Abstract
Biallelic mutations in the human breast cancer susceptibility gene, BRCA2, are associated with Fanconi anemia, implying that some persons who inherit 2 deleterious variants of BRCA2 are able to survive even though it is well established that BRCA2 is indispensable for viability in mice. One such variant, IVS7 + 2T > G, results in premature protein truncation because of skipping of exon 7. Surprisingly, the persons who are either IVS7 + 2T > G homozygous or compound heterozygous are born alive but die of malignancy associated with Fanconi anemia. Using a mouse embryonic stem cell–based functional assay, we found that the IVS7 + 2T > G allele produces an alternatively spliced transcript lacking exons 4-7, encoding an in-frame BRCA2 protein with an internal deletion of 105 amino acids (BRCA2Δ105). We demonstrate that BRCA2Δ105 is proficient in homologous recombination-mediated DNA repair as measured by different functional assays. Evaluation of this transcript in normal and leukemia cells suggests that BRCA2Δ105 may contribute to the viability of persons inheriting this mutation. In this study, we have also characterized 5 other BRCA2 variants and found 3 of these (p.L2510P, p.R2336H, and p.W2626C) to be deleterious and 2 (p.I2490T and p.K2729N) probably neutral. Such studies are important to understand the functional significance of unclassified BRCA2 variants.
Introduction
Fanconi anemia (FA) is a rare and predominantly autosomal recessive disorder characterized by defective DNA repair and chromosomal instability.1,2 Studies of this rare disorder have elucidated the general mechanisms of DNA repair, bone marrow failure, and cancer pathogenesis. Homozygous or biallelic mutations of 14 genes (FANCA, FANCB, FANCC, FANCD1, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCJ, FANCL, FANCM, FANCN, and FANCP) are associated with the development of FA.1-3 Recently, biallelic mutations in RAD51C have also been found in patients with FA-like disease.4 Because FANCB is located on X-chromosome, affected males harbor a single mutant allele. Many of the proteins encoded by these genes exist in a macromolecular complex involved in the recognition and repair of damaged DNA.2,4-6 FANCD1/BRCA2 interacts directly with FANCN, FANCD2, and FANCG.7-9 A number of genes involved in the FA pathway have been found to be inactivated in a variety of cancers. Four genes (FANCD1, FANCN, FANCJ, and RAD51C) in this pathway have been shown to be breast cancer susceptibility genes.2,10 Mutations in FANCD1, FANCC, FANCN, and FANCG have been found also to be associated with pancreatic cancer.2,11,12
Among the different FA-complementation groups, the clinical phenotypes of FANCD1/BRCA2 (FA-D1) and FANCN/PALB2 (FA-N) are indistinguishable and also most severe in terms of very early age at onset, as well as high risks of leukemia, and specific spectrum of solid tumors.1,13 Patients in the FA-D1 subgroup have been identified to carry biallelic mutations in BRCA2, including 2 clearly deleterious or a clearly deleterious mutation in one allele and a missense mutation or variants of unknown clinical significance in the other allele. Interestingly, several of these FA-associated variants are located near the C-terminus of BRCA2 protein, or more specifically, between positions 2336 and 2729 (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).1 It is important for appropriate genetic counseling and risk factor assessment of mutation carriers to determine whether these variants lead to functional inactivation or retain normal BRCA2 activity. Limited availability of family history and mutation linkage data have been barriers in the classification of these missense mutations. A recently reported functional assay using mouse embryonic stem (ES) cells has now made it feasible to characterize any variant identified in human BRCA2.14
In this study, we have functionally evaluated 5 variants of unknown significance (p.R2336H [c.7235G > A], p.I2490T [c.7697T > C], p.L2510P [c.7757T > C], p.W2626C [c.8106G > C], and p.K2729N [c.8415G > T]) and 1 splice-site mutation c.864 + 2T > G (for simplicity referred to as IVS7 + 2T > G) and examined their effect on BRCA2 function using the ES cell–based functional assay (Table 1). In addition to IVS7 + 2T > G, p.R2336H is also known to affect splicing and both are considered to be deleterious.22-24 The other 4 (p.I2490T, p.L2510P, p.W2626C, and p.K2729N) are listed as variants of unknown clinical significance in the Breast Cancer Information database (http://research.nhgri.nih.gov/bic; supplemental Table 1). We have also examined the effect of these variants on the structural integrity of BRCA2, using structural modeling to support the functional data. Our findings demonstrate the usefulness of the ES cell–based assay to understand the functional effect of BRCA2 variants associated with FA that are currently considered to be of unknown clinical significance because of limited functional and family linkage data.
Patient malignancies and BRCA2 mutations
| Patient no. ID . | Cancer (age, y) . | Allele 1 . | Allele 2 . | References . |
|---|---|---|---|---|
| EUFA579 | AML | 7235G > A (p.R2336H)* | 5837TC > AG (p.F1807X) | 1,15 |
| AP37P | AML M2 (2) | 8415G > T (p.K2729N)* | 8732C > A (p.S2835X) | 1,15,16 |
| 3 | Brain (2.5) | 5301insA | 7690T > C (p.1249T)* | 1,17 |
| K2S1 | Wilms (0.5), AML (2) | 4876G > T (p.E1550X) | 7757T > C (p.L2510P)* | 1,18 |
| K2S2 | T-ALL (4.9) | 4876G > T (p.E1550X) | 7757T > C (p.L2510P)* | 1,18 |
| 129/1 | AML (2.2) | IVS7 + 2T > G* | IVS7 + 2T > G* | 1,19,20 |
| 357/1 | AML (1.9) | 8106G > C (p.W2626C) | 2041insA | 1,19 |
| 800/1 | AML (0.9) | IVS7 + 2T > G* | 5164del4 | 1,19 |
| 800/2 | Wilms (0.8) | IVS7 + 2T > G* | 5164del4 | 1,19 |
| SB1685CB | AML (2.1) | IVS7 + 2T > G* | 3827delGT | 1,21 |
| Patient no. ID . | Cancer (age, y) . | Allele 1 . | Allele 2 . | References . |
|---|---|---|---|---|
| EUFA579 | AML | 7235G > A (p.R2336H)* | 5837TC > AG (p.F1807X) | 1,15 |
| AP37P | AML M2 (2) | 8415G > T (p.K2729N)* | 8732C > A (p.S2835X) | 1,15,16 |
| 3 | Brain (2.5) | 5301insA | 7690T > C (p.1249T)* | 1,17 |
| K2S1 | Wilms (0.5), AML (2) | 4876G > T (p.E1550X) | 7757T > C (p.L2510P)* | 1,18 |
| K2S2 | T-ALL (4.9) | 4876G > T (p.E1550X) | 7757T > C (p.L2510P)* | 1,18 |
| 129/1 | AML (2.2) | IVS7 + 2T > G* | IVS7 + 2T > G* | 1,19,20 |
| 357/1 | AML (1.9) | 8106G > C (p.W2626C) | 2041insA | 1,19 |
| 800/1 | AML (0.9) | IVS7 + 2T > G* | 5164del4 | 1,19 |
| 800/2 | Wilms (0.8) | IVS7 + 2T > G* | 5164del4 | 1,19 |
| SB1685CB | AML (2.1) | IVS7 + 2T > G* | 3827delGT | 1,21 |
Alleles are shown as nucleotide changes. Where the change results in a missense or nonsense mutation, the resulting (amino acid) change is indicated below the nucleotide.
ALL indicates acute lymphoblastic leukemia.
Missense mutation.
Methods
Reagents
Human B-lymphocyte cell lines (GM05920 and GM14805) were obtained from Coriell Cell Repositories. Human EBV-lymphoblastoid cell lines (AVO35), leukemia cell line (SB1685CB), and nonimmortilized fibroblasts (AC389) were maintained as described previously.21 All oligonucleotides were obtained from Invitrogen, and their sequences are listed in supplemental Table 3. Antibodies used are: c-myc tag (ab18185, Abcam), actin (Ab-5, NeoMarkers), Rad51 (H92, Santa Cruz Biotechnology, sc8349), and γ-H2AX (clone JBW103, Upstate Biotechnology).
Generation of mutations in BRCA2 in a bacterial artificial chromosome clone
Generation of Brca2-null ES cells carrying a mutant transgene
BRCA2 functional assays
BRCA2 functional assays were performed as described previously14 (supplemental Methods).
Splicing minigene reporter assay
Splicing minigene reporter assay was done using RHCglo plasmid as described before.27 Ratios of exon inclusion/exclusion were quantified using ImageQuant TL Version 2005 software (GE Healthcare).
Crystal structure modeling
Homology-based modeling was performed using UCSF Chimera Version 1.5 software (supplemental Methods).
Coimmunoprecipitation
Coimmunoprecipitation experiments were performed as described previously28 (supplemental Methods).
Statistical analyses
All data were expressed as mean ± SD. Differences between 2 groups were compared using 2-tailed unpaired Student t test (Microsoft Excel for Mac). P < .05 was considered significant.
Results
Rescue of mouse Brca2KO/KO ES cells by human BRCA2 variants
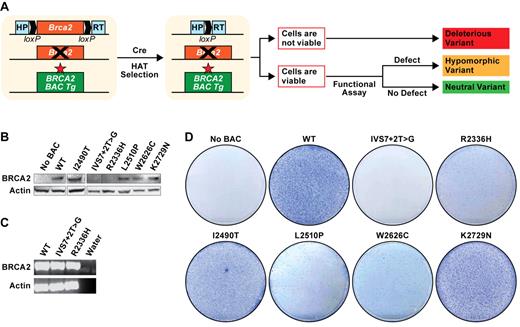
The mouse ES-cell based assay to analyze BRCA2 variants is based on the functional complementation of the lethality of Brca2-null (Brca2KO/KO) ES cells by human BRCA2 variants. For this assay, the ES cell line PL2F7, carrying one null allele of Brca2 and a conditional allele flanked by 2 loxP sites, is used.14 Furthermore, to express the transgene at physiologic levels, the variants are generated in a BAC containing the entire region of the genome encoding the BRCA2 gene. Variants that fail to rescue the lethality of Brca2KO/KO ES cells, or are deficient in any of the known functions of BRCA2 are considered to be deleterious, whereas those that are functionally similar to WT BRCA2 are neutral14 (Figure 1A).
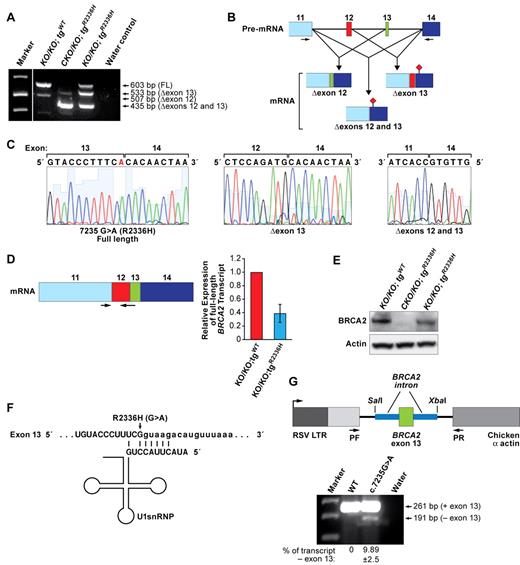
ES cell–based assay for functional analysis of BRCA2 variants. (A) Schematic representation of ES cell–based functional assay. Human BRCA2 BAC DNA with any mutation is introduced into mouse PL2F7 ES cells containing a conditional allele of Brca2. After CRE-mediated recombination of the conditional allele and HAT selection, the cells may or may not be viable, depending on the impact of the mutation on BRCA2 function. The viable HATr ES cells can be functionally similar to the WT cells or may be defective in BRCA2 functions, depending on the impact of the mutations. The neutral variants are functionally indistinguishable from WT. The hypomorphic variants with severe defect in BRCA2 function are probably deleterious. Star in the BAC construct indicates the mutation in BRCA2. Boxes with HP and RT indicate the 2 halves of the human HPRT minigene. loxP sites are indicated by solid arrows. (B) Expression of BRCA2 variant transgenes as detected by WB in the cells containing BRCA2 BAC before Cre-mediated deletion of Brca2. BRCA2 was detected using c-myc antibody. β-actin was used as a loading control. WB did not detect expression of some variants because of truncated protein product. (C) RT-PCR analysis showing the expression of IVS7 + 2T > G and p.R2336H variants. (D) Methylene blue staining of the plates of HATr ES cell colonies with no BAC (PL2F7), WT, IVS7 + 2T > G, p.R2336H, p.I2490T, p.L2510P, p.W2626C, and p.K2729N BRCA2 BAC transgenes.
ES cell–based assay for functional analysis of BRCA2 variants. (A) Schematic representation of ES cell–based functional assay. Human BRCA2 BAC DNA with any mutation is introduced into mouse PL2F7 ES cells containing a conditional allele of Brca2. After CRE-mediated recombination of the conditional allele and HAT selection, the cells may or may not be viable, depending on the impact of the mutation on BRCA2 function. The viable HATr ES cells can be functionally similar to the WT cells or may be defective in BRCA2 functions, depending on the impact of the mutations. The neutral variants are functionally indistinguishable from WT. The hypomorphic variants with severe defect in BRCA2 function are probably deleterious. Star in the BAC construct indicates the mutation in BRCA2. Boxes with HP and RT indicate the 2 halves of the human HPRT minigene. loxP sites are indicated by solid arrows. (B) Expression of BRCA2 variant transgenes as detected by WB in the cells containing BRCA2 BAC before Cre-mediated deletion of Brca2. BRCA2 was detected using c-myc antibody. β-actin was used as a loading control. WB did not detect expression of some variants because of truncated protein product. (C) RT-PCR analysis showing the expression of IVS7 + 2T > G and p.R2336H variants. (D) Methylene blue staining of the plates of HATr ES cell colonies with no BAC (PL2F7), WT, IVS7 + 2T > G, p.R2336H, p.I2490T, p.L2510P, p.W2626C, and p.K2729N BRCA2 BAC transgenes.
To evaluate the functional significance of selected variants, BACs containing myc-tagged BRCA2 with the desired mutations were electroporated into PL2F7 ES cells. Expression of full-length BRCA2 was confirmed by Western blot (WB) analysis for each variant except IVS7 + 2T > G and p.R2336H (Figure 1B). Because expression of the BRCA2 transcript was confirmed by RT-PCR in cells expressing the IVS7 + 2T > G and p.R2336H variants, a defect was predicted in the translation or stability of the full-length proteins (Figure 1C). After Cre-mediated deletion of the conditional Brca2 allele, no hypoxanthine-aminopterin-thymidine-resistant (HATr) colonies were observed in IVS7 + 2T > G variant expressing ES cells, supporting its deleterious nature (Figure 1D). In contrast, ES cells expressing p.I2490T and p.K2729N variants resulted in similar number of HATr colonies as those expressing WT: P = .88 (p.I2490T) and .80 (p.K2729N; Figure 1D). At least 3 independent BAC-expressing ES cell clones were tested for all variants, and each showed similar results. Southern analysis of the HATr colonies confirmed lack of endogenous Brca2 (supplemental Figure 2). The p.R2336H, p.L2510P, and p.W2626C variants rescued the ES cell lethality, but there was 70%-80% reduction in the numbers of HATr colonies (Figure 1D). Furthermore, the mutant ES cells grew considerably more slowly in vitro and had reduced plating efficiency (supplemental Figures 3, 6A; and data not shown).
Effect of IVS7 + 2T > G on alternative splicing of BRCA2
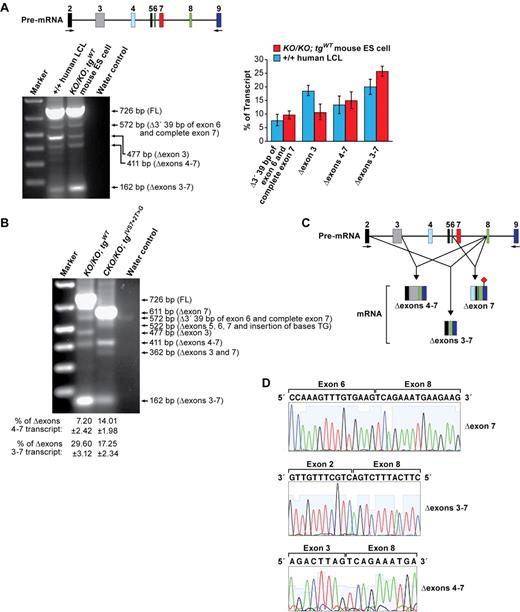
IVS7 + 2T > G allele of BRCA2 is a known deleterious mutation that alters a splice site, which results in skipping of exon 7 and premature protein truncation.23 On the other hand, a few patients who were either homozygous for this mutation or had a clearly deleterious mutation in the other allele of BRCA2 are born alive and survived for a few years with severe FA phenotype (Table 1). We hypothesized that there may be an alternatively spliced transcript encoding a partially functional BRCA2 protein that may be responsible for fetal viability. To identify this splicing variant, we first compared the splicing patterns of BRCA2 in EBV-transformed lymphoblastoid cell line of an apparently healthy person with wild-type BRCA2 and Brca2KO/KO;tgWT mouse ES cells by RT-PCR using the primers specific to exons 2 and 9 (Figure 2A; supplemental Table 2). The expression of alternate transcripts was found to be similar between human LCL and mouse transgenic ES cells, although the level of expression of some transcripts varies (Figure 2A). We detected 3 alternatively spliced transcripts (Δexon3, Δexons 4-7, and Δexons 3-7) that are predicted to code for BRCA2 proteins with an internal deletion of a few amino acids (supplemental Table 2). Next, we evaluated the transcripts encoded by BRCA2 carrying the IVS7 + 2T > G mutation in mouse ES cells. Other than the predominant fragment (611 bp) that represents the transcript lacking exon 7 (Figure 2B-D) and predicted to result in a premature stop codon in exon 9 (supplemental Figure 1), we identified 4 alternatively spliced transcripts (Figure 2B; supplemental Table 2). Three of them (522 bp, 411 bp and 362 bp) were up-regulated in cells expressing the IVS7 + 2T > G allele (Figure 2B). Among those, 411-bp and 162-bp fragments represent transcripts that are predicted to encode BRCA2 proteins, each with an internal deletion (supplemental Table 2). The 162-bp fragment represents a transcript that lacks exons 3-7 (Δexons 3-7; Figure 2B-D) and encodes BRCA2 protein with an internal deletion of 188 amino acids. The deleted amino acids include 17 that are encoded by the 5′ end of exon 3 (amino acids 24-40 of BRCA2) and are essential for binding to PALB2 (FANCN; supplemental Figure 1).9 PALB2 is required for BRCA2 function as PALB2 loss affects homologous recombination (HR), cell survival, and proliferation similar to BRCA2-depleted cells.9 To examine the importance of exon 3 and the PALB2 binding domain of BRCA2, we generated 2 exon 3 deletion mutants. The first mutant had a deletion in the 5′ region of exon 3 that encodes amino acids 25-40, which are a part of the PALB2 binding domain: deletion (BRCA2Δ25-40). The second mutant lacked the 3′ portion of exon 3 (encoding amino acids 41-104) that is not involved in PALB2 binding (BRCA2Δ41-104; supplemental Figure 4A,C-E). After Cre expression, the cells expressing BRCA2Δ25-40 did not yield any HATr colonies, but BRCA2Δ41-104 rescued the ES cell lethality similar to WT BAC (supplemental Figure 4B). Furthermore, the rescued ES cells expressing BRCAΔ41-104 were similar to WT cells in various functional assays (supplemental Figure 4F-G). These results suggest that the PALB2 binding domain is essential for BRCA2 function and the deletion of exon 3, as in the Δexons 3-7 transcript, will be deleterious for BRCA2 function. However, because humans with biallelic PALB2 mutation are born alive, we cannot rule out the possibility that the transcripts with exon 3 deletion may provide some BRCA2 function in humans.29
Alternative splicing events in IVS 7 + 2T > G variant. (A) RT-PCR analysis of EBV-transformed lymphoblastoid cell line (GM05920) and mouse ES cells expressing WT and human BRCA2 transgene. FL indicates fragment of a full-length product. Schematic representation of the genomic region analyzed for alternate transcript expression is shown at the top. Numbers above the colored boxes represent exon numbers. Arrows indicate the primers used for RT-PCR analysis. Quantification of different transcripts is shown on right panel. (B) RT-PCR analysis of mouse ES cells expressing either WT or IVS7 + 2T > G human BRCA2 transgenes. (C) Schematic representation of the alternative splicing that deletes exon 7. Exon numbers are indicated above the colored boxes representing the different exons. Arrows correspond to the position of the primers used for RT-PCR. (♦) represents the stop codon. (D) Sequence analysis of RT-PCR product reveals the deletion of exon 7, exons 3-7, and exons 4-7.
Alternative splicing events in IVS 7 + 2T > G variant. (A) RT-PCR analysis of EBV-transformed lymphoblastoid cell line (GM05920) and mouse ES cells expressing WT and human BRCA2 transgene. FL indicates fragment of a full-length product. Schematic representation of the genomic region analyzed for alternate transcript expression is shown at the top. Numbers above the colored boxes represent exon numbers. Arrows indicate the primers used for RT-PCR analysis. Quantification of different transcripts is shown on right panel. (B) RT-PCR analysis of mouse ES cells expressing either WT or IVS7 + 2T > G human BRCA2 transgenes. (C) Schematic representation of the alternative splicing that deletes exon 7. Exon numbers are indicated above the colored boxes representing the different exons. Arrows correspond to the position of the primers used for RT-PCR. (♦) represents the stop codon. (D) Sequence analysis of RT-PCR product reveals the deletion of exon 7, exons 3-7, and exons 4-7.
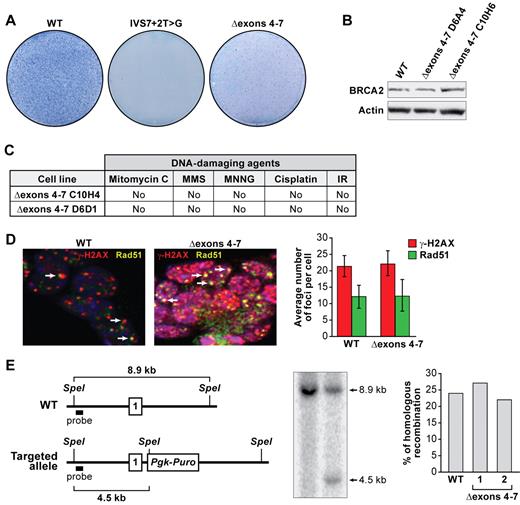
The 411-bp fragment (Figure 2B) represents a transcript lacking exons 4-7 (Δexons 4-7; Figure 2D). These deleted exons encode 105 amino acids of BRCA2 (supplemental Figure 1) that are evolutionarily conserved but of unknown functional significance (supplemental Figure 5A). To examine the effect of a loss of these105 amino acids, we deleted exons 4-7 in a BAC and expressed the mutant BRCA2 in PL2F7 ES cells. BRCA2Δ105 was able to rescue the lethality of Brca2KO/KO ES cells (supplemental Figure 4A-B). Although BRCA2Δ105 was expressed at levels comparable with WT BRCA2, we obtained 60%-70% fewer HATr (Figure 3A-B; supplemental Figure 5C) colonies, suggesting that BRCA2Δ105 may not be fully functional. Interestingly, the rescued ES cells exhibited no difference in growth or plating efficiency compared with control cells (supplemental Figure 4D; and data not shown). Furthermore, these cells were not hypersensitive to DNA-damaging agents (Figure 3C; supplemental Figure 5E-I), formed RAD51 foci in response to DNA-damaging agents similar to the WT cells (Figure 3D), were proficient in HR (Figure 3E), and exhibited no genomic instability (supplemental Figure 5H). Taken together, these results suggest that BRCA2Δ105 retains the known DNA repair functions of BRCA2. Our findings are supported by a recent report showing a chimeric BRCA2 lacking several amino acids, including the region coded by exons 4-7 to be proficient in DNA double-strand break repair in vitro.30 We postulate that IVS7 + 2T > G variant failed to rescue the Brca2KO/KO ES cell lethality because of the low abundance of the Δexons 4-7 transcript.
BRCA2Δexons 4-7 variant is efficient in DNA-repair function. (A) Methylene blue staining of HATr ES cell colonies expressing WT BRCA2, IVS7 + 2T > G BRCA2, and BRCA2Δexons 4-7. (B) Expression of BRCA2Δexons 4-7 variant analyzed by WB. Two independent clones were used for the mutant cell line. Actin was used as a control. (C) Sensitivity of 2 independent ES cell clones expressing BRCA2Δexons 4-7 to different DNA-damaging agents: mitomycin C (MMC); methyl-methanesulfonate (MMS); methyl-N′-nitro-N-nitrosoguanidine (MNNG); cisplatin; and ionizing radiation (IR). No, same as control (ES cells expressing WT BRCA2). (D) RAD51 foci formation 6 hours after ionizing radiation. RAD51 foci are shown in yellow, γ-H2AX foci marking DNA damage are shown in red, nuclei are stained with 4,6-diamidino-2-phenylindole (blue). Arrows point to RAD51 foci. Graphs on the right represent the quantification of RAD51 and γ-H2AX foci after ionizing radiation. Thirty nuclei were counted in each case, and their mean values are shown. Error bars represent the mean ± SD. (E) HR efficiency test. A scheme for targeting intron 1 of the Rosa26 locus (top panel). Number 1 indicates the first exon of Rosa26. WT indicates wild-type. Southern blot shows the targeted allele and WT allele. The bar graph on the right shows the quantification of the homologous recombination efficiency. Two independent clones were tested for BRCA2Δexons 4-7, and they are numbered as 1 and 2.
BRCA2Δexons 4-7 variant is efficient in DNA-repair function. (A) Methylene blue staining of HATr ES cell colonies expressing WT BRCA2, IVS7 + 2T > G BRCA2, and BRCA2Δexons 4-7. (B) Expression of BRCA2Δexons 4-7 variant analyzed by WB. Two independent clones were used for the mutant cell line. Actin was used as a control. (C) Sensitivity of 2 independent ES cell clones expressing BRCA2Δexons 4-7 to different DNA-damaging agents: mitomycin C (MMC); methyl-methanesulfonate (MMS); methyl-N′-nitro-N-nitrosoguanidine (MNNG); cisplatin; and ionizing radiation (IR). No, same as control (ES cells expressing WT BRCA2). (D) RAD51 foci formation 6 hours after ionizing radiation. RAD51 foci are shown in yellow, γ-H2AX foci marking DNA damage are shown in red, nuclei are stained with 4,6-diamidino-2-phenylindole (blue). Arrows point to RAD51 foci. Graphs on the right represent the quantification of RAD51 and γ-H2AX foci after ionizing radiation. Thirty nuclei were counted in each case, and their mean values are shown. Error bars represent the mean ± SD. (E) HR efficiency test. A scheme for targeting intron 1 of the Rosa26 locus (top panel). Number 1 indicates the first exon of Rosa26. WT indicates wild-type. Southern blot shows the targeted allele and WT allele. The bar graph on the right shows the quantification of the homologous recombination efficiency. Two independent clones were tested for BRCA2Δexons 4-7, and they are numbered as 1 and 2.
Expression of Δexons 4-7 splice variant in human cells
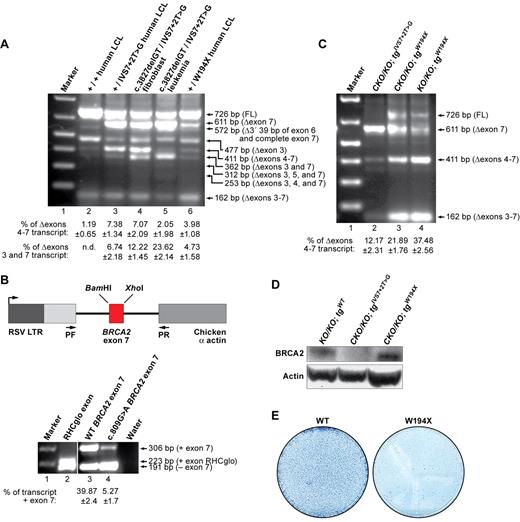
We next examined whether the Δexons 4-7 splice variant is expressed in human cells. We found this splice variant to be expressed in an EBV-transformed lymphoblastoid cell line (AVO35) established from a healthy IVS7 + 2T > G carrier21 (Figure 4A lane 3). We then tested the expression level of this alternatively spliced transcript in the normal fibroblasts (AC389) and the leukemia cells (SB1685CB) of this carrier's son who was affected by FA and developed acute myeloid leukemia (AML) at the age of 2 years.21 The patient was a BRCA2 compound heterozygote, inheriting the IVS7 + 2T > G mutation from the father and a 2-bp deletion in exon 11 (c.3827delGT) from the mother. Interestingly, although the same transcripts were present in the child's normal fibroblast and leukemia cells as those detected in the father's cells, there was a marked quantitative difference in the expression of 2 transcripts. The first transcript, which lacked exons 3 and 7 (Δexons 3 and 7) was expressed at higher levels in the child's cells (Figure 4A lanes 4 and 5). This transcript is predicted to result in a nonfunctional protein because of premature protein truncation. In contrast, the second transcript, the Δexons 4-7 splice variant, exhibited a marked reduction only in the leukemia cells (Figure 4A lane 5). A second leukemia cell line from the same patient (SB1690CB) exhibited similar pattern of expression of these transcripts (data not shown).
Expression of BRCA2 Δexons 4-7 transcript in human cell lines carrying IVS7 + 2T > G or another truncation mutation at exon 7. (A) RT-PCR analysis of different human cell lines: GM05920B (+/+ LCL), AVO35 (+/IVS7 + 2T > G LCL), AC389 (c.3827delGT/IVS7 + 2T > G fibroblast), SB1685CB (c.3827delGT/IVS7 + 2T > G leukemia), and GM14805 (+/W194X LCL). (B) RHCglo minigene reporter construct is shown in the left panel. BRCA2 exon7 variants replaced the RHCglo exon. Wild-type (WT) and W194X (c.809G > A) mutated were PCR-amplified using overlapping oligonucleotides containing WT or mutated sequences and BamHI and XhoI flanking restriction sites. Primers used for RT-PCR are marked by arrows and indicated as pF and pR. RT-PCR analysis of BRCA2 exon 7 alternative splicing in RHCglo minigene is shown at right. (C) Expression of Δexon 7 and Δexons 4-7 transcripts in ES cells carrying the W194X BRCA2 transgene. (D) WB showing the expression of BRCA2Δexons 4-7 variant in W194X ES cells. Actin was used as a loading control. (E) Methylene blue staining of HATr colonies obtained after Cre expression into the ES cells expressing either WT or W194X BRCA2.
Expression of BRCA2 Δexons 4-7 transcript in human cell lines carrying IVS7 + 2T > G or another truncation mutation at exon 7. (A) RT-PCR analysis of different human cell lines: GM05920B (+/+ LCL), AVO35 (+/IVS7 + 2T > G LCL), AC389 (c.3827delGT/IVS7 + 2T > G fibroblast), SB1685CB (c.3827delGT/IVS7 + 2T > G leukemia), and GM14805 (+/W194X LCL). (B) RHCglo minigene reporter construct is shown in the left panel. BRCA2 exon7 variants replaced the RHCglo exon. Wild-type (WT) and W194X (c.809G > A) mutated were PCR-amplified using overlapping oligonucleotides containing WT or mutated sequences and BamHI and XhoI flanking restriction sites. Primers used for RT-PCR are marked by arrows and indicated as pF and pR. RT-PCR analysis of BRCA2 exon 7 alternative splicing in RHCglo minigene is shown at right. (C) Expression of Δexon 7 and Δexons 4-7 transcripts in ES cells carrying the W194X BRCA2 transgene. (D) WB showing the expression of BRCA2Δexons 4-7 variant in W194X ES cells. Actin was used as a loading control. (E) Methylene blue staining of HATr colonies obtained after Cre expression into the ES cells expressing either WT or W194X BRCA2.
In conclusion, we have detected 9 splice variants in both the IVS7 + 2T > G carrier and the FA-derived AML cells (Figure 4A; supplemental Table 2). However, among all these different splice variants, the Δexons 4-7 transcript is the only one that is predicted to generate a functional protein and is highly expressed in cells with IVS7 + 2T > G mutation (Figure 4A; supplemental Table 2). In addition, the same transcript showed marked reduction in leukemia cells. Based on these observations, we speculate that the child with 2 deleterious BRCA2 mutations was born and survived for 2 years because of the partial BRCA2 function provided by this transcript. Subsequently, because of reasons currently unknown, in some cell(s) a change in splicing may have occurred that caused reduction in the Δexons 4-7 transcript and up-regulation of the Δexons 3 and 7 transcript, which may have contributed to leukemogenesis.
To determine whether such alternatively spliced transcripts are a common feature and not unique to the IVS7 + 2T > G variant, we examined another variant p.W194X (c.809 G > A), which is predicted to result in a premature stop codon in exon 7. We detected the presence of the Δexons 4-7 splice variant in an EBV-lymphoblastoid cell line expressing this allele (Figure 4A lane 6). Surprisingly, we also detected the presence of Δexon 7 transcript in these cells (Figure 4A lane 5). To further examine the effect of this mutation on exon 7 exclusion, we cloned the WT and mutant exon 7 sequences in a RHCglo minigene reporter plasmid (Figure 4B left panel).27 We examined the transcripts in COS7 cells by RT-PCR (Figure 4B right panel). In the case of the c.809G > A mutation, the average rate of exon 7 inclusion in 3 independent experiments was 5.27 ± 1.7 compared with 39.87 ± 2.4 in WT exon (Figure 4B right panel). These results suggest that the c.809 G > A mutation affects a potential cis-regulatory element that causes exclusion of the exon 7. Interestingly, we identified a potential exonic splicing silencer sequence (CTTAGT) in exon 7 with c.809G > A mutation.31
Next, we examined the effect of this mutation on ES cell survival and HR-mediated DNA repair function in PL2F7 cells. Interestingly, compared with the IVS7 + 2T > G expressing ES cells, the relative level of Δexons 4-7 was higher in W194X ES cells (Figure 4C), and we also detected BRCA2 protein by WB (Figure 4D). Furthermore, this relatively higher level of Δexons 4-7 transcript was sufficient to rescue the lethality of ES cells, albeit at a 4- to 5-fold reduced frequency (Figure 4E). It is remarkable that, despite being a clearly deleterious variant, W194X resulted in viable Brca2KO/KO ES cells and the rescued ES cells were functionally indistinguishable from WT BRCA2 expressing ES cells (data not shown).
p.R2336H is a splice site mutation
The mutation (c.7235G > A) resulting in an p.R2336H variant is located at the consensus splice donor site in exon 13 and has been shown to disrupt normal splicing.22 It results in 2 splice variants: one skipping exon 13 (Δexon 13) and the other skipping exons 12 and 13 (Δexons 12 and 13). Both transcripts result in frameshift, generating a premature stop codon in exon 14.22
Brca2CKO/KO ES cells expressing the p.R2336H variant (Brca2CKO/KO;tgR2336H) show the presence of these 2 transcripts (Δexon 13 and Δexons 12 and 13, Figure 5A-C) as reported in heterozygous carriers.22 After Cre expression, we obtained 800-1000 HATr colonies, which is 75%-80% fewer than the number obtained with WT BRCA2-expressing cells (Figure 1D). These HATr colonies were confirmed to be Brca2KO/KO (supplemental Figure 2). Because we had predicted that no full-length BRCA2 would be expressed in these cells, we reexamined the transcripts expressed in these cells to determine the cause of their rescue. Surprisingly, in addition to the Δexon 13 and Δexons 12 and 13 transcripts, these viable cells (Brca2KO/KO;tgR2336H) also expressed a full-length transcript retaining exon 13 (Figure 5A-C), which may account for their rescue. Next, we quantified the level of full-length transcript in WT and Brca2KO/KO;tgR2336H cells. Real-time RT-PCR using primers specific to exon 11 and the junction of exons 12 and 13 showed 2.5-fold reduction of full-length transcript production in the rescued cells compared with WT (Figure 5D). WB analysis confirmed that the p.R2336H variant results in expression of the full-length BRCA2 but at reduced levels (Figure 5E). It is possible that a small fraction of Brca2CKO/KO;tgR2336H ES cells may be expressing full-length BRCA2 and such cells are able to survive when the conditional Brca2 allele is deleted. Alternatively, some regulatory changes may occur that enable the expression of full-length protein in response to the deletion of the endogenous Brca2.
Alternative splicing because of c.7235G > A (p.R2336H) mutation. (A) RT-PCR analysis of the Brca2KO/KO ES cells expressing WT transgene, and Brca2CKO/KO and Brca2KO/KO ES cells expressing p.R2336H variant human BRCA2 transgene. FL indicates fragment of a full-length product; Δexon12, transcript with deletion of exon 12 (this is a natural, alternatively spliced form of BRCA232 ); Δexon 13, transcript with deletion of exon 13; and Δexons 12 and 13, transcript with deletion of exons 12 and 13. (B) Schematic diagram of the effect of p.R2336H (c.7235G > A) mutation on splicing. Exon numbers are indicated above the boxes representing the different exons. Arrows indicate the positions of the primers. (♦) represents the stop codon generated because of exon skipping. (C) Sequence analysis of different RT-PCR products from Brca2KO/KO ES cells expressing p.R2336H mutation. Left panel: Full-length transcript with the presence of mutation marked in red. Middle panel: Skipping of exon 13. Right panel: Skipping of exons 12 and 13. (D) Real-time RT-PCR analysis of the full-length transcript in mouse ES cells expressing either WT or p.R2336H BRCA2 variant. Left panel: Scheme of mature RNA quantified by real-time RT-PCR. Arrows indicate the primers. (E) WB showing the expression of WT and p.R2336H BRCA2 transgenes. c-myc antibody was used to detect BRCA2. For loading control, β-actin blot is shown. (F) Binding of U1 snRNA to the 5′ splice site. Uppercase letters correspond to the exon, and lowercase letters represent intron. Arrow indicates the change because of p.R2336H mutation. (G) RHCglo minigene reporter construct containing BRCA2 exon 13 variants (WT or p.R2336H) along with the flanking intron segments shown on top. Genomic fragments were PCR amplified using the primers containing flanking SalI and XbaI sites from the corresponding BACs and cloned into RHCglo reporter construct. After transient transfection into COS7 cells, RT-PCR was done using the primers pF and pR as indicated by arrows. RT-PCR analysis of BRCA2 exon 13 exclusion/inclusion in RHCglo minigene is shown below.
Alternative splicing because of c.7235G > A (p.R2336H) mutation. (A) RT-PCR analysis of the Brca2KO/KO ES cells expressing WT transgene, and Brca2CKO/KO and Brca2KO/KO ES cells expressing p.R2336H variant human BRCA2 transgene. FL indicates fragment of a full-length product; Δexon12, transcript with deletion of exon 12 (this is a natural, alternatively spliced form of BRCA232 ); Δexon 13, transcript with deletion of exon 13; and Δexons 12 and 13, transcript with deletion of exons 12 and 13. (B) Schematic diagram of the effect of p.R2336H (c.7235G > A) mutation on splicing. Exon numbers are indicated above the boxes representing the different exons. Arrows indicate the positions of the primers. (♦) represents the stop codon generated because of exon skipping. (C) Sequence analysis of different RT-PCR products from Brca2KO/KO ES cells expressing p.R2336H mutation. Left panel: Full-length transcript with the presence of mutation marked in red. Middle panel: Skipping of exon 13. Right panel: Skipping of exons 12 and 13. (D) Real-time RT-PCR analysis of the full-length transcript in mouse ES cells expressing either WT or p.R2336H BRCA2 variant. Left panel: Scheme of mature RNA quantified by real-time RT-PCR. Arrows indicate the primers. (E) WB showing the expression of WT and p.R2336H BRCA2 transgenes. c-myc antibody was used to detect BRCA2. For loading control, β-actin blot is shown. (F) Binding of U1 snRNA to the 5′ splice site. Uppercase letters correspond to the exon, and lowercase letters represent intron. Arrow indicates the change because of p.R2336H mutation. (G) RHCglo minigene reporter construct containing BRCA2 exon 13 variants (WT or p.R2336H) along with the flanking intron segments shown on top. Genomic fragments were PCR amplified using the primers containing flanking SalI and XbaI sites from the corresponding BACs and cloned into RHCglo reporter construct. After transient transfection into COS7 cells, RT-PCR was done using the primers pF and pR as indicated by arrows. RT-PCR analysis of BRCA2 exon 13 exclusion/inclusion in RHCglo minigene is shown below.
We evaluated the strength of this splice site using the Splice Site Prediction program33 and found that the score for donor site prediction is approximately 2.5-fold reduced because of the c.7235G > A mutation (0.99 vs 0.4). The c.7235G > A mutation also affects the binding of the U1 snRNP complex to the splice site. The 5′ terminus of the U1 snRNA of this complex has a sequence that base pairs with the splice donor consensus sequence. The c.7235G > A mutation decreases the base pairing of U1 snRNA that may affect the inclusion of exon 13 (Figure 5F). To determine the effect of this mutation on the inclusion of exon 13, we performed a splicing reporter assay using the RHCglo minigene reporter plasmid.27 We cloned WT and mutant exon 13 along with 200-bp flanking intronic sequences into the vector and examined the transcript expression in COS7 cells by RT-PCR. We found the majority of the transcripts in c.7235G > A to include exon 13 similar to the WT (Figure 5G). Less than 10% of the transcripts from the reporter plasmid with the mutation skipped exon 13. These observed differences in exon 13 inclusion/exclusion suggest that the effect of the mutation on skipping of exon 13 is not merely dependent on the point mutation but is also dependent on the flanking genomic region.
The Brca2KO/KO;tgR2336H ES cells expressing full-length p.R2336H BRCA2 exhibited hypersensitivity to various DNA-damaging agents, formed reduced numbers of IR-induced RAD51 foci, and showed marked reduction in HR efficiency and an increase in genomic instability (supplemental Figures 6, 8). These results show that p.R2336H BRCA2 is defective in HR-mediated DNA repair. It is possible that the phenotypic differences may be the result of the reduced levels of full-length protein. The p.R2336H mutation is located in the FANCD2 and FANCG binding domains of BRCA2 (supplemental Figure 1).7,8 Therefore, it is also possible that this mutation may affect interaction of BRCA2 with these proteins, which may change the organization of the DNA repair complex.
Analysis of missense mutations in the C-terminal domain of BRCA2
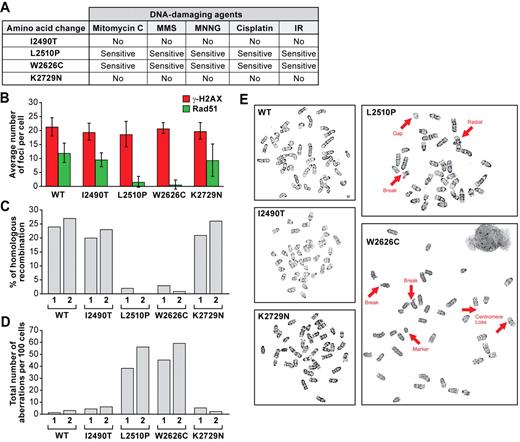
Four of the missense mutations (p.I2490T, p.L2510P, p.W2626C, and p.K2729N) that either fully or partially rescued the lethality of Brca2KO/KO ES cells are located in the evolutionarily conserved domain near the C-terminus of BRCA2 (supplemental Figure 1). We examined the effect of these variants on the DNA double-strand break repair functions of BRCA2. Brca2KO/KO ES cells expressing p.L2510P and p.W2626C variants were hypersensitive to the different DNA-damaging agents we tested and were deficient in IR-induced RAD51 foci formation and HR efficiency (Figure 6A-C; supplemental Figure 7A-E,P-T). We also observed an increase in chromosomal aberrations, such as breaks, gaps, or radial structures, in cells expressing p.L2510P and p.W2626C variants compared with the control cells (Figure 6D-E). In contrast, ES cells rescued by p.I2490T and p.K2729N variants were similar to WT BRCA2-expressing cells in all the functional assays (Figure 6A-E; supplemental Figure 7F-O). These results strongly support the deleterious nature of p.L2510P and p.W2626C variants, whereas p.I2490T and p.K2729N variants are probably neutral because they have no effect on BRCA2 functions.
Functional evaluation of BRCA2 variants located at the C-terminal domain. (A) Chart summarizing the sensitivity of ES cells expressing different mutant BRCA2 to different DNA-damaging agents. (B) Quantification of RAD51 and γ-H2AX foci after ionizing radiation. Error bars represent the mean ± SD. (C) HR efficiency as measured by gene targeting to the Rosa26 locus. For each variant and WT BRCA2, 2 independent clones were used, and they are marked 1 and 2. (D) Total number of chromosomal abnormalities present in WT and different mutant ES cells are represented. Randomly selected 100 metaphase spreads were counted blindly in each case. Two independent clones were used for each mutant and WT cell. (E) Representative metaphase spreads are shown. Thick arrows indicate different chromosomal abnormalities. To rule out the possibility of secondary mutations, each experiment was conducted using at least 2 independent clones of each mutant and they behaved similarly.
Functional evaluation of BRCA2 variants located at the C-terminal domain. (A) Chart summarizing the sensitivity of ES cells expressing different mutant BRCA2 to different DNA-damaging agents. (B) Quantification of RAD51 and γ-H2AX foci after ionizing radiation. Error bars represent the mean ± SD. (C) HR efficiency as measured by gene targeting to the Rosa26 locus. For each variant and WT BRCA2, 2 independent clones were used, and they are marked 1 and 2. (D) Total number of chromosomal abnormalities present in WT and different mutant ES cells are represented. Randomly selected 100 metaphase spreads were counted blindly in each case. Two independent clones were used for each mutant and WT cell. (E) Representative metaphase spreads are shown. Thick arrows indicate different chromosomal abnormalities. To rule out the possibility of secondary mutations, each experiment was conducted using at least 2 independent clones of each mutant and they behaved similarly.
Homology-based molecular modeling of variants present in the C-terminal region
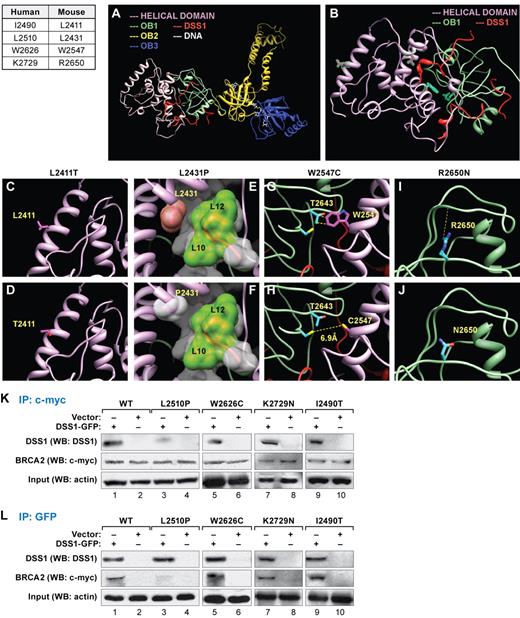
Four of the missense mutants examined in this study involve residues that map to the C-terminal domain (supplemental Figure 1). The crystal structure of the mouse C-terminal domain is known, and we used it to model the effect of human BRCA2 variants (Figure 7A-B).34 Leucine at position 2411 (corresponding to human p.I2490) is located at the first helix of the helical domain and found to be completely exposed to the solvent (Figure 7C). This residue is absent from any interface with cofactors and does not have any crucial contacts with the rest of the protein. Therefore, a threonine residue at this position (p.I2490T) is not expected to disrupt any crucial contacts or lose structure or function (Figure 7D). In contrast, leucine at position 2431 (corresponding to human L2510) is present at a core helix of the helical domain, which contacts DSS1, which is essential for BRCA2 stability (Figure 7E).35,36 One of the residues critical for BRCA2/DSS1 interaction is L2431 of BRCA2, which locks onto a hydrophobic patch between Leu10-Leu12 of DSS1 (Figure 7E). The p.L2431P (human p.L2510P) mutant may lose this contact because proline is low in hydrophobicity (Figure 7F). Moreover, because proline residues have unique dihedral angles, the secondary structure at the interface of BRCA2 and DSS1 is predicted to result in misfolding and loss of function.
Effect of BRCA2 variants on the structure of the C-terminal domain of mouse BRCA2 and analysis of BRCA2-DSS1 interaction. (A-B) Human BRCA2 mutations in C-terminal region and the corresponding mutations in the mouse BRCA2 are listed in the table. Structure of C-terminal region of BRCA2 that includes 3 oligonucleotide/oligosaccharide-binding (OB) domains OB1, OB2, and OB3, preceded by a helix-rich helical domain, whereas the helical domain (A) and OB1 interact with an essential cofactor DSS1 (B). The mutations studied in this work are present in the OB1 and the helical domain, whose side chains are colored magenta (if present in the helical domain) or cyan (if present in the OB1 domain) and highlighted in green. The effects of variants on the structure of BRCA2 described here are computer predictions and not primary crystallographic data. (C-D) Leucine at position 2411 is present at the first helix of helical domain (C). Its side chain is colored magenta, and its mutant is shown in Thr2411 (D). (E) Leucine at position 2431 is present at the core of the helical domain and interacts with DSS1. The side chain of Leu2431 is displayed, and its surface is colored pink. The protein DSS1 surface is displayed and colored white, except the hydrophobic patch between Leu10 and Leu12 of DSS1 is colored green. (F) The mutant of the same residue, Pro2431, surface is displayed in white. (G) Tryptophan at position 2547 is present within the helical domain and crucially placed at the interface of this domain and OB1. Trp2547 makes several van-der Waals interactions with Thr2643 of the OB1 domain, shown at yellow dotted lines. (H) A cysteine mutant Cys2547 side chain is shown as stick diagram, which might have the tendency to form a disulfide bond with a cysteine residue Cys2610. (I) Arginine at position 2650 is present at the surface of the OB1 domain (blue sticks), which forms a hydrogen bond with the backbone of Asp2693, whereas mutant Asn2650 loses the hydrogen bond (J). (K) Coimmunoprecipitation experiments of extracts after transient expression of DSS1-GFP construct in ES cells expressing either WT or mutant BRCA2. DSS1-GFP was detected by WB in immunoprecipitations (IP) with c-myc antibody. BRCA2 is tagged with c-myc, and DSS1 is tagged with GFP. Note that, in p.L2510P mutants, DSS1-GFP IP was reduced (lane 3), but in p.W2626C (lane 5), p.K2729N (lane 7), and p.I2490T (lane 9) mutants, the amount of DSS1 immunoprecipitated with BRCA2 is comparable to the WT (lane 1). Actin was used for input control. (L) IP using GFP antibody. WB using c-myc antibody after performing IP with GFP antibody detected BRCA2. DSS1-GFP was detected by DSS1 antibody. The amount of immunoprecipitated BRCA2 is reduced in p.L2510P mutants (lane 3), but not in p.W2626C (lane 5), p.K2729N (lane 7), and p.I2490T (lane 9) compared with the WT cells (lane 1). Actin was used to show the same amount of protein used in the IP experiments.
Effect of BRCA2 variants on the structure of the C-terminal domain of mouse BRCA2 and analysis of BRCA2-DSS1 interaction. (A-B) Human BRCA2 mutations in C-terminal region and the corresponding mutations in the mouse BRCA2 are listed in the table. Structure of C-terminal region of BRCA2 that includes 3 oligonucleotide/oligosaccharide-binding (OB) domains OB1, OB2, and OB3, preceded by a helix-rich helical domain, whereas the helical domain (A) and OB1 interact with an essential cofactor DSS1 (B). The mutations studied in this work are present in the OB1 and the helical domain, whose side chains are colored magenta (if present in the helical domain) or cyan (if present in the OB1 domain) and highlighted in green. The effects of variants on the structure of BRCA2 described here are computer predictions and not primary crystallographic data. (C-D) Leucine at position 2411 is present at the first helix of helical domain (C). Its side chain is colored magenta, and its mutant is shown in Thr2411 (D). (E) Leucine at position 2431 is present at the core of the helical domain and interacts with DSS1. The side chain of Leu2431 is displayed, and its surface is colored pink. The protein DSS1 surface is displayed and colored white, except the hydrophobic patch between Leu10 and Leu12 of DSS1 is colored green. (F) The mutant of the same residue, Pro2431, surface is displayed in white. (G) Tryptophan at position 2547 is present within the helical domain and crucially placed at the interface of this domain and OB1. Trp2547 makes several van-der Waals interactions with Thr2643 of the OB1 domain, shown at yellow dotted lines. (H) A cysteine mutant Cys2547 side chain is shown as stick diagram, which might have the tendency to form a disulfide bond with a cysteine residue Cys2610. (I) Arginine at position 2650 is present at the surface of the OB1 domain (blue sticks), which forms a hydrogen bond with the backbone of Asp2693, whereas mutant Asn2650 loses the hydrogen bond (J). (K) Coimmunoprecipitation experiments of extracts after transient expression of DSS1-GFP construct in ES cells expressing either WT or mutant BRCA2. DSS1-GFP was detected by WB in immunoprecipitations (IP) with c-myc antibody. BRCA2 is tagged with c-myc, and DSS1 is tagged with GFP. Note that, in p.L2510P mutants, DSS1-GFP IP was reduced (lane 3), but in p.W2626C (lane 5), p.K2729N (lane 7), and p.I2490T (lane 9) mutants, the amount of DSS1 immunoprecipitated with BRCA2 is comparable to the WT (lane 1). Actin was used for input control. (L) IP using GFP antibody. WB using c-myc antibody after performing IP with GFP antibody detected BRCA2. DSS1-GFP was detected by DSS1 antibody. The amount of immunoprecipitated BRCA2 is reduced in p.L2510P mutants (lane 3), but not in p.W2626C (lane 5), p.K2729N (lane 7), and p.I2490T (lane 9) compared with the WT cells (lane 1). Actin was used to show the same amount of protein used in the IP experiments.
Tryptophan at position 2547 (corresponding to human p.W2626) is present within the helical domain and placed at the interface with the OB1 domain (Figure 7G). This hydrophobic residue with a bulky side chain creates several van der Waals interactions across the interface with T2643 of the OB1 domain (Figure 7G), which is critical for the packing between these 2 domains. The smaller, cysteine side chain at this position (human p.W2626C) will lose these van der Waals contacts and may disrupt the interface packing (Figure 7H). A free cysteine at position 2610 across the interface may also induce disulfide bond formation and may have to come closer to 2.8Å instead of 6.9Å, at the expense of major structural rearrangements at the interface. These structural modifications probably dislodge the proper arrangement of these domains.
Finally, we examined arginine at position 2650 (corresponding to human K2729), which is present at a surface loop of OB1 domain, whose side chain forms a hydrogen bond with an adjacent residue Asp2693 (Figure 7I). The p.R2650N (p.K2729N) variant will lose the hydrogen bond, and it is expected that there may be some local structural modifications (Figure 7J). However, the hydrophilicity of the residue remains intact; and because this region does not interact with other domains or cofactors, the local modifications may not be detrimental to function, which is consistent with our functional data.
Interaction of mutant BRCA2 and DSS1
We tested whether p.I2490T, p.L2510P, p.W2626C, and p.K2729N BRCA2 variants are able to bind to DSS1. Interaction with DSS1 is involved in the stability of BRCA2 protein in insect cells.34,36 Green fluorescent protein (GFP)–tagged DSS1 was transiently expressed in ES cells expressing WT BRCA2 or these variants. Immunoprecipitation analysis from the cell extracts showed reduced binding of p.L2510P BRCA2 to DSS1 compared with the control cells (Figure 7K-L lanes 1-4), but the interaction was not affected in p.W2626C, p.I2490T, or p.K2729N (Figure 7K-L lanes 1,2,5-10).
Discussion
Biallelic mutations in BRCA2 can cause a very severe FA subtype associated with malignancy very early in life. Several of the reported BRCA2 mutations associated with FA are missense variants. To understand the clinical and cellular features of this FA subgroup and determine whether the missense variants contribute to the disease, the functional effects of BRCA2 variants of unknown clinical significance need to be examined carefully. The development of a novel assay by Kuznetsov et al using mouse ES cells has made it possible to analyze different kinds of mutations identified in BRCA2.14 We have now used this assay to examine 5 BRCA2 missense variants and one splice-site mutation (Table 1). Although this assay has been successfully used to analyze several BRCA2 variants14,37 because mouse cells are used to examine the effect of mutations in a human gene, the data should be cautiously analyzed, especially for splicing mutations. Therefore, when possible, we have validated our findings in human cells.
Among all the FANCD1/BRAC2 mutations reported in FA-D1 patients, IVS7 + 2T > G is the only one that has been detected as homozygous.19 This mutation causes skipping of exon 7, and a frameshift leading to a premature protein truncation is predicted.23 The mutation is strongly associated with the development of AML in FA patients very early in life; and according to the Breast Cancer Information database, this mutation has also been found in families with breast and ovarian cancer.1 In our functional assay, it showed the most severe phenotype and failed to rescue the ES cell lethality. We found an alternatively spliced transcript that encodes BRCA2Δ105 is expressed at very low levels in ES cells. Higher expression of this variant in c.809G > A mutants resulted in viable Brca2KO/KO ES cells. However, the inability of BRCA2Δ105 to rescue the lethality of ES cells to the same extent as the WT BRCA2 suggests that some function(s) other than DNA repair may be disrupted. The deleted 105 amino acids do not contain any domain known for HR repair, but a Plk1 phosphorylation site of BRCA2 (Ser,193 Ser205/206, and Thr203/207) is located in this region. Phosphorylation of BRCA2 in this region regulates the interaction with histone acetyltransferase P/CAF.38 The precise role of BRCA2-P/CAF complex has not yet been elucidated, but it was predicted that this phosphorylation of BRCA2 may be associated with entry or control of mitosis.38 Deletion of this region may affect ES cell survival. It is interesting that the cells that do survive do not show any defect in proliferation or plating efficiency. Although we do not know the precise mechanism, it is possible that the surviving cells express BRCA2Δ105 above a threshold level that allows them to overcome this defect.
Here, we have analyzed the splicing of a human gene in mouse ES cells and identified a potential splice variant that is highly expressed in the presence of IVS7 + 2T > G mutation. Our ES cell findings were subsequently validated in human cells. The expression of Δexons 4-7 transcript in a IVS7 + 2T > G mutation carrier and its lower expression in the leukemia cells suggest that the expression of this splice variant may be regulated and depend on several factors, such as tissue-type or developmental stage.39,40 It is probable that the fetal viability with a severe FA phenotype and early malignancy in persons who are homozygous or compound heterozygous for IVS7 + 2T > G may be associated with loss of this splice variant. Although the precise mechanism is not known, this loss may be the result of a change or defect in some splicing factor. It is also possible that the expression of Δexons 4-7 transcript is sufficient to sustain life in humans but is not enough to support normal functions of BRCA2, and the patients have FA and malignancy very early in life.
Interestingly, in this study, we have also demonstrated that the c.809G > A mutation that is predicted to generate an N-terminal truncation (W194X), actually produces a BRCA2 variant (BRCA2Δ105) that is fully proficient in DNA DSBR function. This suggests that, even though some alleles may be predicted to cause premature protein truncation, they may express an alternatively spliced transcript that can generate a functional protein. Our characterization of IVS7 + 2T > G and W194X demonstrates the importance of the careful functional analysis of each variant. To what extent alternative splicing of other FA genes or other human disease genes occurs would be an important question that needs to be addressed before drawing conclusions about the effect of any sequence variants on its biologic functions.
Our structural and functional analysis of 5 other variants has revealed that, although p.R2336H, p.L2510P, and p.W2626C can rescue the ES cell lethality, the rescued cells have significantly attenuated BRCA2 functions. Based on the reduced ES cell viability, these are considered hypomorphic. However, because of significant defect in BRCA2 functions, these are probably deleterious. In contrast, p.I2490T and p.K2729N variants are neutral. Although these 2 variants have been identified in persons with FA (a p.I2490T/5301insA compound heterozygote have FA and medulloblastoma at the age of 3.5 years17 and a person with p.K2729N/S2835X variants was diagnosed with FA and developed AML16 ), they are not likely to be causal. p.I2490T was previously predicted to be a neutral variant by Offit et al17 as well as by Myriad Genetics Laboratories Inc41 based on its prevalence in unaffected persons. p.K2729N variant was classified as neutral using multifactorial likelihood model and a few BRCA2 functional assays.24 Hu et al also showed that p.K2729N is a neutral variant rather than a familial eosophageal cancer-causing mutation in the Chinese population.42 Although we cannot rule out the possibility that these variants may behave differently when present in association with another deleterious allele, our ES cell–based assay and the available evidence strongly suggest that p.I2490T and p.K2729N variants may not be the cause of FA. Therefore, a careful reevaluation may identify the causal mutation in p.I2490T and p.K2729N carriers who had FA that was indistinguishable from those patients with biallelic deleterious mutations, including severe physical findings and cancer. In conclusion, we recognize the value of the epidemiologic analyses of variants; however, when combined with functional analyses, they can be more accurate in predicting the nature of the variants identified in human disease gene, as exemplified here for the rare cases of FA-D1 (∼ 3% of all FA patients).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Mark Greene and Suhwan Chang for helpful discussions and critical review of the manuscript; Billy Fergusson for help with cell culture; Jiro Wada (SAIC-Frederick Inc, Scientific Publications, Graphics & Media Department) for illustrations; Dr Sudhirkumar Yanpallewar for help with the confocal imaging; Dr Maria Jasin for the DR-GFP plasmid and I-SceI expression vector; and Dr Thomas Cooper for the RHCglo plasmid.
This work was supported by the Center for Cancer Research, National Cancer Institute, National Institutes of Health, and a CRUK Clinical Scientist Fellowship (S.M.).
National Institutes of Health
Authorship
Contribution: K.B. conducted most of the experiments, analyzed data, and wrote the manuscript; R.D. and R.A.B. performed structural studies; B.P.A. and L.C.B. conceived the idea and provided epidemiologic data; S.M. provided human cell lines and contributed to the manuscript; S.G.K. performed some experiments; S.S., S.L.N., and S.B. provided technical support; and S.K.S. supervised the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.G.K. is Institute for Molecular Medicine Finland, University of Helsinki, Helsinki, Finland.
Correspondence: Shyam K. Sharan, Mouse Cancer Genetics Program, Center for Cancer Research, National Cancer Institute at Frederick, Bldg 560, Rm 32-31C, 1050 Boyles St, Frederick, MD 21702; e-mail: sharans@mail.nih.gov.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal