Abstract
Mapping the precise determinants of T-cell efficacy against viruses in humans is a public health priority with crucial implications for vaccine design. To inform this effort, we performed a comprehensive analysis of the effective CD8+ T-cell clonotypes that constitute responses specific for the HIV p24 Gag-derived KK10 epitope (KRWIILGLNK; residues 263-272) restricted by HLA-B*2705, which are known to confer superior control of viral replication in HIV-infected individuals. Particular KK10-specific CD8+ T-cell clonotypes, characterized by TRBV4-3/TRBJ1-3 gene rearrangements, were found to be preferentially selected in vivo and shared between individuals. These “public” clonotypes exhibit high levels of TCR avidity and Ag sensitivity, which impart functional advantages and enable effective suppression of HIV replication. The early L268M mutation at position 6 of the KK10 epitope enables the virus to avoid recognition by these highly effective CD8+ T-cell clonotypes. However, alternative clonotypes with variant reactivity provide flexibility within the overall KK10-specific response. These findings provide refined mechanistic insights into the workings of an effective CD8+ T-cell response against HIV.
Introduction
The importance of Ag-specific CD8+ T cells in the control of viral infections is well established. However, the parameters that allow for an effective CD8+ T-cell response have been difficult to elucidate in humans. The magnitude and targeting breadth of antiviral CD8+ T-cell responses in vivo correlate poorly, if at all, with the control of viral replication, thereby demonstrating that not all CD8+ T cells with specificity for a given virus are equally efficacious. Consequently, qualitative rather than quantitative attributes of antiviral CD8+ T cells have received greater attention in recent years, with several studies unveiling functional correlates of protection.1 Technological advances have fostered ever more precise studies of Ag-specific CD8+ T-cell immunity and recent findings highlight the need to integrate fine analyses of individual clonotypes, defined on the basis of specific TCR expression, into our understanding of antiviral CD8+ T-cell efficacy.2 Thus, each Ag-specific T-cell population is constituted from several different clonotypes, which can be considered as the fundamental units of T-cell reactivity. Collectively, the nature of these individual clonotypes determines the qualitative attributes of a given T-cell population. For example, the Ag sensitivity (AgS) of CD8+ T-cell populations, which may be important for antiviral efficacy,3 is likely governed primarily by the structural and biophysical properties of individual TCR interactions with cognate peptide–MHC class I (pMHCI) molecules. Moreover, particular interest surrounds the nature and functional relevance of public clonotypes, which bear Ag-specific TCRs shared between individuals.4,5 Despite the vanishingly small probability of TCR sharing between individuals given the vast potential for combinatorial diversity during the process of V(D)J gene rearrangement, public clonotypes can be identified in the majority of Ag-specific T-cell populations6 ; furthermore, their presence can be associated with distinct biologic outcomes.7-9
In this study, we aimed to unravel the forces that dictate the selection and maintenance of virus-specific CD8+ T-cell clonotypes associated with effective control of HIV replication in vivo. To this end, we performed detailed parallel ex vivo and in vitro analyses of CD8+ T cells specific for the p24 Gag-derived KK10 epitope (KRWIILGLNK; residues 263-272) restricted by HLA-B*2705. The KK10-specific CD8+ T-cell response is immunodominant in HLA-B*2705+ individuals infected with HIV clade B and linked with slower disease progression rates.10,11 Moreover, the emergence of viral escape mutations in this epitope during late infection has been associated with progression to AIDS.12-14 Here, we report that KK10-specific clonotypes with TRBV4-3/TRBJ1-3 gene rearrangements exhibit high levels of AgS, suppress HIV replication effectively, and tend to be public. Despite such functional advantages, however, these cells were typically subdominant in vivo, a phenomenon that could be linked to their inability to recognize the early L268M mutation that frequently occurs within the KK10 epitope.
Methods
Patients
Samples were obtained from untreated HIV-1–infected HLA-B*2705+ patients enrolled in cohorts in France, Australia, and Spain. All patients were asymptomatic with CD4+ T-cell counts > 300 cells/mm3 and viral loads ranging from undetectable to 3.5 × 105 copies HIV-1 RNA/mL plasma. The study was approved by the institutional review board and local ethics committee of the Hospital Pitié Salpêtrière. Informed consent was obtained in compliance with the Declaration of Helsinki. PBMCs were separated from citrate anticoagulated blood and cryopreserved for subsequent studies. HIV-1 gag DNA sequencing was performed on whole cellular DNA extracted from PBMCs as described previously.11
Tetramers, Abs, CD8+ T-cell clones, and viruses
Soluble biotinylated KK10/HLA-B*2705 monomers and variants thereof were generated and tetramerized as described previously.15 The D227K/T228A compound mutation was introduced into the α3 domain of HLA-B*2705 to generate CD8-null monomers based on extrapolation from studies with HLA-A*0201.16,17 Loss of soluble CD8 binding and maintenance of TCR docking integrity were verified for these novel reagents using surface plasmon resonance (SPR) as described previously with minor modifications16 (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). mAbs were obtained from the following vendors: (1) αCD4-APCCy7, αCD107a-Cy5PE, αIL-2-APC, αIFNγ-Alexa 700, and αTNFα-PECy7 (BD Biosciences); (2) αCD8-Alexa405 (Caltag Laboratories); (3) αMIP-1β-FITC (LIVE DEAD Aqua Systems); and (4) αp24-PE (Beckman Coulter). The viability dye LIVE DEAD Aqua (Molecular Probes) was used to eliminate dead cells from the analysis. HIV-specific CD8+ T-cell clones were isolated from PBMC samples obtained from 3 different HIV-1–infected HLA-B*2705+ patients and characterized as detailed previously.18 The HIV strains used for infection of HLA-B*2705+ CD4+ T cells were HIVNL4-3 and HIVNL4-3 ΔNef, and a strain of HIVNL4-3 with a point mutation creating the L268M substitution.
Clonotypic analysis
Molecular analysis of TRB gene expression in KK10-specific CD8+ T-cell populations isolated directly ex vivo by flow cytometry was conducted using a template switch anchored RT-PCR as described previously.7,19 A similar approach was used to characterize TRB and TRA gene expression in KK10-specific CD8+ T-cell clones. In all cases, TCR nomenclature was directly translated from the IMGT database using web-based alignment of molecular TRB or TRA transcripts (IMGT, The ImMunoGeneTics information system; http://imgt.cines.fr). The germline alignments for the TRBV4-3/TRBV1-3 clonotypes shown in supplemental Figure 2 were determined independently of this program to allow for both potential P additions from the TRBD genes and the determination of multiple alignments.
Assessment of Ag sensitivity and TCR avidity
The AgS of CD8+ T cells was assessed by measuring the peptide concentration required to induce half-maximal responses (EC50) in cytolytic Cr51 release (for clones) or IFN-γ ELISPOT assays (for CD8+ T cells within PBMCs), as described previously.11,18 TCR avidity was measured using tetramer dilution assays. CD8+ T-cell clones were incubated with a range of KK10/HLA-B*2705 (standard or CD8-null) tetramer concentrations (30 μg/mL to 0.013 μg/mL in 1/3 dilutions) for 30 minutes at 4°C, and then stained for CD8 expression before fixation. The median fluorescence intensity (MFI) values for tetramer staining and the percentage of tetramer+ CD8+ T cells were evaluated by flow cytometry.
HIV suppression assay
Primary HLA-B*2705+ CD4+ T cells were purified from thawed PBMCs by positive magnetic bead selection, stimulated for 2 days with PHA (1 μg/mL), and then cultured with 100 U/mL rhIL-2. Seven days later, 105 cells/well were infected with virus by spinoculation20 and mixed with CD8+ T-cell clones at different CD8+/CD4+ ratios. To compare HIVNL4-3 versus HIVNL4-3 ΔNef suppression activity, titrated amounts of each virus were used to generate similar levels of infectivity (ie, equivalent intracellular p24 expression 3 days after infection) in the absence of CD8+ T-cell clones, thereby compensating for the potentially attenuated replication of HIVNL4-3 ΔNef compared with HIVNL4-3 in our assays. Typically, we infected with 20 ng and 90 ng of p24/mL for HIVNL4-3 and HIVNL4-3 ΔNef, respectively. Cells were harvested at day 3 postinfection, and stained intracellularly for CD4 and p24 to evaluate the elimination of HIV-infected targets.
Proliferation and polyfunctional assays
For proliferation, CD8+ T-cell clones were stained with CFSE (Molecular Probes, Invitrogen) at 5μM for 10 minutes, then washed and stimulated with HLA-B*2705+ EBV-transformed B-cell lines pulsed with cognate peptide at the indicated concentrations. Five days later, the percentage of CFSE-low cells was evaluated by flow cytometry. For polyfunctional profiling, CD8+ T-cell clones were incubated for 1 hour at 37°C in the presence of αCD107a mAb and HLA-B*2705+ CD4+ T cells infected 3 days earlier with titrated levels of HIVNL4-3 or HIVNL4-3 ΔNef virus; monensin (2.5 μg/mL; Sigma-Aldrich) and brefeldin A (5μg/mL; Sigma-Aldrich) were added for a further 5 hours. Staining for intracellular markers and data analysis were performed as described previously.18
Statistics
Group medians and distributions were compared using the Mann-Whitney U test or the Wilcoxon signed-rank test. Associations between variables were determined using the nonparametric Spearman rank correlation test. The χ2 test was used for evaluation of TRAV/TRBV gene usage associations. P values < .05 were considered significant. Bonferroni correction was used for multiple comparisons, with P values < .05/(number of comparisons) considered significant.
Results
KK10-specific TRBV4-3/TRBJ1-3 clonotypes are highly selected in vivo
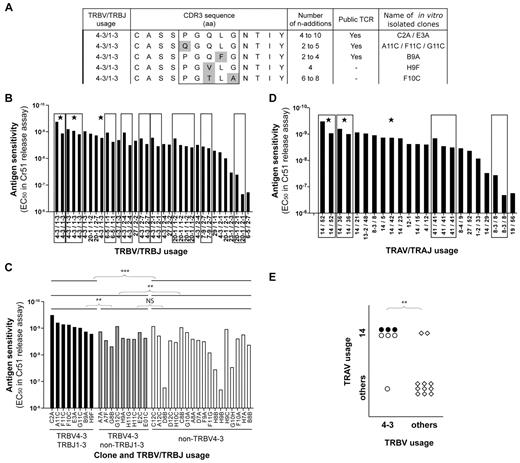
Clonotypic analyses of KK10-specific CD8+ T-cell populations sorted directly ex vivo from 19 HLA-B*2705+ individuals with HIV infection have generated more than 1200 TCRβ sequences comprising nearly 200 distinct clonotypes. Examination of these sequences led to the striking observation that 3 TRBV4-3/TRBJ1-3 clonotypes are public (each found in 2 patients, ie, 6 of 19 patients; Table 1). Furthermore, these public KK10-specific TRBV4-3/TRBJ1-3 clonotypes display closely related CDR3 sequences with a conserved pattern of amino acid usage (Figure 1A). The preferential representation of these clonotypes in KK10-specific CD8+ T-cell populations suggests that they are readily recruited and expand efficiently in vivo. In 2 cases (of the total 19 HLA-B*2705+ donors screened), patients 02.011 and L8146, these clonotypes were even found to be numerically dominant. On this basis, we sought to gain further insight into the public nature of these particular clonotypes and their relevance for the control of HIV replication.
Clonotypic analysis of KK10-specific CD8+ T-cell populations harboring public TRBV4-3/TRBJ1-3 sequences
| Patient/TRBV . | CDR3 sequence, aa . | TRBJ . | Frequency, % . |
|---|---|---|---|
| 02.011, CD4c = 1227, pVL = 1880 | |||
| 4-3*† | CASSPGQLGNTIY*† | 1-3*† | 54 |
| 7-6 | CASRLLGGGQETQY | 2-5 | 35 |
| 4-3 | CASSPGVFGVTQY | 2-3 | 11 |
| 01.01 DOF, CD4c = 716, pVL = 4400 | |||
| 4-3 | CASSQGQYGNEQF | 2-1 | 37 |
| 20-1 | CSARREANYGYT | 1-2 | 22 |
| 4-3 | CASSEGANAYEQY | 2-7 | 9 |
| 3-1 | CASSQDGVYSNQPQH | 1-5 | 6 |
| 4-3 | CASSMGQNSNEQY | 2-7 | 4 |
| 4-3 | CASSQGLSSNEQF | 2-1 | 3 |
| 18 | CASSLGGLDIEQY | 2-7 | 3 |
| 27 | CATSGVTGELF | 2-2 | 3 |
| 4-3*† | CASSPGQLGNTIY*† | 1-3*† | 1 |
| 4-3* | CASSPGVLGNTIY* | 1-3* | 1 |
| 20-1 | CARREANYGYT | 1-2 | 1 |
| 27 | CASAMTGEGYGYT | 1-2 | 1 |
| 27 | CASSGGAHTEAF | 1-1 | 1 |
| 27 | CASSSKTGELF | 2-2 | 1 |
| 27 | CASSSRTGELF | 2-2 | 1 |
| 7-2 | CASSSTRGTEAF | 1-1 | 1 |
| L8146, CD4c = 332, pVL = 202590 | |||
| 4-3*‡ | CASSQGQLGNTIY*‡ | 1-3*‡ | 40 |
| 7-6 | CASSLGGTNHGYT | 1-2 | 13 |
| 7-2 | CASSLVLAIEQY | 2-7 | 7 |
| 9 | CASSPGTGKNIQY | 2-4 | 7 |
| 4-1 | CASSQEGVNTEAF | 1-1 | 4 |
| 4-3 | CASSQAQGLSNSPLH | 1-6 | 4 |
| 5-1 | CASSRTQGPNTDTQY | 2-3 | 4 |
| 7-6 | CASSLDHLAGVNNEQF | 2-1 | 4 |
| 6-1 | CASSGQLLEAF | 1-1 | 2 |
| 7-8 | CASSLEGSRNTEAF | 1-1 | 2 |
| 11-2 | CASSQPPDRGYGYT | 1-2 | 2 |
| 12-4 | CASSLVGSYNEQF | 2-1 | 2 |
| 12-4 | CASSTTSGRYEQY | 2-7 | 2 |
| 18 | CASSFGQGAIEQY | 2-7 | 2 |
| 27 | CASSLSSDEHGYT | 1-2 | 2 |
| 11.007, CD4c = 716, pVL = 4400 | |||
| 20-1 | CSARDLGLAGDTDTQY | 2-3 | 84 |
| 7-9 | CASSLDSYEQY | 2-7 | 13 |
| 4-3*‡ | CASSQGQLGNTIY*‡ | 1-3*‡ | 2 |
| 28 | CASSLGIPGTAQWVYGYT | 1-2 | 2 |
| LTS 12, CD4c = 462, pVL = 17254 | |||
| 28 | CASSLRGGNTDTQY | 2-3 | 61 |
| 24-1 | CATSETGELF | 2-2 | 29 |
| 4-3*§ | CASSPGQFGNTIY*§ | 1-3*§ | 9 |
| 30 | CAWSLTGMNQPQH | 1-5 | 1 |
| LTS 57, CD4c = 1050, pVL = 1900 | |||
| 27 | CASMGGANTEAF | 1-1 | 88 |
| 27 | CASSPTTYGYT | 1-2 | 3 |
| 4-3*§ | CASSPGQFGNTIY*§ | 1-3*§ | 3 |
| 10-3 | CAISEYGNAASPLH | 1-6 | 2 |
| 9 | CASSVLGTSGGAEQF | 2-1 | 2 |
| 20-1 | CSARDWASGLSSYEQY | 2-7 | 2 |
| Patient/TRBV . | CDR3 sequence, aa . | TRBJ . | Frequency, % . |
|---|---|---|---|
| 02.011, CD4c = 1227, pVL = 1880 | |||
| 4-3*† | CASSPGQLGNTIY*† | 1-3*† | 54 |
| 7-6 | CASRLLGGGQETQY | 2-5 | 35 |
| 4-3 | CASSPGVFGVTQY | 2-3 | 11 |
| 01.01 DOF, CD4c = 716, pVL = 4400 | |||
| 4-3 | CASSQGQYGNEQF | 2-1 | 37 |
| 20-1 | CSARREANYGYT | 1-2 | 22 |
| 4-3 | CASSEGANAYEQY | 2-7 | 9 |
| 3-1 | CASSQDGVYSNQPQH | 1-5 | 6 |
| 4-3 | CASSMGQNSNEQY | 2-7 | 4 |
| 4-3 | CASSQGLSSNEQF | 2-1 | 3 |
| 18 | CASSLGGLDIEQY | 2-7 | 3 |
| 27 | CATSGVTGELF | 2-2 | 3 |
| 4-3*† | CASSPGQLGNTIY*† | 1-3*† | 1 |
| 4-3* | CASSPGVLGNTIY* | 1-3* | 1 |
| 20-1 | CARREANYGYT | 1-2 | 1 |
| 27 | CASAMTGEGYGYT | 1-2 | 1 |
| 27 | CASSGGAHTEAF | 1-1 | 1 |
| 27 | CASSSKTGELF | 2-2 | 1 |
| 27 | CASSSRTGELF | 2-2 | 1 |
| 7-2 | CASSSTRGTEAF | 1-1 | 1 |
| L8146, CD4c = 332, pVL = 202590 | |||
| 4-3*‡ | CASSQGQLGNTIY*‡ | 1-3*‡ | 40 |
| 7-6 | CASSLGGTNHGYT | 1-2 | 13 |
| 7-2 | CASSLVLAIEQY | 2-7 | 7 |
| 9 | CASSPGTGKNIQY | 2-4 | 7 |
| 4-1 | CASSQEGVNTEAF | 1-1 | 4 |
| 4-3 | CASSQAQGLSNSPLH | 1-6 | 4 |
| 5-1 | CASSRTQGPNTDTQY | 2-3 | 4 |
| 7-6 | CASSLDHLAGVNNEQF | 2-1 | 4 |
| 6-1 | CASSGQLLEAF | 1-1 | 2 |
| 7-8 | CASSLEGSRNTEAF | 1-1 | 2 |
| 11-2 | CASSQPPDRGYGYT | 1-2 | 2 |
| 12-4 | CASSLVGSYNEQF | 2-1 | 2 |
| 12-4 | CASSTTSGRYEQY | 2-7 | 2 |
| 18 | CASSFGQGAIEQY | 2-7 | 2 |
| 27 | CASSLSSDEHGYT | 1-2 | 2 |
| 11.007, CD4c = 716, pVL = 4400 | |||
| 20-1 | CSARDLGLAGDTDTQY | 2-3 | 84 |
| 7-9 | CASSLDSYEQY | 2-7 | 13 |
| 4-3*‡ | CASSQGQLGNTIY*‡ | 1-3*‡ | 2 |
| 28 | CASSLGIPGTAQWVYGYT | 1-2 | 2 |
| LTS 12, CD4c = 462, pVL = 17254 | |||
| 28 | CASSLRGGNTDTQY | 2-3 | 61 |
| 24-1 | CATSETGELF | 2-2 | 29 |
| 4-3*§ | CASSPGQFGNTIY*§ | 1-3*§ | 9 |
| 30 | CAWSLTGMNQPQH | 1-5 | 1 |
| LTS 57, CD4c = 1050, pVL = 1900 | |||
| 27 | CASMGGANTEAF | 1-1 | 88 |
| 27 | CASSPTTYGYT | 1-2 | 3 |
| 4-3*§ | CASSPGQFGNTIY*§ | 1-3*§ | 3 |
| 10-3 | CAISEYGNAASPLH | 1-6 | 2 |
| 9 | CASSVLGTSGGAEQF | 2-1 | 2 |
| 20-1 | CSARDWASGLSSYEQY | 2-7 | 2 |
Data are shown for 6 of 19 KK10-specific CD8+ T-cell populations sort-purified directly ex vivo from HLA-B*2705+ patients infected with HIV-1. CD4+ T-cell counts (CD4c) and plasma viral load (pVL) are indicated at time of sample.
Bold values represent all TRBV4-3/TRBJ1-3 clonotypes.
First of 3 public TRBV4-3/TRBJ1-3 clonotypes.
Second of 3 public TRBV4-3/TRBJ1-3 clonotypes.
Third of 3 public TRBV4-3/TRBJ1-3 clonotypes.
KK10-specific clonotypes with high levels of Ag sensitivity express TRBV4-3/TRBJ1-3 TCRs. (A) Alignment of observed TCRβ amino acid sequences for TRBV4-3/TRBJ1-3 clonotypes; 3 public and 2 private sequences are shown. Amino acid residues that differ between clonotypes are highlighted in gray. The numbers of n additions required to produce each observed nucleotide sequence and the isolated clone assignments are indicated for each clonotype. (B) KK10-specific CD8+ T-cell clones (n = 35) isolated from 3 patients are grouped by TCRβ sequence, indicated by the box frames, and classified according to mean cognate Ag sensitivity (EC50 for Cr51 release). Public clonotypes are highlighted with a star. (C) Classification of KK10-specific CD8+ T-cell clones according to cognate Ag sensitivity (EC50 for Cr51 release) and TRBV/TRBJ usage. Each bar represents one clone: TRBV4-3/TRBJ1-3 clones are in black, TRBV4-3/non-TRBJ1-3 clones are in gray, and non-TRBV4-3 clones are in white. The clone reference is indicated on the x-axis, and the last letter of the code corresponds to the patient from whom the clone was obtained (A and F for patient 01.01 DOF, B and G for patient 04.064, and C and H for patient 11.007). Statistical analyses were conducted using the Mann-Whitney U test with Bonferroni correction for multiple comparisons (P < .0125 was considered significant). (D) Isolated KK10-specific CD8+ T-cell clones (n = 23) are grouped by TCRα sequence, indicated by the box frames, and classified according to mean cognate Ag sensitivity (EC50 for Cr51 release). Public clonotypes are highlighted with a star. (E) Association between TRAV14 and TRBV4-3 gene usage in KK10-specific CD8+ T-cell clones. Each symbol represents one clone: ●, TRBV4-3/TRBJ1-3 clones; ○, TRBV4-3/non-TRBJ1-3 clones; and ♢, non-TRBV4-3 clones. The χ2 test was used to assess statistical significance. **P < .01 and ***P < .001, respectively.
KK10-specific clonotypes with high levels of Ag sensitivity express TRBV4-3/TRBJ1-3 TCRs. (A) Alignment of observed TCRβ amino acid sequences for TRBV4-3/TRBJ1-3 clonotypes; 3 public and 2 private sequences are shown. Amino acid residues that differ between clonotypes are highlighted in gray. The numbers of n additions required to produce each observed nucleotide sequence and the isolated clone assignments are indicated for each clonotype. (B) KK10-specific CD8+ T-cell clones (n = 35) isolated from 3 patients are grouped by TCRβ sequence, indicated by the box frames, and classified according to mean cognate Ag sensitivity (EC50 for Cr51 release). Public clonotypes are highlighted with a star. (C) Classification of KK10-specific CD8+ T-cell clones according to cognate Ag sensitivity (EC50 for Cr51 release) and TRBV/TRBJ usage. Each bar represents one clone: TRBV4-3/TRBJ1-3 clones are in black, TRBV4-3/non-TRBJ1-3 clones are in gray, and non-TRBV4-3 clones are in white. The clone reference is indicated on the x-axis, and the last letter of the code corresponds to the patient from whom the clone was obtained (A and F for patient 01.01 DOF, B and G for patient 04.064, and C and H for patient 11.007). Statistical analyses were conducted using the Mann-Whitney U test with Bonferroni correction for multiple comparisons (P < .0125 was considered significant). (D) Isolated KK10-specific CD8+ T-cell clones (n = 23) are grouped by TCRα sequence, indicated by the box frames, and classified according to mean cognate Ag sensitivity (EC50 for Cr51 release). Public clonotypes are highlighted with a star. (E) Association between TRAV14 and TRBV4-3 gene usage in KK10-specific CD8+ T-cell clones. Each symbol represents one clone: ●, TRBV4-3/TRBJ1-3 clones; ○, TRBV4-3/non-TRBJ1-3 clones; and ♢, non-TRBV4-3 clones. The χ2 test was used to assess statistical significance. **P < .01 and ***P < .001, respectively.
For a TCR to be public, it must first be present in the naive repertoire of more than one individual. To satisfy this prerequisite, the TCR in question should be produced efficiently during genetic rearrangement from the enormous array of combinatorial possibilities, and survive thymic selection. Recent research has indicated that interindividual TCR sharing within Ag-specific memory CD8+ T-cell populations can be predicted on the basis of relative production frequencies regardless of the selection processes that guide recruitment of individual clonotypes from the naive pool in response to Ag.5 The process by which random V(D)J recombination generates a frequency spectrum of TCR nucleotide and amino acid sequences has been termed convergent recombination21 ; this process operates in all Ag-specific CD8+ T-cell repertoires examined to date21-23 and also shapes clonotypic prevalence in the naive T-cell repertoire, both within and between individuals.24,25 Thus, we assessed the likelihood that public KK10-specific TRBV4-3/TRBJ1-3 clonotypes are produced efficiently during V(D)J recombination. Indicators of the efficiency with which TCRs can be produced by V(D)J recombination include the extent of germline encoding, the number of recombination events that can produce each nucleotide sequence and the variety of nucleotide sequences that can encode each amino acid sequence.5 Analyses of the multiple nucleotide sequences encoding each observed TRBV4-3/TRBJ1-3 clonotype showed that the conserved amino acids in the CASSXGXXXNTIY motif are predominantly germline-encoded (see supplemental Figure 2). Two of the public TRBV4-3/TRBJ1-3 clonotypes could be made with as few as 2 nucleotide additions and 1 required a minimum of 4 nucleotide additions; this compares with a median of 8 nucleotide additions across all observed KK10-specific sequences (data not shown). Moreover, each of the public TRBV4-3/TRBJ1-3 clonotypes was encoded by at least 3 different nucleotide sequences, several of which could have been produced by different recombination events (ie, different contributions from the germline genes and different nt additions; supplemental Figure 2). Overall, these data suggest that public TRBV4-3/TRBJ1-3 clonotypes can be produced with reasonable efficiency.
KK10-specific TRBV4-3/TRBJ1-3 clonotypes exhibit high levels of Ag sensitivity
In addition to genetic considerations, a public TCR must be efficiently recruited from the naive T-cell pool to be detected in the Ag-specific memory T-cell pool of multiple individuals. To assess the functional attributes of TRBV4-3/TRBJ1-3 clonotypes that might impinge on Ag-driven selection processes, we used a bank of KK10-specific CD8+ T-cell clones generated in vitro from HLA-B*2705+ patients infected with HIV. A total of 35 individual clones (from 3 individuals) were derived and characterized. Of note, we were able to isolate and study distinct clones bearing each of the 3 different public TRBV4-3/TRBJ1-3 sequences, as well as 2 clones with private TRBV4-3/TRBJ1-3 sequences (Figure 1A). Functional measurements using target cells loaded with a gradient of KK10 peptide concentrations revealed a broad range of AgS profiles across the bank of clones.18 However, clones with identical TCR sequences displayed very similar levels of AgS (Figure 1B), thereby supporting the premise that the primary determinants of AgS are TCR-dependent. Notably, high levels of AgS were apparent for clones bearing public TRBV4-3/TRBJ1-3 sequences. Further analysis revealed that TRBV4-3 usage (n = 17) conferred significantly higher AgS (P = .004) relative to clones with other TRBV segments (n = 18). Moreover, concomitant TRBJ1-3 usage was largely responsible for this difference (P = .005 for TRBV4-3/TRBJ1-3 [n = 8] vs TRBV4-3/non-TRBJ1-3 [n = 9]); indeed, the TRBV4-3/non-TRBJ1-3 clones did not differ significantly from the non-TRBV4-3 clones with respect to AgS (Figure 1C, supplemental Figure 3A). These differences were not conditional on the presence of weakly sensitive clones among the non-TRBV4-3 group (supplemental Figure 3B). Thus, TRBV4-3/TRBJ1-3 TCRs seem to furnish KK10-specific CD8+ T cells with high levels of AgS. A variety of TCRα sequences were also expressed among these KK10-specific clones, and TRAV14 usage was particularly common (Figure 1D). Of note, we found a significant association between TRAV14 and TRBV4-3 usage (Figure 1E); indeed, all TRBV4-3/TRBJ1-3 clones also expressed rearranged TRAV14 gene products.
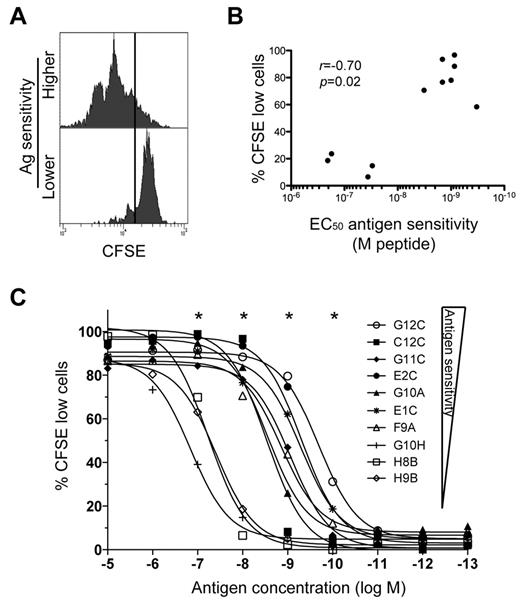
We hypothesized that the high AgS levels displayed by KK10-specific TRBV4-3/TRBJ1-3 clonotypes could confer the ability to proliferate vigorously in response to cognate Ag encounter. To test this possibility, we measured clonal proliferation in response to stimulation with exogenous KK10 peptide at a concentration of 10−8M, which reflects KK10 epitope densities on the surface of HIV-infected cells in vitro as inferred from functional comparisons conducted with 2 different CD8+ T-cell clones.18 Clones with higher levels of AgS divided more readily than clones with lower levels of AgS (Figure 2A). The extent of proliferation, measured as the percentage of cells undergoing division over a period of 5 days, correlated with AgS (Figure 2B). This correlation was maintained across different peptide concentrations. The peptide concentrations required to trigger proliferation were substantially higher (> 100-fold in some cases) for clones with low AgS levels compared with clones with high AgS levels (Figure 2C). Thus, clones with high AgS levels likely proliferate more effectively under physiologic conditions, thereby providing these cells with an expansion advantage. These data support the preferential selection of TRBV4-3/TRBJ1-3 clonotypes in vivo.
Differential induction of CD8+ T-cell proliferation according to Ag sensitivity. (A) Representative examples of CD8+ T-cell proliferation measured by dilution of CFSE fluorescence at 10−8M peptide for clones with higher (C2A) and lower (D8B) AgS levels. Cells were labeled with CFSE and then stimulated with KK10 peptide-loaded EBV-transformed HLA-B*2705+ B cells for 5 days. (B) Proliferation induced by 10−8M cognate peptide is plotted as a function of AgS (EC50 for Cr51 release). Each dot represents a distinct clone. Minimal proliferation was observed in the absence of exogenous cognate peptide. The correlation was determined using the Spearman rank test. (C) Proliferation (% of cells with diluted CFSE fluorescence) across a gradient of peptide concentrations for 10 different KK10-specific CD8+ T-cell clones with different AgS levels. *A significant correlation between AgS and proliferation at a given concentration of peptide.
Differential induction of CD8+ T-cell proliferation according to Ag sensitivity. (A) Representative examples of CD8+ T-cell proliferation measured by dilution of CFSE fluorescence at 10−8M peptide for clones with higher (C2A) and lower (D8B) AgS levels. Cells were labeled with CFSE and then stimulated with KK10 peptide-loaded EBV-transformed HLA-B*2705+ B cells for 5 days. (B) Proliferation induced by 10−8M cognate peptide is plotted as a function of AgS (EC50 for Cr51 release). Each dot represents a distinct clone. Minimal proliferation was observed in the absence of exogenous cognate peptide. The correlation was determined using the Spearman rank test. (C) Proliferation (% of cells with diluted CFSE fluorescence) across a gradient of peptide concentrations for 10 different KK10-specific CD8+ T-cell clones with different AgS levels. *A significant correlation between AgS and proliferation at a given concentration of peptide.
TCR-pMHCI interactions determine Ag sensitivity and HIV-suppressive capacity
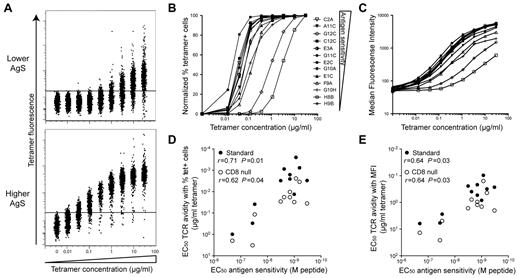
We have recently shown that AgS is a major determinant of the qualitative CD8+ T-cell attributes associated with effective control of HIV replication (ie, polyfunctionality and HIV suppressive capacity) and may be considered a robust correlate of CD8+ T-cell efficacy.18 However, the factors that determine acquisition of high AgS levels are not fully defined. The results reported above suggest a strong relationship between specific TCR sequences and high levels of AgS. We therefore performed tetramer dilution assays, which focus specifically on the TCR-pMHCI interaction, to measure TCR avidity for a series of KK10-specific CD8+ T-cell clones with a range of AgS levels. For this purpose, we used both standard and CD8-null tetramers. These latter reagents, which incorporate a compound D227K/T228A mutation in the α3 domain that abrogates CD8 coreceptor binding without affecting the fidelity of TCR docking (supplemental Figure 1), were produced to enable the assessment of intrinsic TCR avidity in the absence of CD8 compensation.15,17 The percentage of tetramer+ cells and median fluorescence intensity (MFI) were quantified by flow cytometry as a function of decreasing tetramer concentrations26-28 for different clones (Figure 3). Marked differences between CD8+ T-cell clones were observed according to their AgS. Increases in tetramer MFI were apparent for clones with high AgS levels at the most diluted tetramer concentrations used (down to 1.3 × 10−2 μg/mL). In contrast, substantially higher (> 10-fold) tetramer concentrations were required to observe MFI increases in clones with lower AgS levels; these clones also required very high tetramer concentrations to attain 100% staining (Figure 3A). Normalized percentages of tetramer+ cells (Figure 3B) and MFI (Figure 3C) were plotted as a function of tetramer concentration, and the tetramer concentrations required to obtain half maximal values (EC50) were calculated to quantify TCR avidity. EC50 values were generally higher with CD8-null tetramers compared with standard tetramers, consistent with the role of CD8 in stabilization of the TCR-pMHCI interaction.29 However, regardless of the tetramers used (ie, standard or CD8-null), this calculated measure of clonal TCR avidity correlated with the AgS of KK10-specific CD8+ T cells (Figure 3D-E). These data indicate that TCR avidity, determined by the TCR-pMHCI interaction, is a principal determinant of AgS in this set of clones.
Ag sensitivity correlates with TCR avidity. Clones with different AgS levels were labeled with KK10/HLA-B*2705 tetramer across a range of concentrations. (A) Representative tetramer titrations for 1 clone with lower (H8B) and 1 clone with higher (C2A) levels of AgS. (B-C) Representative tetramer titration curves for several clones displayed as a percentage (B) or MFI (C) of tetramer+ cells. The symbol key shown in panel B applies to both panels. (D-E) Correlation between AgS (EC50 for Cr51 release) and TCR avidity displayed as tetramer concentration EC50 for percentage (D) or MFI (E) of tetramer+ cells. Data are shown for both standard (●) and CD8-null (○) tetramer titration assays. Correlations were determined using the Spearman rank test.
Ag sensitivity correlates with TCR avidity. Clones with different AgS levels were labeled with KK10/HLA-B*2705 tetramer across a range of concentrations. (A) Representative tetramer titrations for 1 clone with lower (H8B) and 1 clone with higher (C2A) levels of AgS. (B-C) Representative tetramer titration curves for several clones displayed as a percentage (B) or MFI (C) of tetramer+ cells. The symbol key shown in panel B applies to both panels. (D-E) Correlation between AgS (EC50 for Cr51 release) and TCR avidity displayed as tetramer concentration EC50 for percentage (D) or MFI (E) of tetramer+ cells. Data are shown for both standard (●) and CD8-null (○) tetramer titration assays. Correlations were determined using the Spearman rank test.
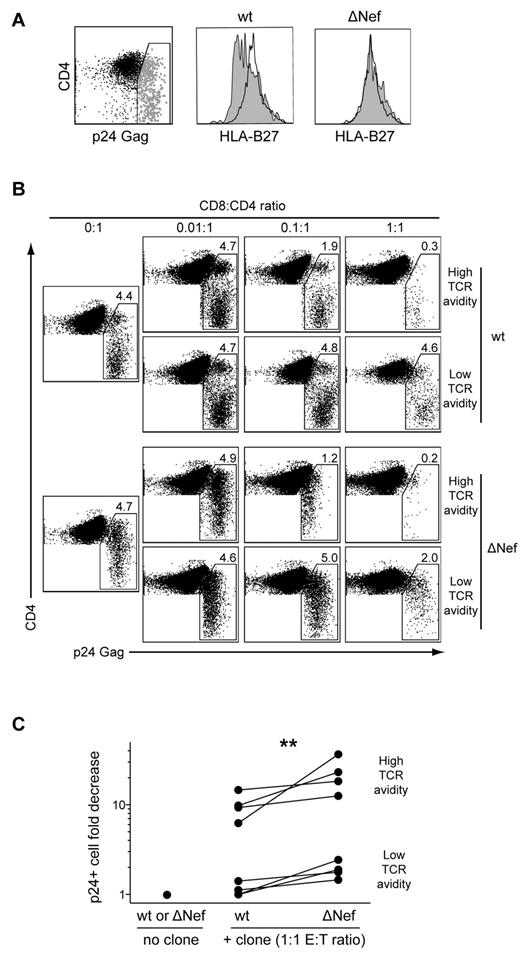
By extension, we reasoned that if TCR-mediated avidity for Ag is a primary determinant of AgS, then clones with high levels of TCR avidity should require fewer TCR-pMHCI ligations to kill infected cells and suppress HIV replication. To evaluate this, we compared the ability of CD8+ T-cell clones with high or low levels of AgS to suppress HIV replication in HLA-B*2705+ CD4+ T cells infected either with wild-type (wt) or Nef-deleted viruses. The HIV Nef protein is known to down-regulate HLA class I molecule expression on the surface of infected cells,30 thus altering the number of cognate pMHCI molecules available for TCR ligation. Indeed, CD4+ T cells infected with a wt HIV strain exhibited significant, although not complete, down-regulation of HLA-B*27 expression on the cell surface; in contrast, CD4+ T cells infected with a ΔNef virus displayed cell surface HLA-B*27 levels similar to those observed on the surface of uninfected cells (Figure 4A). Suppression of HIV replication, calculated as the fold reduction in the percentage of p24 Gag Ag-expressing CD4+ T cells detected by flow cytometry, was evaluated after 3 days of infected CD4+ T-cell coculture with different CD8+ T-cell clones. In line with our previous findings,18 clones with high levels of TCR avidity eliminated infected CD4+ T cells more efficiently than clones with low levels of TCR avidity. Regardless of TCR avidity, the ability to suppress HIV replication was more pronounced for ΔNef compared with wt virus (Figure 4B-C). This suggests stronger CD8+ T-cell activation, likely because of higher levels of HLA class I expression on the surface of CD4+ T cells infected with the ΔNef virus. Consistent with this possibility, representative high- and low-avidity clones displayed marginally more polyfunctional profiles when tested in the presence of CD4+ T cells infected with ΔNef compared with wt virus (supplemental Figure 4). Overall, TCR avidity emerges as a major determinant of AgS and HIV suppressive capacity in CD8+ T cells. Taken together, these results indicate that TCRs constructed with the TRBV4-3/TRBJ1-3 pairing confer KK10-specific CD8+ T cells with functional properties that enable preferential control of HIV replication.
TCR avidity determines the efficacy of HIV suppression by CD8+ T cells. (A) HLA-B*2705+ CD4+ T cells infected with wt (HIVNL4-3) or ΔNef (HIVNL4-3ΔNef) viruses were assayed for HLA-B*27 expression by flow cytometry. Surface expression of HLA-B*27 is shown for infected p24+ cells (gray) and uninfected p24− cells (black). (B) Representative flow cytometry plots showing p24 expression in CD4+ T cells infected with titrated amounts of wt or ΔNef viruses after 3 days of coculture with high- or low-avidity CD8+ T-cell clones. Percentages of p24+ cells and E:T ratios are indicated. (C) Fold decrease in the percentage of p24+ CD4+ T cells infected with titrated amounts of wt or ΔNef viruses after 3 days of coculture with higher (n = 4) or lower (n = 4) avidity CD8+ T-cell clones at E:T ratios of 1:1. The Wilcoxon signed-rank test was used to compare suppression of wt versus ΔNef viruses. **P < .01.
TCR avidity determines the efficacy of HIV suppression by CD8+ T cells. (A) HLA-B*2705+ CD4+ T cells infected with wt (HIVNL4-3) or ΔNef (HIVNL4-3ΔNef) viruses were assayed for HLA-B*27 expression by flow cytometry. Surface expression of HLA-B*27 is shown for infected p24+ cells (gray) and uninfected p24− cells (black). (B) Representative flow cytometry plots showing p24 expression in CD4+ T cells infected with titrated amounts of wt or ΔNef viruses after 3 days of coculture with high- or low-avidity CD8+ T-cell clones. Percentages of p24+ cells and E:T ratios are indicated. (C) Fold decrease in the percentage of p24+ CD4+ T cells infected with titrated amounts of wt or ΔNef viruses after 3 days of coculture with higher (n = 4) or lower (n = 4) avidity CD8+ T-cell clones at E:T ratios of 1:1. The Wilcoxon signed-rank test was used to compare suppression of wt versus ΔNef viruses. **P < .01.
HIV escapes recognition by public TRBV4-3/TRBJ1-3 clonotypes through the L268M mutation
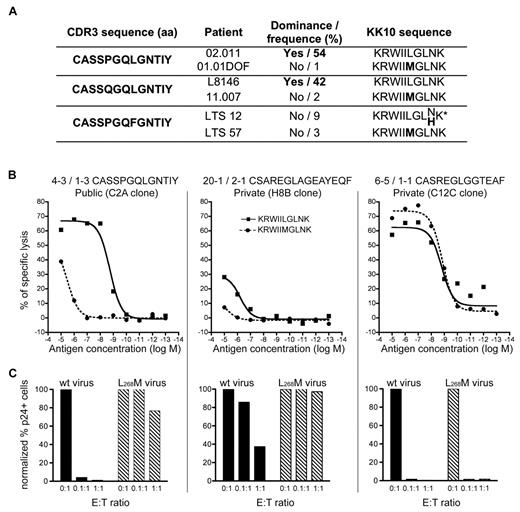
Although public TRBV4-3/TRBJ1-3 clonotypes could proliferate vigorously, they were found to dominate the KK10-specific CD8+ T-cell population in only 2 of 6 patients (02.011 and L8146) in whom they were detected (Table 1). This may reflect the replacement of senescent clonotypes, as we suggested previously11 ; however, the most noticeable feature that was shared between these 2 individuals emerged at the level of the dominant autologous viral sequence. Specifically, the prevalent viral species in patients with dominant TRBV4-3/TRBJ1-3 clonotypes contained wt KK10 epitope sequences. In contrast, individuals with subdominant TRBV4-3/TRBJ1-3 clonotypes harbored viruses with KK10 epitope mutations, in particular the common L268M substitution (Figure 5A). We reasoned that these public clonotypes, although apparently highly efficient, might actually be narrowly constrained and unable to recognize the L268M variant effectively, thereby mitigating their selection advantage in vivo in the presence of mutant viruses.
Inefficient recognition of the L268M mutant by public TRBV4-3/TRBJ1-3 clonotypes. (A) In vivo dominance of public TRBV4-3/TRBJ1-3 clonotypes and prevalent viral variants at the time of sampling. *Codominant viral KK10 sequences. (B) Ag sensitivity of representative CD8+ T-cell clones for the wt KK10 and L268M mutant epitopes was measured in Cr51 cytotoxicity assays using EBV-transformed HLA-B*2705+ B cells pulsed with the corresponding peptides across a range of concentrations. (C) Suppression of HIV replication in HLA-B*2705+ CD4+ T cells infected with wt (HIVNL4-3) virus (■) or a mutant virus encoding the L268M epitope mutation ( ) by representative CD8+ T-cell clones at 3 different E:T ratios. The percentage of p24+ CD4+ T cells was measured by flow cytometry at day 3 postinfection.
) by representative CD8+ T-cell clones at 3 different E:T ratios. The percentage of p24+ CD4+ T cells was measured by flow cytometry at day 3 postinfection.
Inefficient recognition of the L268M mutant by public TRBV4-3/TRBJ1-3 clonotypes. (A) In vivo dominance of public TRBV4-3/TRBJ1-3 clonotypes and prevalent viral variants at the time of sampling. *Codominant viral KK10 sequences. (B) Ag sensitivity of representative CD8+ T-cell clones for the wt KK10 and L268M mutant epitopes was measured in Cr51 cytotoxicity assays using EBV-transformed HLA-B*2705+ B cells pulsed with the corresponding peptides across a range of concentrations. (C) Suppression of HIV replication in HLA-B*2705+ CD4+ T cells infected with wt (HIVNL4-3) virus (■) or a mutant virus encoding the L268M epitope mutation ( ) by representative CD8+ T-cell clones at 3 different E:T ratios. The percentage of p24+ CD4+ T cells was measured by flow cytometry at day 3 postinfection.
) by representative CD8+ T-cell clones at 3 different E:T ratios. The percentage of p24+ CD4+ T cells was measured by flow cytometry at day 3 postinfection.
To confirm this hypothesis, we evaluated the AgS of the 3 public TRBV4-3/TRBJ1-3 CD8+ T-cell clones for both the wt and L268M mutant KK10 epitopes in Cr51 cytotoxicity assays. The AgS of these clones for the L268M mutant was found to be at least 2 orders of magnitude lower than that observed for the wt KK10 epitope (Figure 5B representative results are displayed in the left panel). HIV suppression experiments using HLA-B*2705+ CD4+ T cells infected with either wt virus or a strain presenting the L268M substitution corroborated these results. A public TRBV4-3/TRBJ1-3 clone efficiently eliminated CD4+ cells infected with wt virus but was almost completely ineffective against the L268M variant strain (Figure 5C left panel). For comparison, 2 private non-TRBV4-3/TRBJ1-3 clones (with low or high AgS levels for both the wt KK10 epitope and the L268M variant) were also studied (Figure 5B-C middle and right panels). These results demonstrate that the public TRBV4-3/TRBJ1-3 clonotypes, although very effective at recognizing the wt KK10 epitope, are functionally disabled (ie, consigned to low AgS) by the L268M mutant. Thus, the L268M mutation enables HIV to escape recognition and suppression by otherwise highly effective TRBV4-3/TRBJ1-3 clonotypes. Several other KK10-specific CD8+ T-cell clones also displayed weaker sensitivity for the L268M variant compared with the wt peptide (supplemental Figure 5). Although this may reflect the use of wt KK10/HLA-B*2705 tetramers to isolate these cells for cloning, it does indicate that HIV L263M escape is likely not exclusive to TRBV4-3/TRBJ1-3 clonotypes.
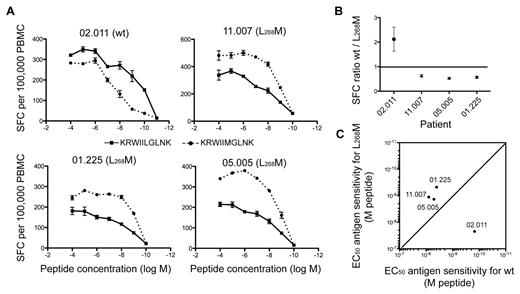
Nonetheless, in contrast to the R264K mutation, the emergence of the L268M mutation in vivo is usually not associated with loss of viral control and HIV disease progression,31 thereby suggesting that KK10-specific CD8+ T-cell responses remain effective in the presence of this variant. Direct ex vivo assessment of KK10 reactivity revealed that CD8+ T cells from 3 patients (11.007, 05.005, and 01.225) harboring a predominant L268M virus were able to recognize this variant efficiently. In contrast, cells from patient 02.011, in whom the circulating virus carried the wt KK10 epitope and the cognate CD8+ T-cell population was dominated by a public TRBV4-3/TRBJ1-3 clonotype, recognized the wt KK10 epitope with higher levels of AgS and greater overall magnitude compared with the L268M variant (Figure 6A-C). These observations may reflect adaptation of the immune response to the appearance of L268M mutants, characterized by the replacement of highly effective but wt epitope-constrained cells, such as those with TRBV4-3/TRBJ1-3 TCRs, by L268M-reactive cells. This process could ensure the maintenance of viral control despite the emergence of L268M escape variants.
Reactivity of KK10-specific CD8+ T-cell populations to emerging L268M mutants. (A) Ag sensitivity of KK10-specific CD8+ T-cell populations from 4 HLA-B*2705+ individuals infected with HIV-1 was measured directly ex vivo using IFN-γ ELISPOT analysis across a range of wt KK10 and mutant L268M peptide concentrations. The patients harbored predominantly either wt (02.011) or L268M (11.007, 01.225, and 05.005) viruses at the time point studied. (B) SFC ratio of wt/L268M-specific responses are plotted for each patient. Ratios were calculated for each peptide concentration and are shown as the mean ± SEM for all peptide concentrations. (C) Ag sensitivity (EC50 values) for the wt KK10 and mutant L268M peptides in each individual. SFC indicates spot-forming cell.
Reactivity of KK10-specific CD8+ T-cell populations to emerging L268M mutants. (A) Ag sensitivity of KK10-specific CD8+ T-cell populations from 4 HLA-B*2705+ individuals infected with HIV-1 was measured directly ex vivo using IFN-γ ELISPOT analysis across a range of wt KK10 and mutant L268M peptide concentrations. The patients harbored predominantly either wt (02.011) or L268M (11.007, 01.225, and 05.005) viruses at the time point studied. (B) SFC ratio of wt/L268M-specific responses are plotted for each patient. Ratios were calculated for each peptide concentration and are shown as the mean ± SEM for all peptide concentrations. (C) Ag sensitivity (EC50 values) for the wt KK10 and mutant L268M peptides in each individual. SFC indicates spot-forming cell.
Discussion
The Ag-mobilized TCR repertoire is a likely determinant of many critical qualitative parameters that contribute to CD8+ T-cell efficacy. Thus, individual clonotypes specific for a given pMHCI structure within an Ag-specific CD8+ T-cell population can potentially exhibit diverse properties, which in sum define the composite nature of the overall response. To dissect this complexity in the setting of a protective HIV-specific response, we performed extensive TCR repertoire analyses together with functional assessments of isolated CD8+ T-cell clones specific for the p24 Gag KK10 epitope restricted by HLA-B*2705. Thus, we were able to assess the role of specific TCRs in relation to CD8+ T-cell efficacy without the confounding variables of differential epitope targeting and HLA restriction.
Initially, we observed that 3 closely related TRBV4-3/TRBJ1-3 clonotypes within the KK10-specific CD8+ T-cell population were public, that is, shared in vivo between different individuals. Several theories have been put forward to explain the existence of public clonotypes,5 which appear to be a pervasive feature of Ag-specific T-cell responses despite the extremely low theoretical probability of recurrence.6 Sequence-based explanations propose that public TCRs are produced by near-germline recombination of V(D)J segments, with some deletion of nucleotides but no or minimal nucleotide additions.32-34 We evaluated this possibility in our public TRBV4-3/TRBJ1-3 clonotypes and found that a minimum of 2-4 nucleotide additions was required to produce each of the observed TCRβ sequences. Nonetheless, these TCRβ amino acid sequences were also encoded by different nucleotide sequences, many of which required greater numbers of nucleotide additions. Thus, as shown for other public TCRs,21,35,36 germline likeness in isolation cannot explain the shared nature of such sequences. Recently, “convergent recombination” has been proposed as a mechanism that could explain the existence of public TCRs.5 In brief, it is known that many different V(D)J recombination events can converge to produce identical nucleotide sequences and that many different nucleotide sequences can converge to encode identical amino acid sequences or motifs. Thus, the number of different ways in which any given TCR amino acid sequence can be constructed will contribute to its public or private nature. The observation that multiple nucleotide sequences can encode each of the public TRBV4-3/TRBJ1-3 amino acid sequences, some of which could be produced by different V(D)J recombination events, suggests that convergent recombination plays a role in the interindividual sharing of these clonotypes. Moreover, in our in vitro experiments, all KK10-specific CD8+ T-cell clones bearing TCRβ amino acid sequences constructed from TRBV4-3/TRBJ1-3 gene segments displayed high levels of cognate AgS. Such TRBV4-3/TRBJ1-3 clones were also characterized by TRAV14 usage, in contrast to the majority of non-TRBV4-3 clones. Thus, these specific TRA and TRB gene segments appear to generate a TCR structure that is particularly Ag-reactive. Given that such high levels of AgS also confer a proliferative advantage, it seems likely that avidity-based selection of TRBV4-3/TRBJ1-3 clonotypes in vivo also contributes to the observed sharing of such TCR sequences within the KK10-specific memory CD8+ T-cell pools of HIV-1 infected HLA-B*2705+ individuals.
The high levels of AgS displayed by TRBV4-3/TRBJ1-3 clonotypes conferred not only extensive proliferative capacity, but also potent HIV-suppressive activity. These findings suggest that TRBV4-3/TRBJ1-3 clonotypes could be major players in the KK10-specific CD8+ T-cell response, which is a characteristic immunologic feature of HLA-B*2705+ individuals who control HIV replication in vivo. In line with this possibility, our data indicate that the virus can mutate specifically to escape recognition by these highly effective clonotypes. Indeed, subdominance of TRBV4-3/TRBJ1-3 clonotypes in vivo, which was observed in 4 of 6 patients in whom these TCR sequences were detected (of 19 patients in total), was associated with the preponderance of circulating viral strains carrying the L268M mutant epitope. The L268M mutation is known to occur early during the course of HIV infection. It is generally not considered a “true” escape mutation because no increase in viral load occurs after its appearance. Instead, it has been proposed that the L268M substitution is a compensatory change required for the appearance, late in infection, of the R264K mutation, which results in increased viral loads and clinical progression.12-14 The R264K mutation dramatically decreases peptide binding to HLA-B*27; in contrast, the L268M mutation does not coincide with an anchor residue and appears to have little or no impact on HLA-B*27 binding.13,14,37 However, the emergence of L268M variants is associated with TCR repertoire changes within KK10-specific CD8+ T-cell populations, suggesting a differential impact on clonotypic recognition.31 Consistent with this possibility, we observed that AgS was reduced by at least 2 orders of magnitude when TRBV4-3/TRBJ1-3 clonotypes were confronted with the L268M variant; furthermore, the HIV-suppressive capacity of these clonotypes was almost entirely abrogated, rendering them ineffective in the face of an L268M variant virus. Thus, it is likely that the L268M mutation enables the virus to escape from the intense immune pressure exerted by such highly efficacious clonotypes early in HIV infection with no apparent fitness cost.38 The appearance of this variant is likely followed by the expansion of L268M-specific clonotypes that supersede the intially mobilized repertoire and thus maintain control of HIV replication.39 Nonetheless, because reversion to the wt epitope rarely occurs in vivo,40 we hypothesize that the L268M variant may ultimately favor the virus in the tight equilibrium between HIV replication and immune control. For instance, newly generated clonotypes may not attain the levels of AgS and anti-HIV efficacy displayed by TRBV4-3/TRBJ1-3 clonotypes for wt virus.
In addition to reinforcing the importance of AgS for CD8+ T-cell selection and efficacy against HIV, our data provide further insights into the determinants of AgS. Several factors contribute to AgS, effectively making it a composite parameter. These factors include the density and topography of HLA class I molecules, the quality of Ag presentation, costimulatory, and coinhibitory receptors on the CD8+ T-cell and target cell, pMHCI decay kinetics, membrane flexibility and TCR-dependent parameters such as monomeric affinity for Ag.29,41-46 The present work highlights the central role of TCR avidity as a determinant of AgS. TCR avidity, measured in the current study by pMHCI tetramer titrations to eliminate the confounding influences of other molecular interactions that can occur in bi-membrane domains, takes into account the intrinsic binding strength (affinity) of the TCR and the role of the CD8 coreceptor, as well as the density, topography, and coordinate relationship of these Ag-binding receptors within the constraints of cell-surface mobility. There was no relationship between AgS and CD8 density, as measured by flow cytometry at the time of assay (data not shown), on the surface of the KK10-specific CD8+ T-cell clones used in this study. A dominant role for the CD8 coreceptor was also excluded by the measurement of intrinsic TCR avidity using CD8-null pMHCI tetramer titrations. Furthermore, studies in mice have shown that the relationship between TCR density and the response to Ag is nonexponential.47 Thus, it seems unlikely that differential cell-surface densities of TCR and CD8 determine the range of clonal Ag avidities observed in this study. This is consistent with the cosegregation of TCR avidities and AgS levels with distinct TCR sequences. Altogether, these data highlight the importance of TCR-pMHCI interactions for CD8+ T-cell efficacy against HIV. This is further exemplified by the increased HIV-suppressive activity of CD8+ T-cell clones in assays using a ΔNef virus, which does not down-regulate HLA class I molecules from the surface of infected target cells. This effect may contribute, at least in part, to the establishment of long-term nonprogressive disease status in patients infected with viruses carrying deletions in the Nef/LTR region, as described previously in the Sydney Blood Bank cohort.48,49 In addition to the attenuated fitness of such viruses, control of HIV may be more easily achieved as lesser constraints are placed on the selection of CD8+ T-cell clonotypes with high levels of AgS.
In summary, the present study of a protective HIV-specific CD8+ T-cell response at the clonotypic level illustrates the importance of TCR avidity as a determinant of AgS and emphasizes that the selection of cognate clonotypes with high AgS levels may be critical for CD8+ T-cell efficacy. Controlling this selection process may be a key consideration for the rational design of effective T cell–based vaccines,50 although further studies with other HIV epitope-specific CD8+ T-cell populations are required to confirm and extend our findings. While the present observations may only represent one of several potential scenarios that could occur during the T-cell response to HIV, the demonstration herein that HIV can escape from specific clonotypes with high levels of AgS highlights the intricacy of the host-pathogen equilibrium and the level of complexity that may be necessary to design an effective T cell–based vaccine.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are very grateful to the staff and patients who participated in this study, to the Agence Nationale de la Recherche sur le SIDA (ANRS) Cohorts Asymptomatique à Long Terme group and to the National Centre in HIV Epidemiology and Clinical Research long-term nonprogressor cohort. They thank Catherine Blanc for sorting viable infected cells at the Pitié-Salpêtrière Flow Cytometry Platform. They are indebted to Zaïna Aït Arkoub and Henri Agut for help with HIV-1 gag DNA sequencing.
This work was supported by the ANR (project ANR-09-JCJC-0114-01), Sidaction, the Inserm AVENIR grant, the French ANRS, the National Institutes of Health via the Intramural Program of the Vaccine Research Center (National Institute of Allergy and Infectious Diseases [NIAID]) and R01 067077 (C.B.), the Australian Research Council (ARC), the Australian National Health and Medical Research Council (NHMRC), and the UK Medical Research Council (MRC). M.C.I. is supported by a Sidaction Fellowship. J.R.A. is supported by a Fundação para a Ciência e Tecnologia Fellowship.
V.V. is an ARC Future Fellow. P.G.W. is an NHMRC C. J. Martin Fellow. J.R. is an ARC Federation Fellow. M.P.D. is a Sylvia and Charles Viertel Senior Medical Research Fellow. D.A.P. is an MRC (UK) Senior Clinical Fellow.
National Institutes of Health
Authorship
Contribution: M.C.I. designed the study, performed research, analyzed data, and wrote the manuscript; J.R.A. designed the study, performed research, and analyzed data; S.F. performed research; D.J.v.B. performed research and analyzed data; M.H. performed research; V.V. analyzed data and wrote the paper; E.G. performed research and contributed vital new reagents; A.U. performed research; L.W. performed research; M.C. performed research; S.G. performed research; P.G.W. performed research; B.A. contributed vital reagents; A.M. contributed vital reagents; J.R. contributed vital analytical tools; M.P.D. contributed vital analytical tools; M.T. contributed vital new reagents; C.B. performed research and analyzed data; D.C.D. designed the study and contributed vital analytical tools; A.D.K. designed the study and analyzed data; D.A.P. designed the study, contributed vital new reagents, and wrote the manuscript; and V.A. designed the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Victor Appay, Inserm UMR S 945, Infections and Immunity, Avenir Group, Université Pierre et Marie Curie-Paris6, Hôpital Pitié-Salpêtrière, Paris, France; e-mail: victor.appay@upmc.fr.
References
Author notes
M.C.I. and J.R.A. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal