Abstract
Myeloproliferative neoplasms (MPNs) are clonal disorders characterized by excessive production of mature blood cells. In the majority of classic MPN—polycythemia vera, essential thrombocythemia, and primitive myelofibrosis—driver oncogenic mutations affecting Janus kinase 2 (JAK2) or MPL lead to constitutive activation of cytokine-regulated intracellular signaling pathways. LNK, c-CBL, or SOCSs (all negative regulators of signaling pathways), although infrequently targeted, may either drive the disease or synergize with JAK2 and MPL mutations. IZF1 deletions or TP53 mutations are mainly found at transformation phases and are present at greater frequency than in de novo acute myeloid leukemias. Loss-of-function mutations in 3 genes involved in epigenetic regulation, TET2, ASXL1, and EZH2, may be early events preceding JAK2V617F but may also occur late during disease progression. They are more frequently observed in PMF than PV and ET and are also present in other types of malignant myeloid diseases. A likely hypothesis is that they facilitate clonal selection, allowing the dominance of the JAK2V617F subclone during the chronic phase and, together with cooperating mutations, promote blast crisis. Their precise roles in hematopoiesis and in the pathogenesis of MPN, as well as their prognostic impact and potential as a therapeutic target, are currently under investigation.
Introduction
The term “myeloproliferative disorders” (MPDs) was first introduced by William Dameshek in 1951 to describe 4 different diseases with clinical and biologic similarities: polycythemia vera (PV), essential thrombocythemia (ET), primary myelofibrosis (PMF), and chronic myeloid leukemia (CML).1 Later on, chronic neutrophil leukemia, hypereosinophilic syndromes, systemic mastocytosis, atypical chronic myeloid leukemia, and other rare chronic hemopathies were added. Certain disorders that combine myeloproliferation and myelodysplasia features, such as chronic myelomonocytic leukemia, are closely related to MPDs. In 2008, the World Health Organization recommended changing the term myeloproliferative disorders to myeloproliferative neoplasms (MPNs).2 In this review, we will focus on the current molecular knowledge of the BCR-ABL–negative MPNs PV, ET, and PMF.
All MPNs are clonal disorders with an initial hit in the HSCs resulting in an excessive production of blood cells because of hypersensitivity or independence from normal cytokine regulation.3,4 This myeloproliferation results from the absence of feedback regulation by mature cells, decreasing cytokine levels. MPNs involve the 3 main myeloid lineages but predominate in one of them: the erythroid lineage for PV, the megakaryocyte (MK)/platelet lineage for ET, and the MK/granulocytic lineages for PMF. MPNs are associated with a normal maturation but increased numbers of mature blood cells, except in PMF, in which abnormalities in MK differentiation might be responsible for the marrow fibrosis. A continuum exists between the BCR-ABL–negative MPN and myelodysplastic syndromes (MDS), as exemplified by chronic myelomonocytic leukemia (CMML), classified as MPN/MDS by the World Health Organization.
Several MPNs are the result of a genetic event leading to the constitutive activation of a tyrosine kinase that mimics the intracellular signaling pathways induced by hematopoietic growth factors. In CML, oligomerization of the BCR-ABL fusion protein via the BCR coiled-coil domain induces the constitutive activation of ABL that triggers many signaling pathways and drives the disease. Inhibition of the ABL kinase activity in patients results in disease regression, demonstrating that BCR-ABL is the oncogenic event responsible for the disease. Additional fusion proteins with constitutive tyrosine kinase activity involving PDGFRβ, fibroblast growth factor receptor, and PDGFRα, and more recently Janus kinase 2 (JAK2), have been described in hypereosinophilic syndromes (FIP1L1-PDGFRα) and in atypical MPNs.3 In addition, systemic mastocytosis involves gain-of-function mutations in the tyrosine kinase receptor c-KIT. Hence, it was likely that classic MPNs are also “kinase diseases.”
The founding mutations
JAK2V617F mutation
The JAK2 locus was found affected by a 9p loss of heterozygosity in 30% of PV patients (Table 1, Figure 1).5 In 2005 a major advance in the understanding of the pathogenesis of MPN was made by the discovery of the JAK2V617F mutation.6-9 The JAK family comprises 4 kinases (JAK1, 2, and 3 and TYK2) that attach to cytokine receptor cytosolic domains. JAK kinases possess 2 highly homologous domains at the carboxyl terminus: an active kinase domain (JAK homology, JH1) and a catalytically “inactive” pseudokinase domain (JH2). The JH2 domain is a negative regulator of the JH1 kinase activity.10 At the N-terminus, the JH5-JH7 domains contain a FERM (Band-4.1, ezrin, radixin, and moesin)–like motif, which plays a role in the binding to the cytosolic domain of cognate cytokine receptors. JAK2 plays a central role in the signaling from “myeloid” cytokine receptors. It binds to the 3 homodimeric “myeloid” receptors (erythropoietin receptor [EPO-R], myeloproliferative leukemia [MPL; TPO-R], G-CSF receptor [G-CSF-R]), to the prolactin and growth hormone receptors, to heterodimeric receptors (GM-CSF-R, IL-3-R, and IL-5-R, which share the common β chain of IL-3-R and the gp130 family of receptors), and to IFN-γ R2. JAK2 is the only JAK capable of mediating the signaling of EPO-R and MPL. JAK2 also functions as a chaperone for trafficking of these 2 receptors to the cell surface and their stability.43 More recently, JAK2 was also shown to promote G-CSF-R cell-surface localization.44 Therefore, JAK2 and the 3 “myeloid receptors” form functional units and have been shown to be required for the promotion of JAK2V617F signaling.45
Genes involved in the pathogenesis of MPN
| Gene . | Localization . | Function . | Comment . | Disease (frequency) . | References . |
|---|---|---|---|---|---|
| JAK2 | 9p24 | Tyrosine kinase, signaling | Gain of function | PV (95%-99%), ET (50%-70%), PMF (40%-50%) | 6,,–9,11,12 |
| MPL | 1p34 | Receptor, signaling | Gain of function | ET (4%), PMF (11%) | 13,,–16 |
| LNK | 12q24 | Adaptor, signal regulation | Loss of function | ET (< 5%), PMF (< 5%), JAK2 mutation-negative erythrocytosis (25%), post-MPN AML (13%) | 17,–19 |
| CBL | 11q23 | Adaptor, E3 ubiquitin ligase, signal regulation | Dominant negative | PMF (6%), post-MPN AML | 20,21 |
| SOCS1 | 16p13.2 | E3 ubiquitin ligase, signal regulation | Methylation | ET (14%-25%), PV (11%-13%), PMF (17%) | 22,23 |
| SOCS2 | 12q22 | E3 ubiquitin ligase, signal regulation | Methylation | All MPN (28%) | 24 |
| SOCS3 | 17q25.3 | E3 ubiquitin ligase, signal regulation | Methylation, mutation | ET (10%), PV (22%) | 22,25,–27 |
| NRAS | 1p13.2 | GTPase, signaling | Gain of function | post-MPN AML (7%-13%) | 21 |
| NF1 | 17q11.2 | RAS signaling regulation | Deletion | PMF (0%-6%), post ET/PV MF (14%) | 28,29 |
| TET2 | 4q24 | DNA hydroxymethylation | Loss of function | PV (15%), ET (4%-11%), PMF (19%), post-MPN AML (26%) | 30,,–33 |
| ASXL1 | 20q11.21 | Chromatin modifications | Loss of function | PV and ET (< 7%) PMF (19%-40%), post-MPN AML (19%) | 34,35 |
| EZH2 | 7q35 | Chromatin methylation | Loss of function | PV (3%), PMF (13%) | 36,–38 |
| IKZF1 | 7p12 | Transcription factor, lymphopoiesis | Deletion | post-MPN AML (21%) | 39 |
| RUNX1 | 21q22.3 | Transcription factor, hematopoiesis | Loss of function | post-MPN AML (37%) | 21,40 |
| RB | 13q14 | Cell cycle, apoptosis | Deletion | PMF (19%) | 29 |
| TP53 | 17p13.1 | Cell cycle, apoptosis | Loss of function | post-MPN AML (20%) | 21,41 |
| IDH1 | 2q33.3 | Metabolism | Neomorphic enzyme | PMF (2%), post-MPN AML (8%) | 42 |
| IDH2 | 15q26.1 | Metabolism | Neomorphic enzyme | PMF (2%), post-MPN AML (18%) | 42 |
| Gene . | Localization . | Function . | Comment . | Disease (frequency) . | References . |
|---|---|---|---|---|---|
| JAK2 | 9p24 | Tyrosine kinase, signaling | Gain of function | PV (95%-99%), ET (50%-70%), PMF (40%-50%) | 6,,–9,11,12 |
| MPL | 1p34 | Receptor, signaling | Gain of function | ET (4%), PMF (11%) | 13,,–16 |
| LNK | 12q24 | Adaptor, signal regulation | Loss of function | ET (< 5%), PMF (< 5%), JAK2 mutation-negative erythrocytosis (25%), post-MPN AML (13%) | 17,–19 |
| CBL | 11q23 | Adaptor, E3 ubiquitin ligase, signal regulation | Dominant negative | PMF (6%), post-MPN AML | 20,21 |
| SOCS1 | 16p13.2 | E3 ubiquitin ligase, signal regulation | Methylation | ET (14%-25%), PV (11%-13%), PMF (17%) | 22,23 |
| SOCS2 | 12q22 | E3 ubiquitin ligase, signal regulation | Methylation | All MPN (28%) | 24 |
| SOCS3 | 17q25.3 | E3 ubiquitin ligase, signal regulation | Methylation, mutation | ET (10%), PV (22%) | 22,25,–27 |
| NRAS | 1p13.2 | GTPase, signaling | Gain of function | post-MPN AML (7%-13%) | 21 |
| NF1 | 17q11.2 | RAS signaling regulation | Deletion | PMF (0%-6%), post ET/PV MF (14%) | 28,29 |
| TET2 | 4q24 | DNA hydroxymethylation | Loss of function | PV (15%), ET (4%-11%), PMF (19%), post-MPN AML (26%) | 30,,–33 |
| ASXL1 | 20q11.21 | Chromatin modifications | Loss of function | PV and ET (< 7%) PMF (19%-40%), post-MPN AML (19%) | 34,35 |
| EZH2 | 7q35 | Chromatin methylation | Loss of function | PV (3%), PMF (13%) | 36,–38 |
| IKZF1 | 7p12 | Transcription factor, lymphopoiesis | Deletion | post-MPN AML (21%) | 39 |
| RUNX1 | 21q22.3 | Transcription factor, hematopoiesis | Loss of function | post-MPN AML (37%) | 21,40 |
| RB | 13q14 | Cell cycle, apoptosis | Deletion | PMF (19%) | 29 |
| TP53 | 17p13.1 | Cell cycle, apoptosis | Loss of function | post-MPN AML (20%) | 21,41 |
| IDH1 | 2q33.3 | Metabolism | Neomorphic enzyme | PMF (2%), post-MPN AML (8%) | 42 |
| IDH2 | 15q26.1 | Metabolism | Neomorphic enzyme | PMF (2%), post-MPN AML (18%) | 42 |
AML indicates acute myeloid leukemia; ET, essential thrombocythemia; JAK2, Janus kinase 2; MPN, myeloproliferative neoplasms; PMF, primary myelofibrosis; and PV, polycythemia vera.
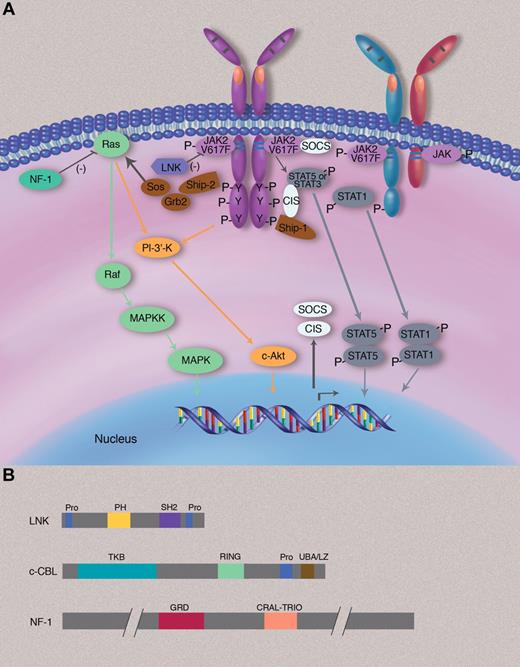
Signaling pathways involved in the pathogenesis of MPNs. (A) JAK2V617F is attached to the cytosolic juxtamembrane region of dimeric cytokine receptors, such as EpoR or MPL (TpoR) but can also be attached to the cytosolic region of other JAK2-using cytokine receptors, such as the IFN-γ receptor 2 chain. When bound to homodimeric cytokine receptors, JAK2V617F induces constitutive signaling via STAT5, STAT3, RAS-MAPK, and PI-3′K-Akt pathways, which all regulate gene expression and promote survival, proliferation, and differentiation of committed myeloid progenitors. Constitutive STAT5 and STAT3 signaling is frequently described in MPN myeloid cells, and constitutive STAT1 signaling has been found in certain cases such as erythroid colonies of ET patients. Negative regulators such as CIS (cytokine-inducible SH2) and SOCS proteins (such as SOCS3) are transcriptionally induced by the activated JAK2 but in general appear to be overwhelmed and cannot efficiently block constitutive JAK2V617F signaling. LNK exerts a negative effect on JAK2V617F signaling and constrains the MPN phenotype. Only a minority of patients have LNK mutations. Activation of RAS signaling is counteracted by NF1 (neurofibromatosis 1), a protein that stimulates the GTPase activity of RAS and that is deleted in a minority of MPN patients. (B) Domain structure of negative signaling regulators LNK, c-CBL, and NF-1. Pro indicates proline-rich domain; PH, plekstrin homology domain; SH2, src-homology 2; TKB, tyrosine kinase binding domain; RING, really interesting new gene finges domain; GRD, GAP (GTPase-activating domain)–related domain; and CRAL-TRIO, cellular retinaldehyde and TRIO domain.
Signaling pathways involved in the pathogenesis of MPNs. (A) JAK2V617F is attached to the cytosolic juxtamembrane region of dimeric cytokine receptors, such as EpoR or MPL (TpoR) but can also be attached to the cytosolic region of other JAK2-using cytokine receptors, such as the IFN-γ receptor 2 chain. When bound to homodimeric cytokine receptors, JAK2V617F induces constitutive signaling via STAT5, STAT3, RAS-MAPK, and PI-3′K-Akt pathways, which all regulate gene expression and promote survival, proliferation, and differentiation of committed myeloid progenitors. Constitutive STAT5 and STAT3 signaling is frequently described in MPN myeloid cells, and constitutive STAT1 signaling has been found in certain cases such as erythroid colonies of ET patients. Negative regulators such as CIS (cytokine-inducible SH2) and SOCS proteins (such as SOCS3) are transcriptionally induced by the activated JAK2 but in general appear to be overwhelmed and cannot efficiently block constitutive JAK2V617F signaling. LNK exerts a negative effect on JAK2V617F signaling and constrains the MPN phenotype. Only a minority of patients have LNK mutations. Activation of RAS signaling is counteracted by NF1 (neurofibromatosis 1), a protein that stimulates the GTPase activity of RAS and that is deleted in a minority of MPN patients. (B) Domain structure of negative signaling regulators LNK, c-CBL, and NF-1. Pro indicates proline-rich domain; PH, plekstrin homology domain; SH2, src-homology 2; TKB, tyrosine kinase binding domain; RING, really interesting new gene finges domain; GRD, GAP (GTPase-activating domain)–related domain; and CRAL-TRIO, cellular retinaldehyde and TRIO domain.
The JAK2V617F mutation results from a guanine to thymine change at nucleotide 1849 of the cDNA, in exon 14 of the gene. This valine is located at one of the predicted interfaces between JH1 and JH2 domains,46 and the change to a phenylalanine appears to relieve the inhibition of the JH2 domain on the kinase domain. There is evidence that constitutive signaling by JAK2V61F requires the homodimeric receptor, explaining why the JAK2V617F-induced proliferation affects 3 myeloid lineages.45 Indeed, the JAK2V617F mutation has been found in the majority of BCR-ABL–negative MPNs (∼ 95% of patients with PV, 50%-70% with ET, and 40%-50% with PMF), as well as in some cases of atypical MPN (30%-50% splanchnic vein thrombosis and sideroblastic anemia associated with a thrombocytosis).47
Beside its role in the cytokine receptor signaling cascade (Figure 1), JAK2 has been shown to influence chromatin structure (Figure 2).48,49 In hematopoietic cells, nuclear JAK2 phosphorylates histone H3Y41, thereby blocking recruitment of the repressor heterochromatin protein 1α and allowing increased expression of several genes, including the LMO2 oncogene.50 In the same line, JAK2V617F has been recently described to interact with and phosphorylate the protein arginine methyltransferase PRMT5 with a much greater affinity than wild-type JAK2.51 This property is specific (or enhanced) for the mutant protein and has been shown to disrupt the interaction between PRMT5 and its cofactor MEP50, leading to a decreased methyltransferase activity. The knockdown of PRMT5 increases colony formation and erythroid differentiation of primary cells.51 This emerging nuclear role of mutated JAK2 may reveal mutant-specific chromatin effects that may open a novel therapeutic window.
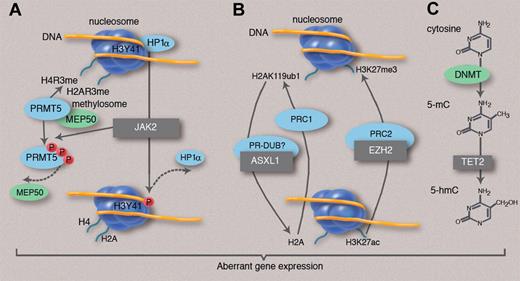
New mutations and epigenetic modifications in MPNs. Several genes mutated in MPN (black boxes) are implicated either in histone modifications or in DNA methylation control. (A) JAK2 activation phosphorylates histone H3Y41, leading to the exclusion of heterochromatin protein 1α (HP1α) from chromatin. In addition, mutant JAK2 phosphorylates PRMT5, thus impairing the methylation of histone H4 and H2A arginine residues. Both phosphorylations are expected to facilitate gene transcription or reduce gene repression. (B) EZH2 belongs to the PRC2 complex, which methylates H3K27 and may also recruit DNMTs. ASXL1 may belong to a complex (PR-DUB in Drosophila melanogaster) that deubiquitinates histone H2AK119, a function that antagonizes the effect of the PRC1 complex. Inactivation of both genes is expected to prevent transcriptional repression mediated by the PRC complexes. (C) TET2 converts DNA 5mC to 5hmC, thus playing a role in active DNA demethylation that would be associated with gene expression. Activation mutations of JAK2 and mutations impairing EZH2, ASXL1, and TET2 functions may result in the deregulation of both DNA methylation and chromatin structure, resulting in aberrant gene expression, gene activation, or failure of repression.
New mutations and epigenetic modifications in MPNs. Several genes mutated in MPN (black boxes) are implicated either in histone modifications or in DNA methylation control. (A) JAK2 activation phosphorylates histone H3Y41, leading to the exclusion of heterochromatin protein 1α (HP1α) from chromatin. In addition, mutant JAK2 phosphorylates PRMT5, thus impairing the methylation of histone H4 and H2A arginine residues. Both phosphorylations are expected to facilitate gene transcription or reduce gene repression. (B) EZH2 belongs to the PRC2 complex, which methylates H3K27 and may also recruit DNMTs. ASXL1 may belong to a complex (PR-DUB in Drosophila melanogaster) that deubiquitinates histone H2AK119, a function that antagonizes the effect of the PRC1 complex. Inactivation of both genes is expected to prevent transcriptional repression mediated by the PRC complexes. (C) TET2 converts DNA 5mC to 5hmC, thus playing a role in active DNA demethylation that would be associated with gene expression. Activation mutations of JAK2 and mutations impairing EZH2, ASXL1, and TET2 functions may result in the deregulation of both DNA methylation and chromatin structure, resulting in aberrant gene expression, gene activation, or failure of repression.
The discovery of JAK2V617F mutation is an important breakthrough in the understanding of BCR-ABL–negative MPN and has demonstrated the role of pathologic signaling by the JAK/STAT pathway in MPN. Two other mutations directly affecting this pathway have subsequently been described.
JAK2 exon 12 mutations
In the rare JAK2V617F-negative PV, different somatic gain-of-function mutations in exon 12 of JAK2 have been found.11,12 These mutations span the linker region between the SH2 and JH2 domains. Although not located in the pseudokinase domain, these mutations may modify the structure of the JH2 domain in a very similar fashion as V617F. Along these lines, residue F595, located in the helix C of the pseudokinase domain, was shown to be required for both V617F and K539L mutants but not for cytokine-induced JAK2 activation.52 However, in contrast to JAK2V617F, exon 12 mutations are not associated with ET and PMF, although JAK2 exon 12 PV may progress to a secondary myelofibrosis.53
MPL mutations
Several gain-of-function mutations of MPL have been found in exon 10, which result in the substitution of a tryptophan 515 to a leucine, lysine, asparagine, or alanine.13-16 Amino acid 515 is located in a stretch of 5 amino acids (K/RWQFP) found inside the cytoplasm just after the transmembrane domain. These 5 amino acids play a major role in the cytosolic conformation of MPL and prevent spontaneous activation of the receptor.54,55 In addition, the MPLS505N mutation, initially described in familial ET, was also found in sporadic MPN.13,16 These MPL mutations have been found in up to 15% of JAK2 V617F-negative ET or PMF.56
Subsequent modeling of the disease has shown that JAK2 and MPL mutations were able to drive the disease in mice. In mouse models of JAK2V617F, both retroviral transplantation assays and transgenic models, including constitutive or inducible knockin approaches, demonstrated the development of a MPN, usually a PV progressing to myelofibrosis.57-63 However, in some models, an ET-like disorder, usually transient, was observed.57,61,64 This phenotype corresponds to a weak JAK2V617F expression, supporting the hypothesis that phenotypic heterogeneity of the JAK2V617F-induced diseases might be because of the intensity of JAK2 signaling, especially to the number of JAK2V617F copies.61 This assumption fits with the human situation in which most patients with ET have a single JAK2V617F mutation, whereas in most PV cases, 2 copies of JAK2V617F are present because of uniparental disomy.65,66 In addition, strain-specific variations were observed in the retroviral mouse model, suggesting that host modifier genes were involved in the phenotype of the disease.57,58,67,68
This finding also fits with recent results in humans in which JAK2V617F-heterozygous erythroid cells from PV and ET patients exhibit different gene expression profiles, with erythroid cells from ET patients displaying an IFN-γ–stimulated gene expression pattern.69 This is due to the fact that ET and not PV erythroid colonies are characterized by constitutive activation of STAT1, possibly via activation of IFN-γ receptor 2 or another STAT1-activating receptor.69 This finding plus other data suggest the existence of a modifier gene in the human disease. In MPL, BM transplantation assays with W515L or W515A mutations led to an ET-like disorder rapidly progressing to myelofibrosis.13,55 Thus, BCR-ABL–negative MPN resembles CML with some significant differences.
Genes involved in intracellular signaling
LNK
LNK, also called SH2B3, is a member of the SH2B family, which contains 2 other members (Table 1, Figure 1).70 This family of adaptor proteins is characterized by a conserved structure with 3 main domains (a proline-rich amino terminus, a plekstrin homology domain, an SH2 domain) and a conserved tyrosine at the C-terminus. LNK plays an important role in hematopoiesis by negatively regulating JAK2 activation through its SH2 domain, thus inhibiting EPO-R and MPL signaling.71,72 In addition, LNK negatively regulates c-KIT and FMS signaling.73 LNK-deficient mice have an increased HSC pool with enhanced self-renewal properties and increased quiescence.74 This phenotype probably results from increased TPO/MPL signaling75 because TPO is required for maintaining HSC quiescence and the HSC reservoir.76,77 In addition, Lnk−/− mice develop MPN with thrombocytosis, splenomegaly, and fibrosis78 and marked B-cell overproduction.
As expected from its negative role in JAK2 signaling, LNK is also capable of attenuating the signaling induced by MPLW515L or JAK2V617F.79 Loss of LNK accelerates the development of MPN induced by JAK2V617F in murine models.80 In JAK2V617F-positive patients, LNK expression is increased and modulates the myeloproliferative process.81 Recently, Oh et al17 identified 2 mutations in LNK exon 2, one in a patient with PMF and the other with ET; both MPNs were JAK2V617F negative. The first mutation leads to a premature stop codon resulting in the absence of the PH and SH2 domains, whereas the second (E208Q) is a missense mutation in the PH domain (Figure 1B). In the first mutation, the capacity to inhibit TPO signaling is lost, whereas in the second mutation, some inhibitory function is maintained.
The frequency of mutations in LNK is low.17 However, other mutations of LNK have been found in leukemic transformation of MPN at a greater frequency (∼13%).18 All mutations, except one, target a hot spot located between codons 208 and 234. Interestingly, some of these mutations appear to be late events involved in disease progression because they were not found in the chronic phase.18 In addition some LNK mutations were associated with JAK2V617F, although it is not known whether LNK mutants and JAK2V617F were present in the same cell. It has been also reported that LNK exon 2 mutations can be found in pure erythrocytosis.19 One mutation (A215V) had been previously described in PMF blast crisis, and another (E208X) leads to absence of the PH and SH2 domains as the mutant described in PMF.17 This finding may suggest that the phenotype of the MPN induced by LNK mutations may depend on different parameters, including the presence of other mutations.
Casitas B-cell lymphoma
The Casitas B-cell lymphoma (CBL) family includes 3 homologs: c-CBL, CBL-b, and CBL-c.82 c-CBL, the founding member, is the cellular counterpart of a murine viral oncogene involved in B-cell and myeloid malignancies. CBL proteins are multifunctional adapter proteins with ubiquitin ligase activity. They are usually involved in negative regulation of receptor tyrosine kinase (RTK) by competitive blocking of signaling and they induce RTK proteosomal degradation by mediating ubiquitination in endosomes. However, CBL may have numerous targets other than RTK, including JAK2 and cytokine receptors such as MPL.83 c-CBL and CBL-b proteins contain several domains: an N-terminal tyrosine kinase binding domain followed by a Ring domain, which is important for the transfer of ubiquitin moieties (Figure 1B). A linker separates these 2 domains. The C-terminus part contains a proline-rich domain involved in the binding of several SH3 proteins. Finally, a C-terminal UBA/LZ domain is implicated in CBL oligomerization and ubiquitin binding.82 CBL-c is the member of the family devoid of the C terminal domains. c-CBL is located at 11q.23.3 and is mutated in a variety of myeloid malignancies.84,85
The greatest frequency of mutations is found in CMML and juvenile myelomonocytic leukemia. In acute myeloid leukemia (AML), the transforming activity of c-CBL may be related to an increased FLT3 signaling.86 Usually variants are missense mutations, which are homozygous because of an acquired uniparental disomy87 or, rarely, because of a deletion of the wild-type copy. For these reasons, CBL has been considered a tumor suppressor gene. In fact, most mutated CBL forms behave as loss-of-function molecules having a dominant-negative effect not only on c-CBL but also on CBL-b, leading to an excessive sensitivity to a variety of growth factors.88 Retroviral transplantation assays with mutated c-CBL induce a mastocytosis phenotype and myeloid leukemia, albeit with a long latency. Similarly, c-CBL knockout mice develop a mild MPN with an increase in HSC. Cbl-b–deficient mice lack a hematologic phenotype.89 The double knockout leads to a rapidly lethal MPN with leukocytosis and excess of monocytes, a phenotype close to myelo-monocytic leukemia.89
In the chronic phase of classic MPN, c-CBL mutations have been found in a low percentage of PMF patients (6%) but were not detected in a small series of PV and ET patients.20 In one case, the c-CBL mutation occured after JAK2V617F. However, during progression of the disease, JAK2V617F was outcompeted by the CBL mutant, suggesting that the 2 mutations had occurred in 2 different cells.20 Similarly a c-CBL mutation has been detected in blasts from a JAK2V617F-positive MPN, which became JAK2V617F negative during transformation.21 Presently, c-CBL seems to be involved more in progression toward myelofibrosis or acute leukemia than in the chronic phase of the disorder, but further studies are required to establish its precise role.
Suppressor of cytokine signaling 1, 2, and 3
Suppressor of cytokine signaling (SOCS) proteins are also important negative regulators of JAK signaling through a classic feedback loop.90 Loss of SOCS activity induces excess signaling by cytokines. SOCS1-inactivating mutations have been described in B-cell lymphoma.91 Some mutations in the different SOCS have been found in MPNs, but they seem rare.25 In contrast, hypermethylation of CpG islands in SOCS1 and SOCS3 associated with a decrease in expression was found in JAK2V617F PV and ET as well as in JAK2V617F and MPLW515-mutation negative ET22,23 Treatment with 2′-deoxyazacytidine restored SOCS1 and SOCS3 expression. SOCS1 not only regulates cytokine signaling90 but may also be implicated in direct activation of p53-dependent senescence.92 It was suggested that the association between SOCS3 promoter methylation and diminution of transcription is found specifically in JAK2V617F-negative patients with PMF.26 However, the role of SOCS3 in regulating JAK2V617F signaling is controversial. On the one hand, it has been reported that SOCS3 inhibits JAK2V617F signaling through proteosomal degradation.93 On the other hand, when JAK2V617F-transformed hematopoietic cell lines were used, it was reported that JAK2V617F mutant kinase escapes SOCS3-negative regulation by inducing hyperphosphorylation of the SOCS box, thereby blocking its interaction with Elongin C and stabilizing JAK2V617F.94 Similar results have been obtained with exon 12 JAK2 mutant kinases.95 SOCS2 may also inhibit JAK2V617F signaling, and its promoter is hypermethylated in some MPN.24 Presently, the role of SOCS proteins in the pathogenesis of MPN remains unclear: Is the epigenetic silencing a cooperating event with JAK2V617F27 or is it at the origin of some MPNs such as ET?
Genes involved in leukemic progression
Leukemic transformation, or blast phase, occurs in approxiamtely 15% of PMF patients and in < 10% of PV and ET patients (Table 1).96-98 To identify the main actors of leukemic transformation in MPN, several teams have conducted comprehensive genotypic analyses of post–MPN-AML cells, with particular attention to both MPN- and AML-related genes.97-99 To fit the definition of a blast phase-transforming event, the candidate event had to be detected in all blast cells but not in the majority of cells from the preceding MPN clone.21
IDH1/2 mutations
IDH1 and IDH2 encode isocitrate dehydrogenase 1 and 2, which are NADP+ enzymes that catalyze the conversion of isocitrate to α-ketoglutarate (αKG).100,101 Heterozygous mutations of IDH1 (R132S, R132G) and IDH2 (codon R140 and R172) have been identified in AML.102,103 Mutated IDH1/2 are neomorphic enzymes that catalyze the reduction of αKG to (R)-2-hydroxyglutarate. A subsequent overproduction of (R)-2-hydroxyglutarate has been proposed to affect the function of αKG-dependent enzymes such as TET2, resulting in a decrease of 5hmc.104 Although the effects of IDH1/2 mutations on HSCs and progenitors remain unknown, these mutations may belong to a set of initiating events of the same nature as the TET2 mutations. The analysis of 1473 patients with MPN revealed a low incidence of IDH1/2 mutations in chronic phase ET, PV, and MF (0.8%, 1.9%, and 4.2%, respectively) contrasting with a 21.6% frequency in blast phase.42 Thus IDH1/IDH2 mutations are associated with MPN transformation, but clonal hierarchy and follow-up studies are needed to better define whether they promote the initiation or the progression of MPN to AML.
IKZF deletion
The IKZF1 gene encodes for the Ikaros transcription factor, which regulates the development of B and T cells.105 IKZF1 deletions are frequent in acute lymphoblastic leukemia, especially in BCR-ABL–positive cases.106 Hemizygous deletions of the IKZF1 gene region on chromosome 7p were observed in 1 of 437 patients with chronic-phase MPN and 6 of 29 patients with post–MPN-AML.39 Clonal hierarchy analysis showed that in some cases IKZF1 deletion was a late event in the progression of the disease and was frequently related to a complex karyotype. Thus, like in post–CML- acute lymphoblastic leukemia, IKZF1 deletions in post–MPN-AML are late events in the progression of MPN to AML.
NRAS/KRAS mutations and NF1 deletion
RAS proteins are membrane-associated GTPases that in their active form trigger a variety of effector signaling pathways, such as the MAPK cascade of serine/threonine kinases. The most frequent mutations in KRAS and NRAS occur at codons 12, 13, and 61, leading to an inhibition of the GTP-ase activity, thus allowing constitutive activation of effector pathways such as MAPK signaling and downstream transcriptional control. It is still unknown whether RAS mutations promote the initiation or the progression of myeloid malignancies to acute leukemia. In MPN, this question remains unresolved because NRAS mutations were found in post–MPN-AML blasts and in cells from the preceding chronic phase, but also, in some patients, exclusively in AML blasts at transformation.21 NF1 deletions were mainly found in myelofibrosis and rarely in PV or ET.28,29 Their frequency in leukemic transformation is unknown.
TP53 mutations
TP53 gene encodes p53, a major tumor suppressor protein involved in various biologic activities, including the control of cell-cycle checkpoints and apoptosis. Germline loss-of-function mutations in TP53 predispose patients to a multiplicity of cancers, and acquired mutations in p53 occur in approximately 10% of AML samples.107 TP53 mutations are not associated with the chronic phase of MPNs. However, mutated TP53 have been found with a 20% frequency in post–MPN-AML patients.21,41,108 In one particular case with both MPL and TET2 mutations during chronic-phase ET, multiple TP53 mutations were identified at the time of post–ET-AML that resulted in an oligoclonal pattern in progenitors, with one selected subclone giving rise to leukemic blasts.109 Thus, the picture of TP53 mutations in post–MPN-AMLs suggests that they play a prominent role in the transformation process.
RUNX1 mutations
The RUNX1/AML1 gene encodes a transcription factor with a major role in hematopoiesis. It was found mutated in MDS and AML.110 In a series of 34 post–MPN-AML patients, RUNX1 mutations were found in leukemic blasts of 11, most of which were localized in the RUNT domain (residues 50-177).18,37 This observation suggests that RUNX1 mutation may represent one of the most frequent genetic events implicated in MPN transformation to AML.
Mutations in “epigenetic regulator genes”: a pre-JAK2 event?
An emerging hypothesis posits that mutations in signaling molecules are not sufficient for disease development in humans and that several cooperating genetic hits might be required to induce disease and allow progression (Table 1, Figure 2). This hypothesis was determined by 4 lines of evidence:
Familial MPN has been described with a dominant-autosomal transmission.111,112 The phenotype of these familial forms correspond either to a classic MPNs (PV, ET) in the absence of germinal transmission of JAK2V617F or MPLW515L, or to a larger spectrum of diseases (CML, systemic mastocytosis and also acute leukemia).111,113
Certain ET and rare PV were clonal as judged by X inactivation assays, whereas JAK2V617F was present only in a minority of cells, suggesting the existence of a pre-JAK2 clone, in which the JAK2V617F mutation has occurred.114,115 In some JAK2V617F MPNs associated with the 20q deletion, this deletion was present in the majority of cells, whereas the JAK2V617F burden was low116 ; furthermore, JAK2V617F was not found in some erythropoietin-independent colonies.115
More surprisingly, it was initially reported that approximately 50% of acute leukemia developing on a JAK2V617F MPNs was JAK2 wild type, even in the absence of previous chemotherapy.97,117 This finding suggests the existence of a pre-JAK2 clone, on the background of which 2 independent genetic events have occurred, one being the JAK2V617F mutation leading to a MPN and another one being a genetic alteration leading to leukemia.
Hematopoiesis of JAK2V617F MPN is quite surprising for a clonal disorder involving a HSC. In most PV and ET at diagnosis, the allele burden is extremely low in HSC, and the clonal dominance only occurs during late stages of hematopoiesis, corresponding to the maximum activity of hematopoietic cytokines (Figure 3).66,118 In contrast, JAK2V617F HSCs predominate in advanced MPNs such as PMF or secondary myelofibrosis.119,120 This finding suggests that JAK2V617F confers only a weak advantage to HSC.
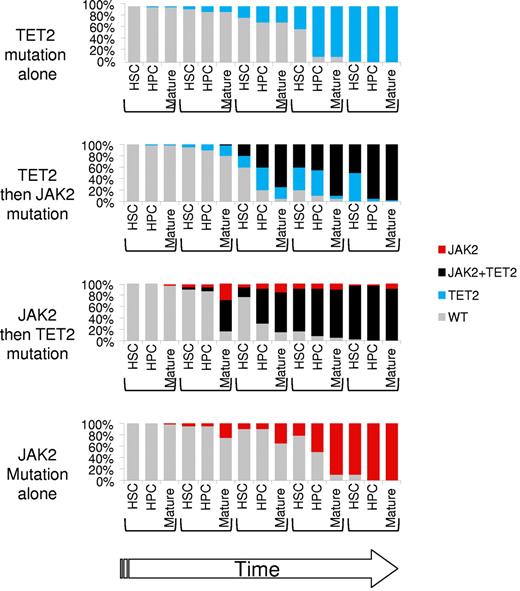
Dynamic representation of clonal dominance in the presence of TET2 and/or JAK2 mutations in the HSC, hematopoietic stem progenitor (HPC), and mature fractions. JAK2V617F would mainly expand the mature fraction, whereas TET2 mutation would mainly expand the HPC fraction. TET2 and JAK2 mutations are synergistic by combining an early and late amplification. The order of mutations would not matter if they occur before the development of a full-blown disorder. All numbers and ratios are arbitrary.
Dynamic representation of clonal dominance in the presence of TET2 and/or JAK2 mutations in the HSC, hematopoietic stem progenitor (HPC), and mature fractions. JAK2V617F would mainly expand the mature fraction, whereas TET2 mutation would mainly expand the HPC fraction. TET2 and JAK2 mutations are synergistic by combining an early and late amplification. The order of mutations would not matter if they occur before the development of a full-blown disorder. All numbers and ratios are arbitrary.
As a consequence of the development of whole genome assays (first comparative genomic hybridization and single nucleotide polymorphism arrays and more recently whole genome sequencing), an increasing number of mutations have been observed in BCR-ABL–negative compared with BCR-ABL–positive MPNs, and these mutations involve genes modifying epigenetic regulation. Mutations in these genes are also found in a great variety of myeloid malignancies, including MDS and AML, suggesting a common pathogenesis in the 3 disorders but implying that these mutated genes are not directly involved in the phenotype of the myeloid malignancies.
Genes that might be mutated before JAK2 in MPNs normally participate in the epigenetic control of transcription. It is not known whether all pre-JAK2 mutations yet to be identified will fall into this category.
EZH2
EZH1 and EZH2 proteins (enhancer of zeste homolog) belong to the polycomb repressive complex 2 (PRC2).121 PRC2 is involved in various cellular processes, including proliferation, differentiation, cell-identity maintenance, aging, and plasticity.121,122 In addition to EZH1 or EZH2, PRC2 includes EED, RbAp46/48, SUZ12, AEBP2, JARID2, and PCL. It is thought that PRC2 contributes to chromatin structure regulation. PRC2 methylates histone H3 at lysine 27, a mark of inactive chromatin, through the SET domain of EZH1 or EZH2.123 Its interaction with DNA in mammals is still elusive but may involve YY1 and long intergenic noncoding (ie, Linc) RNAs. It also associates to DNA-methyltransferase (DNMT) proteins to direct DNA methylation.
EZH2 codes for 1 of the 2 possible catalytic subunits of PRC2. EZH proteins are methyltransferases involved in the di- and trimethylation of K27.123-125 EZH2 overexpression is observed in numerous solid tumors such as breast and prostate cancers and may induce dedifferentiation.126 EZH2 is mutated in B-cell lymphomas, but the recurrent mutation targeting Y641 results in a gain of function: the mutated enzyme preferentially generates trimethylated K27.127 In contrast, in myeloid disorders EZH2 mutations may be associated with deletion of the other copy or preferentially with loss of heterozygosity.36-38 EZH2 mutations are predicted to be inactivating through either the truncation of the protein or modification of essential amino acids. They are not associated with a specific MPN or MDS subtype but might be more frequent in MPD/MPNs and are associated with poor prognosis. EZH2 mutations are not observed in ET, but are seen in 3% of PV and 13% of MF.36
ASXL1
ASXL1, together with ASXL2 and ASXL3, are related to the Drosophila melanogaster additional sex combs (Asx), a polycomb gene required for long-term repression of the HOX genes.128 Asx has recently been identified in a new polycomb complex (PR-DUB) able to deubiquitinate histone H2A through the activity of the BAP1 protein.129 This activity is the converse of the monoubiquitination mediated by the PRC1 complex, and the balance between the 2 activities is important in the regulation of target genes, such as the HOX genes.121 It is not known whether ASXL1 has a similar function in mammals.
Mutations in ASXL1 are frameshifts and stop mutations located within the 12th exon of the gene; they usually affect only one copy of the gene and result in the loss of the carboxyterminal PHD domain.34,130 The frequency is low in low-grade disorders but greater in late MDS, AML, and in proliferative CMML. However, the veracity of the most frequently reported ASXL1 mutation has been recently questioned.131 In MPNs, ASXL1 mutations are rare in ET and PV (< 7%) but frequent in PMF (from 19%-40%).34,35
The function of ASXL1 in hematopoiesis is still poorly understood. Asxl1 knockout mice have a mild defect in hematopoiesis, predominantly in lymphopoiesis.132 A marked decrease in myeloerythroid progenitors was observed but without detectable effect on HSC. Its role in human hematopoiesis is unknown.
TET2
The Ten-Eleven-Translocation2 (TET2) gene codes for a 2-oxoglutarate and Fe(II)-dependent hydroxylase that is able to hydroxylate methylated cytosine (methylcytosine; mC).133-135 This function is shared with the 2 other TET proteins known in mammals, TET1 and TET3.133,135 The function of the resulting modified nucleotide, hydroxymethylcytosine (5hmC), is not clear yet, but it appears to be present in all cell types.136-138 5hmc may have a function by itself and/or represent a step toward cytosine demethylation.136 There is preliminary evidence that the function of TET1 and TET2 is mainly related to the generation of 5hmc.139
The founding member of the TET family is TET1. It has been isolated as a fusion partner of MLL in acute leukemia with the t(10;11)(q21;q32) chromosomal translocation.140,141 TET2 has 9 coding exons. Exon 2 is included in half of the transcripts starting from exon 1 and contains an ATG; a second ATG is located at the beginning of exon 3. The main TET2 protein is 2002 amino acids long (starting from the ATG in exon 3), but a second product is predicted to be translated from a RNA species that is not spliced at the 3′ side of exon 3 and runs on until a polyA site downstream of exon 4.30
TET2 is mutated in a wide range of myeloid malignancies.30-33 Observed mutations are mainly small insertions and deletions and nonsense mutations that are expected to result in the loss of function of the protein. Missense mutations affecting conserved amino acids have been shown also to impair the catalytic activity of the protein. Accordingly, patients with TET2 mutations have lower global 5hmC content than wild type.142 Of note, the global level of 5hmC may not directly impact the mC level as evaluated on a gene-specific base because TET2-mutated patients have lower mC in MDS samples and greater mC in AML samples.104,142
TET2 mutations are found in approximately 14% of MPNs ranging from ET (11%) to PMF (19%).32,143 In roughly 20% of the patients, 2 mutations are observed, suggesting that the inactivation of a single copy of TET2 is sufficient for the transformation process. TET2 mutations are not responsible for the familial MPNs identified to date, although a germline mutation has been described.144 In vitro studies have first shown that TET2 mutations occur before JAK2 mutations during the natural history of sporadic MPNs.32 However, subsequent studies suggest that the converse may happen and also that TET2 mutations may occur when MPNs transform to AML.144-147
One caveat: every mutational event that inactivates TET2 may not have been identified. For example, small deletions outside of the coding sequence or deletion of a single exon would have been missed by the current analyses.
Mutations have been shown to be present in the HSC/progenitor populations.32 In vivo analyses demonstrated that the TET2-mutated cells from MPN samples are able to engraft into immunodeficient mice.32 Clone-amplification studies have shown that TET2 mutations and JAK2V617F act synergistically to induce a clonal dominance at the level of hematopoietic progenitors in PV patients (Figure 3).32,145 TET1 and 2 proteins appear to play a crucial role in stem cell biology as shown in the control of pluripotency of murine embryonic stem cells. Initially, it was suggested that TET1 was involved in the self-renewal of ES cells by controlling Nanog,133 but recent evidence suggests that both TET1 and TET2 are necessary for pluripotency by controlling cell fate through the regulation of distinct sets of genes.139 Thus, TET2 may have a similar role on HSC. Preliminary results show that in the mouse, in vitro knockdown of TET2 by shRNAs increases monocytic differentiation in the presence of GM-CSF, causing an excess of immature forms.105,142 In human hematopoiesis, knocking down TET2 transiently favors monocytic differentiation at the expense of the granulocytic differentiation and decreases the proliferation of erythroid progenitors (F.D., unpublished data, June 2011).
Mutations in EZH2 and in ASXL1 point at deregulation of polycomb activity in MPN. In addition, L3MBTL1, a polycomb gene, may be one of the important lost genes in chromosome 20q deletion frequently seen in MPN.148 It is known that DNMTs interact with EZH2, allowing the coupling between PRC2 and DNA methylation.125 Because DNMT3A interacts with EZH2 and is mutated in AML and MDS,149 mutational analyses of this gene in MPN may identify a new “epigenetic player” in MPN pathogenesis.
Mutations in TET2, ASXL1, and EZH2 genes are observed in a wide range of myeloid malignancies. Furthermore, virtually all the other genes mutated in MPNs are in fact not specific to these disorders. Mutations in JAK2 and MPL have also been described in other diseases, such as acute leukemia,150-153 but they affect different amino acids and their frequency is lower than in MPNs.
Discussion
Initially, MPNs were considered as simple, single-hit diseases that lead to an increase in mature blood cells. ET and PV, disorders with a pure excess of mature blood cells, fit with this hypothesis, whereas PMF, which also has abnormal MK differentiation, is likely more complex. Thus, as with BCR-ABL in CML, a single mutation in a signaling protein would be at the origin of classic MPNs. The discovery of JAK2 and MPL gain-of-function mutations in classic MPN and the recapitulation of the human disorder in mouse models support this hypothesis. However, other lines of evidence indicate that JAK2V617F mutation may not be the initial event in certain MPNs and that other genetic events may be required for disease development. These data led to the concept of a pre-JAK2 event. At least 3 different genes involved in epigenetic regulation may precede JAK2 mutation in MPNs. It is likely that mutations in other genes have yet to be discovered. Thus, instead of being a single-hit disorder, MPN would result from the combination of several genetic events as exemplified by the finding of EZH2, TET2, and JAK2 mutations in the same MPN patient.36 Thus, the pathogenesis of the MPN has evolved from a simple to a complex model. At a first glance, this appears to sharply contrast with CML, where BCR-ABL is thought to fully explain the disease, although this is not the case for all CML patients.154
Why would several genetic events be required for the development of MPN?
The hallmark of these disorders is an increased production of mature blood cells. In some instances, a single germline mutation can induce thrombocytosis, erythrocytosis, or neutrophilia.155-161 In inherited diseases, all HSC carry the mutation, whereas in sporadic MPNs the signaling mutation (JAK2V617F, MPLW515L, or BCR-ABL) is acquired by a HSC that must compete with wild-type HSCs.
Mathematical models predict that at least 1%-2% of the HSC need to carry the JAK2V617F mutation to produce the clinical phenotype.162 Given an estimate of 1 × 106 HSCs in humans the initial JAK2V617F-positive HSC must expand 104 fold more than wild-type HSC before inducing a clinical phenotype.163,164 Evaluation of the JAK2V617F burden at different hematopoietic differentiation steps in humans has shown that the clonal expansion occurs mainly in late differentiation steps in PV and ET.66,118 In addition, CD34+ cells from PV or ET patients at diagnosis generate essentially JAK2 wild-type hematopoiesis in xenograft experiments, showing that JAK2V617F HSC are still in minority in early disease.119,165,166 Furthermore, JAK2V617F HSC did not out-compete JAK2 wild-type HSC in one case of allogenic human BM transplantation.167 This mild effect of JAK2V617F on HSC biology is also observed in mouse models. Transgenic JAK2V617F HSCs displayed only a slight advantage on JAK2 wild-type HSC in transplantation assays.59 Furthermore, in one knockin model, JAK2V617F HSCs were at a disadvantage.64 These results are unexpected because the TPO/MPL axis plays an important role in HSC self-renewal76,77,168-170 and quiescence in the BM niche.77,78 Thus, it is possible that MPL signaling differs between HSC and MKs and/or that JAK2V617F only partially activates normal MPL functions in HSC.
Therefore, a crucial (and presently unknown) factor in MPN pathogenesis is the latency between the mutation of JAK2 and the appearance of the disease. On the basis of the log-log incidence of MPN with age (that increases after 60 years), mathematical models suggested that a single mutation, such as JAK2V617F, although providing a minor advantage to HSC, may cause a MPN with a very long latency (> 45 years).163 Another prediction of the model is that > 95% of the JAK2V617F clones die by exhaustion and disappear.163,164 Therefore, cooperation of JAK2V617F with other genetic events modifying HSC biology will greatly facilitate the development of a clinical phenotype. It cannot be excluded, however, that JAK2V617F may give a significant advantage to HSC in specific conditions such as inflammation, as suggested by preliminary studies.171 It is thus conceivable that JAK2V617F HSC remain harmless during a long period, until genetic or environmental changes such as hematopoietic stress or aging allow clonal dominance and MPN emergence. Of note, clonal dominance occurs at the HSC level in PMF that frequently carry several mutations.119,120
Does a pre-JAK2 event really exist?
There is clear evidence that TET2 mutations, and also ASXL1 and EZH2 mutations, may precede JAK2V617F140 but that the converse may also occur.144-146 This finding suggests that the important issue is not the order of appearance of the mutations but the fact that they all occur in a HSC with the MPN phenotype being mainly driven by the JAK2V617F mutation.172 There is also evidence that secondary acquisition of TET2 mutations may be associated with disease progression and transformation.147 In support of this notion, it has been shown that TET2, EZH2, or ASXL1 mutations are far less frequent in PV and ET than in PMF, which combine at least 2 mutations.32,34-36
Two classes of MPNs may thus exist; a simple one, in which a single JAK2 or MPL mutation induces PV or ET, and a complex class, with 2 or more mutations producing PMF. For this reason, as previously suggested,173 PMF might be the accelerated phase of classic MPN. Next-generation sequencing may help establish whether a large fraction of ET and PV at diagnosis is the consequence of a single genetic event.
In several MPN patients, the coexistence of 2 oncogenic events, such as JAK2V617F, MPLW515L, JAK2 exon 12, BCR-ABL, and c-CBL mutants, has been demonstrated.18,20,174-178 In nearly all these cases, the 2 events have occurred in 2 different cells; moreover, independent acquisitions of JAK2V617F in a single patient have been reported.108,179 In one case it has been demonstrated that the MPLW515L and JAK2V617F subclones were derived from the same ancestral TET2-mutated clone.21 Thus, a reasonable hypothesis is that TET2, ASXL1, and EZH2 mutations favor the occurrence of secondary genetic events. From the original clone, a complex combination of mutations that does not follow a linear dynamics may give rise to different subclones. Depending on the type of dominant clone, different hemopathies evolving in time and appearing in different sequences will be induced. Such a model will explain the occurrence of JAK2 wild-type leukemia in JAK2V617F-positive MPNs, whereas, in contrast, the linear accumulation of mutations in the JAK2V617F-dominant clone will favor the progression of MPN to myelofibrosis and leukemia. This hypothesis has been validated only in one case of wild-type JAK2 AML, which had emerged from an original TET2 mutated clone found in the initial MPN.180
In other examples, TET2 mutations were only found in the leukemic cells.21,147 There are 2 possible scenarios for these patients:
The ancestral clone exhibits a mutation different from a coding sequence mutation on TET2 as exemplified by the EZH2 mutation preceding TET2 mutation36
AML and MPN are independent disorders. This would mean that a “mutator” phenotype exists in some MPN patients. This phenotype may be acquired, for example, related to changes in the hematopoietic environment181,182 or in the germline. This last possibility is supported by a marked increased risk (∼ 5- to 7-fold) among first-degree relatives of MPN patients.183 Such a risk has been at least partly related to 3 SNPs in the JAK2 gene, including one in the exon 14 defining the haplotype 46/1. The haplotype 46/1 is present with an allelic prevalence of approximately10% in the normal populations and confers a 3- and a 1.4-fold increase in the risk of developing a JAK2V617F or exon 12 or a MPLW515 MPN, respectively.184-187 The mechanism of this low penetrance susceptibility is unknown. This predisposition is different from the familial forms of MPN with a Mendelian transmission.112 However, none of the genetic events at the origin of this familial form is presently known, and this could correspond either to a “mutator” phenotype related to an alteration in gene repair or to a haploinsufficiency in a tumor suppressor gene. The precise understanding of the molecular mechanism of these familial forms will be certainly be critical for understanding the pathogenesis of MPN.
What are the consequences for therapy?
New targeted therapies against JAK2 have been developed.188-191 In contrast to the initial results, which showed a targeted effect of JAK2 inhibitors on JAK2V617F cells, including HSC,192,193 there is increasing evidence that they also affect JAK2 wild-type and JAK2V617F cells although having less effect on JAK2V617F HSC.59 This finding supports the clinical results that have shown good clinical responses in constitutional symptoms like asthenia and splenomegaly in myelofibrosis but a weak effect on the JAK2V617F clone.189 However, in cases in which JAK2V617F is not the initiating event, this approach may revert the MPN hematopoiesis to a pre-JAK2 clonal hematopoiesis (pre-JAK2 disorder) with the risk of developing other malignant blood disorders. The proof of this potential was demonstrated in one PV patient treated by pegylated IFN-α, where, after treatment, the JAK2V617F burden was at the threshold of detection, but the hematopoiesis was clonal because of a preexisting TET2 mutation.194 More recently, studies of “epigenetic” therapies in which the authors use either HDAC inhibitors or “demethylating” agents have been initated.195-198 It is unknown whether these therapies will be able to target the alterations induced by ASXL1, EZH2, or TET2 mutations.
Future studies aimed at identifying mutations present in MPN in which the authors use new-generation sequencing will be crucial not only to characterize the many cases of MPNs with no known molecular cause but also to better understand the evolution of the clone and the synergistic effects of the different mutations. This will be very important for understanding MPN pathogenesis and develop new therapies, and also, more generally, to understand the stepwise mechanisms leading to cancer development in humans.
Acknowledgments
The authors thank Françoise Wendling for a critical reading of the manuscript and Christian Pecquet for helping with the figures.
This work was supported by Inserm and by grants from la Ligue Nationale Contre le Cancer, l'Institut National du Cancer, la Cancéropole Ile de France, l'Association Laurette Fugain, la Fondation de France, the MPD Foundation, the Belgian PAI Program, Salus Sanguinis Foundation, the ARC Program of Université Catholique de Louvain, the Fondation contre le cancer, and the FRS-FNRS Belgium.
Authorship
Contribution: W.V. and O.A.B. wrote the manuscript; and F.D. and S.N.C. contributed to the writing of the manuscript and made the figures.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: William Vainchenker, Inserm U1009, Institut Gustave Roussy, PR1, 39 rue Camille Desmoulins, 94805 Villejuif, France; e-mail: verpre@igr.fr.