Abstract
Chronic myeloid leukemia chronic phase (CML-CP) CD34+ cells contain numerous DNA double-strand breaks whose unfaithful repair may contribute to chromosomal instability and disease progression to blast phase (CML-BP). These phenomena are often associated with the appearance of imatinib-resistant BCR-ABL1 kinase mutants (eg, T315I) and overexpression of BCR-ABL1. Here we show that BCR-ABL1 (nonmutated and T315I mutant) promoted RAD51 recombinase-mediated unfaithful homeologous recombination repair (HomeoRR) in a dosage-dependent manner. BCR-ABL1 SH3 domain interacts with RAD51 proline-rich regions, resulting in direct phosphorylation of RAD51 on Y315 (pY315). RAD51(pY315) facilitates dissociation from the complex with BCR-ABL1 kinase, migrates to the nucleus, and enhances formation of the nuclear foci indicative of recombination sites. HomeoRR and RAD51 nuclear foci were strongly reduced by RAD51(Y315F) phosphorylation-less mutant. In addition, peptide aptamer mimicking RAD51(pY315) fragment, but not that with Y315F phosphorylation-less substitution, diminished RAD51 foci formation and inhibited HomeoRR in leukemia cells. In conclusion, we postulate that BCR-ABL1 kinase-mediated RAD51(pY315) promotes unfaithful HomeoRR in leukemia cells, which may contribute to accumulation of secondary chromosomal aberrations responsible for CML relapse and progression.
Introduction
Chronic myeloid leukemia in chronic phase (CML-CP) is a myeloproliferative disorder characterized by the presence of the Philadelphia (Ph) chromosome that results from a (9;22)(q34;q11) reciprocal translocation that juxtaposes the c-abl oncogene 1 (ABL1) gene on chromosome 9 with the breakpoint cluster region (BCR) gene on chromosome 22, generating the BCR-ABL1 fusion oncogene. Most CML-CP patients are currently treated with tyrosine kinase inhibitor (TKI) imatinib designed to block the enzymatic action of the ABL1 tyrosine kinase. Approximately 60%-70% of patients achieve and maintain a complete cytogenetic response (CCyR) 5 years after initiating imatinib treatment. Two “second-generation” TKIs, dasatinib and nilotinib, are effective at inducing or restoring CCyR in 40%-50% of patients who appear to have failed primary treatment with imatinib. However approximately 20% of patients presenting with CML-CP fail to respond to both imatinib and a subsequent second-generation TKI; their prognosis is poor because of a higher risk of disease progression. In addition, because TKIs do not eradicate the disease, the patients in CCyR may carry 106 to 109 leukemia cells, and even BCR-ABL1-negative patients in complete molecular response may have up to 106 leukemia cells.1 These cells may be the “time-ticking bombs” eventually exploding into TKI-resistant clone and/or CML-blast phase (CML-BP) clone on accumulation of additional genetic aberrations.2
Clinical observations and experimental findings clearly demonstrated that genomic instability in CML is driven, at least in part, by BCR-ABL1 kinase.2,3 However, TKI-treated CML patients continue to accumulate chromosomal aberrations probably because of inefficient TKI activity (eg, in low-oxygen bone marrow niche), and/or acquired TKI resistance (TKI-resistant BCR-ABL1 mutants and overexpression of BCR-ABL1).4-6 In concordance, TKI-resistant BCR-ABL1 kinase mutants (eg, T315I) and BCR-ABL1 overexpression are associated with clonal cytogenetic evolution and greater likelihood of disease relapse and progression, which suggests enhanced genomic instability in these cells.6,7
The frequency of additional chromosomal abnormalities is approximately 7% in CML-CP and increases to 40%-70% in the advanced phase of disease CML-BP, as evaluated by standard cytogenetic analysis. However, more sensitive comparative genomic hybridization (CGH) and single nucleotide polymorphism (SNP) analyses detect multiple genetic aberrations already in late CP, but BP patients have much more complex karyotypes.8,9 Numeric chromosomal changes are detected at a 50-fold higher frequency and structural changes at a 12-fold higher frequency in CML-BP, compared with CML-CP.
Genomic instability usually emerges from enhanced DNA damage and/or unfaithful DNA repair. We and others reported that CD34+ CML-CP cells contain elevated numbers of DNA double-strand breaks (DSBs) caused by reactive oxygen species, cytotoxic drugs, and γ-radiation treatment, and that DSB repair mechanisms are stimulated but unfaithful, resulting in accumulation of chromosomal aberrations.10-15
Homologous recombination repair (HomoRR) is one of the most important DSB repair mechanisms.16 It usually involves RAD51 recombinase-dependent single-strand invasion and pairing between allelic sequences on the sister chromatide or homologous chromosome followed by DNA resynthesis. However, when overstimulated and/or not controlled by mismatch repair (MMR) machinery, it can promote pairing between nonallelic DNA sequences that share high sequence identity (divergent sequences) resulting in nonallelic homologous recombination, also called homeologous recombination (HomeoRR).17 Recombination between sequences other than those in equivalently positioned sister chromatids can lead to genome instability by promoting unequal sister chromatid exchanges, which leads to sequence deletions and expansions (intrachromosomal recombination), and by generating translocations when nonhomologous chromosomes are involved (interchromosomal recombination).
We and others reported that BCR-ABL1 and other oncogenic tyrosine kinases (OTKs), such as TEL-ABL1, TEL-JAK2, TEL-PDGFβR, NPM-ALK, ZNF198-FGFR1, FLT3-ITD-TKD, JAK2(V617F), IGF-IR, PDGFβR, EGFR, and MET(M1268T and Y1248H) stimulated the expression and/or tyrosine phosphorylation of RAD51 recombinase.18-24 Thus, an OTK → RAD51 pathway may represent a common mechanism regulating the response to DNA damage not only in CML, but also in other tumor cells.
We show here that BCR-ABL1 interacts with RAD51 to compromise the fidelity of recombination. BCR-ABL1–mediated tyrosine phosphorylation site (pY315) in RAD51 was essential for stimulation of unfaithful HomeoRR and may play an important role in accumulation of chromosomal aberrations and CML progression.
Methods
Cells
The 32Dcl3 murine parental hematopoietic cell line and its counterpart transformed with p210BCR-ABL1 (BCR-ABL1–32Dcl3) were used before and maintained in Iscove modified defined medium supplemented with 1mM glutamine, 10% FBS and IL-3–conditioned medium.18 Human embryonic kidney 293 cells and Phoenix Amphotropic packaging cells (ATCC) were maintained in Dulbecco modified Eagle medium supplemented with 10% FBS. CML-CP patient cells were obtained after receiving informed consent. CD34+ CML cells were isolated using human CD34+ selection cocktail (StemCell Technologies). The research activities involving human samples were approved by the Temple University Institutional Review Board.
RAD51 constructs
The cDNAs of RAD51-Flag wild-type (WT) was cloned into pcDNA 3.1 expression vector and into pMIG-IRES-GFP (obtained from Dr Warren Pear, University of Pennsylvania, Philadelphia, PA) and pMX-puro retroviral vectors. The RAD51(Y315F), RAD51(P286L), RAD51(P321L), and RAD51(P286L+P321L) mutants were generated by site-directed mutagenesis using overlapping oligonucleotides and QuikChange II XL site-directed mutagenesis kit (Stratagene/Agilent Technologies). In addition, cDNAs of RAD51 full length, RAD51-C (amino acids 243-339), RAD51-C(Y315F), RAD51-C(P286L + P321L) and RAD51-C(Y315F + P286L + P321L) were cloned in frame into pGEX-4T2 to generate GST-fusion proteins.
Retroviral infection
Retroviral constructs were transiently transfected into Phoenix Amphotropic cells. Viral supernatants were collected 36 hours later, filtered and concentrated by centrifugation at 5300g for 2 hours at 16°C. BCR-ABL1–32Dcl3 cells (0.5 × 106cells/mL) were infected with 1:1 mixture of freshly concentrated virus and medium, twice a day for 2 days in the presence of 4 μg/mL of Polybrene (Sigma-Aldrich) as described before.18 GFP+ cells and puromycin-resistant cells were sorted and selected, respectively, and used for experiments.
HomeoRR assay
The 32Dcl3 cells were electroporated with 30 μg of pLB4 expression plasmid containing the unfaithful recombination cassette (Figure 1A, obtained from Dr A. Waldman, University of South Carolina)25 and selected with 400 μg/mL hygromycin. The presence of a single copy of genomic integrated HomeoRR cassette was confirmed by Southern blot (Figure 1B) and by polymerase chain reaction (PCR) detecting appropriate AluI-digests (Figure 1D left box). The 32Dcl3-pLB4 cells were electroporated with the pBABE-puro vector encoding p210BCR-ABL1 nonmutated and p210BCR-ABL1(T315I) mutant, and pBABE-puro empty vector (control); expression of BCR-ABL1 in puromycin-resistant clones was confirmed by Western analysis. When indicated, BCR-ABL1–32Dcl3-pLB4 cells were transfected with pMIG-Flag-RAD51(WT)-IRES-GFP and pMIG-Flag-RAD51(Y315F)-IRES-GFP retroviral constructs; expression of Flag-tagged RAD51 proteins in GFP+ cells was confirmed by Western analysis. Cells growing in Iscove modified defined medium supplemented with 1.5% IL-3–conditioned medium were cotransfected with 90 μg of I-SceI and 10 μg of pMIG-IRES-GFP or pDsRed1-Mito (controls for I-SceI transfection). At 36 hours after transfection, GFP+ or Red1+ cells were sorted and individual cells were transferred to 96-well plates and cultured in the presence of G418 for 14 days to detect cells in which recombination did occur. Resistant colonies were counted and selected for further analysis. Genomic DNA was isolated from G418-resistant cells and PCR was performed using the following primers: AW85 (5′-ATACGACTCACTATAGGGCCAGCGTCTTGTCATTGGCG-3′) and AW91 (5′-GATTAGGTGACACTATAGCCAAGCGGCCGGAGAACCTG-3′). An approximately 1.4-kb PCR product was digested with AluI and analyzed by agarose gel electrophoresis.
BCR-ABL1 kinase and the TKI-resistant T315I mutant promote HomeoRR in a “dosage”-dependent manner. (A) Homeologous recombination cassette contains a functional hygromycin gene (hyg), used to select for stably transfected cells, and a 3.9-kb tk-neo fusion gene rendered nonfunctional by the insertion of a 22-bp oligonucleotide containing 18-bp recognition site for endonuclease I-SceI (yellow strip). Three AluI restriction sited (red stripes) are present, too. In addition, a 2.5-kb fragment containing a complete HSV-1 tk gene, which is more than 1% divergent compared with that in the tk-neo fusion gene (0.8% and 1.5% divergent in 5′ and 3′ direction from the DSB site, respectively), is inserted. This inserted fragment serves as a potential recombination template (tk donor). HomeoRR by gene conversion between the I-SceI–induced DSB in the tk-neo fusion gene and the divergent tk donor template restores the function of neo gene encoding resistance to G418 (G418-R) and removes 2 AluI restriction sites closest to the DSB site.25 (B) Southern blot analysis of 32Dcl3-pLB4 parental cell clone (P) and BCR-ABL1–32Dcl3-pLB4 clone (B/A), in which a single copy of the pLB4 cassette was integrated in the genome. Genomic DNA was digested with HindIII and blotted with radiolabeled NcoI-NarI fragment of neo gene as a probe. (C) The 32Dcl3-pLB4 parental cells (P), cells expressing nonmutated BCR-ABL1 (B/A[nm]) and BCR-ABL1(T315I) (B/A[T315I]), and also cells expressing low (B/A-low) and high (B/A-high) levels of BCR-ABL1 were cotransfected with I-SceI (DSB inducer) and GFP (transfection control) expression vectors. HomeoRR activity is shown as percentage of G418-resistant cells in the GFP+ population. (D) Representative analysis of the recombination products by PCR (the primer positions are indicated by blue arrows in panel A) followed by AluI restriction digest. Left box: Nonrecombinant cassette produces 1393-bp product, which generates 653-, 381-, 337-, and 22-bp bands when digested by AluI. Right box: HomeoRR was confirmed in the mixture of G418-resistant clones by PCR amplifying 1371-bp HomeoRR products, which after AluI restriction digest generate characteristic recombination-specific 990-bp band and also 381-bp band (panel A, scheme of the cassette).
BCR-ABL1 kinase and the TKI-resistant T315I mutant promote HomeoRR in a “dosage”-dependent manner. (A) Homeologous recombination cassette contains a functional hygromycin gene (hyg), used to select for stably transfected cells, and a 3.9-kb tk-neo fusion gene rendered nonfunctional by the insertion of a 22-bp oligonucleotide containing 18-bp recognition site for endonuclease I-SceI (yellow strip). Three AluI restriction sited (red stripes) are present, too. In addition, a 2.5-kb fragment containing a complete HSV-1 tk gene, which is more than 1% divergent compared with that in the tk-neo fusion gene (0.8% and 1.5% divergent in 5′ and 3′ direction from the DSB site, respectively), is inserted. This inserted fragment serves as a potential recombination template (tk donor). HomeoRR by gene conversion between the I-SceI–induced DSB in the tk-neo fusion gene and the divergent tk donor template restores the function of neo gene encoding resistance to G418 (G418-R) and removes 2 AluI restriction sites closest to the DSB site.25 (B) Southern blot analysis of 32Dcl3-pLB4 parental cell clone (P) and BCR-ABL1–32Dcl3-pLB4 clone (B/A), in which a single copy of the pLB4 cassette was integrated in the genome. Genomic DNA was digested with HindIII and blotted with radiolabeled NcoI-NarI fragment of neo gene as a probe. (C) The 32Dcl3-pLB4 parental cells (P), cells expressing nonmutated BCR-ABL1 (B/A[nm]) and BCR-ABL1(T315I) (B/A[T315I]), and also cells expressing low (B/A-low) and high (B/A-high) levels of BCR-ABL1 were cotransfected with I-SceI (DSB inducer) and GFP (transfection control) expression vectors. HomeoRR activity is shown as percentage of G418-resistant cells in the GFP+ population. (D) Representative analysis of the recombination products by PCR (the primer positions are indicated by blue arrows in panel A) followed by AluI restriction digest. Left box: Nonrecombinant cassette produces 1393-bp product, which generates 653-, 381-, 337-, and 22-bp bands when digested by AluI. Right box: HomeoRR was confirmed in the mixture of G418-resistant clones by PCR amplifying 1371-bp HomeoRR products, which after AluI restriction digest generate characteristic recombination-specific 990-bp band and also 381-bp band (panel A, scheme of the cassette).
Recombinant proteins
The p185BCR-ABL1 WT and the kinase defective K1172R mutant (KD) proteins were produced using the TNT Coupled Reticulocyte Lysate System (Promega) according to the manufacturer's protocol. GST-fusion proteins were produced in Escherichia coli–competent bacteria DH5α transformed by the pGEX-4T2 constructs.
In vitro kinase assay
The translation mixtures were diluted in kinase buffer (25mM Tris-HCl, pH 7.4, 10mM MgCl2, 1mM dithiothreitol, 30μM adenosine triphosphate) and incubated in 30°C for 30 minutes with GST-RAD51 fusion protein bound to glutathione beads. The beads were washed with kinase buffer containing 1mM phenylmethylsulfonyl fluoride, 0.1mM sodium fluoride, 1mM sodium vanadate, and 10 μg/mL of aprotinin and leupeptin, and the reaction products were resolved by SDS-PAGE, transferred onto nitrocellulose, and analyzed by immunoblotting with the indicated antibodies.
Pull-down assays
GST-fusion proteins immobilized on glutathione-Sepharose beads (Sigma-Aldrich) were incubated for 2 hours at 4°C with cell lysates or rabbit reticulocyte the in vitro transcribed and translated (IVTT) reaction mixtures, washed and boiled for 5 minutes in 2× Laemmli sample buffer. The reaction products were resolved by SDS-PAGE and analyzed by Western blotting using the antibodies recognizing ABL1 (EMD Chemicals and BD Biosciences), phosphotyrosine (Milipore), and Flag (Sigma-Aldrich). GST-fusion proteins were detected by Ponceau red staining.
Immunoblotting
Total cell lysates were obtained by boiling and sonicating cell pellet directly in 1× SDS loading buffer. Cytoplasmic and nuclear extracts were prepared from cultured cells using NE-PER Reagents (Thermo Scientific/Pierce Biotechnology), according to the manufacturer's instructions. Protein fractions were resolved by SDS-PAGE followed by Western analysis using the antibodies specific for: RAD51 (Milipore), RAD51(pY54) and RAD51(pY315) (described before26 ), lamin B1 (Abcam), and α-tubulin (Novus Biologicals).
RAD51 nuclear foci
Cells were irradiated, or not, with 4 Gy from 137Cs source, and 3 hours later cytospins were prepared and fixed as described before.10 Nuclear foci were detected by primary mouse anti-Flag monoclonal antibody (Sigma-Aldrich) and RAD51 rabbit polyclonal antibody (EMD Chemicals/Calbiochem) followed by secondary goat anti–mouse and goat anti–rabbit antibody, respectively, conjugated with FITC (Alexa 488; Invitrogen). DNA was counterstained with the DNA fluorochrome 4,6-diamidino-2-phenylindole (Invitrogen). Staining and images from 25 to 50 cells/group were processed using SlideBook Version 3.0 (Intelligent Imaging Innovations) and Adobe Photoshop Version 6.0 (Adobe Systems) software.
Protein aptamers
Synthetic fusion peptides (aptamers) containing 16 amino acids sequence surrounding RAD51(Y315) were purchased from (Genemed Synthesis). ETRICKIpYDSPCLLEA-GGG-YARAAARQARA (pY315) contained phosphotyrosine (pY) in the position corresponding to Y315; and in ETRICKIFDSPCLLEA-GGG-YARAAARQARA (Y315F), the Y315 was substituted with F. In addition, P321 was substituted with L to disrupt the proline-rich region and avoid entrapment by SH3 domain(s). Three-residue polyglycine linker and protein transduction domain 4 promoting the crossing of lipid bilayers and direct intracellular transduction of peptides were added.27 These fusion peptides were also modified by N-terminal acetylation and C-terminal amidation to reduce proteolytic degradation. In addition, they were linked to rhodamine or for the uptake analysis. The aptamers were purified by high performance liquid chromatography and analyzed by mass spectroscopy.
Results
Nonmutated BCR-ABL1 and TKI-resistant BCR-ABL1(T315I) stimulate HomeoRR
We showed that leukemia cell lines expressing nonmutated and TKI-resistant BCR-ABL1 mutants and CML-CP CD34+ cells contain elevated numbers of DSBs and RAD51 nuclear foci; this effect depends on BCR-ABL1 “dosage”13,28 (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Moreover, BCR-ABL1 (nonmutated and TKI-resistant mutants) promoted the colocalization of RAD51 foci and γ-H2AX foci (recombination sites) and stimulated HomoRR10,19 (supplemental Figure 2). Altogether, these observations support the role of recombination in reparation of numerous DSBs in CML cells.
To determine whether BCR-ABL1 promotes unfaithful HomeoRR between divergent sequences, a reporter cassette (Figure 1A) was integrated in the genome of 32Dcl3 parental cells and BCR-ABL1–positive cells (Figure 1B).25 Leukemia cells expressing nonmutated TKI-sensitive BCR-ABL1 kinase and TKI-resistant BCR-ABL1(T315I) mutant displayed approximately 3-fold increase in HomeoRR activity compared with parental counterparts (Figure 1C upper left box). BCR-ABL1–mediated stimulation of unfaithful recombination depended on the kinase “dosage”: higher levels of BCR-ABL1 caused stronger stimulation of the HomeoRR activity (Figure 1C upper right box). Recombination repair-dependent events in G418-resistant cells were documented by detection of recombination-specific AluI-digested 990-base PCR product (Figure 1D right panel).25
BCR-ABL1 SH3 domain interacts with RAD51 proline-rich regions in a kinase-independent manner
To determine the mechanism of BCR-ABL1–RAD51 interaction, IVTT BCR-ABL1 kinase and the kinase-deficient K1172R mutant were used for the pull-down reaction with the GST-fused full-length RAD51 protein. A relatively weak interaction was detected between BCR-ABL1 kinase and RAD51 (Figure 2A), which resulted in abundant tyrosine phosphorylation of RAD51 (Figure 2B). Interestingly, the interaction was significantly enhanced when BCR-ABL1(K1172R) kinase-deficient mutant was used (Figure 2A) and RAD51 was not phosphorylated (Figure 2B). This observation suggests that BCR-ABL1 initially interacts directly with RAD51 in a kinase-independent manner and that subsequent BCR-ABL1 kinase-mediated RAD51 phosphorylation facilitates dissociation of the complex.
BCR-ABL1 interacts with RAD51 in a kinase-independent manner. BCR-ABL1 nonmutated (nm) and the kinase defective (KD) mutant were IVTT and analyzed by Western blotting with the use of anti-ABL1 (A, top left box) and antiphosphotyrosine (B, left box) antibodies to detect BCR-ABL1 and phosphotyrosine proteins (p-Tyr), respectively. IVTT products were incubated with GST-RAD51 in the kinase buffer supplemented with adenosine triphosphate. The presence of BCR-ABL1 in a GST-RAD51 pull-downs (A, top right box) and tyrosine phosphorylation of GST-RAD51 (B, top right box) was detected by Western analysis with the use of anti-ABL1 and antiphosphotyrosine antibodies, respectively. GST-fusion proteins were detected by Ponceau red staining (bottom boxes).
BCR-ABL1 interacts with RAD51 in a kinase-independent manner. BCR-ABL1 nonmutated (nm) and the kinase defective (KD) mutant were IVTT and analyzed by Western blotting with the use of anti-ABL1 (A, top left box) and antiphosphotyrosine (B, left box) antibodies to detect BCR-ABL1 and phosphotyrosine proteins (p-Tyr), respectively. IVTT products were incubated with GST-RAD51 in the kinase buffer supplemented with adenosine triphosphate. The presence of BCR-ABL1 in a GST-RAD51 pull-downs (A, top right box) and tyrosine phosphorylation of GST-RAD51 (B, top right box) was detected by Western analysis with the use of anti-ABL1 and antiphosphotyrosine antibodies, respectively. GST-fusion proteins were detected by Ponceau red staining (bottom boxes).
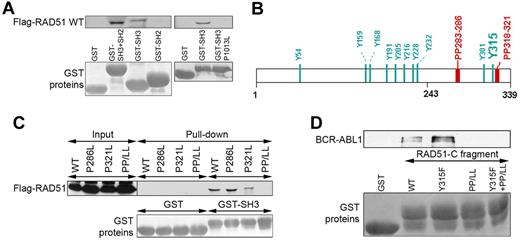
BCR-ABL1 SH3 and SH2 domains may provide a docking site for numerous proteins because of their capability to interact with proteins containing proline-rich region (PP) and phosphotyrosine (pY). In concordance, Flag-RAD51 was pulled down with GST-ABL SH3 + SH2 and GST-ABL SH3, but not with GST-ABL SH2 fusion proteins, implicating a pY-independent interaction (Figure 3A). Disruption of the ability of ABL SH3 domain to interact with PP by P1013L single amino acid substitution prevented the interaction with Flag-RAD51, clearly suggesting the role of RAD51 PPs.29
BCR-ABL1 SH3 domain interacts with RAD51 PPs. (A) GST fusion proteins containing the ABL1 SH3 + SH2, SH2, SH3, and SH3(P1013L) fragments were used for the pull-down assay along with cell lysates from BCR-ABL1–32Dcl3 cells expressing the Flag-RAD51 WT protein. RAD51 was detected by Western analysis (top boxes), and GST proteins were visualized by Ponceau red staining (bottom boxes). (B) Positions of tyrosine residues (Y) and PP in the RAD51 are marked. (C) Cell lysates from 293 cells expressing Flag-RAD51 WT and indicated mutants were used for the pull-down assay with GST and GST-ABL1 SH3 (GST-SH3) fusion protein. Flag-RAD51 was detected by Western analysis with the use of anti-Flag antibody (top box), and GST proteins were visualized by Ponceau red staining (bottom box). (D) GST-RAD51-C fragment containing amino acids 243 to 339 (WT) and the indicated mutants were used for pull-down with cell lysates from BCR-ABL1–32Dcl3 cells. BCR-ABL1 was detected by Western analysis with the use of anti-ABL1 antibody (top box), and GST proteins were visualized by Ponceau red staining (bottom boxes).
BCR-ABL1 SH3 domain interacts with RAD51 PPs. (A) GST fusion proteins containing the ABL1 SH3 + SH2, SH2, SH3, and SH3(P1013L) fragments were used for the pull-down assay along with cell lysates from BCR-ABL1–32Dcl3 cells expressing the Flag-RAD51 WT protein. RAD51 was detected by Western analysis (top boxes), and GST proteins were visualized by Ponceau red staining (bottom boxes). (B) Positions of tyrosine residues (Y) and PP in the RAD51 are marked. (C) Cell lysates from 293 cells expressing Flag-RAD51 WT and indicated mutants were used for the pull-down assay with GST and GST-ABL1 SH3 (GST-SH3) fusion protein. Flag-RAD51 was detected by Western analysis with the use of anti-Flag antibody (top box), and GST proteins were visualized by Ponceau red staining (bottom box). (D) GST-RAD51-C fragment containing amino acids 243 to 339 (WT) and the indicated mutants were used for pull-down with cell lysates from BCR-ABL1–32Dcl3 cells. BCR-ABL1 was detected by Western analysis with the use of anti-ABL1 antibody (top box), and GST proteins were visualized by Ponceau red staining (bottom boxes).
RAD51 has 2 PPs located in the C-terminal portion (Figure 3B). Disruption of both of them by P286L + P321L (PP/LL) was required to abrogate the interaction between ABL1 SH3 domain and full-length RAD51 (Figure 3C). In addition, GST-fused RAD51 C-terminal fragment (RAD51-C, amino acids 243-339) was able to pull down BCR-ABL1; disruption of the PPs in RAD51-C by P286L + P321L amino acid substitutions (PP/LL) abrogated its capability to pull down BCR-ABL1 (Figure 3D). Altogether, these results clearly indicate that BCR-ABL1 SH3 domain interacts with RAD51 PPs. Interestingly, introduction of the Y315F amino acid substitution to the RAD51-C fragment, but not to the PP/LL mutant, enhanced the interaction with BCR-ABL1 in the pull-down assay (Figure 3D), suggesting that phosphorylation on Y315 facilitates the release of RAD51 from the complex with BCR-ABL1.
RAD51 can also bind to Shc and Grb2, but not Gab2, Crkl, and Cbl, which belong to BCR-ABL1 proteome (data not shown). Therefore, we cannot exclude the possibility that BCR-ABL1–RAD51 interaction may also occur via these scaffold proteins.
Phosphorylation of RAD51 on Y315 (pY315) facilitates nuclear foci formation and HomeoRR
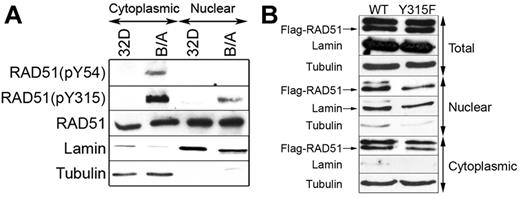
We reported before that BCR-ABL1 kinase phosphorylated RAD51 on Y315 to induce the resistance to cisplatin and mitomycin C, thus implicating its role in stimulation of recombination.19 Using RAD51-specific antibodies recognizing phospho-Y315 (pY315) and phospho-Y54 (pY54), we show that both RAD51(pY54) and RAD51(pY315) were detectable in the cytoplasm of BCR-ABL1–32Dcl3 leukemia cells but not parental counterparts (Figure 4A). Moreover, pY315 was readily detectable in the leukemia cell nuclei and barely detectable in the nuclei from parental cells; pY54 was not detected in both nuclear lysates.
RAD51(pY315) is detected in the nuclei of BCR-ABL1-positive leukemia cells. (A) RAD51(pY54), RAD51(pY315), and total RAD51 were detected by Western blotting with the use of phospho-specific and with anti-RAD51 antibodies, respectively, in the cytoplasmic and nuclear lysates from 32Dcl3 parental cells (32D) and BCR-ABL1–positive counterparts (B/A). The amount of protein lysates run in the gel was adjusted to detect similar levels of RAD51. (B) Flag-RAD51(WT) and Flag-RAD51(Y315F) were expressed in B/A cells, and Flag-tagged RAD51 proteins were detected in total, nuclear, and cytoplasmic cell lysates by Western analysis using anti-Flag antibody (Flag-RAD51 bands are marked by arrow). Lamin and tubulin served as the lysates purity controls.
RAD51(pY315) is detected in the nuclei of BCR-ABL1-positive leukemia cells. (A) RAD51(pY54), RAD51(pY315), and total RAD51 were detected by Western blotting with the use of phospho-specific and with anti-RAD51 antibodies, respectively, in the cytoplasmic and nuclear lysates from 32Dcl3 parental cells (32D) and BCR-ABL1–positive counterparts (B/A). The amount of protein lysates run in the gel was adjusted to detect similar levels of RAD51. (B) Flag-RAD51(WT) and Flag-RAD51(Y315F) were expressed in B/A cells, and Flag-tagged RAD51 proteins were detected in total, nuclear, and cytoplasmic cell lysates by Western analysis using anti-Flag antibody (Flag-RAD51 bands are marked by arrow). Lamin and tubulin served as the lysates purity controls.
To examine whether pY315 facilitates cytoplasmic-nuclear shuttling, Flag-RAD51 WT and Flag-RAD51(Y315F) were expressed in BCR-ABL1–32Dcl3 leukemia cells. Western analysis followed by densitometry analysis showed that nuclear localization of RAD51 was not affected by Y315F phosphorylation-less amino acid substitution (Figure 4B).
To determine the role of RAD51(pY315) in localization on DSBs, BCR-ABL1–32Dcl3 cells expressing Flag-RAD51(WT) and Flag-RAD51(Y315F) mutant (Figure 5A top panel) were untreated or irradiated with 4 Gy; and 3 hours later, they accumulated approximately 14 ± 9 and 50 ± 12 DSBs, respectively, painted by γ-H2AX foci. These experimental conditions are optimal to study RAD51 foci formation in response to DSB induced by γ-irradiation.10 Using fluorescent anti-Flag antibody, we found that Y315F amino acid substitution compromised the ability of RAD51 to form spontaneous and irradiation-induced nuclear foci (Figure 5A bottom panel, B). In concordance, RAD51(Y315F) exerted approximately 2.5-fold reduction of HomeoRR in BCR-ABL1-positive leukemia cells (Figure 5C). In addition, RAD51(Y315F) exerted an inhibitory effect on HomoRR (supplemental Figure 2C).
RAD51(Y315F) mutant displayed reduced nuclear foci formation and inhibited HomeoRR in BCR-ABL1-positive leukemia cells. (A) Top panel: Western blot showing the expression of Flag-tagged RAD51 proteins in BCR-ABL1–32Dcl3-pLB4 cells transfected with Flag-RAD51(WT) and Flag-RAD51(Y315F) expression plasmids or with empty (E) plasmid. Bottom panel: Cells were irradiated with 4 Gy; and 3 hours later, RAD51(WT) and RAD51(Y315F) nuclear foci were detected by immunofluorescence with anti-Flag antibody. Representative nuclei are shown; nuclei borders are marked in blue. (B) Data are number of Flag-RAD51(WT) and Flag-RAD51(Y315F) foci/cell ± SD in nonirradiated (Control) and 4 Gy-irradiated cells. *P < .001 versus corresponding WT group (Student t test). (C) HomeoRR activity in I-SceI transfected DsRed1-Mito+ cells. Data are percentage of G418-resistant clones ± SD. *P < .001 (Student t test).
RAD51(Y315F) mutant displayed reduced nuclear foci formation and inhibited HomeoRR in BCR-ABL1-positive leukemia cells. (A) Top panel: Western blot showing the expression of Flag-tagged RAD51 proteins in BCR-ABL1–32Dcl3-pLB4 cells transfected with Flag-RAD51(WT) and Flag-RAD51(Y315F) expression plasmids or with empty (E) plasmid. Bottom panel: Cells were irradiated with 4 Gy; and 3 hours later, RAD51(WT) and RAD51(Y315F) nuclear foci were detected by immunofluorescence with anti-Flag antibody. Representative nuclei are shown; nuclei borders are marked in blue. (B) Data are number of Flag-RAD51(WT) and Flag-RAD51(Y315F) foci/cell ± SD in nonirradiated (Control) and 4 Gy-irradiated cells. *P < .001 versus corresponding WT group (Student t test). (C) HomeoRR activity in I-SceI transfected DsRed1-Mito+ cells. Data are percentage of G418-resistant clones ± SD. *P < .001 (Student t test).
Targeting RAD51(pY315) reduced HomeoRR in BCR-ABL1-positive leukemias
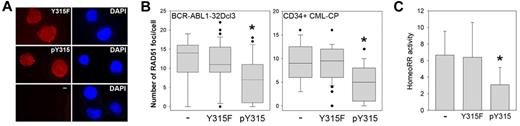
Next, we targeted RAD51(pY315) using synthetic fusion peptide (aptamer) as a decoy. The aptamer containing pY315 and surrounding 16 amino acids of RAD51 Y315 and that containing phenylalanine in the position of Y315 (Y315F) were used. Aptamers were readily detectable in the cytoplasm and nucleus (Figure 6A) in concordance with other studies showing fast and efficient cellular uptake of the modified aptamers.27 pY315, but not Y315F, aptamer reduced RAD51 nuclear foci formation in BCR-ABL1–32Dcl3 cells and CD34+ CML-CP patient cells (Figure 6B), and inhibited unfaithful recombination in BCR-ABL1–32Dcl3 cells by approximately 2-fold (Figure 6C). Moreover, pY315 aptamer reduced also the activity of HomoRR (supplemental Figure 2D).
Peptide aptamer containing RAD51(pY315) inhibits HomeoRR in BCR-ABL1-positive leukemia cells. BCR-ABL1–32Dcl3-pLB4 leukemia cells and CD34+ CML-CP primary cells were incubated with 2μM of rhodamine-labeled Y315F or pY315 aptamer, or left untreated (−). (A) Cellular uptake of the aptamers in BCR-ABL1–32Dcl3-pLB4 leukemia cells was visualized by fluorescence after 4-hour incubation (left boxes). DNA was counterstained with 4,6-diamidino-2-phenylindole (right boxes). (B) RAD51 nuclear foci were detected using anti-RAD51 antibody in BCR-ABL1–32Dcl3-pLB4 cells and CD34+ CML-CP cells as indicated. Data are number of RAD51 foci/cell ± SD. *P < .001 versus untreated cells (Mann-Whitney test). (C) HomeoRR activity in I-SceI transfected GFP+ cells. Data are percentage of G418-resistant clones ± SD. *P < .03 versus untreated cells (Student t test).
Peptide aptamer containing RAD51(pY315) inhibits HomeoRR in BCR-ABL1-positive leukemia cells. BCR-ABL1–32Dcl3-pLB4 leukemia cells and CD34+ CML-CP primary cells were incubated with 2μM of rhodamine-labeled Y315F or pY315 aptamer, or left untreated (−). (A) Cellular uptake of the aptamers in BCR-ABL1–32Dcl3-pLB4 leukemia cells was visualized by fluorescence after 4-hour incubation (left boxes). DNA was counterstained with 4,6-diamidino-2-phenylindole (right boxes). (B) RAD51 nuclear foci were detected using anti-RAD51 antibody in BCR-ABL1–32Dcl3-pLB4 cells and CD34+ CML-CP cells as indicated. Data are number of RAD51 foci/cell ± SD. *P < .001 versus untreated cells (Mann-Whitney test). (C) HomeoRR activity in I-SceI transfected GFP+ cells. Data are percentage of G418-resistant clones ± SD. *P < .03 versus untreated cells (Student t test).
Discussion
RAD51-mediated recombination plays a prominent role in directing DSB repair caused by reactive oxygen species, ionizing radiation and fragile sites in normal and malignant cells. In normal cells, RAD51-dependent HomoRR usually preserves survival and genomic stability; however, when aberrantly regulated in tumor cells, it may promote alternative DSB repair, intrachromosomal and interchromosomal deletions, chromosomal translocations, and contribute to malignant disease progression.30 The mechanisms responsible for an oncogene-induced compromising of the fidelity of RAD51-dependent HomoRR are largely unknown.
We reported before that BCR-ABL1 oncogenic kinase stimulated RAD51-mediated homologous recombination repair (HomoRR).19 However, point mutations may be acquired during otherwise faithful HomoRR in BCR-ABL1 leukemia cells, probably because of abnormal activity of unfaithful DNA polymerase(s), which use homologous fragment in sister chromatide or in homologous chromosome as a template to resynthesize a missing DNA fragment.28 Here we describe how BCR-ABL1 modulates RAD51 recombinase to promote unfaithful HomeoRR.
We found that the SH3 domain of BCR-ABL1 binds to the PPs of RAD51 in a kinase-independent manner, resulting in RAD51 phosphorylation on Y315, which subsequently facilitates disassembly of the complex. Phosphotyrosine-induced dissociation of the protein complexes was described before.31 Although RAD51(pY315) and also RAD51(pY54) were readily detectable in the cytoplasm of leukemia cells, only RAD51(pY315) was detected in the nuclei. Because RAD51(Y315F) phosphorylation-less mutant was present in the nuclear lysates comparable with the RAD51 WT, pY315 does not seem to regulate cytoplasmic-nuclear shuttling (when analyzed by subcellular protein fractionation followed by Western blotting). Instead, our results suggest that pY315 facilitated the self-assembly of RAD51 into nucleoprotein filaments (nuclear foci), which represent DNA repair sites.32 This observation is supported by the biochemical analyses indicating that Y315 regulated for chromatin association and/or filament formation by RAD51.33 In addition, because phosphorylation of Y54 may be secondary to Y315, the role of pY54 in foci formation cannot be excluded.26
RAD51-promoted strand exchange is not stringent and, if overstimulated, can easily bypass up to 3% divergence at the level of DNA sequence. BCR-ABL1-RAD51(pY315)–mediated enhancement of recombination foci formation was accompanied by stimulation of unfaithful HomeoRR which, instead of using a perfect homology sequence in sister chromatide, allows homeologous sequence as a template for DSB repair.17
We show here that unfaithful HomeoRR was triggered by the nonmutated BCR-ABL1 as well as by the TKI-resistant BCR-ABL1(T315I) kinase mutant in the “dosage” -dependent manner. This observation suggests a role of enhanced HomeoRR activity in genomic instability, especially in CML cells resistant to TKIs and in these expressing high levels of BCR-ABL1, such as leukemia stem cell (LSC)-enriched CD34+CD38− CML-CP cells and CML-BP cells. This speculation is supported by the fact that CML clones expressing TKI-resistant BCR-ABL1 kinase mutants and those overexpressing BCR-ABL1 are associated with clonal cytogenetic evolution.6,7
BCR-ABL1–induced RAD51(pY315) –directed HomeoRR may promote recombination involving homeologous sequences sharing high level of homology, for example, Alu elements, long interspersed repetitive elements, and low-copy repeats (also known as segmental duplications).34 Interestingly, high Alu frequency has been detected in various chromosomal breakpoints in CML.35 Unfaithful HomeoRR may be prevented by MMR, which however is inhibited in CML, thus creating an environment facilitating unfaithful recombination.36,37 Altogether, BCR-ABL1–mediated stimulation of RAD51(pY315)-dependent HomeoRR combined with impaired MMR may lead to unfaithful recombination, accumulation of chromosomal translocations, deletions, or inversions in CML.
CML-CP is an LSC-derived but a leukemia progenitor cell (LPC)-driven disease.38 If CML-CP progresses to CML-BP, LPCs also display the ability of self-renewal and to sustain leukemogenesis.39 Therefore, the role of RAD51(pY315) in genomic instability should be considered in different cell lineages. Genomic instability acquired in LSCs is particularly dangerous because all LPCs will eventually carry the aberrations generated in LSCs. In concordance, chromosomal aberrations have been detected in LSC-enriched CD34+CD38− cells and also in LPC-enriched CD34+ cells.40 In addition, the fact that CML-CP can progress to either myeloid or lymphoid blast crisis (sometimes a mix myeloid/lymphoid phenotype is observed), and that chromosomal abnormalities are documented in both phenotypes, suggests that genomic instability occurs at the level of LSCs and/or LPCs.41
Recombination is effective in S and G2/M cell cycle phase when the templates (sister chromatide or homologous chromosome) are available. Therefore, HomeoRR should be more active in TKI-resistant proliferating LPCs than in mostly quiescent LSCs; however, HomeoRR may occur in LSCs undergoing cell cycle progression to replenish their population and/or generate early LPCs. Elevated RAD51 and enhanced recombination was already described in leukemia stem cells displaying dysfunctional MMR, the environment potentially present in CML-CP LSCs.37,42,43 HomeoRR could be very active also in TKI-sensitive CD34+ LPCs because they can undergo up to one to 3 cell cycles and remains alive in the presence of TKI and growth factor.44 Moreover, TKIs may not significantly affect BCR-ABL1 kinase and its signaling pathways in LSCs and also in LPCs in the presence of physiologic concentrations of growth factors or in a protective environment, such as bone marrow niche.5,45
Because up-regulation of RAD51 recombinase may cause genomic instability, several approaches have been developed to inhibit/control its activity, for example, protein aptamer derived from BRC4 motif of BRCA2 to inhibit RAD51 filament formation and DNA aptamer and a chemical compound that inhibit RAD51-dependent pairing and strand exchange.46-48 However, these strategies will also target RAD51-mediated HomoRR in normal cells, which may exert a deleterious effect on cell survival and chromosomal stability.49 Instead, we propose to target the region of RAD51, RAD51(pY315), which is directly phosphorylated by BCR-ABL1 oncogenic kinase, preferentially detected in the nuclei of leukemia, but not normal cells, and involved in HomeoRR.
Interestingly, targeting RAD51(pY315) either by RAD51(Y315F) mutant or by pY315 aptamer exerted much stronger inhibitory effect on HomeoRR than HomoRR (50%-60% and 20%-30% inhibition, respectively). Although we cannot exclude the possibility that a weaker effect of targeting RAD51(pY315) on HomoRR may depend on technical issues (different cassette, integration site, cell clone), our results suggest that RAD51(pY315) may play a more prominent role during HomeoRR than HomoRR. If pY315 stabilizes RAD51 filament formation required for strand exchange, it may enhance stability of the filaments formed at low fidelity homeologous sequences, thus increasing the chances of unfaithful recombination, whereas the requirement for pY315 in the filaments formed at more stable high fidelity homologous sequences is less stringent.33 In support of this hypothesis, nuclear foci formed by RAD51(Y315F) phosphorylation-less mutant were approximately 2 times less likely to colocalize with RAD54 (data not shown), which may promote strand exchange through mismatches.50
In addition to BCR-ABL1, RAD51 recombinase activity could be modulated by tyrosine phosphorylation exerted by other OTKs.18-24 Thus, targeting OTK-mediated phosphotyrosine residues in overstimulated RAD51 recombinase may prevent unfaithful/excessive recombination not only in CML, but also in a variety of other tumors expressing OTKs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health/National Cancer Institute (grant CA123014; T.S.) and Ligue contre le Cancer (Comité 44; F.F.).
National Institutes of Health
Authorship
Contribution: A.S. performed the experiments and evaluated the data; Y.D., S.R., E.G., M.D., and M.N.-S. performed the experiments; F.F. generated anti-phosphoRAD51 antibodies and discussed the results; and T.S. designed research, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tomasz Skorski, Department of Microbiology and Immunology, School of Medicine, Temple University, 3400 N Broad St, MRB 548, Philadelphia, PA 19140; e-mail: tskorski@temple.edu.

![Figure 1. BCR-ABL1 kinase and the TKI-resistant T315I mutant promote HomeoRR in a “dosage”-dependent manner. (A) Homeologous recombination cassette contains a functional hygromycin gene (hyg), used to select for stably transfected cells, and a 3.9-kb tk-neo fusion gene rendered nonfunctional by the insertion of a 22-bp oligonucleotide containing 18-bp recognition site for endonuclease I-SceI (yellow strip). Three AluI restriction sited (red stripes) are present, too. In addition, a 2.5-kb fragment containing a complete HSV-1 tk gene, which is more than 1% divergent compared with that in the tk-neo fusion gene (0.8% and 1.5% divergent in 5′ and 3′ direction from the DSB site, respectively), is inserted. This inserted fragment serves as a potential recombination template (tk donor). HomeoRR by gene conversion between the I-SceI–induced DSB in the tk-neo fusion gene and the divergent tk donor template restores the function of neo gene encoding resistance to G418 (G418-R) and removes 2 AluI restriction sites closest to the DSB site.25 (B) Southern blot analysis of 32Dcl3-pLB4 parental cell clone (P) and BCR-ABL1–32Dcl3-pLB4 clone (B/A), in which a single copy of the pLB4 cassette was integrated in the genome. Genomic DNA was digested with HindIII and blotted with radiolabeled NcoI-NarI fragment of neo gene as a probe. (C) The 32Dcl3-pLB4 parental cells (P), cells expressing nonmutated BCR-ABL1 (B/A[nm]) and BCR-ABL1(T315I) (B/A[T315I]), and also cells expressing low (B/A-low) and high (B/A-high) levels of BCR-ABL1 were cotransfected with I-SceI (DSB inducer) and GFP (transfection control) expression vectors. HomeoRR activity is shown as percentage of G418-resistant cells in the GFP+ population. (D) Representative analysis of the recombination products by PCR (the primer positions are indicated by blue arrows in panel A) followed by AluI restriction digest. Left box: Nonrecombinant cassette produces 1393-bp product, which generates 653-, 381-, 337-, and 22-bp bands when digested by AluI. Right box: HomeoRR was confirmed in the mixture of G418-resistant clones by PCR amplifying 1371-bp HomeoRR products, which after AluI restriction digest generate characteristic recombination-specific 990-bp band and also 381-bp band (panel A, scheme of the cassette).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/4/10.1182_blood-2010-09-307256/4/m_zh89991175380001.jpeg?Expires=1769091794&Signature=K7G85urMBbqNgy01yVOngtX0qmNoH-ECh~iXl9iAtfyCJjI-7Rwy2~plVno2n8RCs1v16lbE6b2wE-DirjiBIitXLrdVIHpmnhPZjs5iET-lyz5CFW48WXJ5pGPQf~uU7KOApcV-tcaowpQ5SHN73eQac~K64ZmlJ-Xtphp70fhbVwOKJAh1N83irEVFyNQaJOyeTonMrhpEAVDpqs6u5zsd0--N-H6vArSu7D~hp1WCiGEcUeIYq4RGsitzIm-F89NRhnm1k51Qok2tVOF90QMm9dPJJK~TLjq6FzRms~PKUhoHo7PUO75sRV-Hn3Ju-uX8ZgstiQndzMx-tjM86A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal