Abstract
IFNα, a cytokine with multiple functions in innate and adaptive immunity and a potent inhibitor of HIV, exerts antiviral activity, in part, by enhancing apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3 (APOBEC3) family members. Although IFNα therapy is associated with reduced viral burden, this cytokine also mediates immune dysfunction and toxicities. Through detailed mapping of IFNα receptor binding sites, we generated IFNα hybrids and mutants and determined that structural changes in the C-helix alter the ability of IFN to limit retroviral activity. Selective IFNα constructs differentially block HIV replication and their directional magnitude of inhibition correlates with APOBEC3 levels. Importantly, certain mutants exhibited reduced toxicity as reflected by induced indoleamine 2,3-dioxygenase (IDO), suggesting discreet and shared intracellular signaling pathways. Defining IFN structure and function relative to APOBEC and other antiviral genes may enable design of novel IFN-related molecules preserving beneficial antiviral roles while minimizing negative effects.
Introduction
Highly active antiretroviral therapy (HAART) may not eradicate all long-lived and productive reservoirs of HIV, including macrophages,1-3 and additional approaches to viral suppression continue to center on host cell-derived molecules that antagonize the virus life cycle. Although hundreds of host cell factors are required by HIV for successful infection4 and may serve as potential intervention targets, considerable effort has focused on those endogenous factors that contribute to cellular resistance to HIV. Among the innate intracellular HIV antagonists are the apolipoprotein-B mRNA-editing enzyme-catalytic polypeptide-like 3 (APOBEC3) family of cytidine deaminases,5,6 and members of the tripartite motif family (TRIM),7 but HIV has the potential to effectively eliminate or neutralize these resistance factors.
In vitro studies demonstrated that IFNα enhances innate antiviral molecules including APOBEC family members as a counter-maneuver to HIV viral infectivity factor (VIF).8,9 Importantly, treatment of HIV and HCV coinfected patients with pegylated-IFNα-2a (peg-IFNα) and Ribavirin or HIV mono-infected patients with peg-IFNα only10 resulted in enhanced APOBEC expression in PBMCs along with other IFN stimulated genes (ISGs).10 These studies illustrate the potential to alter the virus-host imbalance and suppression of de novo synthesis of HIV. Despite these promising findings, the use of IFNα therapeutically is fraught with potential side effects, including cytotoxicity, immune dysregulation, neuropathicity and apoptosis.11-14 Although the use of peg-IFNα with its longer half-life has enabled reductions in levels of administered cytokine on a less frequent regimen, toxicity remains a deterrent.
Based on the potential utility of IFNα in the arsenal of anti-HIV therapies, we have attempted to determine whether its antiviral capacities can be dissociated from its toxic functions. Dissection of the IFNα molecule through generation of IFNα hybrids and mutants enabled us to examine whether there are structural determinants that differentially favor signaling pathways with regard to generation of molecules linked to antiviral activity (APOBEC, TRIM, PKR)7,9,15 compared with IFN-inducible agents of toxicity, as exemplified by indoleamine 2,3-dioxygenase (IDO) and TNFα.13,14,16 Alterations in the C-helix (amino acids 75-95) of IFN molecule, specifically amino acid residues 86 and 90, previously associated with changes in its antiproliferative activity in Daudi cell line,17 modified antiviral properties of the intact cytokine. Here we show that exposure of CD4+CCR5+ human macrophages to mutated IFN constructs results in differential STAT phosphorylation and suppression of HIV production. Using IDO as an indicator of toxicity, based on its recognition as a marker for IFN-therapy–induced psychiatric disorders and for disease progression and negative prognosis in HIV infection,13,18-20 we demonstrate that depending on the structural modifications of the IFN amino acid sequence, IDO and APOBEC could be differentially regulated. These findings provide insight into how structural alterations in IFNα may modify its functional repertoire to enhance the antiviral benefit-to-risk ratio.
Methods
Peripheral blood monocyte isolation and differentiation to macrophages
Human PBMCs from leukapheresis of healthy volunteers (Department of Transfusion Medicine, National Institutes of Health, Bethesda, MD) were diluted in endotoxin-free–PBS without Ca2+/Mg2+ (BioWhittaker), and separated on lymphocyte sedimentation medium (Organon-Teknika) before counterflow centrifugal elutriation to obtain T lymphocytes and monocytes without monocyte activation.15 Cells were phenotypically characterized as previously described.8 For differentiated macrophages, monocytes were adhered in serum-free DMEM supplemented with 2mM l-glutamine, 50 μg/mL gentamicin (Mediatech Inc) at 37°C and 5% CO2 in 6-well plates or 96-well plates (Corning) for 2-4 hours, 10% heat-inactivated FBS added and the cells cultured for 6-7 days.9
Infection with HIV-1
Macrophages were incubated with the R5 strain HIV-1BaL (25μl TCID50 = 104/mL; Advanced Biotechnologies Inc) for 2 hours at 37°C. After infection, cells were washed and cultured with DMEM 10% FCS for 10-14 days and 50% supernatant collected and replaced with fresh DMEM twice-weekly. IFNα and IFN constructs were added at indicated concentrations once after removal of excess virus.8,9 In indicated experiments, anti-CD118 (0.1 μg/mL); anti-IFNα/β receptor-chain 2 (IFNAR2; PBL Biologic Laboratories) was added to macrophage cultures after infection and before addition of IFN. Viral replication in macrophages was measured by supernatant p24 levels by ELISA (Perkin-Elmer Life Sciences) 10-14 days postinfection. Infection of activated (anti-CD3/anti-CD28) human T lymphocytes was determined as described.15
IFN and IFN constructs
Expression and purification of IFN proteins were previously described,17,21 purity evaluated by SDS-PAGE, and concentrations determined using Bradford protein quantification assay (Pierce) and absorbance at 280 nm (Nanodrop spectrophotometer, ThermoScientific). Biologic activities of IFN were tested in a cell line assay as described.21 IFN hybrids and mutant constructs containing amino-terminal from parental IFNα21b and the carboxy-terminal from parental IFNα2c (Figure 1A) were designated as follows: Hybrid1 (HY1):IFNα21b(1-75)/IFNα2c(76-166), HY2:IFNα21b(1-95)/IFNα2c(96-166) and HY4:IFNα21b(1-75)/IFNα2c(76-81)/IFNα21b(82-95)/IFNα2c(96-166). From HY4, site-directed mutant-1 (SDM1):IFNα21b(1-75)/IFNα2c(76-81)/IFNα21b(82-95)/IFNα2c,(96-166) S86- > Y, SDM2: IFNα21b(1-75)/IFNα2c(76-81)/IFNα21b(82-95)/IFNα2c, (96-166) N90- > Y; and Cassette mutant (CM3): IFNα21b(1-75)/IFNα2c(76-81)/IFNα21b(82-95)/IFNα2c(96-166), S86- > K17,21 . All IFN constructs were tested for endotoxin levels (EU/mL < 1 at 0.6mg/mL).
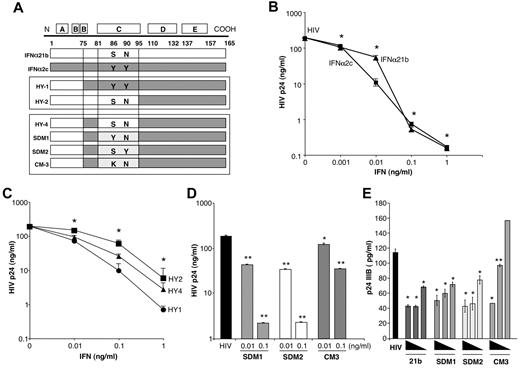
IFNα constructs and antiviral activity. (A) Diagram illustrating construction of IFNα hybrids and mutants. HY1 and HY2 were generated by combining amino acids (aa)1-75 (HY1) or aa1-95 (HY2) from the amino terminus of IFNα21b and carboxy terminus from IFNα2c. Parental IFNα2c tyrosines at aa86 and 90 were present in HY1, while HY2 contains serine and asparagine from IFNα21b. HY4 was generated using HY2 and inserting aa75-81 from IFNα2c retaining serine (aa86) and asparagine (aa90) from IFNα21b. SDM1 differs from HY4 only at aa86 containing a tyrosine. SDM2 derived from HY4 has serine and tyrosine at position 86 and 90, respectively. CM3 differs from SDM1 only at position 86 (S86K). (B) Macrophages (4 × 105) were exposed to HIV for 2 hours, washed and treated once with the indicated concentrations of IFNα (1, 0.1, 0.01, 0.001 ng/mL) parental molecules (*P < .003) or (C) IFN hybrids (HY1, HY2, and HY4 at 0.1-0.1ng/mL *P < .05). Viral replication on day 10 after infection as determined by p24 viral Ag (n = 3). (D) Macrophages (4 × 105) were exposed to HIV for 2 hours, washed, and treated once with SDM1, SDM2, and CM3 (0.01-0.1ng/mL) or media. Ten-day supernatants were tested for p24 by ELISA. Representative donor in duplicates (n = 3). Mutant IFNα molecules compared with cultures infected with HIV in the absence of IFN 2-tailed t test, **P < .001 or *P < .01. SDM1 and SDM2 at 0.1 or 0.01ng/mL compared with CM3 at 0.1 (P < .001) or 0.01 ng/mL (P < .05). (E) Activated (anti-CD3/CD28) T lymphocytes were exposed to HIV IIIB and then incubated in the presence or absence of parental IFN or mutants. (1.0-0.01 ng/mL). *P < .01 HIV alone vs IFN constructs. **P < .05 HIV alone vs CM3. IFNα21b, SDM1, and SDM2 (at 0.1 and 0.01ng/mL) were more effective in controlling HIV replication compared with CM3, P < .02. Data represent mean + SEM.
IFNα constructs and antiviral activity. (A) Diagram illustrating construction of IFNα hybrids and mutants. HY1 and HY2 were generated by combining amino acids (aa)1-75 (HY1) or aa1-95 (HY2) from the amino terminus of IFNα21b and carboxy terminus from IFNα2c. Parental IFNα2c tyrosines at aa86 and 90 were present in HY1, while HY2 contains serine and asparagine from IFNα21b. HY4 was generated using HY2 and inserting aa75-81 from IFNα2c retaining serine (aa86) and asparagine (aa90) from IFNα21b. SDM1 differs from HY4 only at aa86 containing a tyrosine. SDM2 derived from HY4 has serine and tyrosine at position 86 and 90, respectively. CM3 differs from SDM1 only at position 86 (S86K). (B) Macrophages (4 × 105) were exposed to HIV for 2 hours, washed and treated once with the indicated concentrations of IFNα (1, 0.1, 0.01, 0.001 ng/mL) parental molecules (*P < .003) or (C) IFN hybrids (HY1, HY2, and HY4 at 0.1-0.1ng/mL *P < .05). Viral replication on day 10 after infection as determined by p24 viral Ag (n = 3). (D) Macrophages (4 × 105) were exposed to HIV for 2 hours, washed, and treated once with SDM1, SDM2, and CM3 (0.01-0.1ng/mL) or media. Ten-day supernatants were tested for p24 by ELISA. Representative donor in duplicates (n = 3). Mutant IFNα molecules compared with cultures infected with HIV in the absence of IFN 2-tailed t test, **P < .001 or *P < .01. SDM1 and SDM2 at 0.1 or 0.01ng/mL compared with CM3 at 0.1 (P < .001) or 0.01 ng/mL (P < .05). (E) Activated (anti-CD3/CD28) T lymphocytes were exposed to HIV IIIB and then incubated in the presence or absence of parental IFN or mutants. (1.0-0.01 ng/mL). *P < .01 HIV alone vs IFN constructs. **P < .05 HIV alone vs CM3. IFNα21b, SDM1, and SDM2 (at 0.1 and 0.01ng/mL) were more effective in controlling HIV replication compared with CM3, P < .02. Data represent mean + SEM.
Homology modeling
Homology models were obtained by submitting sequences for IFNα constructs to I-TASSER homology modeling server22 and choosing models with best consensus scores. Models were examined for steric clashes and undesirable geometries (PROCHECK). Color coding based on secondary structure was performed using Visual Molecular Dynamics program. Models were subjected to energy minimization followed by short-simulated annealing molecular dynamics under isobaric-isothermal conditions to obtain a lower energy conformation. Homology models were explicitly solvated with TIP3P water molecules and Na+ and Cl- counterions using the VMD program. Simulations were performed using NAMD23 program (Version 2.7) on Biowulf Linux cluster at National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov) and included periodic boundary conditions. Electrostatic interactions were handled using a Particle-Mesh Ewald summation. CHARMM2724 forcefield was used with CHARMM atom types and charges. Initially, a system was energy minimized, followed by slow warming to 310 K in 10 K increments. Each increment ran for 5 picosecond sufficient equilibration at a given temperature. Once the 310 K target temperature was reached, a system was further equilibrated for 50 psec. The simulation was heated 3 times to 450 K in 10 K increments and cooled to 40 K in 10 K decrements with system held at each temperature for 5 picosecond. For all simulations, a 1 femtosecond integration timestep was used along with a 12Å cutoff. Langevin dynamics were used to maintain temperature and a modified Nosé-Hoover Langevin piston was used to control pressure.
IFNAR2/IFN docking
Rosetta program (Version 2.3) was used to perform all-atom, perturbation docking of the interferon-binding ectodomain (EC) of the human IFNAR (PDB-ID-1N6U) to the homology model of IFNα2c.25 Backbones were held rigid while sidechains were allowed to flex. Before docking, IFNα2c was placed 25Å from the side of the receptor thought to interact with IFN based on available 3-D structure information and computational studies. During the run, IFN was able to change its orientation before docking by translating, rotating and rocking relative to the receptor. Docking solutions (20 000) were generated and the top 100 scoring solutions (lowest energy) were clustered based on an all-atom root mean square deviation (RMSD) of 5Å. Representative solutions from the top 10 scoring clusters were retained and of these, the solution with the lowest energy was used for further analysis. Calculations were performed using the high-performance computational resources of Biowulf Linux cluster.
IFNAR2-EC
The EC domain of human IFNα receptor subunit 2 (IFNAR2-EC) was expressed and purified from baculovirus expression vector system (BEVS) cultured supernatant. Recombinant baculovirus containing IFNAR2-EC gene under control of the polyhedrin promoter was prepared by transfection of bacmid DNA in Sf-9 insect cells. The resulting virus was amplified, its titer determined and virus stock stored at 4°C in the dark. One liter of High 5 cells was grown at 27°C in a 3-liter flask at shaker speed of 110 rpms. When cell count reached 1.88 × 106/mL the culture was infected with IFNAR2-EC virus at a multiplicity of infection of 3. Cultures were grown at 21°C for 48 hours and supernatant collected after centrifugation at 1700g. Nondenaturing immobilized metal affinity chromatography (IMAC) purification from culture supernatant, dialysis and sample concentration and SDS-PAGE for quality control was performed.21
RNA isolation and real time PCR
Total RNA was isolated from macrophages or monocytes and DNase digested using Qiagen-RNeasy Mini kit and RNase-Free DNase Set (QIAGEN). For real-time PCR, 1 μg total RNA was reverse-transcribed using oligodeoxythymidylic acid primer (Invitrogen). Amplification of cDNA was performed with TaqMan expression arrays for APOBEC3A (Hs00377444_m1), APOBEC3G (Hs00222415_m1), PKR-EIF2AK2 (Hs00169345_m1), TRIM22 (Hs00232319_m1), IDO (Hs00158032_m1), TNFα (Hs00174128_m1) and GAPDH (Hs99999905_m1; Applied Biosystems). Data were analyzed using the 2−ddCT method and results reported as fold change15 or by conventional PCR9 with primers for TNFα 5′-TGAGCACTGAAAGCATGATCCG-3′(forward) and 5′ AGAGGGCTGATTAGAGAGAGGTCC-3′(reverse).
Western blots
Macrophages were treated or not with IFNα parental molecules and indicated hybrids/mutants (0.1-1 ng/mL) and harvested at indicated time points after treatment. Cell lysates were prepared using protein extraction reagent with protease/phosphatase inhibitor cocktail (Pierce) and cell debris removed by centrifugation (20 000g, 20 minutes, 4°C). Total protein concentration in lysates was determined using Bio-Rad DC Protein Assay (Bio-Rad). Samples were analyzed by SDS-PAGE followed by Western blot as described.21 Antibodies for STAT1 and pSTAT1 (Y701) were obtained from BDTransduction Lab and Cell Signaling Technology. Antibody for STAT2 was obtained from Upstate Biotechnology. Antibodies for pSTAT2 (Y689), STAT3, pSTAT3 (Y705), pSTAT3 (S727) pSTAT1 (Ser727), pSTAT5 (Y694), pSTAT6 (Y641), P-IκBα, IκBα, and p-JAK1(Y1022-1023) were from Cell Signaling. Antibodies for actin were obtained from ABCAM and tubulin from Sigma-Aldrich. In the indicated experiments, macrophages were pretreated with anti-CD118 or isotype-matched control antibody (eBiosciences), Ly294002 (Cell Signaling), Bay 11-7082, KN93, W7 or JAK inhibitor I (Calbiochem) for 1 hour, or Polymyxin B (Sigma-Aldrich) before addition of IFN or soluble IFNAR2 (30 minutes). Signal was developed using Super Signal West-Femto Maximum Sensitivity Kit (Pierce).
ELISA TNFα
Macrophage culture supernatants were collected after parental IFN, SDM1 or SDM2 mutant treatment and evaluated for TNFα by ELISA (R&D Systems).
Statistical analyses
The data are expressed as mean + SEM. Statistical significance between 2 groups was determined using Student t test. Values of P < .05 were considered significant.
Results
Antiviral functions of parental IFN and hybrids
To determine structural properties of IFN molecules responsible for antiviral activity, we used parental IFNs and hybrid constructs generated by recombinant methodology that have previously been shown to have differential biologic properties relative to cell growth.21 Components of parental IFNα21b and IFNα2c (Figure 1A), which both suppress R5 HIV with only a single treatment after the macrophages have been exposed to virus (Figure 1B, day 10 p24 levels shown), similar to recombinant IFNα2a,8,9 were compared with engineered hybrid molecules generated by combining the most active components of these 2 IFN molecules (Figure 1A-C).21 One hybrid molecule (HY1), containing N-terminal of IFNα21b combined with amino acids 75-165 that include the C-helix (amino acids 75-95) of IFNα2c, was most active at inhibiting HIV infection of macrophages (Figure 1C). By comparison, HY2 containing N-terminal and C-helix regions of IFNα21b and COOH-terminus of IFNα2c, while inhibitory at 0.1-1 ng/mL, was less active than parental IFN or the other hybrids at 0.01 ng/mL (Figure 1C). HY4 that differed at amino acids 75-81, but shared C-helix with HY2, had intermediate activity, suggesting that the C-helix configuration is key to triggering an optimal cellular response to HIV.
IFN mutants and HIV suppression
To further define structural determinants for antiviral activity, we generated mutants retaining the N-terminus of IFNα21b and COOH-terminus of IFNα2c, inserting mutations in the C-helix region of IFNα21b specifically at positions 86 and 90 which are involved in the interaction with the IFNAR1 chain17 (Figure 1A). Amino acids 75-81, originating from IFNα2c in HY1 with optimal antiviral activity, were included in all new constructs with further dissection of this sequence through site-directed mutagenesis (SDM) and cassette mutagenesis (CM).17,21 Furthermore, each of the mutants contains both the D and E helices of IFNα2c (amino acids 95-165; Figure 1A) using the N terminal region derived from IFNα21b. Macrophage cultures were treated with a single application of these mutant IFNα constructs at 0.01-0.1 ng/mL, concentrations which enabled differential effects to be seen, after exposure to HIV8,9 and culture supernatants collected every 2-3 days for HIV p24 analysis (Figure 1D, day 10 shown). Two mutants generated by site-directed mutagenesis, SDM1 (S86Y) and SDM2 (N90Y) effectively inhibited HIV at concentrations similar to parental IFNα21b and IFNα2c (Figure 1B-D), and importantly, even exceeded the activity of the hybrid from which they were derived. In contrast, cassette mutant 3 (CM3), distinct from SDM1 only at amino acid position (S86K) or from SDM2 at position 90, was a considerably less potent inhibitor of HIV replication (Figure 1D) with similar results observed in activated T-lymphocytes exposed to HIV IIIB (Figure 1E), indicating that preservation of tyrosine at position 86 and/or 90 is functionally relevant, and perhaps that a serine at position 86 can also confer functional competence.
Induction of IFNα-induced antiviral genes by parental and mutant IFN
Based on their differential effects on HIV suppression, we examined hybrid and mutant molecules for induction of macrophage genes linked with antiviral activity, such as TRIM22, PKR and APOBEC3 family members, based on our recent studies.15 By RT-PCR, all IFNα constructs tested induced significant elevations in PKR gene expression compared with untreated macrophages (2-6 fold; P < .05; Figure 2A). However, PKR levels did not directly correspond to the observed differential activity against HIV in that SDM1, SDM2 and CM3 induced comparable PKR to parental IFNα2c, yet parental IFNs, SDM1 and SDM2 exhibit greater antiviral activity than CM3 (S86K). Parallel analyses for TRIM22 showed a modest, but indistinguishable increase (∼ 2-3 fold) in gene expression induced by IFNα21b, IFNα2c, SDM1, SDM2, HY4 and CM3 (Figure 2B).
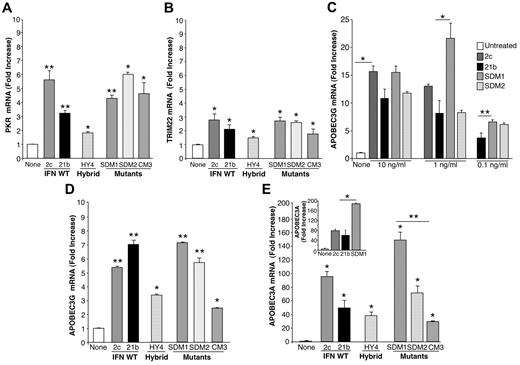
Antiviral gene induction by parental IFNα, hybrid, and mutants. After treatment of macrophages (6 × 106) with IFNα21b, IFNα2c, their hybrids or mutants (0.1 ng/mL) for 4 hours, total mRNA was isolated and examined for (A) PKR and (B) TRIM22 gene transcription using real-time PCR. Representative donor; n = 3. For PKR 2-tailed t test for IFNα-treated compared with control cells (None), **P < .01 (SDM1, SDM2, IFNα21b, IFNα2c) and *P < .05 (CM3, HY4). For TRIM22, *P < .05 for IFN constructs compared with untreated macrophages. Induction of PKR and TRIM22 by SDM1, SDM2, or parental IFN is not significantly different from that of CM3. (C) APOBEC3G gene expression in macrophages incubated with IFN constructs at 0.1-10 ng/mL. *P < .03 untreated vs IFN-treated constructs, *P < .03 parental IFN vs SDM1, **P < .05 IFNα21b vs SDM1. (D) APOBEC3G gene expression correlates with antiretroviral activity. Macrophages (6 × 106) were treated in parallel with IFNα mutants, hybrid (HY4), or parental IFNα (1 ng/mL) for 4 hours and mRNA analyzed for APOBEC3G by RT-PCR. IFNα21b, IFNα2c, SDM1, and SDM2 significantly enhance APOBEC3G gene expression. **P < .001. For HY4 and CM3, *P < .01. APOBEC3G induction by SDM1, SDM2, IFNα2c, and IFNα21b is statistically higher than CM3, *P < .01. Representative donor, n = 3. (E) Macrophages were examined for APOBEC3A transcription after IFNα treatment (1 ng/mL). IFNα parent, hybrid, and mutants induced APOBEC3A (*P < .01), with maximal levels induced by mutant SDM1, *P < .01. Parental IFN vs SDM1, P < .05 and SDM1 vs CM3, **P = .01. Inset: APOBEC3A gene expression in monocytes exposed to IFN constructs. Untreated compared with IFN-treated constructs, *P < .05. Representative donor in duplicate n = 3. Data represent mean + SEM.
Antiviral gene induction by parental IFNα, hybrid, and mutants. After treatment of macrophages (6 × 106) with IFNα21b, IFNα2c, their hybrids or mutants (0.1 ng/mL) for 4 hours, total mRNA was isolated and examined for (A) PKR and (B) TRIM22 gene transcription using real-time PCR. Representative donor; n = 3. For PKR 2-tailed t test for IFNα-treated compared with control cells (None), **P < .01 (SDM1, SDM2, IFNα21b, IFNα2c) and *P < .05 (CM3, HY4). For TRIM22, *P < .05 for IFN constructs compared with untreated macrophages. Induction of PKR and TRIM22 by SDM1, SDM2, or parental IFN is not significantly different from that of CM3. (C) APOBEC3G gene expression in macrophages incubated with IFN constructs at 0.1-10 ng/mL. *P < .03 untreated vs IFN-treated constructs, *P < .03 parental IFN vs SDM1, **P < .05 IFNα21b vs SDM1. (D) APOBEC3G gene expression correlates with antiretroviral activity. Macrophages (6 × 106) were treated in parallel with IFNα mutants, hybrid (HY4), or parental IFNα (1 ng/mL) for 4 hours and mRNA analyzed for APOBEC3G by RT-PCR. IFNα21b, IFNα2c, SDM1, and SDM2 significantly enhance APOBEC3G gene expression. **P < .001. For HY4 and CM3, *P < .01. APOBEC3G induction by SDM1, SDM2, IFNα2c, and IFNα21b is statistically higher than CM3, *P < .01. Representative donor, n = 3. (E) Macrophages were examined for APOBEC3A transcription after IFNα treatment (1 ng/mL). IFNα parent, hybrid, and mutants induced APOBEC3A (*P < .01), with maximal levels induced by mutant SDM1, *P < .01. Parental IFN vs SDM1, P < .05 and SDM1 vs CM3, **P = .01. Inset: APOBEC3A gene expression in monocytes exposed to IFN constructs. Untreated compared with IFN-treated constructs, *P < .05. Representative donor in duplicate n = 3. Data represent mean + SEM.
Comparison of APOBEC3G transcription revealed higher (∼ 5-7 fold; P < .001) induction with IFNα21b and IFNα2c than with the hybrid construct (Figure 2D). However, the SDM1 mutant was uniquely equal or more effective in relative concordance with its anti-HIV activity (Figure 1C), compared with CM3 mutant with the least activity against HIV. Induction of elevated levels of APOBEC3G by SDM1 were detected optimally at 1 ng/mL (P < .03; Figure 2C). Although a similar pattern emerged with APOBEC3A, reported to suppress adeno-associated virus, retrotransposons and HIV,8,9,15,26 this cytidine deaminase was much more dramatically increased in response to parental IFNs (50- > 100-fold; Figure 2E), as reported.8,9 In addition to differentiated macrophages, monocyte levels of APOBEC3A were also up-regulated by intact IFN (Figure 2E inset). Of considerable interest, the engineered SDM1 mutant triggered levels of APOBEC3A in macrophages and monocytes that significantly exceeded levels evident for parental IFNs, hybrids or the CM3 mutant (Figure 2E; P < .01). CM3 with limited antiviral activity was a much less effective inducer of APOBEC, supporting a connection between determinants of IFN that signal APOBEC transcription and HIV antagonism. Once again, these data point to the tyrosine (Y) and likely, asparagine (N) amino acid residues at positions 86 and 90, respectively, as being important for regulating antiviral activity and APOBEC induction. To understand how modifications in amino acids 86/90 alter IFN function, we generated computational structural models (Figure 3). IFN homology models show that these constructs are very similar and sequence alignments illustrate that major changes that would affect how IFN binds to its receptor occur at the C-terminus. Therefore, it appears that the changes in positions 86 and 90 are likely not altering binding through IFNAR2, but rather altering the complex binding interactions between ligand, IFNAR1 and IFNAR2 leading to signal transduction.
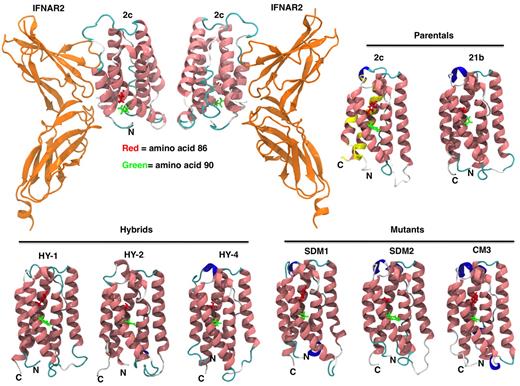
IFNα constructs 3-D models. IFN homology models for parental IFN (IFNα2c and IFNα21b), hybrids, and mutants were generated as described in “Homology modeling.” The ribbon diagram renderings of IFN homology models are colored by secondary structure (pink = α helix, blue = 310 helix, cyan = turn, white = coil). Mutations in the C-loop of the molecule in amino acid positions 86 and 90 are shown in red and green, respectively. Overall the structures are very similar with the mean root mean square deviation (RMSD) between the C-α carbons being 2.5 Å (median RMSD = 2.4 Å). Most of the structural differences between the models occur in loop regions, specifically positions 21-31, 45-51, and 100-110. Docking of IFN-binding ectodomain of the homology model of IFNα2c to IFNAR2 is shown (top left panel). Yellow color on IFNα2c model illustrates IFNAR2 interacting region (top right panel). Coloring on secondary structure was performed using Visual Molecular Dynamics (VMD) program.
IFNα constructs 3-D models. IFN homology models for parental IFN (IFNα2c and IFNα21b), hybrids, and mutants were generated as described in “Homology modeling.” The ribbon diagram renderings of IFN homology models are colored by secondary structure (pink = α helix, blue = 310 helix, cyan = turn, white = coil). Mutations in the C-loop of the molecule in amino acid positions 86 and 90 are shown in red and green, respectively. Overall the structures are very similar with the mean root mean square deviation (RMSD) between the C-α carbons being 2.5 Å (median RMSD = 2.4 Å). Most of the structural differences between the models occur in loop regions, specifically positions 21-31, 45-51, and 100-110. Docking of IFN-binding ectodomain of the homology model of IFNα2c to IFNAR2 is shown (top left panel). Yellow color on IFNα2c model illustrates IFNAR2 interacting region (top right panel). Coloring on secondary structure was performed using Visual Molecular Dynamics (VMD) program.
Signaling pathways engaged by IFN and IFN mutants
The distinct functional properties of mutant SDM1 focused our attention on dissecting targeted signaling pathways engaged to orchestrate its antiretroviral activities. First, we used an anti–human CD118 antibody that selectively blocks IFNα receptor chain-2 (IFNAR2) ligand binding to establish that its signal transduction was mediated solely by IFNAR. As evident in Figure 4A, ISG induction by SDM1, as represented by APOBEC3A, was abrogated in parallel with that of wild-type IFN by pre-exposure of macrophages to the receptor antibody. In concert with blockade of ISG, anti-CD118 largely reversed the ability of IFN and IFN mutant to suppress HIV replication relative to control cultures (no Ab) and isotype (Figure 4B). Interestingly, anti-CD118 enhanced HIV replication in the absence of exogenously added IFN, consistent with endogenous IFN in the infected cultures.27 Further confirmation of the receptor specificity of the SDM1 construct was obtained when we added excess soluble recombinant IFNAR2 (sIFNAR2) with IFN and the SDM1 mutant, which blocked their induction of APOBEC3A/G gene expression by ∼ 90% (Figure 4C), documenting that interaction with IFNAR2 by the engineered constructs is preserved and essential for activation of the receptor associated kinase JAK1 (Figure 4C inset).
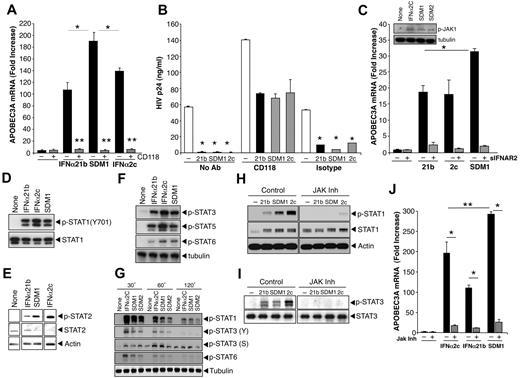
IFNα and mutant signaling. (A) Macrophages were pretreated or not with anti-IFNAR (CD118) for 20 minutes and treated with parental IFNα or SDM1 for 4 hours. Levels of APOBEC3A for SDM1 compared with parental IFNs *P < .01. APOBEC3A transcription by RT-PCR shows that blockade of IFNAR2 reduces APOBEC3A gene expression, **P < .01. (B) Macrophages were exposed to HIV 2 hours, washed and incubated with CD118 antibody or isotype control for 20 minutes. IFNα21b, IFNα2c SDM1 were then added once at 1 ng/mL and supernatants collected at day 13 after infection and tested for p24 (*P < .01, HIV alone vs IFN-treated). (C) Macrophages were treated with 1 ng/mL of IFNα21b, IFNα2c, or SDM1 in the presence or absence of excess soluble IFNAR2. *P < .05 SDM1 vs 21B and 2C. Inset: Whole cell lysates from macrophages treated with IFNα2c, SDM1 and SDM2 were analyzed for P-JAK1 by Western blot. Representative assay n = 2. (D) Whole cell protein lysates collected after 15-minute exposure to IFNα (0.1 ng/mL) were examined for STAT activation. Levels of STAT1 phosphorylation were analyzed by Western blot using an anti-pSTAT1 antibody. Membranes were then probed with STAT1 antibody to confirm equivalent protein loading. Representative data, n = 4. (E) STAT2 activation was detected 15 minutes after IFN (1 ng/mL) by immunoblotting using pSTAT2 and STAT2 antibodies or β-actin (n = 3). (F) Enhanced phosphorylation of STAT3, STAT5, and STAT6 after treatment with indicated IFNα constructs (1 ng/mL) for 20 minutes (n = 3). (G) Whole cell lysates from macrophages exposed to medium alone, IFNα2c, SDM1, and SDM2 for the indicated time points were analyzed by Western blot for pSTAT1 (Y), pSTAT3 (Y), pSTAT3 (S), pSTAT6 (Y) and tubulin. (H) Macrophages were treated with JAK inhibitor I for 1 hour before addition of IFNα and cell lysates analyzed for pSTAT1 or (I) pSTAT3 by Western blot. Interference with IFNAR-associated JAK suppresses STAT1 and STAT3 phosphorylation when compared with macrophages not treated with inhibitor. Representative data, n = 3. (J) Total RNA analyzed by RT-PCR for APOBEC3A transcription in macrophages pretreated with JAK inhibitor 1 hour prior to 4-hour IFNα. Inhibition of JAK almost completely abrogates APOBEC3A transcription. Representative data, n = 3, *P < .003. Levels of APOBEC3A for SDM1 compared with parental IFNs, **P < .01. Data represent mean + SEM.
IFNα and mutant signaling. (A) Macrophages were pretreated or not with anti-IFNAR (CD118) for 20 minutes and treated with parental IFNα or SDM1 for 4 hours. Levels of APOBEC3A for SDM1 compared with parental IFNs *P < .01. APOBEC3A transcription by RT-PCR shows that blockade of IFNAR2 reduces APOBEC3A gene expression, **P < .01. (B) Macrophages were exposed to HIV 2 hours, washed and incubated with CD118 antibody or isotype control for 20 minutes. IFNα21b, IFNα2c SDM1 were then added once at 1 ng/mL and supernatants collected at day 13 after infection and tested for p24 (*P < .01, HIV alone vs IFN-treated). (C) Macrophages were treated with 1 ng/mL of IFNα21b, IFNα2c, or SDM1 in the presence or absence of excess soluble IFNAR2. *P < .05 SDM1 vs 21B and 2C. Inset: Whole cell lysates from macrophages treated with IFNα2c, SDM1 and SDM2 were analyzed for P-JAK1 by Western blot. Representative assay n = 2. (D) Whole cell protein lysates collected after 15-minute exposure to IFNα (0.1 ng/mL) were examined for STAT activation. Levels of STAT1 phosphorylation were analyzed by Western blot using an anti-pSTAT1 antibody. Membranes were then probed with STAT1 antibody to confirm equivalent protein loading. Representative data, n = 4. (E) STAT2 activation was detected 15 minutes after IFN (1 ng/mL) by immunoblotting using pSTAT2 and STAT2 antibodies or β-actin (n = 3). (F) Enhanced phosphorylation of STAT3, STAT5, and STAT6 after treatment with indicated IFNα constructs (1 ng/mL) for 20 minutes (n = 3). (G) Whole cell lysates from macrophages exposed to medium alone, IFNα2c, SDM1, and SDM2 for the indicated time points were analyzed by Western blot for pSTAT1 (Y), pSTAT3 (Y), pSTAT3 (S), pSTAT6 (Y) and tubulin. (H) Macrophages were treated with JAK inhibitor I for 1 hour before addition of IFNα and cell lysates analyzed for pSTAT1 or (I) pSTAT3 by Western blot. Interference with IFNAR-associated JAK suppresses STAT1 and STAT3 phosphorylation when compared with macrophages not treated with inhibitor. Representative data, n = 3. (J) Total RNA analyzed by RT-PCR for APOBEC3A transcription in macrophages pretreated with JAK inhibitor 1 hour prior to 4-hour IFNα. Inhibition of JAK almost completely abrogates APOBEC3A transcription. Representative data, n = 3, *P < .003. Levels of APOBEC3A for SDM1 compared with parental IFNs, **P < .01. Data represent mean + SEM.
Interaction of IFNα with its cognate receptors triggers canonical IFN-induced signaling, including Janus protein kinase (JAK) phosphorylation and activation of cytoplasmic STAT1/2, culminating in ISG transcription. Phosphorylation of STAT1 in response to SDM1 occurred rapidly, comparable with parental IFNα21b (Figure 4D-H), whereas IFNα2c often appeared more active. Induction of STAT2 phosphorylation followed a similar pattern, and interestingly, STAT3 and STAT5 phosphorylation were also engaged and to a lesser extent, STAT6 (Figures 4E-F). Since STAT phosphorylation by less active constructs CM3 and HY4 was lower (data not shown), the extent of STAT phosphorylation appears to correspond to regulation of APOBEC and antiviral activity. Additional kinetics studies reveal that phosphorylation of STATs was transient for all IFN constructs, detectable by 30 minutes and declining by 2 hours. However, while the pSTAT1(Y), pSTAT3(Y) and pSTAT6(Y) signal was declining by 60 minutes the pSTAT3(ser) was optimal at 1 hour (Figure 4G). Addition of an upstream JAK inhibitor (1-100nM) 1 hour before IFN and SDM1, not only blocked STAT1/3 phosphorylation (Figure 4H-I), but also significantly suppressed induction of APOBEC3 expression (Figure 4J). Our data provide evidence for a differential level of signal transduction that may underlie the unique ability of the SDM1 mutant to orchestrate an enhanced cellular response against HIV.
Dissociation between antiviral and toxicity-related genes
We next investigated whether or not the ability of SDM1 to promote antiviral properties in macrophages was also associated with enhanced expression of molecules linked to IFN toxicity. Among the multiple IFN-induced molecules that have been linked with its toxicity is IDO,13 and exposure of differentiated macrophages to parental IFNα2c and IFNα21b triggered substantive increases in IDO mRNA expression within 4 hours. (Figure 5A-B). Since IDO is also inducible via endotoxin interaction with TLR, we added Polymyxin B to neutralize any potential contaminating endotoxin which did not alter IFN-induced IDO. More importantly, in surprising contrast, SDM1 and SDM2 induced significantly less IDO than parental molecules (P < .05) whether in presence or absence of Polymyxin B, providing a useful clue that it may be possible to dissociate IFN antiviral functions from its toxic properties. Whether tested at 1 ng/mL (Figure 5A-B) or at reduced concentrations (Figure 5C), SDM1 induced significantly less IDO than the parental IFN (P < .01). Confirming the specificity of the signal the antibody to IFNAR anti-CD118 blocked both IFNα2c and SDM1 mediated up-regulation of IDO (Figure 5C inset). As anticipated, regulation of IDO triggered by parental or mutant SDM1 was also blocked by a JAK inhibitor (Figure 5D) albeit induction of IDO by parental IFNα2c was not totally suppressed by the JAK inhibitor, implicating potential accessory signaling networks.
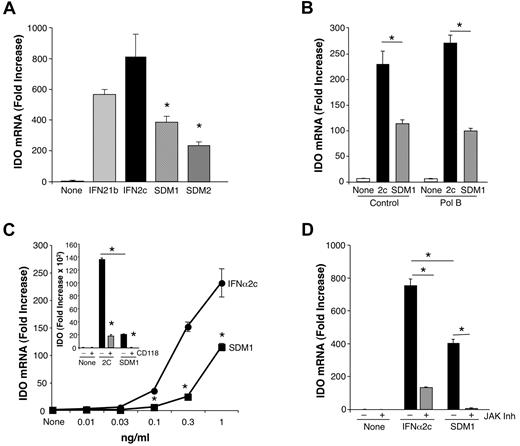
Comparison of IFN constructs for induction of IDO. (A) Total mRNA was isolated from macrophages (6 × 106) treated with IFNα21b, IFNα2c, SDM1 or SDM2. SDM1 (S86- > Y) and SDM2 (N90- > Y) induced reduced IDO gene expression in comparison to parental IFN molecules at 1 ng/mL (*P < .02). Representative data, n = 3. (B) Total mRNA analyses of macrophages incubated in the presence or absence of Polymyxin B and exposed to IFNα2c or SDM1 as indicated for 4 hours (*P < .05). (C) Real-time PCR of monocytes treated with indicated concentrations of IFNα2c, SDM1 for 4 hours confirms limited IDO transcription for mutant IFN, *P < .01. Inset: CD118 antibody blocks parental IFN and SDM1 IDO gene induction. Representative data, n = 3, *P < .01 IFNα2c vs SDM1. *P < .01 IFN alone compared with anti-CD118/IFN-treated. (D) Real-time PCR analyses comparing levels of transcriptional activity for IDO in macrophages cultures pretreated with JAK inhibitor (100nM) 60 minutes before 4-hour treatment with 1 ng/mL of IFNα2c or SDM1. *P < .01 for levels of IDO for SDM1 vs IFNα2c and IFN constructs versus inhibitor/IFN-treated. Data represent mean + SEM.
Comparison of IFN constructs for induction of IDO. (A) Total mRNA was isolated from macrophages (6 × 106) treated with IFNα21b, IFNα2c, SDM1 or SDM2. SDM1 (S86- > Y) and SDM2 (N90- > Y) induced reduced IDO gene expression in comparison to parental IFN molecules at 1 ng/mL (*P < .02). Representative data, n = 3. (B) Total mRNA analyses of macrophages incubated in the presence or absence of Polymyxin B and exposed to IFNα2c or SDM1 as indicated for 4 hours (*P < .05). (C) Real-time PCR of monocytes treated with indicated concentrations of IFNα2c, SDM1 for 4 hours confirms limited IDO transcription for mutant IFN, *P < .01. Inset: CD118 antibody blocks parental IFN and SDM1 IDO gene induction. Representative data, n = 3, *P < .01 IFNα2c vs SDM1. *P < .01 IFN alone compared with anti-CD118/IFN-treated. (D) Real-time PCR analyses comparing levels of transcriptional activity for IDO in macrophages cultures pretreated with JAK inhibitor (100nM) 60 minutes before 4-hour treatment with 1 ng/mL of IFNα2c or SDM1. *P < .01 for levels of IDO for SDM1 vs IFNα2c and IFN constructs versus inhibitor/IFN-treated. Data represent mean + SEM.
IFNα-mediated NFκB signaling
Since APOBEC3 and IDO appeared to be differentially induced by the IFN mutant containing S86Y, which is linked to the IFNAR binding site, we explored potential cross-talk pathways that might influence disparate regulation. When we monitored parameters of NFκB activation, considered an accessory Type I IFN signaling pathway,28-30 we determined by Western blots that an increase in phosphorylation of IκBα in response to parental IFN was seen relative to SDM1 (Figure 6A inset). Parental IFNα substantially increased IκBα phosphorylation (Figure 6A inset), consistent with activation of the NFκB pathway.28-30 Since NFκB modulates IDO gene expression in conjunction with STAT,31 heightened engagement of this pathway could favor IDO. SDM1 was a much less active trigger for phosphorylation of IκBα, but interestingly a NFκB pathway inhibitor was still partially suppressive to the limited levels of SDM1-driven IDO, as well as IFN-induced IDO gene expression (Figure 6A).
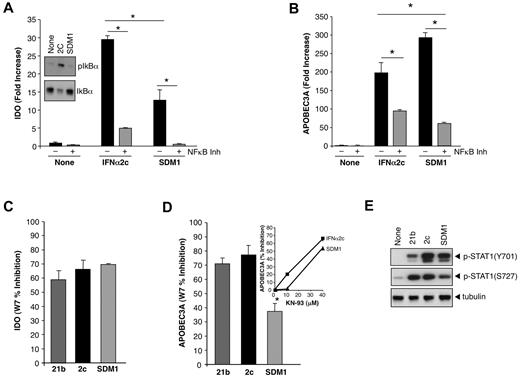
Regulation of IFNα-induced APOBEC3 and IDO by NFκB and calmodulin/CaMK. (A) Whole cell protein lysates collected after treatment with parental IFN or SDM1 (1 ng/mL, 20 minutes) were examined for IκBα phosphorylation by Western blot using an anti–P-IκBα antibody (inset). Transcription of IDO mRNA in macrophages that were pretreated with Bay 11-7082 (10μM) for 1 hour followed by 4 hours of parental or SDM1 mutant is diminished by 82% and 96%, respectively *P < .05 for IDO levels 2C compared with SDM1 and IFN treated compared with inhibitor/IFN treated. Representative data, n = 3. (B) Transcription of APOBEC3A in cells that were pretreated with Bay 11-7082 inhibitor for 1 hour followed by IFNα2c or SDM1 (4 hours), *P < .05 for control compared with IFN-treated; IFNα2c vs SDM1; and IFN constructs treated versus inhibitor/IFN-treated. Representative data, n = 3. (C, D) Macrophage cultures were pretreated for 1 hour with the calmodulin inhibitor W7 (40μM) or (D inset) CaMKII inhibitor KN93 (10-40μM) before 4-hour treatment with parental IFNs and SDM1 mutant. Total mRNA was examined for APOBEC3A and IDO gene transcription, in C P = not significant; in D *P < .05. (E) Whole cell lysates were analyzed for pSTAT1(Y701)(S727) and tubulin (30 minutes). Representative data, n = 3.
Regulation of IFNα-induced APOBEC3 and IDO by NFκB and calmodulin/CaMK. (A) Whole cell protein lysates collected after treatment with parental IFN or SDM1 (1 ng/mL, 20 minutes) were examined for IκBα phosphorylation by Western blot using an anti–P-IκBα antibody (inset). Transcription of IDO mRNA in macrophages that were pretreated with Bay 11-7082 (10μM) for 1 hour followed by 4 hours of parental or SDM1 mutant is diminished by 82% and 96%, respectively *P < .05 for IDO levels 2C compared with SDM1 and IFN treated compared with inhibitor/IFN treated. Representative data, n = 3. (B) Transcription of APOBEC3A in cells that were pretreated with Bay 11-7082 inhibitor for 1 hour followed by IFNα2c or SDM1 (4 hours), *P < .05 for control compared with IFN-treated; IFNα2c vs SDM1; and IFN constructs treated versus inhibitor/IFN-treated. Representative data, n = 3. (C, D) Macrophage cultures were pretreated for 1 hour with the calmodulin inhibitor W7 (40μM) or (D inset) CaMKII inhibitor KN93 (10-40μM) before 4-hour treatment with parental IFNs and SDM1 mutant. Total mRNA was examined for APOBEC3A and IDO gene transcription, in C P = not significant; in D *P < .05. (E) Whole cell lysates were analyzed for pSTAT1(Y701)(S727) and tubulin (30 minutes). Representative data, n = 3.
Although the NFκB pathway has not been associated with APOBEC3A/G gene transcription, blocking NFκB in IFNα2c or SDM1 stimulated macrophages (Figure 6B) partially reduced APOBEC3 transcription. These data, together with the known JAK/STAT signaling requirement, infer a functional bifurcation of intracellular pathways leading to induction of APOBEC3A and IDO. Engagement of NFκB appears to contribute to a greater extent in the transcriptional activation of IDO, but to a lesser extent for cytidine deaminases (Figure 6A-B) and a potential shifting of the balance between these signaling events may underlie the ability of SDM1 to favor antiviral over toxicity sequelae.
Regulation of IFN-induced gene expression by Calmodulin/CaMK
To further explore this altered signaling balance, we examined STAT1 phosphorylation. A key step in STAT complex formation allowing its translocation into the nucleus is tyrosine phosphorylation by activated JAK1/TYK2, whereas serine phosphorylation of STAT1, although not required for nuclear translocation or binding to ISG promoters, is important for full transcriptional activation.32 Because calmodulin and calcium-activated calmodulin kinase (CaMK) have been reported to regulate Type I and Type II IFN signaling in macrophages by both tyrosine and serine phosphorylation on STAT133,34 and to regulate another cytidine deaminase, activation-induced cytidine deaminase (AID), we investigated their potential involvement in IFN mutant signaling. In this regard, the calmodulin inhibitor W7 reduced gene expression of IDO for both parental IFNs and mutant SDM1 (Figure 6C). W7 similarly inhibited APOBEC3 in the case of parental IFN, but its effect on SDM1 mediated APOBEC expression was significantly less (Figure 5D).
Since treatment of macrophage cultures with W7 reveal a differential suppression only for APOBEC3A we decided to explore CaMKII, a calmodulin downstream mediator. KN93, an antagonist of CaMKII, partially interferes (50%-60%) with the induction of APOBEC3A by parental IFN and SDM1, as well as APOBEC3G (data not shown) at 40μM (Figure 6D inset). Suppression of parental IFN-induced APOBEC3A, but not that triggered by SDM1, occurred even at concentrations as low as 10μM, whereas at 40μM the kinase inhibitors were maximally inhibitory for both stimuli (Figure 6D inset). These data are consistent with a lesser dependence of SDM1 on calmodulin kinase/CaMK than parental IFN. In keeping with this signaling cascade, the reduced phosphorylation of STAT1 (S727), but not STAT1 (Y701) by SDM1 compared with both IFNα2c and IFNα21b (Figure 6E) favors a more substantive involvement of this pathway for IDO, indicative of further uncoupling of these molecular pathways.
Differential TNFα induction modulates toxicity
Since IFN is known to synergize with TNFα for IDO transcription31 and TNFα can facilitate HIV infection,35 we considered the potential for TNFα involvement as an intermediate link in IFN toxicity. Parental IFN triggered rapid and abundant levels of TNFα as determined by real time PCR and by protein detection (Figure 7A-B), which was abolished in the presence of the IFNAR antibody (Figure 7C). Quite the contrary, SDM1 and SDM2 initiated limited TNFα by PCR compared with IFNα2c and IFNα21b correlating with reduced cytokine levels detected by ELISA (Figure 7B). Furthermore, direct addition of exogenous TNFα to macrophages resulted in significantly enhanced IDO expression, even at 1 ng/mL (Figure 7D) confirming the ability of this cytokine to regulate IDO transcription. Consistent with dissociable pathways, TNFα was not an effective stimulus for APOBEC3A (Figure 7D), regardless of concentration. The early and substantial IFNα2c and IFNα21b induced TNFα can augment and sustain IFN-driven IDO transcription in a positive feedback loop, without influencing APOBEC generation. This sequence of events likely exacerbates IFN's toxicity-related pathways, and/or potentially favors HIV replication in opposition to IFN. In contrast, SDM1, which does not readily support TNFα generation, will engender less TNFα and IDO relative to its ability to enhance antiviral APOBEC3 and likely other antiviral molecules.
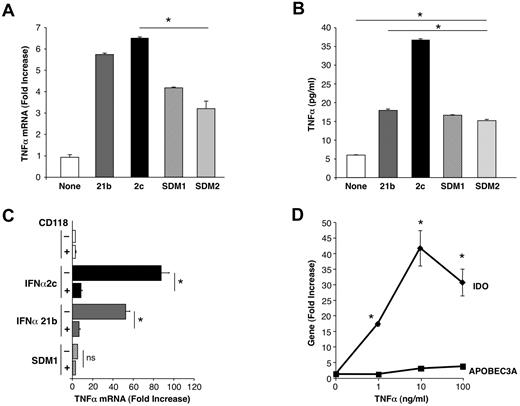
Differential TNFα induction after exposure to IFN constructs. (A) Macrophages were examined for TNFα gene expression (*P < .003 SDM1 and SDM2 vs IFNα2c; P < .02 SDM1 and SDM2 vs IFNα21b (4 hours) by RT-PCR. P < .002 untreated vs IFN constructs treated macrophages. (B) Supernatant TNFα protein (4 hours) was determined by ELISA (*P < .001 IFN constructs compared with untreated and SDM1 or SDM2 vs parental IFNα2c; mutants vs IFNα21b P < .02). Representative data n = 3. (C) Total mRNA was isolated from macrophages that were incubated with anti-CD118 before treatment with parental IFNs or SDM1 mutant for 4 hours. TNFα gene transcription was examined by RT-PCR. (*P < .01; ns, not significant; P < .01 for parental IFN vs SDM1). (D) Macrophages were treated with rhTNFα (1-100 ng/mL) for 4 hours and total mRNA examined for IDO or APOBEC3A gene expression by RT-PCR. Data represent mean + SEM * P < .05, untreated compared with TNFα-treated.
Differential TNFα induction after exposure to IFN constructs. (A) Macrophages were examined for TNFα gene expression (*P < .003 SDM1 and SDM2 vs IFNα2c; P < .02 SDM1 and SDM2 vs IFNα21b (4 hours) by RT-PCR. P < .002 untreated vs IFN constructs treated macrophages. (B) Supernatant TNFα protein (4 hours) was determined by ELISA (*P < .001 IFN constructs compared with untreated and SDM1 or SDM2 vs parental IFNα2c; mutants vs IFNα21b P < .02). Representative data n = 3. (C) Total mRNA was isolated from macrophages that were incubated with anti-CD118 before treatment with parental IFNs or SDM1 mutant for 4 hours. TNFα gene transcription was examined by RT-PCR. (*P < .01; ns, not significant; P < .01 for parental IFN vs SDM1). (D) Macrophages were treated with rhTNFα (1-100 ng/mL) for 4 hours and total mRNA examined for IDO or APOBEC3A gene expression by RT-PCR. Data represent mean + SEM * P < .05, untreated compared with TNFα-treated.
Discussion
A renewed interest in type-I interferons for treatment of infectious diseases, particularly HIV,10 has been tempered by recurring toxicity issues associated with its use as a therapeutic agent.12-14 In this study, we approached this problem through a structure-function analysis of IFNα to identify the minimal requirements for antiviral function with the intent to dissociate this activity from its less desirable functions. We have uncovered structural determinants in the IFNα molecule critical for differentially controlling HIV replication that appear to correlate with induction of host cell innate antiviral APOBEC3 family members. After construction of hybrids containing components of IFNα21b and IFNα2c, which retained antiviral activity, we generated site-specific mutants to identify requisite structural determinants for antiviral function. IFN hybrids with mutations at positions 86 and/or 90, notably SDM1, containing a tyrosine at position 86 and asparagine at position 90 exhibit enhanced anti-HIV activity in concert with elevated APOBEC3 transcription. SDM2, containing a serine at position 86 and tyrosine at 90, had activity, whereas CM3 differing from SDM1 only at 86 (Y to K) did not, narrowing down the activity focal point.
Using SDM1 as our model agonist, we demonstrated that its activity was IFNAR and JAK/STAT dependent and that phosphorylation of STATs was better achieved when macrophages were treated with SDM1 relative to the least efficient antiviral IFNα constructs. Type I IFNs engage the same IFNAR1 and IFNAR2 cellular receptors leading to a similar transcription profile but can exhibit variations in kinetics of certain genes and immunomodulatory activities. Some of these variations can be independent of receptor affinity or attributed to differences in amino acid residues interacting with IFNAR that influence the nature of ternary structure, receptor turnover expression, stability of signaling complex binding to ISRE/GAS elements and rate of phosphatase activity. In competition binding studies with Daudi cells, we demonstrated that SDM containing IFNα21b N-terminus and tyrosine (aa 86 or 90) compete well with I125IFNα21 binding sites.17 Our study suggests that structural amino acid changes in the C-helix known to interact with IFNΑR1 may modify the overall IFNΑR signaling dynamics leading to elevated APOBEC3 and lower IDO by SDM1 which may involve additional transcriptional elements, posttranslational modifications or subcellular localization36 of signaling mediators responsible for differential levels of gene transcription.
How SDM1 preferentially steers macrophages to an antiviral phenotype while dampening toxicity-related IDO and TNFα provides an opportunity for segregating these functions of IFN. In this regard we demonstrate that SDM1 induction of IDO and APOBEC3 transcription was equally inhibited by anti-CD118, sIFNAR2 and JAK inhibitor I, authenticating that SDM1 signaling requires IFNAR and receptor associated tyrosine kinases for both genes. In contrast to parental IFNs, limited SDM1 induction of IDO transcription is the result of differential engagement of the auxiliary signaling network NFκB, known to participate and synergize with the STAT pathway. However, transcription of APOBEC3 seems less influenced by this pathway. SDM1 induces higher APOBEC3A despite lower pSTAT1(S727) activation pointing to a more complex transcriptional regulation involving multiple STATs, accessory signaling and recruitment of additional gene specific coactivators. pSTAT6 has been reported to be inducible by IFNα in cells of lymphoid and endothelial origin.30,37 However, additional studies are necessary to determine whether pSTAT6 induction by IFN constructs leads to functional formation/translocation of ISGF3-like complex (STAT6:STAT2:IRF9)37 and differential binding to ISRE to impact IFNα-induced IDO or APOBEC3 gene expression in macrophages. Furthermore, CAMKII known to modulate JAK-STAT33,34 regulates both APOBEC3 and IDO gene expression pathways, implicating NFκB and PI3K in regulation of APOBEC. Nonetheless, the diminished NFκB response induced by SDM1 in comparison with parental IFN illustrates how structural components of IFNs differ in the regulation of antiviral and toxic ISG. IFNs, besides activating the classic JAK/STAT signaling cascade,30 engage accessory pathways involving the interaction of TRAF2 with the IFNAR1 subunit and also activate NFκB transcription factors regulating expression of genes involved in cell survival and inflammatory responses.28,29,38 Although at this point, it is not known whether binding of SDM with IFNAR1 leads to TRAF2 interaction, our evidence favors an apparent signaling cascade for antiviral activity, with both STAT and NFκB involvement in generation of genes associated with antiviral activity and yet to be determined signaling intermediates.
Concomitant with enhanced APOBEC and antiviral activity, SDM1-mediated TNFα transcription much less than parental IFNs. We find that exacerbation of the elevated levels of IDO attributable to intact IFNα2c or IFNα21b may be the consequence of coinduction of TNFα. TNFα, by synergizing with IFNγ via NFκB activation, enhancing STAT1 and IRF1 binding to GAS and ISRE sites upstream, sustains IDO in a cooperative fashion with other IFN-induced transcriptional elements.31,39-41 The heme containing enzyme IDO catalyzes the initial rate limiting step in tryptophan degradation to form N-formylkynurenine derived catabolites and has been highlighted as one of the genes linked to interferon-therapy mediated toxicity,13,14 and IDO is expressed in many cells including astrocytes, microglia, DC, monocytes, and macrophages.42 Additional complications are seen in HCV43,44 and HIV infected patients because of virus induced IDO-mediated catabolism, exemplified in HIV-associated dementia (HAD), disease progression and persistence.18,20 Moreover, increased IDO in tonsils of HIV-infected patients not responding to antiretroviral therapy involves T regulatory lymphocytes.45 In this regard, future studies will help determine how the differential antiviral effects observed with IFN constructs with T lymphocytes correlate with IDO and APOBEC3. Augmented IDO expression is seen after exposure of PBMCs to either infectious or noninfectious HIV46 and discrepant regulation of IDO by different retroviral clades has been correlated with prevalence of HAD.47 In addition to IFNα2 enforced TNFα generation, HIV-induced macrophages produce TNFα,48 and elevated levels of this inflammatory mediator are detected in HAD patient CNS, further contributing to HIV entry into the brain.49 Although detection of CaMKII activation was more challenging to assess in our cultures, this pathway can potentially enhance NFκB signaling,50 contributing to a self-perpetuating response for parental IFNs with a lesser role of CaMKII in the case of SDM1, which may explain lower phosphorylation levels of STAT1(S727). Increased STAT1(S727) phosphorylation, known to be important for most IFN transcriptional activity, may favor IDO by parental IFN compared with SDM1.
In summary, we have identified structural components of IFNα optimized in SDM1, an engineered mutant derived from the amino-terminal region of IFNα21b and the COOH-terminus from IFNα2c that preferentially support macrophage antiviral activity with reduced engagement of pathways responsible for certain toxicity related sequelae. This mutant possesses tyrosine at position 86 preserving asparagine at amino acid 90 as a critical determinant imparting novel differential signaling and consequently, dissociable induction of ISG. While only a first step, these observations provide a template for further dissection and engineering of IFN molecules to reduce adverse-related gene induction and shifting the balance in favor of antiviral activity. Besides providing a better understanding of the normal physiologic, pharmacologic and pathologic functions of IFN, these newly customized IFNs, in addition to impacting the innate resistance repertoire to HIV, may suggest a more global benefit in other infectious and neoplastic diseases.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Wenwen Jin and Bhavna Gupta for technical assistance and to Calley Grace for editorial assistance. The authors acknowledge William Gillette, Dominic Esposito, and Ralph Hopkins (SAIC, NCI, Frederick, MD) for expression and purification of IFNs and IFNAR2-EC.
This research was supported by Intramural Research Program of National Institutes of Health, National Institute of Dental and Craniofacial Research (NIDCR) and National Institute of Allergy and Infectious Diseases (NIAID).
National Institutes of Health
Authorship
Contribution: N.V. designed and performed experiments, collected, analyzed and interpreted data, and wrote the manuscript; H.S. performed selected experiments, provided reagents, interpreted data and reviewed manuscript; J.B. and K.C.Z. provided reagents, interpreted data and reviewed manuscript; M.A.D. performed homology modeling dynamics and IFNAR2-IFN docking simulations; and S.M.W. designed experiments, interpreted data and contributed to writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Nancy Vázquez, Bldg 30, Rm 332, 30 Convent Dr, MSC 4352, NIDCR, National Institutes of Health, Bethesda, MD 20892-4352; e-mail: nvazquez@mail.nih.gov.
References
Author notes
K.C.Z. and S.M.W. contributed equally to this article.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal