Abstract
Numerous molecular markers have been recently discovered as potential prognostic factors in acute myeloid leukemia (AML). It has become of critical importance to thoroughly evaluate their interrelationships and relative prognostic importance. Gene expression profiling was conducted in a well-characterized cohort of 439 AML patients (age < 60 years) to determine expression levels of EVI1, WT1, BCL2, ABCB1, BAALC, FLT3, CD34, INDO, ERG and MN1. A variety of AML-specific mutations were evaluated, that is, FLT3, NPM1, N-RAS, K-RAS, IDH1, IDH2, and CEBPADM/SM (double/single). Univariable survival analysis shows that (1) patients with FLT3ITD mutations have inferior overall survival (OS) and event-free survival (EFS), whereas CEBPADM and NPM1 mutations indicate favorable OS and EFS in intermediate-risk AML, and (2) high transcript levels of BAALC, CD34, MN1, EVl1, and ERG predict inferior OS and EFS. In multivariable survival analysis, CD34, ERG, and CEBPADM remain significant. Using survival tree and regression methodologies, we show that CEBPADM, CD34, and IDH2 mutations are capable of separating the intermediate group into 2 AML subgroups with highly distinctive survival characteristics (OS at 60 months: 51.9% vs 14.9%). The integrated statistical approach demonstrates that from the multitude of biomarkers a greatly condensed subset can be selected for improved stratification of intermediate-risk AML.
Introduction
It is widely accepted that certain cytogenetic abnormalities consistently associate with particular subsets of acute myeloid leukemia (AML) that carry distinct responses to therapy.1,2 Approximately 40% of all AML patients are currently classified into distinct groups with variable prognosis based on the presence or absence of specific recurrent cytogenetic abnormalities. AML without favorable and particular unfavorable cytogenetic aberrations is classified as intermediate prognosis. The intermediate-risk cytogenetic subclass of AML includes cytogenetically normal (CN) and AML with other cytogenetic abnormalities and accounts for approximately 60% of all AML patients; and accordingly, recent gene-mutation and gene-expression studies represent a mixture of leukemias with favorable and unfavorable prognoses.
In recent years, a variety of novel molecular markers have refined the risk stratification of intermediate-risk AML. For instance, mutations in FLT3,3-5 NPM1,6-9 and CEBPA10-16 all carry variable prognostic value. Recently, IDH1 and IDH2 mutations were identified; but for the time being, the prognostic value of these mutations appears to be controversial.17-20
Besides acquired mutations, a number of individual genes have been proposed as important prognostic expression markers (ie, specific gene expression levels were shown to be associated with treatment outcome in AML). For instance, expression of EVI1,21-23 BAALC,24,25 ERG,26,27 and MN128,29 was proposed as indicators for treatment outcome in AML. Some expression markers, such as WT1,30-33 BCL2,34 INDO,35 CD34,36 ABCB1,37 and FLT3,38 have been put forward as clinical markers, but their applicability has been less well established or has been controversial.
Previous studies have often assessed the prognostic value of various biomarkers on an individual basis or in a limited collective context. For the purpose of risk stratification and understanding of the relative prognostic importance, it has become crucial to integrate them in a joint analysis. In the present study, we investigate the role of gene expression markers EVI1, WT1, BAALC, ERG, BCL2, ABCB1, INDO, CD34, BCL2, and MN1 (evaluated using standardized microarray analysis) as well as somatic gene mutations in FLT3, N-RAS, CEBPASM,CEBPADM, NPM1, IDH1, and IDH2 in survival prognosis in cytogenetically defined intermediate-risk AML. In addition to univariate and multivariate analyses, we adopted a statistical approach that is capable of deriving a simplified prognostic index that can be used for the risk stratification of the intermediate-risk group.
Methods
Patients, cell samples, and molecular analyses
We investigated a cohort of 439 patients (age < 60 years) with a diagnosis of primary AML or refractory anemia with excess blasts(-t) (n = 17; Table 1). All patients were treated according to the HOVON (Dutch-Belgian Hematology-Oncology Cooperative group) protocols between 1987 and 2006 (www.hovon.nl).39-41 This study was approved by the Medical Ethical Committee of the Erasmus University Medical Center.
Clinical and molecular characteristics of the 439 patients
| Variables . | Range . | Mean/median or % . | No. of patients . |
|---|---|---|---|
| Clinical variables | |||
| White blood cell count, × 109/L | 0.3-278 | 52.0/29.8 (mean/median) | |
| Bone marrow blast count, % | 0-98 | 62.1/66 (mean/median) | |
| Platelet count, × 109/L | 3-998 | 78.9/52 | |
| Patient characteristics | |||
| Age, y | 15-60 | 42.11/43 (mean/median) | |
| Sex (female) | 219 | ||
| FAB classification | |||
| M0 | 3.6 | 16 | |
| M1 | 19.1 | 84 | |
| M2 | 23.2 | 102 | |
| M3 | 5 | 22 | |
| M4 | 18.5 | 81 | |
| M4Eo | 6.2 | 27 | |
| M5 | 23.7 | 104 | |
| M6 | 1.1 | 5 | |
| RAEB | 0.9 | 4 | |
| Not determined | 4.8 | 21 | |
| Cytogenetics | |||
| t(8;21) | 8 | 35 | |
| inv (16) | 8.2 | 36 | |
| t(15;17) | 5.7 | 25 | |
| CN | 43.5 | 192 | |
| CA | 28.7 | 126 | |
| MK* | 5.7 | 25 | |
| Mutations | |||
| NPM1+ | 29.6 | 130 | |
| FLT3ITD+ | 26.9 | 118 | |
| FLT3TKD+ | 10.7 | 47 | |
| N-RAS+ | 987 | 43 | |
| K-RAS+ | 0.9 | 4 | |
| CEBPASM+ | 1.6 | 7 | |
| CEBPADM+ | 5.2 | 23 | |
| IDH1+ | 7.2 | 32 | |
| IDH2+ | 8.2 | 36 | |
| NPM1+ FLT3ITD+ | 15.3 | 67 | |
| NPM1+ FLT3ITD− | 14.4 | 63 | |
| NPM1− FLT3ITD+ | 11.6 | 51 | |
| NPM1− FLT3ITD− | 58.8 | 258 |
| Variables . | Range . | Mean/median or % . | No. of patients . |
|---|---|---|---|
| Clinical variables | |||
| White blood cell count, × 109/L | 0.3-278 | 52.0/29.8 (mean/median) | |
| Bone marrow blast count, % | 0-98 | 62.1/66 (mean/median) | |
| Platelet count, × 109/L | 3-998 | 78.9/52 | |
| Patient characteristics | |||
| Age, y | 15-60 | 42.11/43 (mean/median) | |
| Sex (female) | 219 | ||
| FAB classification | |||
| M0 | 3.6 | 16 | |
| M1 | 19.1 | 84 | |
| M2 | 23.2 | 102 | |
| M3 | 5 | 22 | |
| M4 | 18.5 | 81 | |
| M4Eo | 6.2 | 27 | |
| M5 | 23.7 | 104 | |
| M6 | 1.1 | 5 | |
| RAEB | 0.9 | 4 | |
| Not determined | 4.8 | 21 | |
| Cytogenetics | |||
| t(8;21) | 8 | 35 | |
| inv (16) | 8.2 | 36 | |
| t(15;17) | 5.7 | 25 | |
| CN | 43.5 | 192 | |
| CA | 28.7 | 126 | |
| MK* | 5.7 | 25 | |
| Mutations | |||
| NPM1+ | 29.6 | 130 | |
| FLT3ITD+ | 26.9 | 118 | |
| FLT3TKD+ | 10.7 | 47 | |
| N-RAS+ | 987 | 43 | |
| K-RAS+ | 0.9 | 4 | |
| CEBPASM+ | 1.6 | 7 | |
| CEBPADM+ | 5.2 | 23 | |
| IDH1+ | 7.2 | 32 | |
| IDH2+ | 8.2 | 36 | |
| NPM1+ FLT3ITD+ | 15.3 | 67 | |
| NPM1+ FLT3ITD− | 14.4 | 63 | |
| NPM1− FLT3ITD+ | 11.6 | 51 | |
| NPM1− FLT3ITD− | 58.8 | 258 |
Mutation present (or absent) groups denoted with (+) or (−).
RAEB indicates refractory anemia with excess blasts; FAB, French-American-British; CN, normal cytogenetics or -X or -Y as the sole abnormality; CA, cytogenetically abnormal; MK, monosomal karyotype; and M4Eo, M4 category with inv(16).
MK category contains 12 AML patients classified as complex karyotype and 13 other cases with complex karyotypes are in the CA category.
All AML cases in this study were also included in other studies,16,17,22,23,42 and subsets of cases have also been investigated in other studies35,43-45 (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The earlier studies had different study objectives (ie, dealing with individual markers or selected subsets of leukemia; for instance, CN AML).
AML was cytogenetically classified into the following prognostic categories: (I) favorable: t(8;21) and inv(16); (II) very unfavorable: monosomal karyotypes as defined earlier46 ; (III) intermediate-risk I: CN; and (IV) intermediate-risk II: the remaining AML cases (cytogenetically abnormal).
After informed consent was given in accordance with the Declaration of Helsinki, bone marrow aspirates or peripheral blood samples were taken at diagnosis. Blasts and mononuclear cells were purified by Ficoll-Hypaque (Nygaard) centrifugation and cryopreserved. The AML samples contained 80%-100% blast cells after thawing, regardless of the blast count at diagnosis. Mutational analyses were all performed as described previously.9,12,17,47,48
Gene profiling and quality control for assessment of gene expression variations
A total of 439 AML samples were analyzed using Affymetrix U133Plus2.0 GeneChips (Affymetrix) that contains 54 675 probe sets, representing 20 650 unique genes. The methods have been reported in detail elsewhere.45,49 The differences between the scaling/normalization factors of the GeneChips in complete cohort was < 3-fold (0.62 ± 0.20). All additional measures of quality (percentage genes present, [39.8 ± 3.5]; GAPDH 3′ to 5′ ratio, [1.08[ ± 0.15]; and actin 3′to 5′ ratio, [1.30 ± 0.26]), indicated high overall sample and assay quality in the complete AML cohort.
Informative probe sets detecting expression of various genes were selected. Only those probe sets with accurate annotation and genomic localization using the ENSEMBL genome browser (www.ensembl.org) were included: ABCB1: 209993_at and 209994_s_at; WT1: 206067_s_at and 216953_s_at; BCL2: 203684_s_at and 203685_at; BAALC: 218899_s_at and 222780_s_at; ERG: 213541_s_at and 241926_s_at; EVI1: 221884_at and 226420_at; FLT3: 206674_at; CD34: 209543_s_at; and MN1: 205330_at and INDO: 210029_at.
Data preparation
Each of the mutation markers is coded as a binary variable (ie, mutation present [+] or absent). The gene expression of each selected gene was determined from either a single probe or a combination of multiple probes linked to that gene. Probe sets for each expression marker were selected from the Affymetrix U133Plus2.0 GeneChip, based on an accurate annotation and localization using the ENSEMBL genome browser. If one probe per gene was available (MN1, CD34, FLT3, and INDO1), the probe expression intensity was log2-transformed and scaled so that the minimal value equals 0 and the maximal value equals 1. In case multiple probe sets were annotated for a single gene (BAALC, BCL2, ABCB1, EVI1, WT1, and ERG), we reduced the number of variables by performing a factor analysis per gene using the log2-transformed expression data of all 439 AML patient samples. This resulted in a factor score, composed of the expression values from all the representative probe sets, for each individual expression marker. The factor scores were also rescaled so that the minimal value of the score for each gene is 0 and the maximal value is 1.
Statistical analysis
Statistical analyses were performed with R (Version 2.9.2). Both overall survival (OS; with failure defined as death because of any cause) and event-free survival (EFS; with failure defined as no complete remission [set at day 1], relapse, or death in first complete remission) were considered as endpoints for survival analyses.
To determine the prognostic value of the markers, the Cox proportional hazard regression model was used in univariable and multivariable analyses. To further inspect the prognostic importance and/or redundancy of the markers, we applied a variable selection in the Cox proportional hazards model, namely, the Akaike information criterion-based stepwise variable selection and the Least Absolute Shrinkage and Selection Operator (LASSO),50 where the optimal penalty parameter was chosen so that it maximizes the cross-validated partial log-likelihood (20-fold cross-validation).51 To further evaluate the hierarchy of the prognostic importance, we used tree-structured survival modeling (unbiased recursive partitioning approach of Hothorn et al52 ). Estimated probabilities of OS and EFS were calculated using the Kaplan-Meier method. Partial likelihood ratio test was used to evaluate differences between survival distributions.
The bimodal shape of the EVI1 expression distribution (supplemental Figure 3) suggests that there are 2 populations of patients with high and low EVI1 expression. A mixture model fit with normally distributed components52 supports the evidence for this observation (supplemental Figure 3). The intersection point of the 2 superimposed densities naturally suggests a threshold (c = 1.15) to decide whether or not the EVI1 was overexpressed. EVI1 expression based on quantitative reverse-transcribed polymerase chain reaction was treated as a categorical variable in previous reports.21,22 Here we also use a categorical EVI1 in survival analyses using the reference value of 1.15. A penalized spline fit in Cox proportional hazard regression suggests a nonlinear behavior of EVI1 (P value of a test for linearity .04, best degrees of freedom 2.1 determined by Akaike information criterion). Despite a piecewise constant, transformation might not be the best approximation of the true relationship it manages to separate the distinctive survival characteristic of the small group of patients (8.8%) with high EVI1, which would be masked if we treated EVI1 as linear. The remaining markers are treated on a continuous scale in accordance with their actual distribution pattern. Unless otherwise stated, with “high expression” we refer to high values extreme with respect to the distribution of each marker.
Pair-wise associations between binary markers were assessed by means of χ2 test (or Fisher [Halton-Freeman] exact tests when the expected count number in at least one of the cells dropped to < 5). The direction of the observed associations was measured by a φ coefficient. Spearman correlation coefficient was used to assess the pair-wise correlations between gene expression markers. Differential gene expression across patient subcategories was tested by means of Wilcoxon sum-rank test (2 categories only) and Kruskal-Wallis test (> 2 categories). The level .05 has been used as a threshold to declare the statistical significance.
Results
Recurrent mutations and expression levels in cytogenetically defined AML subsets
Details on the molecular and clinical characteristics of the investigated cohort of 439 patients are summarized in Table 1.
The distribution of the recurrent mutations among the cytogenetically defined AML subsets is summarized in supplemental Table 2. We note increased frequencies of FLT3ITD and NPM1 mutations in CN AML as well as the common occurrence of FLT3ITD and FLT3TKD in AML with t(15;17). The prevalence of FLT3TKD and N-RAS mutations is higher in AML with inv(16). IDH1 and both CEBPASM and CEBPADM were observed exclusively in intermediate-risk cytogenetic categories (CN and cytogenetically abnormal). K-RAS mutations are relatively rare in AML and were not considered in further analyses.
The majority of expression markers genes show a differential expression in the cytogenetically defined AML subsets (supplemental Figure 1). Expression marker genes INDO1 and FLT3 do not have distinctive expression patterns in relationship to the cytogenetically defined subgroups (P values of the overall Kruskal-Wallis test: .301 and .204, respectively). Compared with the normal karyotype group, significantly higher expression of WT1 is associated with t(15;17) (P < .001), relatively low BCL2 expression is observed in AML with t(8;21) (P < .001), and high expression of both BAALC and CD34 was detected in t(8;21) and inv(16) groups (P < .001 for both comparisons). We further noticed elevated MN1 expression in AML with inv(16) compared with the CN group (P < .001).
Associations between recurrent mutation and expression markers
The summary of pair-wise associations between the binary mutation markers is given in supplemental Table 3. FLT3ITD, FLT3TKD, and IDH1 mutations appear significantly overrepresented in NPM1 mutant group (φ coefficients 0.36, 0.15, and 0.28, respectively). On the other hand, FLT3ITD are more prevalent in AML without FLT3TKD (φ = −0.11), N-RAS (φ = −0.2), or IDH2 mutations (φ = −0.11).
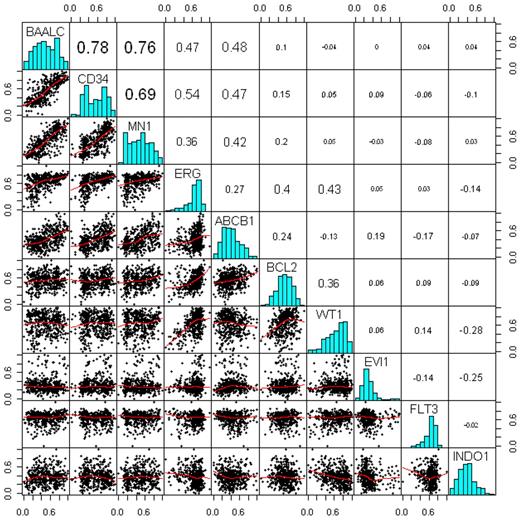
Spearman correlation analysis between the gene expression markers in Figure 1 revealed the following associations: (1) the expression of the marker genes BAALC, CD34, MN1, ERG, and ABCB1 is relatively strongly associated; (2) BAALC exhibits the strongest positive correlation with CD34 expression (correlation coefficient [R] = 0.78) and MN1 (R = 0.76); and (3) moderate associations are also observed between ERG and WT1 (R = 0.43), ERG and BCL2 (R = 0.4), and BCL2 and WT1 (R = 0.36); (4) INDO1 appears to be inversely associated with EVI1 (R = −0.25), WT1 (R = −0.28), and ERG (R = −0.14).
Associations between the gene expression markers. Bottom triangle represents a scatter-plot matrix of the markers, where the red lines are the lowess smoothing curves. Top triangle represents pair-wise Spearman correlation coefficients. On the diagonal, there are histograms of each of the markers.
Associations between the gene expression markers. Bottom triangle represents a scatter-plot matrix of the markers, where the red lines are the lowess smoothing curves. Top triangle represents pair-wise Spearman correlation coefficients. On the diagonal, there are histograms of each of the markers.
The summary of the association analysis between the mutation and gene expression markers is given in supplemental Table 4. The NPM1 mutant patient group is significantly associated with higher WT1 expression. In contrast, the expression of BCL2, BAALC, ERG, ABCB1, CD34, and MN1 is elevated in NPM1 wild-type AML. Other associations that we observe are, for example, decreased BAALC, CD34, and MN1 expression as well as increased FLT3 and WT1 expression in FLT3ITD AML. Increased ABCB1 expression associates with CEBPADM AML. Likewise, BAALC and CD34 expression is higher in IDH1 wild-type AML, and BCL2 expression is higher in IDH2 mutant AML.
Survival analyses in intermediate- risk AML
Univariable survival analysis (Table 2) indicated inferior OS in intermediate-risk AML patients with FLT3ITD mutations (hazard ratio [HR] = 1.41; P = .017), whereas CEBPADM (OS: HR = 0.38, P = .004; EFS: HR = 0.45, P = .007) and NPM1 mutations (OS: HR = 0.73, P = .03; EFS: HR = 0.69; P = .006) were found indicative of favorable OS and EFS. The positive prognostic impact of NPM1 mutations becomes even more pronounced in FLT3ITD-negative AML (OS: HR = 0.63, P = .022; EFS: HR = 0.64, P = .018). Univariable analysis of the gene expression markers demonstrates that increased expressions of BAALC, CD34, MN1, EVI1, and ERG are significant negative indicators for OS and EFS (all HR > 1.5, P < .01). Univariable survival analysis for CN AML is given in supplemental Table 5. The negative predictive effect of FLT3ITD, BAALC, CD34, EVI1, and ERG is retained in CN AML.
Univariable survival analysis in the intermediate-risk group
| Variable . | . | OS . | EFS . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Hazard ratio . | Lower . | Upper . | P . | Hazard ratio . | Lower . | Upper . | P . | ||
| NPM1 | + | 0.73 | 0.55 | 0.97 | .03 | 0.69 | 0.53 | 0.9 | .006 |
| FLT3ITD | + | 1.41 | 1.06 | 1.86 | .017 | 1.3 | 0.99 | 1.7 | .059 |
| FLT3TKD | + | 0.82 | 0.51 | 1.32 | .418 | 0.74 | 0.47 | 1.16 | .192 |
| N-RAS | + | 0.94 | 0.57 | 1.54 | .798 | 1.23 | 0.77 | 1.94 | .386 |
| CEBPASM | + | 1.01 | 0.38 | 2.72 | .984 | 0.8 | 0.3 | 2.16 | .662 |
| CEBPADM | + | 0.38 | 0.19 | 0.74 | .004 | 0.45 | 0.25 | 0.81 | .007 |
| IDH1 | + | 0.83 | 0.52 | 1.31 | .414 | 0.97 | 0.64 | 1.47 | .877 |
| IDH2 | + | 0.74 | 0.47 | 1.17 | .199 | 0.79 | 0.51 | 1.21 | .273 |
| FLT3ITD × NPM1 | +− | 1.67 | 1.13 | 2.46 | .01 | 1.76 | 1.21 | 2.58 | .003 |
| −+ | 0.63 | 0.42 | 0.94 | .022 | 0.64 | 0.45 | 0.93 | .018 | |
| ++ | 1.03 | 0.72 | 1.47 | .875 | 0.9 | 0.64 | 1.27 | .549 | |
| EVI1* | + | 1.78 | 1.17 | 2.7 | .007 | 2.01 | 1.34 | 3.02 | < .001 |
| BAALC | 3.16 | 1.74 | 5.72 | < .001 | 2.9 | 1.63 | 5.16 | < .001 | |
| CD34 | 3.81 | 2.17 | 6.67 | < .001 | 3.57 | 2.11 | 6.05 | < .001 | |
| MN1 | 2.41 | 1.37 | 4.23 | .002 | 2.51 | 1.46 | 4.32 | < .001 | |
| ERG | 3.69 | 1.65 | 8.26 | .001 | 3.48 | 1.63 | 7.43 | .001 | |
| ABCB1 | 0.99 | 0.51 | 1.93 | .983 | 0.92 | 0.49 | 1.73 | .798 | |
| BCL2 | 1.07 | 0.5 | 2.3 | .861 | 1.19 | 0.57 | 2.46 | .644 | |
| INDO1 | 0.65 | 0.32 | 1.36 | .254 | 0.69 | 0.35 | 1.38 | 0.3 | |
| Variable . | . | OS . | EFS . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Hazard ratio . | Lower . | Upper . | P . | Hazard ratio . | Lower . | Upper . | P . | ||
| NPM1 | + | 0.73 | 0.55 | 0.97 | .03 | 0.69 | 0.53 | 0.9 | .006 |
| FLT3ITD | + | 1.41 | 1.06 | 1.86 | .017 | 1.3 | 0.99 | 1.7 | .059 |
| FLT3TKD | + | 0.82 | 0.51 | 1.32 | .418 | 0.74 | 0.47 | 1.16 | .192 |
| N-RAS | + | 0.94 | 0.57 | 1.54 | .798 | 1.23 | 0.77 | 1.94 | .386 |
| CEBPASM | + | 1.01 | 0.38 | 2.72 | .984 | 0.8 | 0.3 | 2.16 | .662 |
| CEBPADM | + | 0.38 | 0.19 | 0.74 | .004 | 0.45 | 0.25 | 0.81 | .007 |
| IDH1 | + | 0.83 | 0.52 | 1.31 | .414 | 0.97 | 0.64 | 1.47 | .877 |
| IDH2 | + | 0.74 | 0.47 | 1.17 | .199 | 0.79 | 0.51 | 1.21 | .273 |
| FLT3ITD × NPM1 | +− | 1.67 | 1.13 | 2.46 | .01 | 1.76 | 1.21 | 2.58 | .003 |
| −+ | 0.63 | 0.42 | 0.94 | .022 | 0.64 | 0.45 | 0.93 | .018 | |
| ++ | 1.03 | 0.72 | 1.47 | .875 | 0.9 | 0.64 | 1.27 | .549 | |
| EVI1* | + | 1.78 | 1.17 | 2.7 | .007 | 2.01 | 1.34 | 3.02 | < .001 |
| BAALC | 3.16 | 1.74 | 5.72 | < .001 | 2.9 | 1.63 | 5.16 | < .001 | |
| CD34 | 3.81 | 2.17 | 6.67 | < .001 | 3.57 | 2.11 | 6.05 | < .001 | |
| MN1 | 2.41 | 1.37 | 4.23 | .002 | 2.51 | 1.46 | 4.32 | < .001 | |
| ERG | 3.69 | 1.65 | 8.26 | .001 | 3.48 | 1.63 | 7.43 | .001 | |
| ABCB1 | 0.99 | 0.51 | 1.93 | .983 | 0.92 | 0.49 | 1.73 | .798 | |
| BCL2 | 1.07 | 0.5 | 2.3 | .861 | 1.19 | 0.57 | 2.46 | .644 | |
| INDO1 | 0.65 | 0.32 | 1.36 | .254 | 0.69 | 0.35 | 1.38 | 0.3 | |
Mutation present (or absent) groups are denoted with (+) or (−). The reference category for all binary mutation markers is mutation absent (−). The reference category for the combined aberration in FLT3ITD and NPM1 is both mutations absent.
EVI1 expression categorized with the reference category EVI1 < 1.15.
The multivariable Cox regression analysis (Table 3) shows that CD34, ERG, and CEBPADM remain significant predictors for OS and EFS after the correction for the remaining markers (respective P values for OS, P = .004, P = .036, and P < .001; and EFS, P = .005, P = .032, and P < .001), whereas neither increased BAALC, increased MN1, nor EVI1 expression or the presence of FLT3ITD is no longer indicative of adverse OS and EFS in intermediate-risk AML. The multivariable survival analysis for CN AML is summarized in supplemental Table 6. When we control for the remaining prognostic markers, only CEBPADM and CD34 remain significant in CN AML.
Multivariable survival analysis of the intermediate-risk group
| Variable . | . | OS . | EFS . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Hazard ratio . | Lower . | Upper . | P . | Hazard ratio . | Lower . | Upper . | P . | ||
| FLT3ITD × NPM1 | + − | 1.23 | 0.79 | 1.92 | .37 | 1.54 | 1 | 2.37 | .05 |
| −+ | 0.73 | 0.43 | 1.25 | .25 | 0.7 | 0.42 | 1.17 | .17 | |
| ++ | 1.09 | 0.66 | 1.79 | .74 | 0.94 | 0.58 | 1.52 | .81 | |
| FLT3TKD | + | 1.13 | 0.67 | 1.93 | .64 | 0.99 | 0.6 | 1.63 | .95 |
| N-RAS | + | 0.99 | 0.59 | 1.68 | .99 | 1.28 | 0.79 | 2.08 | .32 |
| CEBPASM | + | 0.99 | 0.35 | 2.81 | .99 | 0.84 | 0.3 | 2.36 | .75 |
| CEBPADM | + | 0.21 | 0.1 | 0.44 | < .001 | 0.26 | 0.14 | 0.51 | < .001 |
| IDH1 | + | 1.09 | 0.67 | 1.79 | .73 | 1.31 | 0.83 | 2.06 | .25 |
| IDH2 | + | 0.66 | 0.4 | 1.1 | .11 | 0.79 | 0.49 | 1.27 | .32 |
| EVI1* | + | 1.15 | 0.72 | 1.83 | .56 | 1.3 | 0.83 | 2.04 | .26 |
| BAALC | 1.43 | 0.43 | 4.72 | .56 | 1.07 | 0.35 | 3.28 | .91 | |
| CD34 | 5.09 | 1.73 | 15.01 | < .001 | 4.47 | 1.62 | 12.32 | < .001 | |
| MN1 | 0.59 | 0.21 | 1.67 | .32 | 0.7 | 0.25 | 1.95 | .49 | |
| ERG | 4.13 | 1.01 | 16.86 | .05 | 3.93 | 1.05 | 14.62 | .04 | |
| ABCB1 | 0.84 | 0.32 | 2.22 | .72 | 0.67 | 0.26 | 1.68 | .39 | |
| BCL2 | 0.34 | 0.12 | 0.96 | .04 | 0.42 | 0.15 | 1.15 | .09 | |
| WT1 | 0.68 | 0.28 | 1.64 | .39 | 0.66 | 0.29 | 1.53 | .34 | |
| FLT3 | 0.75 | 0.28 | 2 | .56 | 0.66 | 0.25 | 1.74 | .41 | |
| INDO1 | 0.7 | 0.3 | 1.61 | .4 | 0.69 | 0.32 | 1.51 | .36 | |
| Variable . | . | OS . | EFS . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Hazard ratio . | Lower . | Upper . | P . | Hazard ratio . | Lower . | Upper . | P . | ||
| FLT3ITD × NPM1 | + − | 1.23 | 0.79 | 1.92 | .37 | 1.54 | 1 | 2.37 | .05 |
| −+ | 0.73 | 0.43 | 1.25 | .25 | 0.7 | 0.42 | 1.17 | .17 | |
| ++ | 1.09 | 0.66 | 1.79 | .74 | 0.94 | 0.58 | 1.52 | .81 | |
| FLT3TKD | + | 1.13 | 0.67 | 1.93 | .64 | 0.99 | 0.6 | 1.63 | .95 |
| N-RAS | + | 0.99 | 0.59 | 1.68 | .99 | 1.28 | 0.79 | 2.08 | .32 |
| CEBPASM | + | 0.99 | 0.35 | 2.81 | .99 | 0.84 | 0.3 | 2.36 | .75 |
| CEBPADM | + | 0.21 | 0.1 | 0.44 | < .001 | 0.26 | 0.14 | 0.51 | < .001 |
| IDH1 | + | 1.09 | 0.67 | 1.79 | .73 | 1.31 | 0.83 | 2.06 | .25 |
| IDH2 | + | 0.66 | 0.4 | 1.1 | .11 | 0.79 | 0.49 | 1.27 | .32 |
| EVI1* | + | 1.15 | 0.72 | 1.83 | .56 | 1.3 | 0.83 | 2.04 | .26 |
| BAALC | 1.43 | 0.43 | 4.72 | .56 | 1.07 | 0.35 | 3.28 | .91 | |
| CD34 | 5.09 | 1.73 | 15.01 | < .001 | 4.47 | 1.62 | 12.32 | < .001 | |
| MN1 | 0.59 | 0.21 | 1.67 | .32 | 0.7 | 0.25 | 1.95 | .49 | |
| ERG | 4.13 | 1.01 | 16.86 | .05 | 3.93 | 1.05 | 14.62 | .04 | |
| ABCB1 | 0.84 | 0.32 | 2.22 | .72 | 0.67 | 0.26 | 1.68 | .39 | |
| BCL2 | 0.34 | 0.12 | 0.96 | .04 | 0.42 | 0.15 | 1.15 | .09 | |
| WT1 | 0.68 | 0.28 | 1.64 | .39 | 0.66 | 0.29 | 1.53 | .34 | |
| FLT3 | 0.75 | 0.28 | 2 | .56 | 0.66 | 0.25 | 1.74 | .41 | |
| INDO1 | 0.7 | 0.3 | 1.61 | .4 | 0.69 | 0.32 | 1.51 | .36 | |
Mutation present (or absent) groups denoted with (+) or (−). The reference category for binary mutation markers is mutation absent (−). The reference category for the combined aberration in FLT3ITD and NPM1 is both mutations absent.
EVI expression categorized with the reference category EVI < 1.15.
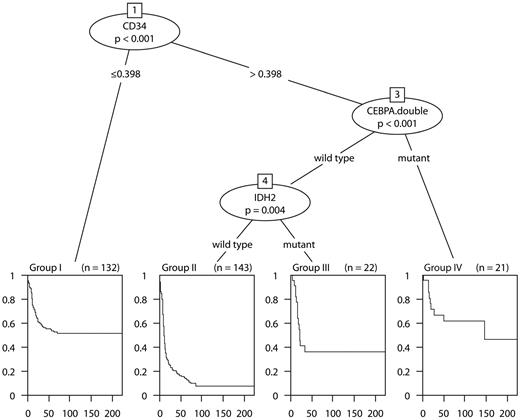
To investigate which minimal subset/combination of markers is sufficient for assessing prognosis, variable selection in Cox proportional hazards models was performed. The LASSO variable selection with an optimal value of 8.7 of the penalization parameter identified the following markers: CD34, CEBPADM, IDH2, BCL2, ERG, NPM1, EVI1, FLT3ITD, and INDO1. Estimated regression coefficients of the penalized Cox proportional hazards model for different values of the penalization parameter for OS are shown in supplemental Figure 2. The plot indicates that, among the considered series of markers, CD34 and CEBPADM play a predominant role in survival prognosis. The Akaike information criterion-based stepwise selection identified a similar set of markers (ie, CD34, CEBPADM, IDH2, BCL2, and ERG). The variables recognized as important by the recursive binary partitioning in the survival tree methodology were CD34, CEBPADM, and IDH2 (Figure 2). Similar results were obtained for EFS. The tree model is in accordance with the penalized Cox regression approach in that CD34 and CEBPADM were again identified as the most important predictors.
The survival tree model (OS). The tree shows the partitioning of the 318 intermediate-risk AML into 4 groups with more similar survival characteristics. Kaplan-Meier estimates of the survival curves for each of the groups attached at the bottom of the tree. Group I (n = 132) consists of patients with CD34 expression ≤ 0.398. Group II (n = 143) are patients with CD34 expression > 0.398, IDH2 wild type CEBPADM absent. Group III is characterized by CD34 expression > 0.398, IDH2 mutation present, and no CEBPADM. Group IV includes patients with CD34 expression > 0.398 and CEBPADM.
The survival tree model (OS). The tree shows the partitioning of the 318 intermediate-risk AML into 4 groups with more similar survival characteristics. Kaplan-Meier estimates of the survival curves for each of the groups attached at the bottom of the tree. Group I (n = 132) consists of patients with CD34 expression ≤ 0.398. Group II (n = 143) are patients with CD34 expression > 0.398, IDH2 wild type CEBPADM absent. Group III is characterized by CD34 expression > 0.398, IDH2 mutation present, and no CEBPADM. Group IV includes patients with CD34 expression > 0.398 and CEBPADM.
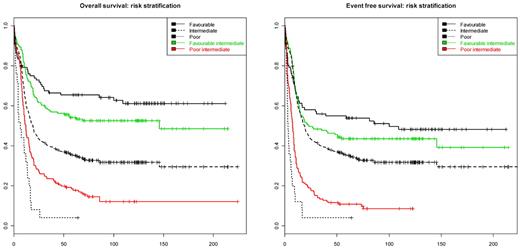
The survival tree model in Figure 2 naturally suggests stratification of the intermediate-risk AML into subgroups with more homogeneous survival characteristics. According to the model, the intermediate-risk group could be divided into 4 categories: (I) “low CD34” (defined as CD34 < 0.398); (II) “high CD34” (defined as CD34 > 0.398), IDH2 wild-type and CEBPADM absent; (III) “high CD34” (> 0.398), IDH2 mutated, and no CEBPADM; and (IV) “high CD34” (> 0.398) and CEBPADM. Of the 4 categories, the groups (I, III, and IV) have statistically indistinguishable survival characteristics (P value of a 2 df partial likelihood ratio test: 0.298 [OS] and 0.333 [EFS]). The 3 groups (I, III, and IV) together could be aggregated as the favorable intermediate-risk group (estimated OS and EFS at 60 months, 51.9% and 41.5%, respectively). In contrast, the estimated OS and EFS at 60 months in the group (II) is 14.9% and 8.3%, respectively, which indicates unfavorable prognosis. The latter group has been designated: poor intermediate-risk group. The survival characteristics in the proposed strata compared with the survival profile of the established cytogenetic prognostic stratification (as described in “Patients, cell samples, and molecular analyses”) is given in Figure 3. The difference in survival between favorable and intermediate favorable prognostic groups is not statistically significant (P value of 1 df likelihood ratio test: 0.153 [OS], 0.44 [EFS]). The survival characteristics between poor and poor intermediate group are significantly different (P value of 1 df likelihood ratio test: .012 for both OS and EFS).
Risk stratification of intermediate-risk AML. The left and right panels presents Kaplan-Meier survival curve estimates for the OS and EFS in 5 AML subsets. Black lines indicate survival curves for favorable (solid line), intermediate (dashed line), and unfavorable (dotted line) cytogenetic risk subgroups of AML as defined in “Patients, cell samples, and molecular analyses.” The red curve represents the poor intermediate group defined as CD34 expression > 0.398, no IDH2 mutation, and no CEBPADM. The green line represents the favorable intermediate group defined as (1) CD34 expression < 0.398, (2) CD34 expression > 0.398 and CEBPADM, or (3) CD34 expression > 0.398, no CEBPADM, and IDH2 mutant.
Risk stratification of intermediate-risk AML. The left and right panels presents Kaplan-Meier survival curve estimates for the OS and EFS in 5 AML subsets. Black lines indicate survival curves for favorable (solid line), intermediate (dashed line), and unfavorable (dotted line) cytogenetic risk subgroups of AML as defined in “Patients, cell samples, and molecular analyses.” The red curve represents the poor intermediate group defined as CD34 expression > 0.398, no IDH2 mutation, and no CEBPADM. The green line represents the favorable intermediate group defined as (1) CD34 expression < 0.398, (2) CD34 expression > 0.398 and CEBPADM, or (3) CD34 expression > 0.398, no CEBPADM, and IDH2 mutant.
Discussion
AML is a group of neoplasms characterized by a variety of genetic and epigenetic aberrations and variable responses to therapy.1,2 The pretreatment karyotype of leukemic blasts is currently a key determinant for therapy decision-making in AML. Usually, the largest cytogenetic subclass of AML (ie, those patients with a normal karyotype and patients with prognostically noninformative cytogenetic aberrations) is categorized as intermediate risk. In recent years, a number of novel markers have been identified as putative classifiers for these AML patients. These markers include a wide variety of acquired mutations as well as expression changes in specific genes.
In previous studies, prognostic risk assessments were put forward based on various expression markers BAALC,24,25,53 ERG,26,27 MN1,28,29 and EVI1.21-23 These studies have postulated risk algorithms mainly for CN AML and included only few of the wide variety of mutations and expression markers. Studies addressing the relative importance of the various postulated mutations and expression markers are limited.54
In this study, we investigated the role of a wide series of genomic biomarkers that included mutations in FLT3, CEBPA, NPM1, NRAS, IDH1, IDH2, and WT1 genes as well as high expression of EVI1, WT1, BCL2, ABCB1, BAALC, FLT3, CD34, INDO, ERG, and MN1 in the risk stratification of intermediate-risk AML. The results reveal particular associations between some of these markers that may strongly affect the collective use of these markers in risk assessment. For instance, we demonstrate an inverse association between NPM1 mutations and Affymetrix HGU133 Plus2.0-derived CD34 expression, as was shown by others.9,55,56 Importantly, relatively strong associations exist between expression levels of CD34, BAALC, MN1, ERG, and ABCB1. Consequently, expression values of all these markers inversely correlate with the presence of mutant NPM1. These interactions indicate that these markers will have similar value in risk stratification of AML and should therefore be taken into account when prognostic scores based on selected markers are constructed.
By univariable analyses, we confirmed the prognostic ability of previously established markers in intermediate-risk AML: CEBPADM and NPM1 mutations as indicators for favorable OS and EFS6-12 and FLT3ITD mutations as markers for poor response to therapy.3-5,57 High expression of BAALC, CD34, MN1, and ERG all express unfavorable prognostic value with regard to OS and EFS, which is in line with earlier publications.24-29,53 Importantly, expression of CD34 mRNA strongly associates with poor OS and EFS.
In multivariable analyses, CEBPADM independently predicts favorable outcome, whereas CD34 and ERG are independent predictors for inferior OS and EFS. ERG expression has emerged as a strong negative predictor in multivariate analyses previously54 ; however, in this model, CD34 expression is the strongest expression marker for poor outcome. By conducting a model selection in both Cox proportional hazards regression models and survival trees, it becomes evident that CEBPADM and CD34 expression stands out as the most prominent predictors for treatment outcome. Although the value of CD34 protein expression has been controversial,36 CD34 mRNA appears to be notably valuable in AML risk stratification.
Although stratification based on expression levels is challenging, the usage of standardized protocols and Affymetrix GeneChips may facilitate the implementation of gene expression level analyses. Indeed, because many laboratories currently use Affymetrix GeneChips, the results of these types of analyses may be relatively easily implemented.
We developed a simplified stratification rule of intermediate-risk AML, which identifies 2 distinctive groups of patients with survival characteristics being similar to the generally established favorable and poor risk cytogenetic subgroups, respectively. We acknowledge that the proposed stratification needs further validation in future studies and will probably be improved with new emerging knowledge. Nevertheless, the model presented here discloses several particularly interesting associations with respect to the hierarchy of the prognostic importance of a scale of molecular biomarkers and adds to the understanding of the heterogeneity of intermediate-risk AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Dutch Cancer Society (Koningin Wilhelmina Fonds) and Center for Translational Molecular Medicine. This study was performed within the framework of the Center for Translational Molecular Medicine (Leukemia BioCHIP project, grant 03O-102).
Authorship
Contribution: V.R. designed research, analyzed data, and wrote the paper; S.A., B.J.W., C.A.J.E., H.B.B., and R.D. performed research; W.L.J.v.P. analyzed data and wrote manuscript; B.L. designed research and wrote the paper; and P.J.M.V. designed and performed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: R.D., B.L., and P.J.M.V. have declared ownership interests in Skyline Diagnostic, a start-up and spin-off company of Erasmus University Medical Center. The remaining authors declare no competing financial interests.
Correspondence: Peter J. M. Valk, Erasmus University Medical Center Rotterdam, Department of Hematology, Ee1391a, Dr Molewaterplein 50, 3015 GE Rotterdam Z-H, The Netherlands; e-mail: p.valk@erasmusmc.nl.
References
Author notes
B.L. and P.J.M.V. contributed equally to this study as co-senior authors.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal