Abstract
Notch1 signaling is absolutely essential for steady-state thymic lymphopoiesis, but the role of other Notch receptors, and their potential overlap with the function of Notch1, remains unclear. Here we show that like Notch1, Notch3 is differentially expressed by progenitor thymocytes, peaking at the DN3 progenitor stage. Using mice carrying a gene-trapped allele, we show that thymic cellularity is slightly reduced in the absence of Notch3, although progression through the defined sequence of TCR-αβ development is normal, as are NKT and TCRγδ cell production. The absence of a profound effect from Notch3 deletion is not explained by residual function of the gene-trapped allele because insertion mapping suggests that the targeted allele would not encode functional signaling domains. We also show that although Notch1 and Notch3 are coexpressed on some early intrathymic progenitors, the relatively mild phenotype seen after Notch3 deletion does not result from the compensatory function of Notch1, nor does Notch3 function explain the likewise mild phenotype seen after conditional (intrathymic) deletion of Notch1. Our studies indicate that Notch1 and Notch3 carry out nonoverlapping functions during thymocyte differentiation, and that while Notch1 is absolutely required early in the lymphopoietic process, neither receptor is essential at later stages.
Introduction
T lymphocytes are lost throughout life because of a variety of causes, and therefore must be continuously replaced. Steady-state T lymphopoiesis is the primary function of the thymus. Given this fact, it is somewhat counterintuitive that the thymus contains no self-renewing lymphoid progenitors. Instead, the thymus relies on the semicontinuous importation of BM-derived multipotent progenitors that circulate in the blood.1-3 Once inside the thymus, microenvironmental cues unique to this organ specify the T-lineage fate in these multipotent progenitors, and simultaneously induce and control several other complex processes, including a million-fold proliferative expansion, sequential somatic rearrangements of TCR loci, positive and negative selection, and functional T-lineage asymmetry (reviewed in Petrie and Zuniga-Pflucker4 ). Only a relatively small number of the signals that the thymic microenvironment provides to induce these functions have been identified; the essential ones are mainly limited to Notch ligands that induce T-lineage specification and other functions (for examples, see Krueger et al,5 Schmitt and Zuniga-Pflucker,6 Schmitt et al,7 and Feyerabend et al8 ), the cytokines IL-7 and kit ligand that support proliferation and/or survival,9-12 and MHC proteins that drive the positive- and negative-selection processes (reviewed in von Boehmer13 ). In addition, several chemokines have been shown to be important in controlling the directional migration of progenitor cells within the thymus (for examples, see Uehara et al,14 Ueno et al,15 Janas et al,16 and Plotkin et al17 ), which appears to be the primary mechanism by which temporal order is imposed during steady-state differentiation (reviewed in Petrie and Zuniga-Pflucker4 ).
The relatively small number of essential signals attributed to the thymic microenvironment seems insufficient to explain the complexity of the lymphopoietic process found there. In particular, identification of stromal signals that are stratified between different signaling microenvironments has been challenging because of inherent difficulties in isolation of stable stromal cells, as well as a paucity of region-specific markers. To identify other microenvironmental (stromal) signals that might play a role in thymocyte differentiation, we developed a high-throughput approach for reverse identification of stromal signals. This approach is based on the logical assumption that if lymphoid progenitors respond to stromal signals, they should express specific receptors for those signals. In particular, receptors that are restricted to one stage of development (or a few adjacent stages), or that change substantially during developmental progression, might be predictive of stratified stromal signals, because the histological location corresponding to each progenitor stage has been mapped.18,19 This approach has the further advantage that all stages of the lymphoid isolation process can be carried out on ice, and thus, minimal changes in gene expression are expected to occur.
In this manuscript, we applied this approach to all stages of progenitor development that occur in the thymic cortex. Using cDNA microarrays, we identified several hundred receptor genes expressed at one or more stages of progenitor differentiation in the cortex. We then applied a variety of prioritization criteria, including the magnitude of dynamic changes in gene expression, known roles in relevant biologic processes (lineage specification, proliferation, survival), and/or the availability of existing genetic models to identify a short list of novel candidate receptors that could interpolate signals from the thymic microenvironment. We show that one of these receptors, Notch3, undergoes dramatic changes in expression at both the RNA and protein levels during lymphoid differentiation in the thymus. Notch3 appears to be functional in thymocyte differentiation because the thymuses of Notch3 mutant mice are slightly smaller, but this change does not appear to be attributable to any specific phase of development. Because Notch3 and Notch1 exhibit overlapping expression patterns, we tested the possibility that Notch1 might compensate for the absence of Notch3, but this does not appear to be the case, nor does Notch3 appear to compensate for the absence of Notch1. Our findings indicate that while Notch3 is dynamically regulated during postnatal thymocyte differentiation, its function is nonessential under normal biologic conditions.
Methods
Mice
All procedures involving animals were reviewed and approved by The Scripps Research Institute Institutional Animal Care and Use Committee. Mice carrying a gene-trapped Notch3 allele (Notch3lacZ/lacZ) were originally generated by a large-scale secretory gene trap screen,20 and were obtained from R. Kopan (Washington University, St Louis, MO) on a partial C56BL/6 background. Mice used in the experiments described here were at least another 8 generations of backcrossing onto the C57BL/6 background. C57BL/6 mice carrying a loxP-flanked Notch1 allele21 and control C57BL/6 mice were obtained from The Jackson Laboratory. Mice transgenic for Lck[Cre]22 were obtained from Taconic. CD45.1 (Ly-5.1) allelic mice were purchased from The Jackson Laboratory. Strains were maintained and animals were bred at The Scripps Research Institute.
Flow cytometric phenotyping and cell sorting
All steps were performed at 4°C or on ice. For microarray analysis of early progenitor cells, thymuses from 10 male C57BL/6 mice at 5 weeks were pooled. Small cortical lymphocytes (CD4+8+ cells) were depleted by buoyant centrifugation in iso-osmolar Opti-Prep density centrifugation medium (Sigma-Aldrich). Depleted cells were stained with a cocktail of lineage Abs recognizing mature T-lineage cells, B cells, granulocytes, monocytes, macrophages, dendritic cells, and erythrocytes, followed by a second depletion of lineage-positive cells using anti–Ig-coated paramagnetic beads (Dynal). The remaining cells were stained with a fluorochome-conjugated anti-lineage Ab, as well as fluorochrome-conjugated Abs recognizing CD24, CD25, or CD44. DN1 (lin−CD24+CD25−CD44hi), DN2 (lin−CD24+CD25+CD44hi), DN3 (lin−CD24+CD25+CD44lo), or pre-DP (lin−CD24+CD25−CD44lo) populations were sorted to > 98% purity. For DP cells (CD4+8+), an aliquot of the thymocyte pools described above was removed before depletion and stained with fluorochrome-conjugated Abs recognizing CD4 or CD8; DP cells were sorted to > 99% purity. Each of these procedures was repeated until sufficient mRNA could be isolated to serve as template for microarray (> 1 μg). PE-conjugated Notch3 mAb was obtained from BioLegend. NK1.1 Ab was purchased from eBioscience. Most other Abs, including those used for phenotypic analysis, were prepared and conjugated in our laboratory. All microarray data are available on the Gene Expression Omnibus (GEO) under accession number GSE30631.
Generation of competitive hematopoietic chimeras
CD45.1 recipient mice (5-6 weeks of age) were given 2 4.5 Gy doses of γ-irradiation spaced 2 hours apart, and were then injected in the retro-orbital plexus with a combination of either CD45.1 plus C57BL/6 (CD45.2+), or CD45.1 plus Notch3lacZ/lacZ (CD45.2+), sex-matched lineage-negative BM (∼ 1 million donor cells total). After 6 to 8 weeks, the proportions of donor (CD45.2+) cells were determined by flow cytometry, as described in the preceding section.
Analysis of cell death by the TUNEL assay
Thymocytes were first stained with the appropriate cocktail of fluorescent Abs as described in “Flow cytometric phenotyping and cell sorting.” Stained cells were washed once in PBS and then resuspended in a solution of 2% CH2O/PBS for 2 hours on ice. Fixed cells were washed once in PBS and resuspended in PBS containing 5% FBS and 0.5% Tween 20 for 30 minutes on ice, followed by 2 washes in PBS. Fixed, permeabilized cells were then subjected to end-labeling of DNA strand breaks using FITC-dUTP and TdT, exactly as described by the manufacturer in the BD Pharmingen Apo-Direct kit, except that 4′6-diamidino2-phenylindole (DAPI; 10 μg/mL) was used for DNA counterstaining instead of propidium iodide.
Analysis of proliferative index by DNA staining
The methods for cell-cycle analysis were essentially as previously described.23 Briefly, cells were stained using the appropriate fluorescent Abs and fixed in CH2O as for the TUNEL assay, followed by resuspension in PBS containing 5% FBS, 0.5% Tween 20, and DAPI at 10 μg/mL. After data acquisition, DNA content analysis was carried out using the cell-cycle algorithms available in FlowJo 8.8.7 (TreeStar). Mitotic index is represented as the mean ± SD of the hyperdiploid singlet component of each cell type (ie, S + G2 + M phase).
RNA isolation, microarray analysis, and PCR
Pools of mRNA thus represented at least 70 mice corresponding to 7 independent cell-sorting events. mRNA was purified using micro RNAeasy columns (QIAGEN). In vitro transcription, probe synthesis and labeling, hybridization to U74Av2 gene chips (Affymetrix), and chip imaging were performed by the Genomics Core Laboratory at the Sloan-Kettering Institute according to the manufacturer's specifications. All quality control metrics were well within the manufacturer's recommendations for quality data. PCR primers for mapping were as follows: Notch3-forward (exon 16), gatcaagacattgacgactgtgac; Notch3-reverse (exon 17), gtcgaggcaagaacaggaaaag; gene trap-forward, gcagggagagttgagatggaagg; gene trap-reverse, ccgtcactccaacgcagcaccatcac. PCR primers for detection of 3′ Notch3 transcripts were as follows: forward (exon 32), tgggaaatctgccttacactgg; reverse (exon 33), agcagcttggcagcctcatag.
Western blotting
Western blotting was performed as previously described.24 Sorted cells were directly lysed in SDS-PAGE loading buffer at 95°C. DNA was sheared by several passages through a 27-gauge needle. Proteins were separated on a 4% SDS–polyacrylamide gel, and transferred to a PVDF membrane. The membrane was incubated with an anti-Notch3 C-terminal Ab (Santa Cruz Biotechnology) overnight at 4°C. An Ab recognizing β-actin (GeneScrips) was used to monitor lane loading. Ab binding was detected by chemoluminescence using the Pierce ECL Western blotting substrate (Thermo Scientific).
Results
Notch3 is differentially expressed during steady-state thymocyte differentiation
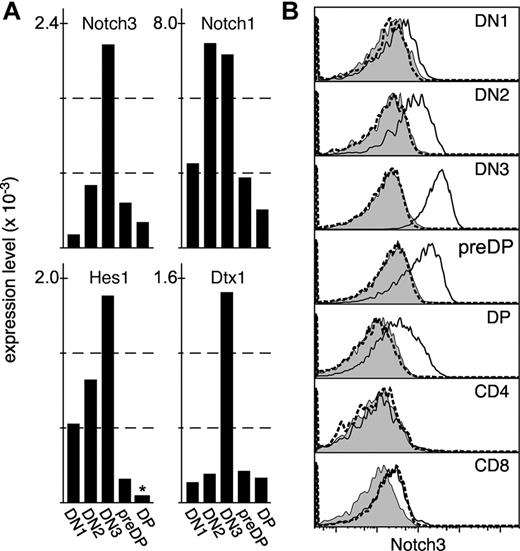
To identify receptors on lymphoid progenitors that might interpolate signals from the thymic microenvironment, we performed several independent microarray series using mRNA from purified thymocyte progenitor stages as a template. In this manuscript, we present data derived from a single Affymetrix gene chip (series U74Av2) for DN1 (lineage−CD24+25−44+), DN2 (lineage−CD24+25+44+), DN3 (lineage−CD24+25+44lo), pre-DP (lineagelowCD24+25−44lo), and DP (CD4+8+) thymocytes. Original datasets can be found at the NCBI Gene Ontology Omnibus. Genes expressed (present) in one or more progenitor stages were identified using the Microarray Suite 5 (MAS5) detection call algorithm (Affymetrix). From these, 398 probesets mapping to the molecular function gene ontology “receptor activity” (GO:0004872) were identified. As a final criterion, receptor gene expression that changed significantly (signal ratio < 0.5 or > 2 and significant MAS5 change call) at any individual developmental transition was identified. The resulting gene list was manually examined for those receptors with known roles in developmental processes relevant to the thymus, such as lineage specification, proliferation, or cell migration (reviewed in Petrie and Zuniga-Pflucker4 ). One such result was Notch3. Notch3 was one of the most-changed receptors expressed during T-cell development, exhibiting an approximately 20-fold change between DN1 and DN3 stages (Figure 1). However, expression was significant at all progenitor stages (probability that differences between matched and mismatched probes for Notch3 is because of chance < 0.04, Tukey range test), suggesting that some level of Notch3 was expressed in all stages tested. In contrast to another study,25 no significant expression was found for Notch2 or Notch4 in this microarray series (not shown); several other microarray series (different gene chip versions) performed by us gave ambiguous results regarding Notch2, with significant expression at all stages for one probeset on these chips, and no significant expression with another (not shown). In any case, Notch2 is unlikely to explain the findings presented here because the patterns of expression found for Notch225 (DN1/DN2 cells) precedes the point at which Notch3 is up-regulated in thymocytes (DN3, see Figure 1). In contrast, we found that Notch1 was coexpressed at all stages where Notch3 was expressed (Figure 1A). Furthermore, most Notch-related gene expression peaked at the DN3 stage, as indicated by the expression patterns of Notch1, Hes1, and Deltex1 (Figure 1A), suggesting that a focus of Notch activity coincides with the coexpression of high levels of both Notch3 and Notch1 at the DN3 stage.
Notch3 is expressed early in thymocyte differentiation. (A) Gene expression as determined by microarray (Affymetrix U74Av2), where the global median (MAS5) is equal to 500. Only data for high-confidence probesets (at) are shown. Expression was statistically significant (MAS 5 detection call of Present) for all genes at all stages except for Hes1 in DP cells, as indicated by an asterisk. Expression of most Notch-related genes, including Notch3, peaks at the DN3 stage. (B) mAb staining of wild-type (solid line) or Notch3-mutant (dashed line) thymocytes; protein expression directly correlates with gene expression data, with the highest levels of expression on DN3 cells. Filled histograms represent isotype-matched control Ab staining on wild-type cells.
Notch3 is expressed early in thymocyte differentiation. (A) Gene expression as determined by microarray (Affymetrix U74Av2), where the global median (MAS5) is equal to 500. Only data for high-confidence probesets (at) are shown. Expression was statistically significant (MAS 5 detection call of Present) for all genes at all stages except for Hes1 in DP cells, as indicated by an asterisk. Expression of most Notch-related genes, including Notch3, peaks at the DN3 stage. (B) mAb staining of wild-type (solid line) or Notch3-mutant (dashed line) thymocytes; protein expression directly correlates with gene expression data, with the highest levels of expression on DN3 cells. Filled histograms represent isotype-matched control Ab staining on wild-type cells.
To confirm this expression at the protein level, we used a commercially available mAb (BioLegend) to screen for surface expression of Notch3 on viable thymocytes. Protein expression at all progenitor stages correlated exactly with message levels (Figure 1B), with low levels on DN1 cells increasing on DN2 and peaking by the DN3 stage, then decreasing but remaining detectable through the DP stage. While we did not perform microarray analysis on mature CD4 or CD8 SP cells in the U74Av2 series, we did screen for protein levels, and found that unlike progenitor cells, mature cells do not appear to express Notch3. The specificity of the Notch3 Ab was confirmed by performing similar staining on thymocytes from mice homozygous for a gene-trapped allele of Notch3,20 which was generally indistinguishable from the staining seen with an isotype-matched control Ab on wild-type thymocytes. Together, these findings indicate that Notch3 is expressed on all thymocyte progenitor stages in a manner that peaks at the DN3 stage, and further, that Notch3 and Notch1 are coexpressed on progenitor thymocytes. Our gene expression data are consistent with previously finding on unfractionated progenitor thymocytes, as described by others.26,27
Characterization of T-lineage development in Notch3 gene-trapped mice
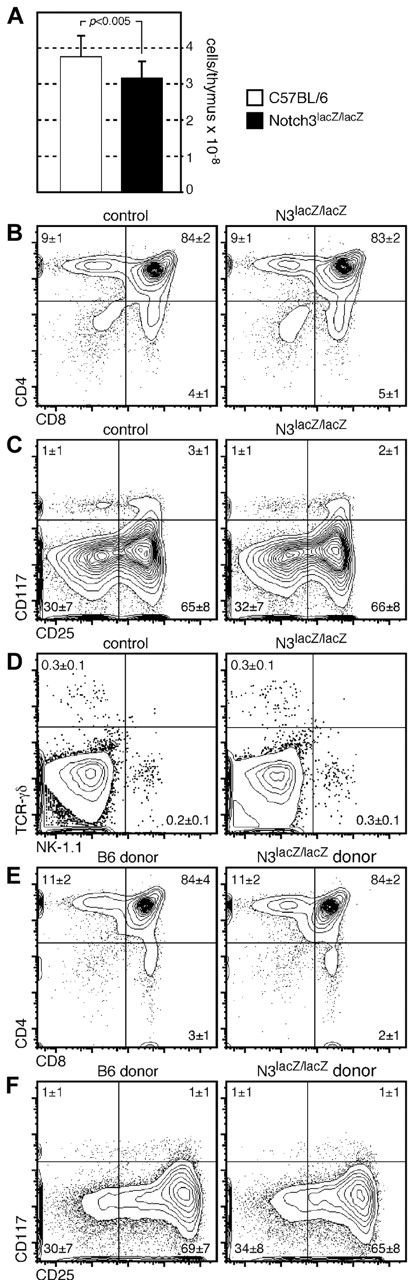
Previous studies with a different strain of Notch3-deficient mice indicated no obvious phenotype in young mice.28 However, data from just one mouse was shown, and because evaluation of CD4−8− subsets indicated an unusual level of DN1 cells in these mice (∼ 30%, vs the more conventional 1%-3%), we decided to more thoroughly evaluate thymocyte differentiation in mice where Notch3 had been inactivated by insertion of a gene trap.20 Mice homozygous for the Notch3 gene trapped allele (referred to as Notch3lacZ/lacZ) were superficially normal and were born at normal Mendelian ratios, as previously described.20,28 We found that thymuses from young (4-5 week old) Notch3lacZ/lacZ mice exhibited a ∼ 15% reduction in cellularity from that found in wild-type controls (Figure 2A). This differs somewhat from the previous study,28 which indicated no change in thymic size in young mice. Despite this difference in cellularity, no obvious phenotypic differences were seen between Notch3lacZ/lacZ and control mice, either at the early progenitor stages, or at more mature stages (Figure 2B-C). The use of CD117 (c-kit) in place of CD44, as was used in the previous study,28 also revealed the presence of normal proportions of CD4−8−cells (particularly DN1 cells) in Notch3lacZ/lacZ mice (Figure 2C), suggesting that these cells do not accumulate abnormally. Absolute numbers of cells at each stage of development in control of N3lacZ/lacZ mice are shown in Table 1; like relative proportions, no significant differences (Student 2-tailed t test for unpaired samples, P < .1) were found. Both NKT cells and TCRγδ lineage cells were found in normal proportions in N3lacZ/lacZ mice (Figure 2D). Together, these findings indicate that Notch3 is not absolutely required for either these alternate lineages, or for mainstream TCRαβ cell development in the thymus.
Thymocytes from mice with a gene-trapped Notch3 allele exhibit slightly reduced cellularity, but otherwise develop normally. (A) Absolute cell counts in thymuses from mice homozygous for a targeted (lacZ gene trapped) Notch3 allele, or wild-type control mice; a ∼ 15% reduction P < .005, Student 2-tailed t test for independent samples) in cellularity is seen in Notch3lacZ/lacZ mice (n = 15 for each type of mice). Nonetheless, the overall proportion of cells at each developmental stage, as defined by CD4 versus CD8 (B), or CD117 versus CD25 on lineage-negative cells (C) was not different in targeted versus control mice. The frequency of cells in the TCR-γδ or NK lineages was also nearly identical in Notch3lacZ/lacZ and control mice (D). (E-F) The phenotype of cells developing in the thymus of competitive chimeras constructed from N3lacZ/lacZ or wild-type donor cells mixed with allelically marked (CD45.1+) donor cells. Again, no substantial differences in phenotype were noted.
Thymocytes from mice with a gene-trapped Notch3 allele exhibit slightly reduced cellularity, but otherwise develop normally. (A) Absolute cell counts in thymuses from mice homozygous for a targeted (lacZ gene trapped) Notch3 allele, or wild-type control mice; a ∼ 15% reduction P < .005, Student 2-tailed t test for independent samples) in cellularity is seen in Notch3lacZ/lacZ mice (n = 15 for each type of mice). Nonetheless, the overall proportion of cells at each developmental stage, as defined by CD4 versus CD8 (B), or CD117 versus CD25 on lineage-negative cells (C) was not different in targeted versus control mice. The frequency of cells in the TCR-γδ or NK lineages was also nearly identical in Notch3lacZ/lacZ and control mice (D). (E-F) The phenotype of cells developing in the thymus of competitive chimeras constructed from N3lacZ/lacZ or wild-type donor cells mixed with allelically marked (CD45.1+) donor cells. Again, no substantial differences in phenotype were noted.
Absolute numbers of thymocytes (in millions)
| . | DN1 . | DN2 . | DN3 . | Pre-DP . | DP . | CD4 . | CD8 . |
|---|---|---|---|---|---|---|---|
| Wild type | 0.13 ± 0.07 | 0.32 ± 0.13 | 7.8 ± 1.9 | 3.1 ± 0.6 | 287 ± 37 | 30 ± 4 | 13 ± 2 |
| Notch3lacz | 0.09 ± 0.04 | 0.21 ± 0.07 | 7.1 ± 2.0 | 3.4 ± 1.2 | 264 ± 48 | 29 ± 4.5 | 15 ± 4.8 |
| . | DN1 . | DN2 . | DN3 . | Pre-DP . | DP . | CD4 . | CD8 . |
|---|---|---|---|---|---|---|---|
| Wild type | 0.13 ± 0.07 | 0.32 ± 0.13 | 7.8 ± 1.9 | 3.1 ± 0.6 | 287 ± 37 | 30 ± 4 | 13 ± 2 |
| Notch3lacz | 0.09 ± 0.04 | 0.21 ± 0.07 | 7.1 ± 2.0 | 3.4 ± 1.2 | 264 ± 48 | 29 ± 4.5 | 15 ± 4.8 |
The appearance of a slight reduction in cellularity in the thymuses of N3lacZ/lacZ mice (Figure 2A) led us to consider that such cells might simply lack competitive fitness, and that the full phenotype would not be revealed in the absence of competition. To more stringently test the qualitative nature of N3-targeted cells, we generated competitive hematopoietic chimeras in CD45.1 allelic mice (Figure 2E-F), by transplanting a mixture of either N3lacZ/lacZ or wild-type lineage-negative BM (CD45.2+) together with lineage-negative CD45.1+ BM. Even in this competitive environment, where wild-type (CD45.1 donor) cells would compete with N3lacZ/lacZ donor cells for niche space and nutrient availability, no differences could be found in the phenotype of N3lacZ/lacZ progeny and that of wild-type controls.
Reduction in thymic cellularity in N3lacZ/lacZ mice is not explained by differences in proliferation or cell death
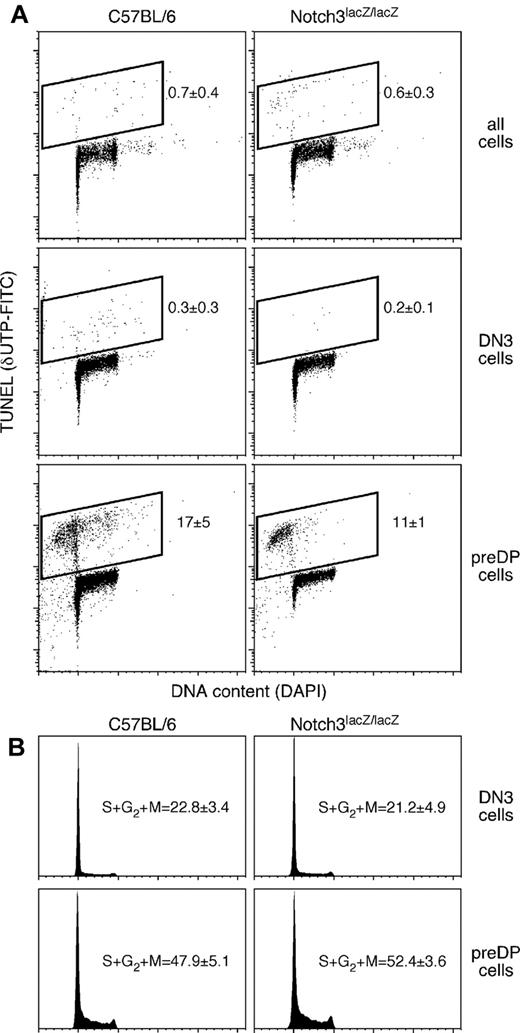
The presence of a slightly smaller thymus in N3lacZ/lacZ mice (Figure 2) suggests that either differentiation, proliferation, or cell survival are impaired in the absence of Notch3. The data shown in Figure 2 indicate that thymocytes from mice homozygous for the N3lacZ allele are capable of normal differentiation. To further probe the mechanism leading to a smaller thymus in N3lacZ/lacZ mice, we assessed proliferation and cell survival in control or N3lacZ/lacZ mice. The data are summarized in Figure 3. Because most thymocytes are poised to die, even isolation procedures can affect assays involving membrane permeability or polarity (dye exclusion, annexin V staining). However, DNA double strand breaks require specific enzymatic activity, and in cells that are kept cold during isolation, are not generated spontaneously. Consequently, as a stringent ex vivo assay for cell death occurring in vivo, we performed DNA end labeling using TdT and FITC-labeled d-UTP (TUNEL). No detectable differences were seen between thymocytes from mice homozygous for the lacZ allele and control (C57BL/6) mice among in DN3 cells (the highest Notch3-expressing cells in the thymus, see Figure 1) or among total thymocytes. Even in pre-DP thymocytes, which exhibit the highest detectable levels of DNA strand breaks among all thymocytes, were not different between N3lacZ/lacZ and control mice. These findings indicate that neither stage-specific nor overall cell death are affected by the absence of Notch3. Similarly, cell-cycle status, as assessed by quantitative assessment of nuclear DNA, was not affected by the absence of Notch3 (Figure 3B). Thus, our findings do not pinpoint a molecular mechanism for a slightly smaller thymus in N3lacZ/lacZ mice, although we cannot rule out the possibility (without performing an exceptionally large number of experiments) that differentiation, proliferation, or cell survival are affected by Notch3, but at levels that are below our ability to measure. It should be noted that even very small changes could lead to a 15% reduction in thymus cellularity, particularly if spread across several stages and/or serial cell divisions.
Thymocytes from mice with a gene-trapped Notch3 allele exhibit normal levels of cell death and cell proliferation. (A) Labeling of DNA strand breaks as an indicator of cell death, using dUTP and TdT (TUNEL). Control (C57BL/6) and Notch3lacZ/lacZ mice showed indistinguishable levels of cell death (Student t test, P > .05) among total thymocytes (top), DN3 thymocytes (representing Notch3 peak expression, see Figure 1), or pre-DP thymocytes (the immediate progeny of Notch3-expressing DN3 cells). Statistics indicate mean ± SD for TUNEL+ cells in 3-5 independent experiments. (B) Cell-cycle status (DNA content, assessed by DAPI staining) in DN3 or pre-DP cells from control or Notch3lacZ/lacZ thymuses; again, no significant differences could be distinguished (Student t test P > .05; statistics indicate mean ± SD for hyperdiploid cells in 5 or more independent experiments). Cell-cycle statistics include both TUNEL-negative cells from cell death experiments (A) and additional experiments where TUNEL was not performed, because there was no difference in the measured proliferative index with these 2 protocols.
Thymocytes from mice with a gene-trapped Notch3 allele exhibit normal levels of cell death and cell proliferation. (A) Labeling of DNA strand breaks as an indicator of cell death, using dUTP and TdT (TUNEL). Control (C57BL/6) and Notch3lacZ/lacZ mice showed indistinguishable levels of cell death (Student t test, P > .05) among total thymocytes (top), DN3 thymocytes (representing Notch3 peak expression, see Figure 1), or pre-DP thymocytes (the immediate progeny of Notch3-expressing DN3 cells). Statistics indicate mean ± SD for TUNEL+ cells in 3-5 independent experiments. (B) Cell-cycle status (DNA content, assessed by DAPI staining) in DN3 or pre-DP cells from control or Notch3lacZ/lacZ thymuses; again, no significant differences could be distinguished (Student t test P > .05; statistics indicate mean ± SD for hyperdiploid cells in 5 or more independent experiments). Cell-cycle statistics include both TUNEL-negative cells from cell death experiments (A) and additional experiments where TUNEL was not performed, because there was no difference in the measured proliferative index with these 2 protocols.
The gene-trapped Notch3 allele is a null allele
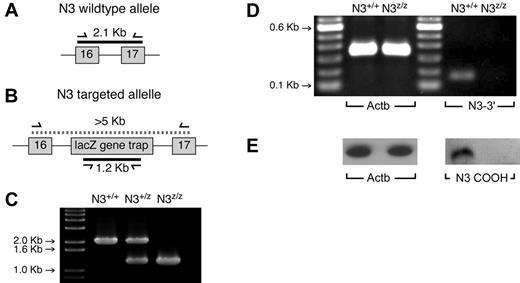
The experiments described above indicate that differentiation, proliferation, and survival occur normally in homozygous N3lacZ mice, and that unlike Notch1, Notch3 is not absolutely essential for these processes, even though it is expressed (Figure 1). One alternative explanation was that because the Notch3 strain we used was generated randomly by gene trap insertion,20 the targeted allele might retain partial function. The intron between exons 16 and 17 of Notch3 was preliminarily identified as the likely insertion site by a BLAST search of 5′ RACE sequencing results.20 Insertion of the gene trap sequence at this point, including a stop codon after the lacZ reporter gene,20 would result in a lacZ fusion protein containing only the secretory signal peptide and limited number of (extracellular) EGF repeats from Notch3. To confirm this, we designed primers spanning parts of exons 16 and 17 and the intronic sequence between them, which would generate a 2.1-kb amplicon using genomic DNA as template (Figure 4A). Disruption of this region by insertion of the gene trap vector20 would generate an amplicon too large to be efficiently detected by conventional PCR, resulting in the absence of a genomic band. A second set of primers was designed to specifically amplify a 1.2-kb amplicon corresponding to the lacZ region of the gene trap construct (Figure 4B). As shown in Figure 4C, PCR amplification using both sets of primers on wild-type (Notch3+/+), homozygous Notch3lacz/lacz, or heterozygous (Notch3+/lacZ) DNA template confirms that the location of the gene trap insertion is between Notch3 exons 16 and 17.
The gene-trapped Notch3 allele is a null allele. Random insertion of the lacZ gene trap was suspected to be in the vicinity of exons 16-17 of Notch3. (A) The location of a 2.1-kb amplicon resulting from a forward primer in exon 16 and a reverse primer in exon 17. (B) Insertion of the lacZ gene trap would generate an amplicon too large for conventional PCR using these primers; presence of the gene trap can also be confirmed using gene trap-specific primers. (C) The expected results are found using DNA from wild-type mice, homozygous gene-trapped mice, or heterozygous mice. (D) To exclude the possibility of alternate transcripts that might skip the gene-trap insert, PCR was performed in the 3′ region of the transcript (spanning exons 32 and 33), revealing that no detectable transcripts could be found. (E) An Ab recognizing the C-terminal (intracellular) domain of Notch3 was used to probe extracts from N3lacZ/lacZ or wild-type lineage-negative thymocytes. No detectable protein could be found. Together these experiments confirm that the gene-trapped allele is a null allele.
The gene-trapped Notch3 allele is a null allele. Random insertion of the lacZ gene trap was suspected to be in the vicinity of exons 16-17 of Notch3. (A) The location of a 2.1-kb amplicon resulting from a forward primer in exon 16 and a reverse primer in exon 17. (B) Insertion of the lacZ gene trap would generate an amplicon too large for conventional PCR using these primers; presence of the gene trap can also be confirmed using gene trap-specific primers. (C) The expected results are found using DNA from wild-type mice, homozygous gene-trapped mice, or heterozygous mice. (D) To exclude the possibility of alternate transcripts that might skip the gene-trap insert, PCR was performed in the 3′ region of the transcript (spanning exons 32 and 33), revealing that no detectable transcripts could be found. (E) An Ab recognizing the C-terminal (intracellular) domain of Notch3 was used to probe extracts from N3lacZ/lacZ or wild-type lineage-negative thymocytes. No detectable protein could be found. Together these experiments confirm that the gene-trapped allele is a null allele.
Because alternate splice forms have been reported for other Notch receptors (for an example, see Gomez-del Arco et al29 ), we tested whether the Notch3 gene-trapped allele might encode functional intracellular domains using 2 different approaches. First, we designed primers spanning 2 adjacent exons in the 3′ Notch3 genomic sequence (Figure 4D). Second, we used an Ab raised against the C-terminal domain of Notch3 to probe cell lysates from lineage-negative thymocytes from wild-type or N3lacZ/lacZ mice (Figure 4E). Both of these approaches confirm the predictions of our mapping of the gene trap insert, confirming that the gene-trapped allele represents a null allele (subsequently designated Notch3−/−).
Notch3 function in thymocytes is not masked by the simultaneous function of Notch1
The dynamic regulation of Notch3 during thymocyte differentiation (Figure 1) suggests a function for this receptor in thymic progenitor cells. A function in thymocytes is further substantiated by the findings that expression of a constitutively activated form of Notch3 can alter T-cell development.27 The absence of a significant phenotype in Notch3−/− mice thus seems somewhat surprising. However, Notch1 is also expressed early in thymocyte differentiation,26,30 and is coexpressed with Notch3 in progenitor thymocytes (Figure 1). Because Notch1 can compensate for the function of other Notch family members in other cell types,31-33 we reasoned that it was possible that presence of a functional Notch1 protein could be masking the requirement for Notch3 in developing thymocytes. To test this, we used the same strain of Lck[Cre] previously shown to enforce complete deletion of a loxP-flanked Notch1 allele by the DN3 stage of development.34 Mice carrying various combinations of the Lck[Cre] transgene,22 the Notch1-floxed allele,21 or the Notch3 null allele20 were generated. As was found for mice lacking Lck[Cre] (Figure 2A), thymuses from mice with an intact Notch1 gene but lacking Notch3 (Lck[Cre]+/−Notch1+/+Notch3−/−) were slightly smaller than those found in control mice (Lck[Cre]+/− only; Figure 5A). Conditional deletion of Notch1 in mice with an intact Notch3 gene (Lck[Cre]+/−Notch1fl/flNotch3+/+) resulted in a significant (4-fold) reduction in thymic size, consistent with previously published results.34 However, thymuses from mice that were deficient in both Notch1 and Notch3 (Lck[Cre]+/−Notch1fl/flNotch3−/−) were of virtually the same size as mice where only Notch1 was mutated, showing that deletion of Notch3 did not exacerbate the Notch1 phenotype. Consistent with this data on thymic cellularity, the phenotype in control or Notch3-only mutants were similar, while Notch1-only mutation resulted in an accumulation of CD4−8− cells (consistent with previously published results34 ) that was not changed by simultaneous deletion of Notch3 (Figure 5B-E). These results indicate that the absence of a major thymic phenotype in Notch3−/− mice does not result from compensatory function provided by Notch1. Simultaneously, because the absence of Notch3 does not compound the effects of conditional Notch1 deletion, it can be concluded that the relatively minor phenotype seen after intrathymic deletion of Notch1 (Figure 5, and Wolfer et al34 ) does not indicate a partial rescue of the Notch1 phenotype by the function of Notch3. Together, these findings indicate that while Notch1 has a clear and essential role very early in thymocyte differentiation, both are somewhat dispensable later in progenitor development, despite being expressed at their highest levels.
Nonoverlapping functions for Notch1 and Notch3 in thymocyte differentiation. (A) Absolute cellularity in 5 or more thymuses from mice expressing a conditional deletion of Notch1 (N1−; Notch1fl/fl), germline deletion of Notch3 (N3−; Notch3lacZ/lacZ), mice with mutations in both alleles (N1−N3−), or control mice (N1+N3+). Note that all mice used for experiments shown in this figure were heterozygous for lck[Cre]. (B-E) CD4/CD8 staining on total thymocytes (right) or CD117/CD25 staining on lineage-negative thymocytes (left) from N1+N3+, N1+N3−, N1−N3+, or N1−N3− mice, respectively. Both cellularity and phenotype reflect the status of the Notch1 allele, and other than a small decrease in cellularity (similar to that found in N3− mice lacking Lck[Cre], see Figure 2), mutation of Notch3 does not affect the outcome independently of the status of Notch1.
Nonoverlapping functions for Notch1 and Notch3 in thymocyte differentiation. (A) Absolute cellularity in 5 or more thymuses from mice expressing a conditional deletion of Notch1 (N1−; Notch1fl/fl), germline deletion of Notch3 (N3−; Notch3lacZ/lacZ), mice with mutations in both alleles (N1−N3−), or control mice (N1+N3+). Note that all mice used for experiments shown in this figure were heterozygous for lck[Cre]. (B-E) CD4/CD8 staining on total thymocytes (right) or CD117/CD25 staining on lineage-negative thymocytes (left) from N1+N3+, N1+N3−, N1−N3+, or N1−N3− mice, respectively. Both cellularity and phenotype reflect the status of the Notch1 allele, and other than a small decrease in cellularity (similar to that found in N3− mice lacking Lck[Cre], see Figure 2), mutation of Notch3 does not affect the outcome independently of the status of Notch1.
Deletion of Notch1 in progenitor thymocytes does not result in the absence of Notch3
Various indirect approaches have suggested that the Notch3 gene is a transcriptional target of Notch1.35-38 Consequently, we reasoned that Notch1-deficient thymocytes might also be Notch3-deficient, thus explaining the similarities in phenotype between Notch1-only mutants and Notch1/Notch3 double mutant mice (Figure 5). To test this, we evaluated the surface expression of Notch3 protein on thymocytes in which Notch1 has been conditionally deleted (Lck[Cre]+/−Notch1fl/fl). Figure 6 shows that Notch3 expression on DN3 thymocytes from Lck[Cre]+/−Notch1fl/fl mice was indistinguishable from that found in DN3 cells from control (Lck[Cre] only). Even at the pre-DP stage, which represents cells 2-3 days older than DN3 cells,19,39,40 no differences can be detected, although overall levels of Notch3 are lower in both cell types, as they are in wild-type cells (Figure 1). Similar results can be found for cells a the DP stage (not shown). Overall, these experiments indicate that Notch3 levels are not affected by the presence or absence of Notch1 in thymocytes, and thus, that the similarities between Notch1-only mutation and Notch1/Notch3 double mutation (Figure 5) result from nonoverlapping functions for these 2 proteins.
Notch1-deficient cells continue to express Notch3. Thymocytes from mice carrying a conditionally deleted allele of Notch1 (Lck[Cre]+/−Notch1fl/fl dashed line), control mice (Lck[Cre] only, solid line) were stained using an Ab specific for Notch3. No reduction was found in the levels of surface-expressed Notch3 in thymocytes lacking Notch1, even in pre-DP cells, by which stage Lck[Cre]-mediated deletion is long since finished. These findings show that Notch3 is not regulated by Notch1, and thus the absence of a more severe thymic phenotype in Notch1/3 double mutant mice (Figure 5) is not because of the absence of Notch3 in Notch1-mutant mice. Filled histograms indicate staining of a nonspecific isotype-matched control Ab on control cells.
Notch1-deficient cells continue to express Notch3. Thymocytes from mice carrying a conditionally deleted allele of Notch1 (Lck[Cre]+/−Notch1fl/fl dashed line), control mice (Lck[Cre] only, solid line) were stained using an Ab specific for Notch3. No reduction was found in the levels of surface-expressed Notch3 in thymocytes lacking Notch1, even in pre-DP cells, by which stage Lck[Cre]-mediated deletion is long since finished. These findings show that Notch3 is not regulated by Notch1, and thus the absence of a more severe thymic phenotype in Notch1/3 double mutant mice (Figure 5) is not because of the absence of Notch3 in Notch1-mutant mice. Filled histograms indicate staining of a nonspecific isotype-matched control Ab on control cells.
Discussion
The Notch family of cell-surface receptors (Notch 1-4 in mice and humans) transmits essential signals regulating lineage specification, proliferation, and/or survival in multiple tissue types. It is now well documented that Notch1 plays a critical role in T-cell development in the thymus (for examples, see Krueger et al,5 Schmitt et al,7 Feyerabend et al,8 Radtke et al,30 Wolfer et al,34 Robey et al,41 and Sambandam et al42 ). In contrast, the roles of other Notch receptors in thymocyte differentiation are less clear. In the postnatal thymus, the best evidence for a contribution by Notch receptors other than Notch1 comes from studies on constitutive overexpression of Notch3, which results in developmental defects and T-cell malignancies,43 and rescues T-cell development in mice lacking the pre-TCR.43 These studies strongly implicate Notch3 in normal T-cell development in the thymus.
We independently identified Notch3 as a candidate signal for thymocyte differentiation using an informatic approach. The Notch3 gene was expressed at significant levels in all progenitor populations, but at trace levels early and late (DN1 and DP), while peaking at the DN3 stage. This differential expression was verified at the protein level using a mAb. Interestingly, most Notch gene activity, including Notch1, Deltex1, and Hes1, also peaked at the DN3 stage together with Notch3, implying a focus for Notch activity at this stage. Consequently, we were surprised to find that disruption of Notch3, using a gene-trap insertion,20 did not have a major effect on thymocyte differentiation. We realized that there were several possible alternative explanations for this outcome. Because the Notch3 strain we used was generated in a gene-trap screen, where insertion sites are somewhat imprecise, it was possible that a hypomorphic allele may have been generated. However, precise mapping of the position of the insert to the intron between exons 16 and 17 suggests that even if the upstream regions were transcribed and translated, they would encode only extracellular domains (EGF repeats), and thus could not carry out signaling or transactivation functions. Consequently, the gene-trap allele is a functional knockout allele.
We also realized that because Notch3 peak expression correlates with Notch1 peak expression (see Figure 1), Notch1 function might compensate for the absence of Notch3. This is especially relevant because Notch1 and Notch3 have been shown to have overlapping functions in other tissues (for examples, see Krebs et al,31 Tanigaki et al,32 and Demehri et al33 ). To test this, we generated thymocytes that lacked both Notch1 and Notch3. Because germline inactivation of Notch1 is embryonically lethal, we used a conditional (loxP-flanked) Notch1 allele,21 and induced deletion in thymocytes by using an Lck[Cre] transgenic strain22 that has been previously shown to efficiently target the DN3 stage.34 We found that thymus cellularity and phenotype were not different between thymocytes lacking both Notch1 and Notch3 and those lacking Notch1 only, suggesting that functional Notch1 was not masking a more severe phenotype in Notch3 mutant thymocytes. Simultaneously, these experiments reveal new insights into the relatively mild phenotype resulting from intrathymic (Lck[Cre]-mediated) deletion of Notch1, which could reflect a compensatory function for Notch3, because both Notch1 and Notch3 are expressed in DN1, DN2, and DN3 cells (Figure 1). Our findings indicate that this is not the case, and thus, that the Notch1 and Notch3 have nonoverlapping functions in the thymus. Our findings also indicate that while Notch1 signaling is critical very early in thymocyte differentiation,26,30 neither Notch1 nor Notch3 are absolutely essential later in the process, although both play a more subtle role. Obviously, the reduction in cellularity after conditional intrathymic deletion of Notch1 (∼ 70%) is substantially more than that seen after deletion of Notch3 (∼ 15%), suggesting that although neither receptor is absolutely required, Notch1 remains a more dominant signal. It has been shown that compared with Notch1, the Notch3 intracellular domain is diminished in its ability to activate canonical Notch target genes,44 and the milder thymic phenotype seen after Notch3 deletion may simply reflect this trait.
Although we evaluated virtually all possible functions (differentiation, proliferation, survival) in lymphoid cells, we were not able to identify the cause of the reduced cellularity in Notch3−/− mice, although as noted earlier, given that lymphoid progenitor cells in the thymus undergo multiple rounds of cell division at each stage of development, even miniscule changes in one of these functions could explain a 15% decrease in cellularity. We do not believe that the Notch3 mutation affects stromal cells because we do not find Notch3 to be expressed at significant levels by thymic stromal cells,45 and further, transplantation of wild-type BM into irradiated Notch3-deficient or wild-type mice resulted in normal lymphoid development (data not shown). In any case, this ∼ 15% reduction, and even the ∼ 70% reduction seen after intrathymic deletion of Notch1, is unlikely to be biologically significant, because peripheral T-lymphoid cellularity remains undisturbed despite the reduction in thymic size (not shown). We note that in contrast to our findings, previous characterization of another strain of Notch3 mutant mice28 did not reveal a difference in thymic cellularity in young mice. This may be a function of the number of mice required to achieve statistical significance for this minor phenotype because when we randomly select a similar number of thymuses (5 control, 5 Notch3−/−) from our data, no statistical significance in the differences is achieved.
Previous publications have suggested that Notch3 may be a transcriptional target of Notch1.35-38 Consequently, the absence of a synergistic effect in Notch1/Notch3 double mutant cells could be explained by the generation of a functional double mutant when Notch1 is deleted. Meta-data analysis of Notch1 gene expression also identified Notch3 as one of the best-correlated genes across a broad range of experiments involving mouse developmental systems.46 In thymocytes, we find that Notch3 is expressed at normal levels despite deletion of Notch1, indicating that, at least in this tissue, Notch3 is not a transcriptional target of Notch1 activity.
In this manuscript, we show that although Notch3 is indeed expressed and is dynamically regulated during thymocyte differentiation, it is not absolutely essential for the success of this process. In this context, it would appear that the profound phenotype seen in response to enforced Notch3 expression in thymocytes27,43 may result either from superphysiologic levels of transgene expression, or from the use of a construct encoding a constitutively activated form of the protein. Like those studies, our results indicate that Notch3 does indeed play some role in thymocyte differentiation, although our studies suggest that its influence is rather subtle, and probably not significant at the biologic level. However, we cannot rule out the possibility that other Notch receptors (Notch2 or Notch4) may be up-regulated in the absence of Notch1 and Notch3, and compensate for their absence, although this event seems unlikely.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Spyros Artavanis-Tsakonis (Harvard University) and Kevin Mitchell (Trinity College Dublin) for input regarding the gene trap insertion in Notch3 mutant mice; Dr Rafael Kopan (Washington University, St Louis, MO) for providing Notch3 mutant mice and for helpful conversations; and Dr William Skarnes (Sanger Institute, Hinxton, United Kingdom) for permission to obtain Notch3 mice.
This work was supported by Public Health Service (PHS) grants AG034876 and AI095831. J.S. is supported by a fellowship from the Frenchmen's Creek Women for Cancer Research.
National Institutes of Health
Authorship
Contribution: J.S. designed and performed wet-laboratory experiments, and contributed to the preparation of the manuscript; M.F. performed informatics analysis and contributed to the preparation of the manuscript; J.L. performed wet-laboratory experiments; and H.T.P. performed some of the wet-laboratory experiments, contributed to informatics analysis, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Howard T. Petrie, The Scripps Research Institute, 130 Scripps Way, 2C1, Jupiter, FL 33458; e-mail: hpetrie@scripps.edu.

![Figure 5. Nonoverlapping functions for Notch1 and Notch3 in thymocyte differentiation. (A) Absolute cellularity in 5 or more thymuses from mice expressing a conditional deletion of Notch1 (N1−; Notch1fl/fl), germline deletion of Notch3 (N3−; Notch3lacZ/lacZ), mice with mutations in both alleles (N1−N3−), or control mice (N1+N3+). Note that all mice used for experiments shown in this figure were heterozygous for lck[Cre]. (B-E) CD4/CD8 staining on total thymocytes (right) or CD117/CD25 staining on lineage-negative thymocytes (left) from N1+N3+, N1+N3−, N1−N3+, or N1−N3− mice, respectively. Both cellularity and phenotype reflect the status of the Notch1 allele, and other than a small decrease in cellularity (similar to that found in N3− mice lacking Lck[Cre], see Figure 2), mutation of Notch3 does not affect the outcome independently of the status of Notch1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/9/10.1182_blood-2011-04-346726/4/m_zh89991177600005.jpeg?Expires=1769080787&Signature=vpH36DRgJ8GhFHnW-mkiCpykq33POFGIIsrTE3zewj02N6kIbMf1V1AI3nlKn9b8-FIp2x-ux0Lu0uMBDVBdyXPItoVw804ZU0Ym7ir~jbot19JHRb5vLmMoIT5BkqjbQWNK9EM7SRDw20MtCDxxcMnClfBmvKlrjHYw4fdu6jQL3opyt~GWP16gAh~2bkDWJL1T6nKZvj0dUUj~AAkceeYrBZ9pdSAZy-4ZmKctE~2QCSBR1DXKdHp-ACUoNYic39VFoODBjGxuhpdCimJsYfQb0zZD1t2qXXACIOIVlvRZTNotxJC0KZL1jRexVZNX35zoooQXQdq8WR28r9WZBg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. Notch1-deficient cells continue to express Notch3. Thymocytes from mice carrying a conditionally deleted allele of Notch1 (Lck[Cre]+/−Notch1fl/fl dashed line), control mice (Lck[Cre] only, solid line) were stained using an Ab specific for Notch3. No reduction was found in the levels of surface-expressed Notch3 in thymocytes lacking Notch1, even in pre-DP cells, by which stage Lck[Cre]-mediated deletion is long since finished. These findings show that Notch3 is not regulated by Notch1, and thus the absence of a more severe thymic phenotype in Notch1/3 double mutant mice (Figure 5) is not because of the absence of Notch3 in Notch1-mutant mice. Filled histograms indicate staining of a nonspecific isotype-matched control Ab on control cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/9/10.1182_blood-2011-04-346726/4/m_zh89991177600006.jpeg?Expires=1769080787&Signature=RfgSeeCgR9WpWnpnGDs10swCsBJggALD2bcjE9Z93ecALqwp5jg0CXqnf3tIh2ZQxeYZowCViasM8MXJmzIeW6~pnSg40J6LwKUvkdbszGmLVYSur3uJpk6FKFPDhHEG8sE-cWqYK18lqdIMb8R2urnUZClkkWwYEkQdPfqI2f-W9hSznn~FrRqytNwsVaH6QDzFiJm39gi~kCIuyxy7QqxVTQSSEKkHUnsSss~DOl0tvrp55CYcPLD2il0rrucCszaC3pKKgrP0-5QlQ0UGN4MDJ-7c8JcKdKxummTFwMv714E0zrkSXYW0ANpdEYgflo2xCwu7vmG5othdEP1T0A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal