The pathogenesis of both classic and nodular lymphocyte predominant Hodgkin lymphoma (NLPHL) is still enigmatic. In this issue of Blood, Saarinen and colleagues report the identification of germ line mutations in the NPAT gene as a candidate risk factor for Hodgkin lymphoma.1
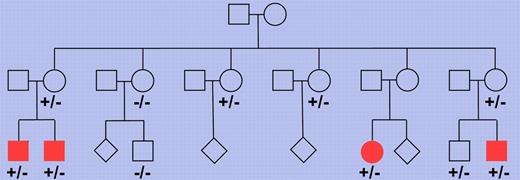
Pedigree of a Finnish family with multiple members affected by nodular lymphocyte predominant Hodgkin lymphoma. Multiple family members were tested for the presence of the truncating NPAT mutation and each of 4 cousins affected by the lymphoma showed this mutation (depicted as filled symbols). −/− indicates wild-type for the 2 base pair deletion in the NPAT gene and +/− indicates presence of the deletion on 1 allele. Where no genotype is indicated, no DNA was available for analysis. The pedigree has been modified for confidentiality.
Pedigree of a Finnish family with multiple members affected by nodular lymphocyte predominant Hodgkin lymphoma. Multiple family members were tested for the presence of the truncating NPAT mutation and each of 4 cousins affected by the lymphoma showed this mutation (depicted as filled symbols). −/− indicates wild-type for the 2 base pair deletion in the NPAT gene and +/− indicates presence of the deletion on 1 allele. Where no genotype is indicated, no DNA was available for analysis. The pedigree has been modified for confidentiality.
Hodgkin lymphoma is one of the most frequent lymphomas in the Western world. Based on differences in tumor cell morphology and phenotype and the histologic picture, this lymphoma entity is subdivided into a classic form and NLPHL, the former accounting for about 95% of cases. We know relatively little about the genetic lesions causing Hodgkin lymphoma, partly because of the rarity of the tumor cells in affected lymph nodes, which hampers the molecular analysis of these cells for somatic alterations. Saarinen and colleagues combined 2 modern genetic methods, single nucleotide polymorphism chips and whole exome sequencing, and applied them to a unique Finnish family with 4 cousins all presenting from 22-26 years of age with NLPHL.1 A germ line mutation in the NPAT (nuclear protein, ataxia-telangiectasia locus) gene was found in all 4 patients (see figure). The mutation was a 2 base pair deletion causing a frame shift and in consequnce a truncation of the protein. Screening of many more Hodgkin lymphoma patients and healthy controls did not reveal additional instances of this mutation but, importantly, a replacement mutation in codon 724 of the gene was found in other NLPHL and classic Hodgkin lymphoma patients at significantly higher frequency than in healthy controls. This identifies NPAT germ line mutations as the first candidate genetic risk factor for NLPHL.
Although it is intriguing that 2 different types of germ line mutations affecting the NPAT protein sequence were found in Hodgkin lymphoma patients, the functional consequences of these mutations remain presently unclear, and deserves future study. NPAT has been reported to be involved in the regulation of the cell cycle, and it is presumably also involved in the regulation of the ATM tumor suppressor gene, which is located directly adjacent to NPAT. Thus, a pathogenetic role of the mutations can be envisioned. However, it is not clear whether the mutated NPAT gene acts as an oncogene or a tumor suppressor gene. If one assumes that the mutations cause a loss of function, it would be important to clarify whether the second allele of the gene is somatically mutated in the lymphocyte predominant (LP) or Hodgkin and Reed-Sternberg cells with germ line mutations on 1 allele. Alternatively, the mutations may have a dominant negative effect (eg, if the protein functions as a dimer or multimer), so that the germ line mutation on 1 allele is sufficient to cause a loss of function. Finally, NPAT may show haploinsufficiency in HL, that is, the amount of remaining wild-type protein encoded from the second allele is not sufficient to sustain normal NPAT function. Cells from patients with NPAT mutations indeed showed reduced expression levels of the gene.1
NPAT germ line mutations were found in NLPHL as well as in classic HL cases, suggesting that such mutations may contribute to the pathogenesis of both subtypes of Hodgkin lymphoma. This is a remarkable finding, because although classic and NLPHL are considered 2 forms of 1 disease, there is presently little indication for common genetic lesions or other shared transforming events in the tumor cells of these lymphomas.2 For example, even though strong constitutive activity of the NF-κB transcription factor signaling pathway is a hallmark of the lymphoma cells in both classic and NLPHL,2,3 the mechanisms for this activation appear to be very distinct: somatic inactivating mutations in the negative NF-κB regulators TNFAIP3 and NFKBIA are frequent in the Hodgkin and Reed-Sternberg tumor cells in classic Hodgkin lymphoma, but they do not play a significant role in the LP tumor cells of NLPHL.2,4 Moreover, infection of the tumor cells by Epstein-Barr virus contributes to NF-κB activation in classic Hodgkin lymphoma, but is not seen in NLPHL.2 Somatic mutations in the SOCS1 gene, encoding for a negative regulator of the JAK/STAT signaling pathway, were, before the study by Saarinen et al, the only known genetic lesion detected in both types of Hodgkin lymphoma.5,6
Besides the genetic lesions in the SOCS1 gene, we know hardly anything about the transforming events in NLPHL. The only other recognized recurrent genetic lesion in these cells are translocations involving the proto-oncogene BCL6.7 BCL6 is a transcription factor that regulates the germinal center B-cell differentiation program. Translocations of BCL6 may thus contribute to NLPHL pathogenesis by freezing the LP tumor cells in the highly proliferative germinal center B-cell differentiation stage.
As Hodgkin lymphoma shows an increased familiar association,8 there is the suspicion that germ line mutations or polymorphisms may represent predisposing factors. With modern genome-wide association studies (GWAS) and next-generation sequencing methods, we now have better tools to identify such predisposing factors. For classic Hodgkin lymphoma, a recent GWAS study identified risk loci at the REL, PVT1, and GATA3 loci,9 and confirmed an association of Hodgkin lymphoma risk with the HLA region.10 Through the work by Saarinen et al, a first candidate predisposing factor for NLPHL has been identified, and a further factor for classic Hodgkin lymphoma. This finding has to be validated in an independent study, but it demonstrates the power of the genome-wide methods and advances our understanding of Hodgkin lymphoma pathogenesis.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal