3 are termed “bivalent domains.” Many are found in embryonic stem cells on genes encoding key developmental transcription factors.52 Similar bivalent domains occur in human primary HSCs/progenitor cells (CD133+) that can differentiate into CD36-expressing erythrocyte precursors.30 In CD133+ cells, promoter regions of 2910 genes bear this bivalent domain mark, including numerous genes involved in development and differentiation. Of these genes, 19% lost the H3K27me3 mark during erythroid differentiation; in the CD133+ cells, a substantial fraction of these genes were enriched with H3K4me1 and H3K9me1 marks as well as bound Pol II, suggesting that these genes are “poised” for activation during erythroid differentiation concomitant with the loss of the repressive H3K27me3 mark.
In addition to trimethylated H3K4me3 and H3K27me3, other histone modifications often mark transcriptionally “poised” genes in erythroid progenitors. Although H3K4me3 and the dimethylated H3K4me2 are concordant at most genes, multipotential hematopoietic cells contain a subset of genes that are marked by H3K4me2 but not H3K4me3. These genes are transcriptionally silent in progenitors and are highly enriched in lineage-specific hematopoietic genes. The H3K4me2 mark is rapidly lost on nonerythroid genes during erythroid differentiation, accompanied by repression of these genes. On the other hand, gain of the H3K4me3 mark correlated with transcriptional activation on erythroid differentiation, Thus the H3K4me2+-H3K4me3− on nonerythroid genes may regulate their repression during erythroid development.53
Several epigenetic regulatory mechanisms control gene induction and repression during erythroid development. Changes in gene expression are not accompanied by significant changes in histone modifications, such as H3K4me3 and H3K27me3, after GATA-1 is reintroduced into a GATA1-null erythroblast cell line to induce differentiation.54 Changes in H4K16ac and H3K79me2 levels, rather than H3K4me3 and H3K27me3, are most predictive of the direction in changes in gene expression during terminal fetal liver erythroid differentiation.55 Because H3K4me3 is usually associated with transcriptional initiation whereas H3K79me2 is tightly correlated with transcription elongation, control of Pol II elongation could be a mechanism for regulating erythroid gene expression. This may be mediated by GATA1 or TAL1 or their associated complexes, especially because nearby regions of genes induced during terminal erythroid development are co-occupied by these factors.54
The precise timing of chromatin switch(es) associated with erythroid differentiation is unclear. Several modifications at the β globin locus (DNA demethylation, formation of DNase I hypersensitive sites, and onset of activation-associated histone modifications) occur during the S phase of an early erythroid cell cycle after stimulation of CFU-E proliferation.56 This raises the possibility that one window of time during DNA replication allows structural changes in chromatin associated with newly synthesized DNA.
Interplay of cell cycle and terminal erythroid differentiation
In many developmental systems, terminal differentiation is tightly coupled with irreversible exit from the cell cycle. Erythroid cells usually undergo 3 to 5 cell divisions during terminal differentiation, preceded by an irreversible exit from the cell cycle. Work on Rb,57 E2F4,58 and cyclin D59 has established that cell division is highly coupled to erythroid differentiation, as defects in those genes result in fetal anemia. Moreover, a crucial step in activating erythroid gene expression occurs during the S phase of an early erythroid cell cycle after CFU-E activation.56 This transition involves repression of a cyclin-dependent kinase (cdk) inhibitor, p57(kip2), which in turn causes the down-regulation of Pu.1, an antagonist of GATA-1 function.56 Much remains to be learned about the interplay of cell-cycle regulation and erythroid gene expression, especially because several agents that slow the cell cycle have been shown to induce γ-globin gene expression in adult erythroid cells.60
Histone deacetylation and erythroblast enucleation
Although in all vertebrates the erythroid precursor nucleus becomes highly condensed and the chromatin transcriptionally inactive as erythropoiesis progresses, the process of enucleation (the budding off and ultimate extrusion of this condensed, inactive erythroid nucleus) is unique to mammals. Our understanding of this elaborate process of mammalian enucleation has advanced significantly since the earliest morphologic documentation of the phenomenon decades ago.61 This intricate developmental process involves multiple molecular and cellular pathways, including histone deacetylation, actin polymerization, vesicle trafficking and cytokinesis, cell-matrix interactions, and even specific miRNAs.62 Importantly, enucleation is not restricted to definitive erythroid cells; primitive yolk- sac derived erythropoiesis also exhibits chromatin condensation and enucleation.63 While nuclear condensation is thought to be critical for mammalian enucleation, chromatin condensation also occurs in many other tissue types, including rather ubiquitous cellular processes, such as cell division and apoptosis. Apoptotic mechanisms, such as caspase activation, are known to be responsible for enucleation of lens epithelia and keratinocytes, although the process is not similar to enucleation of erythroid precursors.62
Certain histone modifications affect not only binding of regulatory proteins but also the stability of chromatin itself. Global levels of several acetylation marks on histones, including H3K9Ac, H4K5Ac, H4K8Ac, and H4K12Ac, are significantly reduced during the terminal stages of erythroid differentiation, concomitant with a decrease in the levels of the histone acetyltransferase GCN5.64 Administration of histone deacetylase (HDAC) inhibitors to human erythroid precursors cultured from CD34+ cells inhibited terminal differentiation.65 Similarly, treatment of Friend virus-infected murine spleen erythroblasts with HDAC inhibitors blocked enucleation.66 The HDAC most important for erythropoiesis is probably HDAC2 because either specific pharmacologic inhibition or shRNA knockdown of HDAC2 blocked chromatin condensation and enucleation of mouse fetal liver erythroblasts in vitro.67 Deacetylation of histones generates a positive charge on the corresponding lysine reside; one plausible hypothesis is that these new positive charges interact with the negative charges on the phosphodiester bonds of DNA to facilitate DNA and chromatin condensation.
Regulation of erythropoiesis by miRNAs
miRNAs are a class of recently identified small regulatory RNAs that down-regulate expression of their target genes by either mRNA degradation or translational inhibition or both.68,69 Specific miRNAs are important regulators of several aspects of erythropoiesis, including erythroid lineage determination, erythroid progenitor proliferation, terminal erythroid differentiation, and enucleation (Table 1). For example, miR-150 is enriched in human umbilical cord blood megakaryocyte-erythrocyte and megakaryocyte progenitors, relative to erythroid progenitors. Overexpression and knockdown assays in MEP cells showed that miR-150 induces differentiation toward the megakaryocytic lineage and inhibits erythroid lineage differentiation through down-regulation of one principal target gene, MYB.70
microRNAs are important regulators for erythropoiesis
| microRNA . | Organism and experimental system . | Function . | Target . | Reference . |
|---|---|---|---|---|
| miR-150 | Human cord blood megakaryocyte-erythrocyte and megakaryocyte progenitor | miR-150 induces differentiation toward the megakaryocytic lineage at the expense of erythroid lineage | MYB | 70 |
| miR-221 and miR-222 | Human cord blood CD34+ cells | Down-regulation of miR-221 and miR-222 is required for terminal erythroid progenitor proliferation | KIT | 71 |
| miR-24 | Human cord blood CD34+ cells and K562 cell line | Down-regulation of miR-24 is required for terminal erythroid differentiation | ALK4 | 72 |
| miR-223 | Human cord blood CD34+ cells | Down-regulation of miR-223 is required for terminal differentiation | LMO2 | 73 |
| miR-144 | Zebrafish embryo | miR-144 is required for zebrafish embryonic α-globin gene expression | Klfd | 76 |
| miR-451 | Zebrafish embryo | miR-451 is required for zebrafish erythropoiesis | Gata2 | 75 |
| Knockout mice | miR-451 is required for steady-state erythropoiesis and resistance to oxidation during stress erythropoiesis | 14–3-3zeta | 78 | |
| miR-15a and miR-16–1 | Human cord blood CD34+ cells | miR-15a and miR-16–1 were identified as potential candidates causing elevated fetal hemoglobin expression; increased expression in elevated fetal hemoglobin gene expression | MYB | 80 |
| miR-191 | Mouse primary erythroid progenitors | The down-regulation of miR-191 is required for erythroblast chromatin condensation and enucleation | Riok3 and Mxi1 | 81 |
| microRNA . | Organism and experimental system . | Function . | Target . | Reference . |
|---|---|---|---|---|
| miR-150 | Human cord blood megakaryocyte-erythrocyte and megakaryocyte progenitor | miR-150 induces differentiation toward the megakaryocytic lineage at the expense of erythroid lineage | MYB | 70 |
| miR-221 and miR-222 | Human cord blood CD34+ cells | Down-regulation of miR-221 and miR-222 is required for terminal erythroid progenitor proliferation | KIT | 71 |
| miR-24 | Human cord blood CD34+ cells and K562 cell line | Down-regulation of miR-24 is required for terminal erythroid differentiation | ALK4 | 72 |
| miR-223 | Human cord blood CD34+ cells | Down-regulation of miR-223 is required for terminal differentiation | LMO2 | 73 |
| miR-144 | Zebrafish embryo | miR-144 is required for zebrafish embryonic α-globin gene expression | Klfd | 76 |
| miR-451 | Zebrafish embryo | miR-451 is required for zebrafish erythropoiesis | Gata2 | 75 |
| Knockout mice | miR-451 is required for steady-state erythropoiesis and resistance to oxidation during stress erythropoiesis | 14–3-3zeta | 78 | |
| miR-15a and miR-16–1 | Human cord blood CD34+ cells | miR-15a and miR-16–1 were identified as potential candidates causing elevated fetal hemoglobin expression; increased expression in elevated fetal hemoglobin gene expression | MYB | 80 |
| miR-191 | Mouse primary erythroid progenitors | The down-regulation of miR-191 is required for erythroblast chromatin condensation and enucleation | Riok3 and Mxi1 | 81 |
Several other miRNAs regulate terminal proliferation and differentiation of erythroid progenitor cells. miR-221 and miR-222 are down-regulated during erythroid differentiation of cultured cord blood CD34+ progenitors. Overexpression of miR-221 and miR-222 in CD34+ progenitor cells impaired proliferation and accelerated differentiation of erythroid precursors. This effect is mediated by down-regulation of one principal target gene, KIT, which encodes the receptor for SCF and is required for proliferation of erythroid progenitors.71 miR-24 functions as a negative regulator of activin signaling by down-regulating the mRNA encoding the activin type I receptor ALK4. In cultured CD34+ cells, miR-24 is down-regulated during erythroid differentiation, which is inversely correlated with the up-regulation of its target gene ALK4. In both CD34+ and K562 erythroleukemia cells, gain- and loss-of-function studies demonstrated that the down-regulation of miR-24 is required for normal erythroid differentiation.72 miR-223 is also down-regulated during erythroid differentiation of cord blood CD34+ progenitor cells. Overexpression of miR-223 impaired erythroid differentiation by down-regulating LMO2, a critical transcription factor that is up-regulated during erythroid differentiation of CD34+ progenitor cells and is required for erythroid differentiation.73 Thus, down-regulation of several miRNAs is essential during terminal erythroid differentiation.
In contrast, 2 cotranscribed miRNAs, miR-144 and miR-451, are highly induced during terminal erythroid differentiation and regulate expression of key erythroid-important genes. miR-144 and miR-451 are highly induced in the erythroid cell line G1E-ER4 when GATA-1 expression is restored by the treatment with estradiol, and ChIP experiments demonstrated that miR-144/451 is a direct transcriptional target of GATA-1.74 Knockdown of miR-451 in zebrafish embryos by injection of antisense morpholinos impaired erythropoiesis,74,75 establishing its importance in erythropoiesis. In zebrafish, gata2 is one important direct target gene of miR-451.75 Up-regulation of miR-144 is required for zebrafish embryonic α-globin gene expression through the down-modulation of its direct target gene Klfd.76
Knockout mice deficient for the miR-144/451 cluster display a cell-autonomous impairment of late erythroblast maturation, resulting in erythroid hyperplasia, splenomegaly, and a mild anemia.77-79 Importantly, these mice exhibit ineffective erythropoiesis in response to oxidative stress. One important direct target gene down-modulated by miR-451 is 14-3-3ζ, a phospho-serine/threonine-binding protein that inhibits nuclear accumulation of the transcription factor FoxO3, a positive regulator of erythroid anti–oxidant genes. In miR-144/451−/− erythroblasts, the excess 14-3-3ζ causes a partial relocalization of FoxO3 from nucleus to cytoplasm with dampening of its transcriptional program, including reduced expression of genes that encode the important anti–oxidant proteins catalase and glutathione peroxidase 1. Importantly, shRNA suppression of 14-3-3ζ protects miR-144/451−/− erythrocytes against peroxide-induced destruction and restores catalase activity.77-79 These studies thus define an important role for miR-144/451 in regulating a subset of erythroid-important genes.
In human trisomy 13, there is delayed switching and persistence of fetal hemoglobin. By analyzing partial trisomy cases, miR-15a and miR-16-1 were identified as potential candidates causing elevated fetal hemoglobin expression. Indeed, increased expression of these miRNAs in primary human erythroid progenitor cells resulted in elevated fetal hemoglobin gene expression. One important direct target of these miRNAs, Myb, plays an important role as a negative regulator of γ-globin gene expression.80
Regulated expression of several other miRNAs is important for erythroblast chromatin condensation and enucleation. The majority of miRNAs present in CFU-E progenitors are down-regulated during terminal erythroid differentiation. Of the predominant developmentally down-regulated miRNAs, ectopic overexpression of miR-191 in mouse fetal liver erythroid progenitors blocked erythroid enucleation but had minor effects on proliferation or erythroid differentiation. Two developmentally up-regulated genes, Riok3 and Mxi1, which are required for chromatin condensation and enucleation, are direct miR-191 targets. Both overexpression of miR-191 and knockdown of Riok3 or Mxi1 impaired the normal down-regulation of histone acetyltransferase Gcn5 (whose down-regulation is required for histone deacetylation and chromatin condensation60 ). Thus, normal down-regulation of miR-191 is essential for erythroid chromatin condensation and enucleation by allowing up-regulation of Riok3 and Mxi1 and down-regulation of Gcn5.81
In addition to the miRNAs listed in the previous 3 paragraphs, many other miRNAs are also abundant and developmentally regulated during erythroid differentiation.81 However, ectopic expression of the majority of miRNAs in cultured CFU-E stage progenitors, effectively preventing their normal down-modulation during terminal differentiation, had subtle or no effects on erythropoiesis.81 This may indicate that these miRNAs are relatively weak regulators of gene expression or indicate that these miRNAs affect differentiation under conditions different from those in culture. This is well illustrated by a recent report showing that miR-144/451−/− mice are only mildly anemic. However, when exposed to phenylhydrazine-induced hemolysis, more than half of miR-144/451−/− mice died whereas all wild-type mice fully recovered.78
In addition to miRNAs, another class of noncoding RNA, lncRNAs (long noncoding RNA) has recently been shown to be crucial for important biologic processes, such as the p53 response82 and stem cell reprogramming.83 It will be interesting to explore the potential function that lncRNAs may play in RBC production.
Stress erythropoiesis and enhanced self-renewal of early committed RBC BFU-E progenitors
RBC levels are normally tightly regulated by Epo, which stimulates erythropoiesis by promoting survival, proliferation, and terminal differentiation of CFU-E cells and more mature erythroblasts. Because normal Epo levels are very low, RBC output from CFU-E cells can be increased more than one order of magnitude by increased Epo production or by injection of recombinant Epo. However, because each Epo-responsive CFU-E cell in the bone marrow can undergo only 3 to 5 terminal cell divisions under maximum Epo stimulation, the number of CFU-E cells limits the maximum Epo-dependent erythrocyte output. Steady-state erythropoiesis alone is therefore not able to correct the RBC deficiency during extreme conditions, such as recovery from bone marrow irradiation or chronic anemia. During such conditions of stress erythropoiesis, new CFU-E are produced from the most immature committed definitive erythroid progenitor cells, the BFU-E cells. Formation of BFU-E and CFU-E progenitors does not require Epo receptor activation.6 One BFU-E progenitor cell can form hundreds of thousands of erythroblasts in vitro; its name is derived from the large burst of red colonies formed in methylcellulose after 7 days of culture. The ability of BFU-E progenitors to undergo limited self-renewal during stress erythropoiesis allows rescue of lethally irradiated mice from anemia, and retransplantation also protects secondary and tertiary recipients.84 In this regard, stress erythropoiesis is similar to definitive fetal liver erythropoiesis and Friend virus-induced erythroleukemia, 2 other situations where similar mechanisms induce erythroid progenitors to undergo self-renewal rather than to differentiate.
Stress erythropoiesis is regulated by SCF, GCs, BMP4, and Hedgehog signaling in addition to tissue hypoxia
Microenvironment
Early observations showed that erythropoiesis moves from the bone marrow to the spleen during stress erythropoiesis (Figure 3).85,86 The importance of factors in the microenvironment for sustaining BFU-E self-renewal was further suggested by the fact that splenectomized mice are resistant to Friend erythroid leukemia virus-induced erythroid leukemia.87 Furthermore, fetal liver and spleen-derived stromal cell lines are superior to cells derived from bone marrow for supporting fetal BFU-E proliferation.88-90 To identify factors that promote BFU-E self-renewal, researchers have therefore studied the spleen and fetal liver milieu, in addition to determining the effect of physiologic responses to stress, such as increased levels of stress hormones.
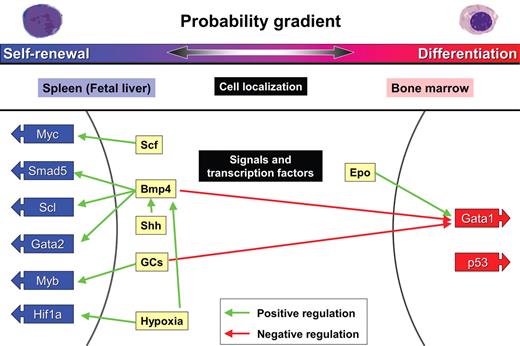
BFU-E self-renewal: The probability of erythroid progenitor self-renewal versus differentiation depends on extrinsic and intrinsic factors. BFU-E progenitors either self-renew or differentiate depending on the body's need for generation of CFU-E and Epo-dependent erythroblasts. Although BFU-E self-renewal is very limited during steady-state erythropoiesis in the bone marrow, it is virtually limitless in the spleen during conditions of stress erythropoiesis. The cytokines Scf, Bmp4, and Shh promote self-renewal in addition, as does stimulation by GCs and hypoxia. These signals activate transcription factors Myc, Smad5, Scl, Gata2, Myb, and Hif1a.
BFU-E self-renewal: The probability of erythroid progenitor self-renewal versus differentiation depends on extrinsic and intrinsic factors. BFU-E progenitors either self-renew or differentiate depending on the body's need for generation of CFU-E and Epo-dependent erythroblasts. Although BFU-E self-renewal is very limited during steady-state erythropoiesis in the bone marrow, it is virtually limitless in the spleen during conditions of stress erythropoiesis. The cytokines Scf, Bmp4, and Shh promote self-renewal in addition, as does stimulation by GCs and hypoxia. These signals activate transcription factors Myc, Smad5, Scl, Gata2, Myb, and Hif1a.
GCs
Release of cortisol from the adrenal glands is increased during conditions of stress erythropoiesis, such as sepsis or severe trauma. The therapeutic effect of the GC analog prednisone in patients with the RBC progenitor disorder Diamond-Blackfan anemia is well documented, although severe side effects limit its use.91,92
Mice that lack the GC receptor or express only a GC receptor defective in DNA binding and transactivation have normal steady-state erythropoiesis, whereas stress erythropoiesis is severely impaired.93,94 In particular, mice with defective or missing GC receptors fail to respond to phenylhydrazine-induced hemolysis by increasing CFU-E numbers in the spleen. For unknown reasons, the GC effect on stress erythropoiesis is antagonized by p53 activation, and p53−/− mice respond to hypoxic stress faster than normal, as shown by the rapid increase of CFU-E and c-Kit+/CD34+ cells in the spleen.95 GCs support stress erythropoiesis by inducing expression of Myb, Kit, and Lmo2 and inhibiting Gata1 expression.95,96 Of those changes in gene expression, only up-regulation of Kit mRNA is detected 4 hours after GC stimulation of BFU-E cells.97
The role of GCs in stress erythropoiesis can be studied in vitro by culturing early erythroid progenitors in medium containing SCF, GCs, and Epo.93,97-99 As during in vivo stress erythropoiesis, in vitro proliferation of fetal liver erythroblasts is severely decreased by a mutation in the GC receptor that disrupts dimerization.93,94 The stimulatory effects of GCs on RBC production therefore probably require GC receptor dimerization, which is required for efficient transactivation of promoters with GC receptor element full sites but may also be necessary for gene repression.93,94 These results suggest that the mechanisms regulating stress erythropoiesis are evolutionarily conserved, and possibly part of an ancient cortisol-mediated stress response to trauma, which also leads to increased blood pressure and glucose levels.100
Using BFU-E and CFU-E cells purified from mouse fetal liver by a new flow cytometric technique (both BFU-E and CFU-E are kit-positive and -negative for Ter119, B220, Mac-1, CD3, Gr-1, CD32/16, Sca-1, CD41, and CD34, whereas BFU-E express low levels of CD71 and CD24a compared with CFU-E) and cultured in a serum-free medium containing only SCF and IGF-1, it was shown that that GCs induce limited self-renewal of BFU-E cells, and not of CFU-E cells or erythroblasts. GCs thereby protect BFU-E cells from exhaustion, and in parallel, over time increase the number of CFU-E cells formed from each BFU-E greater than 10-fold.97 We proposed a physiologic model of stress erythropoiesis where increased levels of GCs help maintain the earliest erythroid progenitors, increase CFU-E output, and at the same time stimulate terminal differentiation, thus promoting both a rapid and long-lasting increase in RBC production. Identification of BFU-E as the target cell of GCs in stress erythropoiesis, together with our novel method to isolate BFU-E, will allow studies toward development of novel erythropoiesis-stimulating agents that act by promoting SE by the same mechanisms used by GCs.
SCF
SCF (Kit ligand) exists both in a soluble and a membrane-bound form. Kit signaling is important not only for erythroid progenitor proliferation but also for HSC growth, mast cell function, melanogenesis, and spermatogenesis. Fetal liver hepatic progenitors that express very high levels of SCF and support expansion of HSCs probably also support BFU-E self-renewal during fetal liver erythropoiesis.101 The stress response of enhanced CFU-E formation in murine spleen on phenylhydrazine treatment is drastically reduced by infusion of anti–Kit antibodies, reiterating the importance of SCF in stress erythropoiesis.102 SCF binding to Kit induces activation of PI3K, and inhibition of PI3K results in decreased numbers of BFU-E and CFU-E cells in vivo and reduced erythroblast proliferation in vitro.103,104 SCF counteracts BFU-E differentiation in part by inducing expression of Myc, which prevents terminal erythroid maturation.64,105
BMP4
Bone morphogenetic protein 4 (BMP4) is also essential for stress erythropoiesis. The flexed-tail mouse strain, which expresses a dominant-negative Smad5 mutant that inhibits BMP4 signaling, exhibits a neonatal anemia that resolves 2 weeks after birth.106-108 This demonstrates that, although BMP4 signaling through Smad5 is not required to maintain adult or fetal liver hematopoiesis, it is essential for stress erythropoiesis.109,110 Indeed, during stress erythropoiesis, BMP4 expression is induced by hypoxia in spleen stromal cells.111,112 For BFU-E cells to be responsive to BMP4, they need to be “primed” with Sonic Hedgehog, a morphogen that is also synthesized and secreted by cells in the spleen microenvironment.113 BMP4, in turn, activates the transcription factor Smad5 as well as Scl and Gata2, which enhances the probability of BFU-E self-renewal over differentiation (Figure 3).84,114,115
Hypoxia
Interestingly, GCs induce expression of genes in BFU-E cells that contain promoter regions highly enriched for hypoxia-induced factor-1α (HIF-1α) binding sites, suggesting that activation of HIF-1α may enhance or replace the effect of GCs on BFU-E self-renewal. Indeed, HIF-1 α activation by a prolyl hydroxylase inhibitor synergized with GCs and enhanced production of CFU-E and then erythroblasts approximately 170-fold.97 Although earlier observations showed that hypoxia supports BFU-E self-renewal through effects on spleen stroma,116 these findings demonstrate an additional effect intrinsic to BFU-E cells.
Prospectus
These studies have provided profound insights into the multiple complex mechanisms by which the body regulates the number of RBCs within a narrow normal range. Equally importantly, they provide novel insights into possible treatments for anemias and other RBC disorders. For example, recent advances in the understanding the regulation of β-hemoglobin switching could lead to better therapies for disorders, such as β-thalassemia and sickle cell disease.117 Increased understanding of how genetic and epigenetic programs regulate different stages of RBC development may enable large-scale production of functional RBCs from hematopoietic or other tissues for clinical blood transfusions. Furthermore, deeper understanding of mechanisms regulating BFU-E self-renewal and thus the output of CFU-E progenitors and mature erythroid cells could result in the development of drugs that stimulate the physiologic mechanisms of stress erythropoiesis. These drugs could be useful in treating relatively erythropoietin-resistant anemias, including patients with bone marrow failure disorders, such as Diamond-Blackfan anemia, trauma, sepsis, and possibly anemia of malaria as well as the approximately 18% of kidney dialysis patients who fail to respond to Epo. One such example is pharmacologically induced HIF-1α activation, which leads to increased erythroblast production at physiologic concentrations of GCs.97
Acknowledgments
The authors thank Vijay Sankaran and Leif Si-Hun Ludwig for useful discussions and Tom DiCesare for graphics rendering.
This work was supported in part by the National Institutes of Health (grants P01 HL 32262, DK068348, and DK067356), the Singapore–Massachusetts Institute of Technology Alliance (grant C-382-641-001-091), and Amgen Inc (research grant). J.F. was supported by a fellowship from the Swedish Research Council and stipends from the Diamond-Blackfan Anemia Foundation, Maja och Hjalmar Leanders Stiftelse, and the Sweden-America Foundation. P.W. was supported by the Croucher Foundation (postdoctoral research grant). S.M.H. was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (K08 DK076848). L.Z. was supported by the Singapore–Massachusetts Institute of Technology Alliance (graduate fellowship).
National Institutes of Health
Authorship
Contribution: S.M.H., P.W., L.Z., J.F., and H.F.L. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Harvey F. Lodish, Whitehead Institute for Biomedical Research, 9 Cambridge Center, Cambridge, MA 02142; e-mail: lodish@wi.mit.edu.
References
Author notes
S.M.H. and P.W. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal